Abstract
Dysfunctional mitochondria are a well-known disease hallmark. The accumulation of aberrant mitochondria can alter cell homeostasis, thus resulting in tissue degeneration. Lysosomal storage disorders (LSDs) are a group of inherited diseases characterized by the buildup of undegraded material inside the lysosomes that leads to autophagic-lysosomal dysfunction. In LSDs, autophagic stress has been associated to mitochondrial accumulation and dysfunction. However, the mechanisms underlying mitochondrial aberrations and how these are involved in tissue pathogenesis remain largely unexplored. In normal conditions, mitochondrial clearance occurs by mitophagy, a selective form of autophagy, which relies on a parkin-mediated mitochondrial priming and subsequent sequestration by autophagosomes. Here, we performed a detailed analysis of key steps of mitophagy in a mouse model of multiple sulfatase deficiency (MSD), a severe type of LSD characterized by both neurological and systemic involvement. We demonstrated that in MSD liver reduced parkin levels resulted in inefficient mitochondrial priming, thus contributing to the accumulation of giant mitochondria that are located outside autophagic vesicles ultimately leading to cytochrome c release and apoptotic cell death. Morphological and functional changes were also observed in mitochondria from MSD brain but these were not directly associated with neuronal cell loss, suggesting a secondary contribution of mitochondria to neurodegeneration. Together, these data shed new light on the mechanisms underlying mitochondrial dysfunction in LSDs and on their tissue-specific differential contribution to the pathogenesis of this group of metabolic disorders.
INTRODUCTION
Macroautophagy (usually referred simply as autophagy) is a catabolic pathway by which parts of the cytoplasm are encapsulated in double-membrane vesicles, named autophagosomes, that are ultimately targeted to lysosomes for degradation (1,2). Autophagy has been described to be an essential regulatory mechanism for mitochondrial quality control (3,4). The term ‘mitophagy’ is widely used to designate this specific form of autophagy. Initially assumed to be a random process, recent studies indicate that mitochondrial degradation is a selective process (5–9).
Mitochondria are recognized to be key regulators of cell death (10,11). Therefore, proper removal of dysfunctional mitochondria is essential to preserve cellular homeostasis. Dysfunctional mitochondria may originate from deleterious effects of specific toxic stimuli or by a series of fusion–fission events that characterize the mitochondrial life cycle (12–15). The recognition of aberrant mitochondria, referred as mitochondrial priming, is based on a PINK1-dependent recruitment of cytosolic parkin to mitochondria (16–19). Upon mitochondrial translocation, parkin exerts its E3 ubiquitin ligase activity by ubiquitinating proteins localized on the outer mitochondrial membrane (OMM) (20,21). Subsequently, parkin substrates are rapidly recognized by the adaptor protein p62/SQSTM1, which binds both the cargo and Atg8/LC3 on the vesicle membrane, thereby targeting mitochondria to autophagosomes for degradation (22–24). Dysfunctional parkin and/or defective cargo recognition prevent proper targeting of irreparably damaged mitochondria for mitophagy resulting in the buildup of toxic, dysfunctional mitochondria, ultimately leading to cell death (25,26).
Lysosomal storage disorders (LSDs) are a group of ∼50 inherited metabolic disorders that are caused by defects in lysosomal function (27,28). The impairment of both endocytic and autophagic pathways, two degradative membrane traffic routes to the lysosomes, is now recognized to play an important role in the pathogenesis of LSDs (29,30). Multiple sulfatase deficiency (MSD) is caused by mutations in the sulfatase modifying factor 1 gene (SUMF1), which is involved in the posttranslational modification of the catalytic site of sulfatases (31,32). The lack of conversion of a cysteine residue into α-formylglycine leads to the absence of all sulfatase activities and the subsequent accumulation of their specific substrates (33). The generation of a mouse model carrying a null mutation in Sumf1 gene (34) has allowed us to study in detail the molecular mechanisms underlying the pathogenesis of this severe type of LSD. We previously demonstrated that defective fusion between autophagosomes and lysosomes results in the buildup of mature unfused autophagic vesicles and in the accumulation of a variety of toxic products (30,35). The presence of dysfunctional mitochondria was also observed in MSD-derived mouse embryonic fibroblasts (MEFs) (30). Altered mitochondria due to autophagic defects have also been described in other LSDs, namely mucolipidosis II (ML II), mucolipidosis III (ML III) (36), mucolipidosis IV (ML IV) (37), GM1-gangliosidosis (GM1) (38,39) and neuronal ceroid-lipofuscinoses or Batten disease (NCL1) (40) among others. However, it is not clear how mitochondrial accumulation is mechanistically linked to autophagic stress. In particular, it has never been explored whether specific defects in mitophagy are the main cause of mitochondrial accumulation in different tissues.
Here, we have analyzed in detail the function of the mitophagic machinery in different tissues from a mouse model of MSD and determined to which extent its deregulation leads to mitochondrial alterations and tissue pathogenesis in LSDs.
RESULTS
Mitochondria release cytochrome c and trigger cell death in a tissue-specific manner in MSD mice
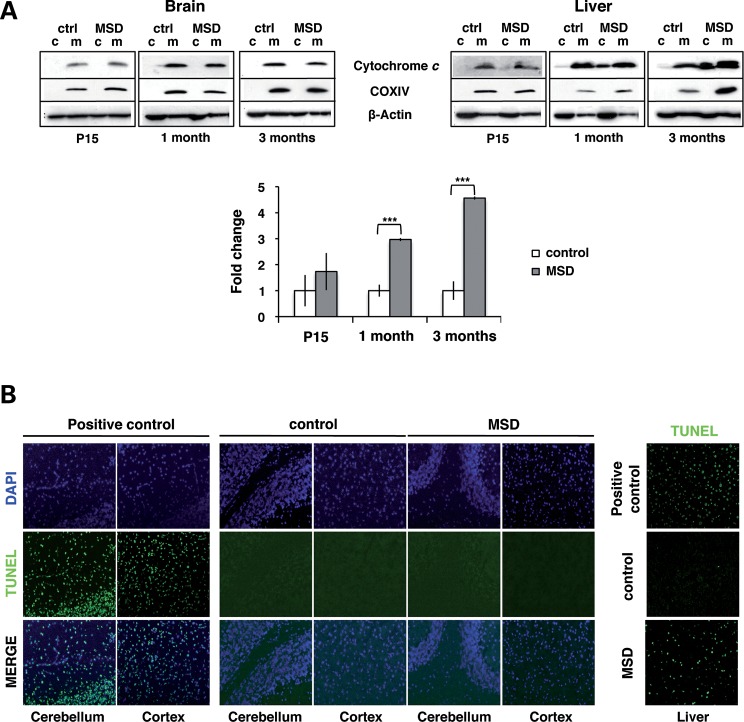
Mitochondria can compromise cell viability leading to tissue damage. An increase in the permeabilization of the mitochondrial membrane, also known as mitochondrial permeability transition, produces mitochondrial swelling, OMM rupture and release of pro-apoptotic factors such as cytochrome c. MSD mice display a multisystemic phenotype that involves both the central nervous system (CNS) and all main organs such as the liver. Glycosaminoglycan (GAG) accumulation, inflammation and cell death are detectable as soon as 15 days after birth and progressively increase with age. At 3 months of age, both the liver and CNS pathology are clearly manifested (30,34,41) (Supplementary Material, Figs S1 and S2). In particular, we evaluated whether mitochondria obtained from the brain and liver of MSD mice contributed to MSD pathology by releasing cytochrome c from the intermembrane space. The presence of cytochrome c was not detected in the cytosolic fractions of MSD brains at any of the time points analyzed (Fig. 1A). On the contrary, we observed high levels of cytochrome c in cytosolic fractions of MSD liver as soon as at 1 month of age.
Figure 1.
Mitochondria release cytochrome c and trigger cell death in MSD liver. (A) Anti-cytochrome c immunoblots of subcellular fractions (c, cytosol; m, mitochondria) obtained from the whole brain and liver from MSD (n = 4) and control mice (n = 4) at P15, 1 month and 3 months. COX IV was used as mitochondrial loading control. In the liver, levels of cytochrome c were quantified in each cytosolic fraction and expressed as the average percentage in terms of fold changes (bottom); **P < 0.01. (B) TUNEL analysis on fixed-paraffin brain and liver sections of MSD and control mice at 3 months. Images were acquired with the 20× magnification. Positive controls were obtained by treating tissue sections with DNAase I. TUNEL-positive cells were stained in green, and nuclei were labeled with 4′,6-diamidino-2-phenylindole (DAPI, blue).
To test whether the release of cytochrome c detected in the liver led to cell death, we performed an in situ apoptosis assay (terminal dUTP nick end labeling—TUNEL) on fixed-paraffin tissue sections of MSD and control mice at 3 months (Fig. 1B). As expected, few or no TUNEL-positive cells were detected in brain slices of MSD mice. This result correlates with the absence of cytochrome c in brain cytosolic fractions (Fig. 1A). On the other hand, MSD liver sections exhibited a high concentration of TUNEL-positive cells per area analyzed, thus suggesting that mitochondria might be the main effectors of cell death in this tissue.
Abnormal mitochondria accumulate in MSD liver in a time-dependent manner
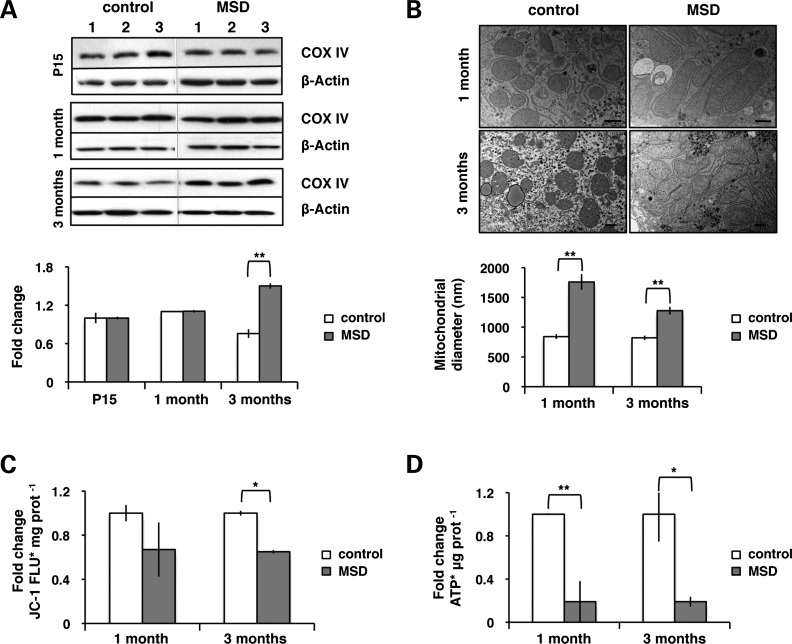
We then evaluated whether the mitochondrial release of cytochrome c was related to morphological and functional changes. We determined the levels of subunit IV of cytochrome oxidase (COX IV), widely used as a marker for quantifying the number of mitochondria (42). COX IV levels were measured in liver homogenates obtained from MSD (n= 3) and littermate control mice (n = 3) by western blot analysis. As shown in Figure 2A, significant changes were only observed at advanced stages, thus indicating that the accumulation of mitochondria is a progressive process in liver pathogenesis.
Figure 2.
Dysfunctional giant mitochondria accumulate in a time-dependent manner in MSD liver. (A) Levels of COX IV in liver total lysates of MSD (n = 3) and control (n = 3) mice at P15, 1 month and 3 months. The COX IV/actin ratio is expressed in terms of fold changes and represents an average value; **P < 0.01. (B) Analysis of mitochondrial size (diameter) using the AnalySIS software in liver ultrathin sections (70 nm) from MSD and control mice at 1 month and 3 months. Average number of mitochondria analyzed: control 1 month = 43, MSD 1 month = 56, control 3 months = 60, MSD 3 months = 127. Values are expressed in nm (scale bar = 400 nm). ANOVA P-value = 1.27 × 10−13, **P < 0.01. (C) Measurement of JC-1 fluorescence (FLU) per milligram (mg) of protein in isolated mitochondria from MSD (n = 3) and control (n = 3) mice at 1 month and 3 months. Values are expressed in terms of fold changes compared with control. ANOVA P-value = 0.048, *P < 0.05. (D) Analysis of ATP content per microgram (μg) of protein in isolated mitochondria from MSD (n = 3) and control (n = 3) mice at 1 month and 3 months. Values are expressed in terms of fold changes compared with control. ANOVA P-value = 4.81 × 10−3, *P < 0.05, **P < 0.01.
Next, we evaluated whether these mitochondria were morphologically and functionally altered. We used the AnalySIS software to measure the diameter of mitochondria in liver ultrathin sections. Mitochondria of MSD appeared significantly larger (giant) than in controls, thus indicating that morphological changes precede the accumulation of mitochondria in MSD liver (Fig. 2B).
Finally, we tested the functionality of liver mitochondria. We assessed the integrity of the mitochondrial membrane by using JC-1, a fluorescent cationic dye that accumulates in mitochondria with intact electrochemical gradient. We isolated mitochondria from livers of MSD (n= 3) and control mice (n= 3) and measured the incorporation of JC-1. A decrease in mitochondrial membrane potential was observed as soon as 1 month and became evident at advanced stages of the pathology (Fig. 2C). As mitochondria are the main source of energy, we measured the amount of adenosine triphosphate (ATP) in mitochondrial fractions. Isolated mitochondria from MSD liver displayed a striking decrease in ATP content (Fig. 2D).
Together, these data indicate that mitochondrial functional alterations and morphological changes appear simultaneously and are both associated to the release of cytochrome c and to liver pathology (Table 1).
Table 1.
Mitochondrial morphology and function in MSD liver
| Mitochondria | Number | Morphology | Functionality | Cytochrome crelease |
|---|---|---|---|---|
| Liver pathology | ||||
| P15 | = | – | – | No |
| 1 month | = | Giant | =JC-1FLU < ATPcontent | Yes |
| 3 months | > | Giant | <JC-1 FLU < ATPcontent | Yes |
Mitochondria accumulate outside autophagosomes
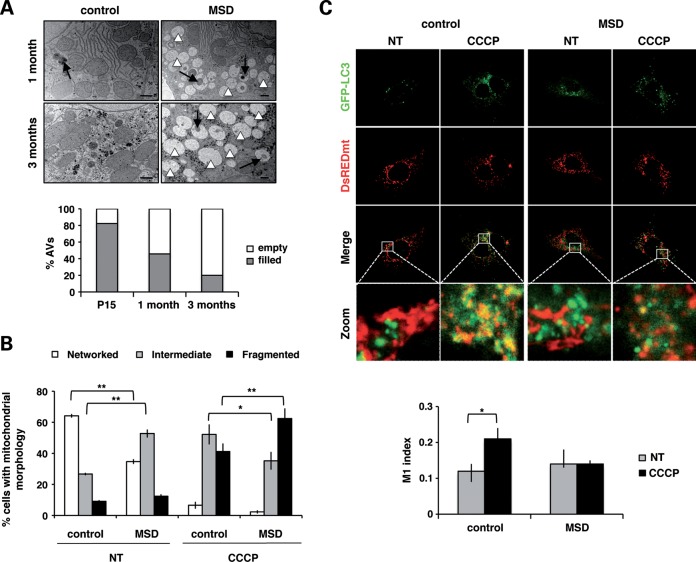
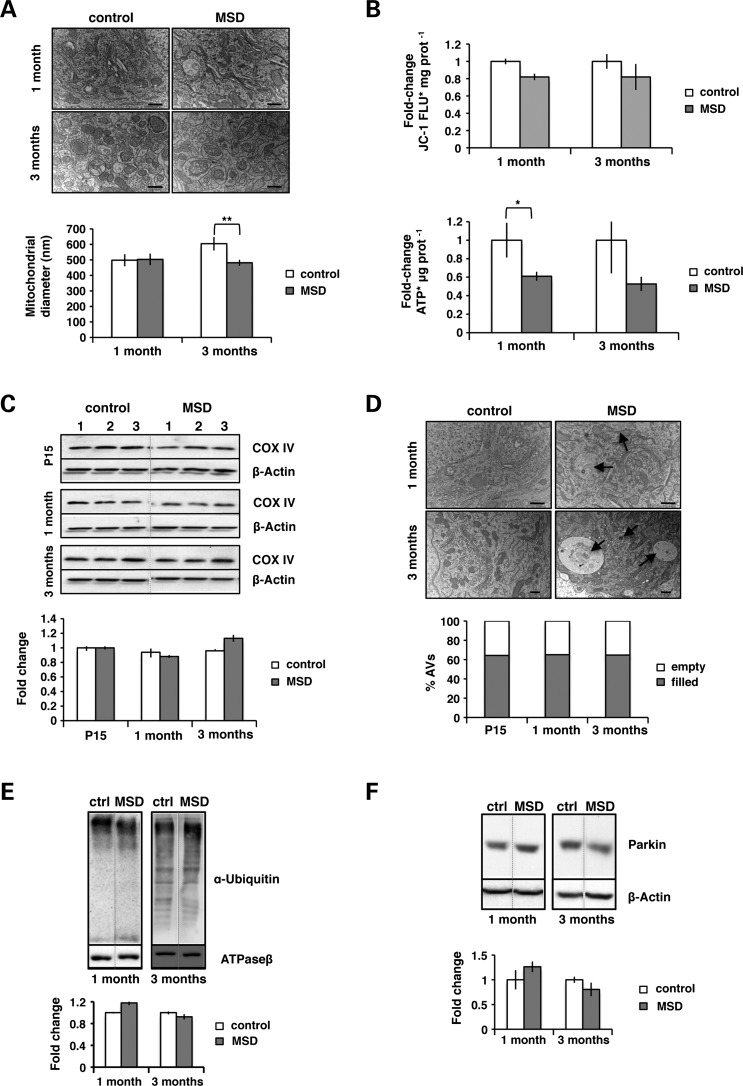
Mitophagy recycles abnormal mitochondria through their targeting and encapsulation by autophagosomes. To assess the functionality of this process, we analyzed the morphology and content of autophagosomes by electron microscopy. In MSD mice, qualitative analysis of liver ultrathin sections revealed the presence of autophagic vesicles along with fuzzy content. The number of vesicles with no-recognizable structures, called ‘empty’ autophagosomes, increased over time, thus suggesting a progressive impairment of autophagic cargo sequestration (Fig. 3A).
Figure 3.
Mitochondria accumulate outside autophagosomes. (A) Electron microscopy analysis of autophagosome (AV) content in liver ultrathin sections (70 nm) at different ages: P15, 1 month and 3 months. Qualitatively, autophagosomes of MSD liver are classified into two categories: empty AVs (white arrowheads) or filled AVs (black arrows) (scale bar = 400 nm). Values are expressed as percentages of the total number of autophagosomes. (B) Analysis of mitochondrial morphology in untreated (NT) and CCCP-treated MSD and control MEFs. Cells were classified into three categories: networked (ANOVA P-value = 3.91 × 10−12), intermediate (ANOVA P-value = 1.24 × 10−3) and fragmented (ANOVA P-value = 6.77 × 10−7). Results are expressed as the percentage of cells in each category; *P < 0.05; **P < 0.01. (C) Confocal images (63×) of untreated (NT) and CCCP-treated (20 μm, 20 h) MSD and control MEFs co-expressing GFP-LC3 (green) and DsREDmito (red) plasmids. The zoomed region is indicated by a white square. Co-localization index is expressed with the M1 (Mander's) coefficient. ANOVA P-value = 0.33, *P < 0.05.
To confirm these results, we assessed mitochondrial sequestration in both control and MSD MEFs that transiently overexpress GFP-LC3 (autophagic marker) and DsREDmito (mitochondrial marker) plasmids. Cells were treated with carbonyl cyanide 3-chlorophenylhydrazone (CCCP; 20 μm, 20 h), a mitochondrial-uncoupling agent that induces membrane depolarization and promotes mitochondrial elimination by autophagy. We analyzed mitochondrial morphology in both treated and untreated cells to ensure that CCCP was able to induce mitochondrial fragmentation in our cell line. Cells were classified in three different categories according to the mitochondrial morphology visualized with DsREDmito: ‘networked’, that included cells with the typical filamentous-elongated mitochondrial morphology; ‘fragmented’, that include cells with mitochondria showing a clear round-shaped morphology; and ‘intermediate’, that include cells with an undefined pattern (some networked and some fragmented). In steady-state conditions, a high percentage of MSD cells presented an intermediate morphology, whereas the great majority of control cells had a networked pattern. Upon CCCP treatment, mitochondria from both MSD and control cells appeared fragmented, thus indicating that CCCP is able to promote membrane depolarization in both types of cells (Fig. 3B).
We next analyzed LC3-mito co-localization in both untreated (NT) and CCCP-treated MSD and control MEFs. In control cells, CCCP treatment resulted in a significant increase in LC3-mito co-localization. Inversely, CCCP treatment did not result in any increase in LC3-mito co-localization in MSD cells, thus confirming a defect in mitochondrial sequestration by autophagosomes (Fig. 3C).
These results indicate that autophagosomes are not able to encircle depolarized mitochondria in MSD.
Impaired mitochondrial priming in MSD is due to insufficient parkin-mediated mitochondrial ubiquitination
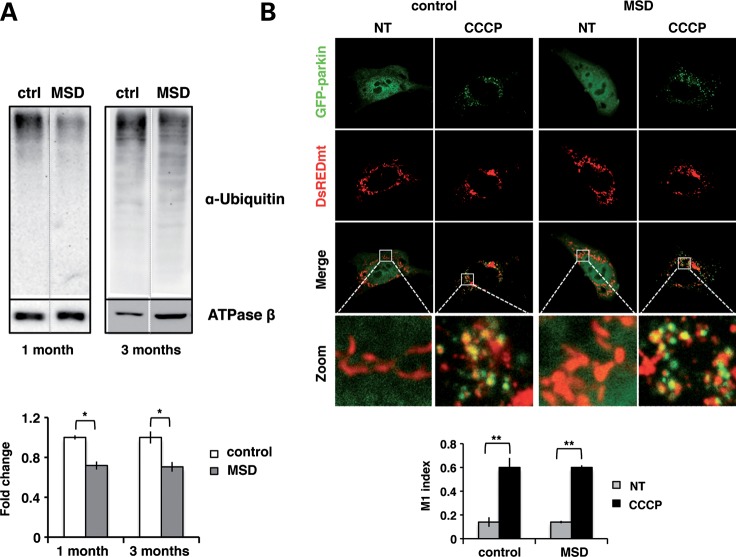
Mitochondrial targeting to autophagosomes relies on parkin-mediated ubiquitination of OMM proteins. This step is crucial for proper mitochondrial recognition and degradation by autophagy. Therefore, we analyzed the efficiency of parkin-mediated ubiquitination in MSD mice. To date, two different OMM proteins have been described as parkin substrates for mitochondrial autophagy: VDAC1 (20) and MFN1 (21). However, the involvement of other OMM proteins cannot be excluded. For this reason, we determined the total amount of ubiquitinated proteins in mitochondrial extracts obtained from livers of MSD (n = 4) and control mice (n = 4) (Fig. 4A). Reduced levels of ubiquitinated proteins per mitochondria (ATPase β content) were detected in MSD livers compared with controls, suggesting that incomplete/partial ubiquitination of mitochondrial proteins leads to inefficient cargo recognition and autophagosome mistargeting.
Figure 4.
Incomplete ubiquitination is not due to impaired parkin translocation to mitochondria. (A) Anti-ubiquitin immunoblots of liver mitochondrial extracts obtained from MSD (n = 4) and control (n = 4) mice at 1 month and 3 months. ATPase β was used as mitochondrial loading control and results were expressed as the fold changes of the ubiquitin/ATPase β ratio; *P < 0.05. (B) Confocal images (63×) of untreated (NT) and CCCP-treated (20 μm, 20 h) MSD and control MEFs co-expressing GFP-parkin (green) and DsREDmito (red) plasmids. The zoomed region is indicated with a white square. Co-localization is expressed as the M1 (Mander's) index. ANOVA P-value = 0.158, **P < 0.01.
We reasoned that incomplete mitochondrial ubiquitination may be due to either inefficient parkin translocation and/or to reduced levels of parkin protein/activity. To address these possibilities, we transiently co-expressed GFP-parkin and DsREDmito plasmids in MSD and control MEFs, which were then incubated in the presence/absence of the mitochondrial uncoupler CCCP (20 μm, 20 h). As shown in Figure 4B, parkin localization was entirely cytoplasmic in both MSD and control untreated cells. Upon CCCP treatment, a massive depolarization of mitochondria induced the translocation of large amounts of parkin in both control and MSD MEFs, indicating that parkin is properly recruited to depolarized mitochondria in MSD cells.
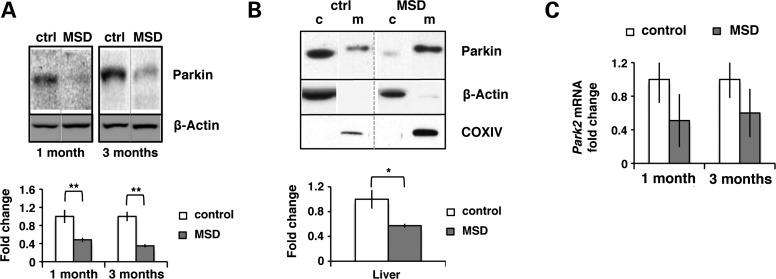
Once we confirmed the proper recruitment of parkin in vitro, we evaluated parkin levels in liver total homogenates obtained from MSD (n = 4) and control mice (n = 4). We found that parkin levels were strongly reduced in MSD liver (Fig. 5A). Similar data were obtained in mitochondrial fractions (Fig. 5B).
Figure 5.
Reduced parkin levels are not a consequence of low Park2 expression. (A) Parkin total levels in liver homogenates from MSD (n= 4) and control (ctrl) mice (n= 4) at 1 month and 3 months. Relative levels are expressed as the Parkin/actin ratio in terms of fold changes; **P < 0.01. (B) Parkin distribution in subcellular fractions (c, cytosol; m, mitochondria) obtained from liver of MSD (n= 4) and control (ctrl) mice (n= 4) at 3 months. Mitochondrial levels were normalized with COX IV (mitochondrial loading control) and the Parkin/COX IV ratio was expressed as fold changes; *P < 005. (C) Park2 mRNA relative levels in MSD liver (n= 3) at 1 month and 3 months obtained by quantitative PCR. Values are expressed as fold changes compared with control; ANOVA P-value = 0.2.
Our results indicate that despite parkin is able to efficiently translocate to mitochondria, its levels are strongly reduced in MSD liver, thus resulting in an incomplete mitochondrial ubiquitination. Reduced parkin levels are likely due to enhanced degradation/decrease stability as no significant differences were observed in Park2 relative expression (Fig. 5C).
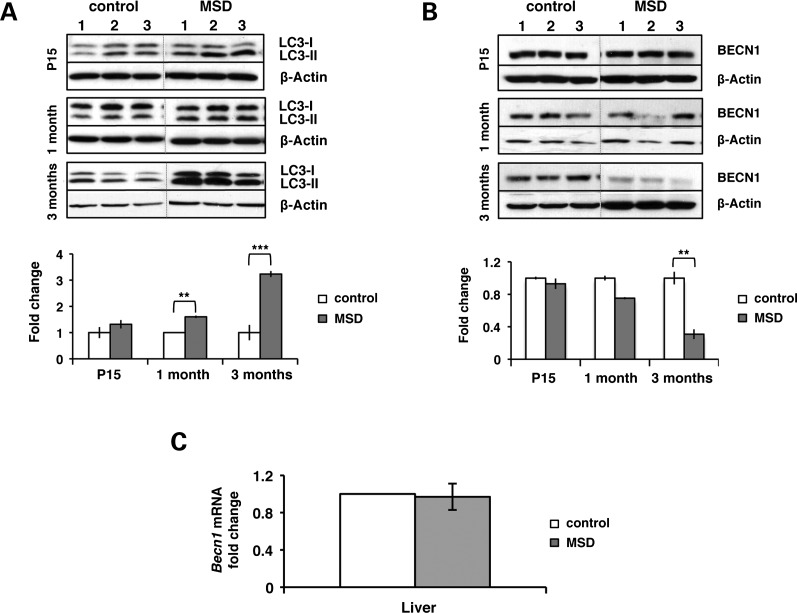
Autophagy induction is inhibited in MSD liver
We monitored the levels of LC3-II in MSD liver at different time points and found that the LC3-II/actin ratio was significantly increased compared with control (Fig. 6A). These data are consistent with the impairment of autophagosome maturation already reported in MSD (30,35). Interestingly, LC3-II accumulation was not associated with the induction of beclin-1 (BECN-1) at this stage of pathology. On the contrary, beclin-1 protein levels were reduced, whereas its mRNA levels remained unaltered (Fig. 6B and C).
Figure 6.
Autophagy is inhibited at advanced stages of MSD liver pathology. (A) LC3 immunoblots on total liver homogenates from MSD (n= 3) and control mice (n= 3) at P15, 1 month and 3 months. Autophagosome accumulation is expressed as the fold change of the LC3-II/actin ratio compared with control; **P < 0.01, ***P < 0.005. (B) Levels of BECN-1 were measured in liver extracts from MSD (n= 3) and control mice (n= 3) at P15, 1 month and 3 months. The BECN-1/actin ratio is expressed as fold changes; **P < 0.01. (C) Becn1 mRNA levels were determined by real-time PCR in liver samples (n= 3) at 3 months.
Therefore, autophagy is downregulated in MSD liver, which might contribute to the mistargeting and subsequent accumulation of mitochondria by limiting the formation of new autophagic vesicles.
Mitochondrial alterations in MSD brain are not associated with defects in mitochondrial priming machinery
As reported in Figure 1, mitochondria from MSD brain do not release cytochrome c. Further morphological and functional analyses revealed that, in contrast to what observed in liver, mitochondria became fragmented (Fig. 7A) and showed reduced membrane integrity and ATP content (Fig. 7B). However, these changes were not as evident as those observed in the liver, indicating a milder mitochondrial phenotype. No alterations in the number of mitochondria were detected at any of the time points analyzed (Fig. 7C), suggesting an efficient mitochondrial removal in this tissue. In fact, the analysis of the content of autophagosomes showed that the percentage of empty mitochondria represented a small fraction and remained constant over time (Fig. 7D). Consistently, we did not find any alteration in the levels of ubiquitinated proteins in brain mitochondrial fractions (Fig. 7E). Moreover, parkin total levels were normal, thus confirming the proper functioning of the mitochondrial priming machinery in MSD brain.
Figure 7.
Altered mitochondria are a late-onset feature of MSD brain pathology. (A) Analysis of mitochondrial size (diameter) using the AnalySIS software in brain (cerebellum area) ultrathin sections (70 nm) from MSD and control mice at 1 month and 3 months. Average number of mitochondria analyzed: control 1 month = 60, MSD 1 month = 79, control 3 months = 62, MSD 3 months = 158. Values are expressed in nm (scale bar = 400 nm). ANOVA P-value = 6 × 10−3, **P < 0.01. (B) Top: measurement of JC-1 fluorescence (FLU) per milligram (mg) of protein in isolated mitochondria from MSD (n = 3) and control (n = 3) mice at 1 month and 3 months, ANOVA P-value = 0.065. Bottom: analysis of ATP content per microgram (μg) of protein in isolated mitochondria from MSD (n = 3) and control (n = 3) mice at 1 month and 3 months, ANOVA P-value = 0.039. Values are expressed in term of fold changes compared with control; *P < 0.05. (C) Levels of COX IV in brain total lysates of MSD (n = 3) and control (n = 3) mice at P15, 1 month and 3 months. The COX IV/actin ratio is expressed as fold changes. (D) Electron microscopy analysis of autophagosome (AV) content in brain ultrathin sections (70 nm) at different ages: P15, 1 month and 3 months. Qualitatively, autophagosomes of MSD tissues are classified into two categories: empty AVs or filled AVs (black arrows) (scale bar = 400 nm). Values are expressed as percentages of the total number of autophagosomes. (E) Anti-ubiquitin immunoblots of brain mitochondrial extracts obtained from MSD (n = 4) and control mice (n = 4) at 1 month and 3 months. ATPase β was used as mitochondrial loading control and values were expressed as the ubiquitin/ATPase β ratio (fold changes). (F) Parkin total levels in brain homogenates from MSD (n = 4) and control mice (n = 4) at 1 month and 3 months. Values are expressed as the Parkin/actin ratio (fold changes).
DISCUSSION
LSDs are metabolic disorders due to mutations in the genes encoding specific proteins involved in lysosomal function, resulting in the accumulation of undegraded material. The degree of severity of the disease depends on the type of storage material and the site of accumulation that generally affects vital organs such as the brain and liver (27). In LSDs, the correlation between enzyme deficiency and the type of storage are well characterized. However, the molecular pathways leading to LSD pathology remain largely unexplored. LSDs have been recently listed as autophagy disorders. In fact, defects in autophagosome formation and maturation have been extensively associated to LSD pathogenesis (30,35–38,40,43). However, whether any of the steps of mitophagy is defective and whether this may be responsible for the mitochondrial phenotype has never been addressed in LSDs. Moreover, how defects in the autophagic pathway are linked to cell death and tissue damage has not been elucidated yet.
In this study, we demonstrate for the first time that the accumulation of mitochondria observed in LSDs is not a mere consequence of defective autophagosome–lysosome fusion but involves a more specific mechanism that mediates mitochondrial priming and targeting to autophagosomes. The analysis of both parkin expression and activity in two affected tissues of MSD mice revealed tissue-specific defects in MSD mice. In fact, reduced parkin protein levels were exclusively detected in MSD liver samples and not in MSD brains at all time points analyzed. In line with this, the reduction of parkin resulted in an incomplete/partial ubiquitination of mitochondrial proteins on the OMM despite the presence of an efficient parkin translocation, as shown in parkin-overexpressing MSD MEFs. As no differences in the Park2 mRNA relative expression were detected, we speculate that low parkin levels in MSD liver may be due to enhanced parkin degradation/reduced stability. Interestingly, it has been reported that parkin degradation is controlled by another E3 ubiquitin ligase called Nrdp1, whose over-expression has been described to significantly reduce the half-life of parkin from 5 to 2.5 h (44). Further studies must be performed in order to understand the real causes of parkin reduction and whether it can be directly correlated to lysosomal dysfunction.
A defective sequestration of mitochondria by autophagosomes was observed as a consequence of impaired mitochondrial priming. A high percentage of empty vesicles was reported in MSD livers, whereas only a small percentage of autophagosomes appeared empty in the brain. Co-localization analysis of GFP-LC3/DsREDmito in MSD and control MEFs confirmed the defective turnover of mitochondria by autophagic vesicles. The pathologic accumulation of autophagosomes was previously reported in MSD brain but not in the liver (30). Here, we show that the accumulation of autophagosomes is a hallmark of liver pathology in MSD mice. Moreover, we observed that the accumulation of autophagic vesicles in this tissue is due to defective autophagosome maturation and not to an enhanced induction of autophagy. Accordingly, unaltered levels of Beclin-1 were observed at P15 and 1-month MSD liver homogenates. Interestingly, at advanced stages of the pathology, Beclin-1 protein levels but not mRNA levels appeared reduced. In fact, it has been previously described that the mitochondrial proteins Bcl-2 and Bcl-XL negatively regulate autophagy by binding to Beclin-1 (45). Therefore, we hypothesize that autophagy inhibition together with a defect in autophagosome maturation may contribute to the phenotype observed in the liver by limiting the availability of new autophagic vesicles.
As a consequence of impaired mitophagy, non-recycled mitochondria accumulate in MSD liver in a time-dependent manner. Further morphological analysis of MSD livers revealed an increase in mitochondrial size as soon as P15. Mitochondrial enlargement can be a consequence of (1) defective fission, (2) a high rate of fusion and (3) impaired mitophagy. However, in the majority of cases, the presence of giant mitochondria is a consequence of defective mitochondrial turnover (15,46). Functional assays based on the measurement of ATP production and membrane integrity showed that mitochondria from MSD liver had reduced ATP content and low membrane potential. Dysfunctional mitochondria are potentially harmful for cell viability. In fact, the release of pro-apoptotic proteins, such as cytochrome c, was observed in MSD livers as soon as at 1 month of age (Table 1). The detection of a high number of TUNEL-positive cells confirmed the contribution of non-removed dysfunctional mitochondria to liver pathology in MSD mice. Contrary to what observed in the liver, mitochondrial alterations in MSD brain were milder and appeared at later stages of the pathology. Furthermore, although loss of Purkinje cells has been described in MSD mice as soon as 2 months (34), neither cytochrome c release nor TUNEL-positive cells were detected at any of the stages analyzed. Therefore, mitochondrial morphological and functional changes cannot be directly correlated with defects in the mitochondrial priming machinery but may be associated to different pathogenic cascades and/or toxic stimuli.
The differences observed between the brain and liver may be explained by different energetic requirements and organ cell composition. Neurons are highly dependent on ATP production and consume more energy than other cells for their metabolism. Moreover, mitochondria play an important role in synaptic maintenance. For this reason, post-mitotic cells are expected to be particularly sensitive to changes in mitochondrial activity and therefore, mitochondrial turnover would reasonably be a very tightly controlled event in neurons through alternative quality control mechanisms. However, in all brain experiments, we used whole brain samples, so it may be possible that some region-specific differences may have been masked. Therefore, we cannot exclude that a more severe mitochondrial phenotype may occur in specific brain areas.
This study demonstrates a tissue-specific impairment of mitochondrial priming machinery in LSDs, resulting in the accumulation of dysfunctional mitochondria ultimately leading to cell death. These tissue-specific pathogenic mechanisms should be taken in consideration when developing therapeutic strategies aimed at treating brain and systemic lesions in LSDs.
MATERIALS AND METHODS
Antibodies, plasmids and reagents
The antibodies used in this work were: rabbit anti-COX IV (IB 1:1000, #4844, Cell Signaling Technology, Boston, MA, USA), mouse anti-Cytochrome c (IB 1:1000, Cat. 556433, BD Pharmingen, NJ, USA), rabbit anti-LC3 (IB 1:1000, NB100-2331, Novus Biologicals, CO, USA), mouse anti-Parkin (IB 1:1000, #4211, Cell Signaling Technology, NJ, USA), rabbit anti-Ubiquitin (IB 1:500, Z0458, Dako Cytomation, CA, USA), mouse anti-ATPase β subunit (IB 1:10 000, ab14730, Abcam, Cambridge, UK), rabbit anti-BECN1 (IB 1:1000, sc11427, Santa Cruz Biotech, CA, USA.) and mouse anti-β-actin (1:10 000, A1978, Sigma-Aldrich, MO, USA). Secondary horseradish peroxidase-conjugated antibodies (anti-rabbit 1:3000 and anti-mouse 1:5000) were from GE Healthcare (UK).
Plasmids GFP-LC3 (37), DsREDmito and GFP-myc-Parkin were a kind gift of Prof. Luca Scorrano (University of Geneva) and Prof. Wolfdieter Springer (University of Tübingen), respectively.
Reagents: CCCP (C2759) was from Sigma-Aldrich. For plasmid transfections, we used Lipofectamine 2000 (Invitrogen).
Animal experiments
Mice were anesthetized with avertin (20 μl/g weight) and then perfused with phosphate-buffered saline (PBS) before tissue collection.
Animal use and analyses were conducted in accordance with the guidelines of the Animal Care and Use Committee of Cardarelli Hospital in Naples and authorized by the Italian Ministry of Health.
Quantification of GAGs
Liver tissue samples were mechanically homogenized in water using the TissueLyser equipment (Qiagen, Germany). Proteins were quantified by the Bradford method (Biorad, CA, USA), and samples were diluted to concentrations equal to 200 μg/ml of total proteins to run on the assay. Samples were then incubated with dimethylmethylene blu–Tris/formate, and colorimetry was immediately measured in an enzyme-linked immunosorbent assay reader plate at 520 nm. Several concentrations of dermatan sulfate were used to calculate the standard curve.
Real-time polymerase chain reaction
Total RNA was extracted from tissue using the RNeasy® Kit and retrotranscribed with QuantiTect® Reverse Transcription Kit (Qiagen) following the manufacturer's instructions. Amplification of cDNA was performed using the SYBR Green I Master for LightCycler® 480 (Roche, Switzerland). Primer sequences were designed using the GenScript Primer Design tool GenScript Primer Design tool (https://www.genscript.com/ssl-bin/app/primer). β-Actin was used as a housekeeping gene.
Protein extraction for immunoblotting
For total protein extracts, tissue samples were homogenized in cold lysis buffer [50 mm Tris–HCl, pH 7.4, 150 mm NaCl, 1 mm ethylenediaminetetraacetic acid (EDTA), 1% Triton X-100, 0.1% sodium dodecyl sulfate (SDS), 0.5% sodium deoxycholate] in the presence of protease inhibitors (Sigma). Mechanical disruption of samples was performed using the TissueLyser equipment (Qiagen). Homogenates were incubated on ice for 30 min and then centrifuged at maximum speed to eliminate cellular debris. Proteins were quantified by the Bradford method (Biorad).
Subcellular fractionation was carried out by homogenization of tissue samples in an ice-cold hypotonic buffer [300 mm sucrose, 10 mm 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), pH 7.4, 200 nm EDTA] in the presence of protease inhibitors (Sigma). Samples were mechanically disrupted with micropestles (Eppendorf, Germany) and then passed through a 20G- and a 26G-needle for several times. The same amount of protein (1 mg) was processed in an equal volume of buffer for all samples. The supernatant of the first centrifugation (5 min, 600 g) was further centrifuged at 16 000g for 15 min. Supernatants were stored as the cytosolic fraction. Pellets were washed in buffer and re-centrifuged. The resultant pellets (mitochondria) were resuspended in the same volume of lysis buffer. For the analysis of cytochrome c release by immunoblotting, same amounts of samples were loaded in a SDS–polyacrylamide gel electrophoresis (PAGE) gel.
Sodium dodecyl sulfate—polyacrylamide gel electrophoresis
Same amounts of protein (20–50 μg) were loaded in an SDS–PAGE gel and run was performed at 80 V for at least 3 h. Blotting was performed at 4°C overnight at a constant voltage (30 V). Membranes were blocked in Tris-buffered saline/T 5% milk for 1 h at room temperature (RT). Primary antibody was then incubated for 1–2 h at RT followed by the incubation with an HPR-conjugated secondary antibody (1 h at RT). Detection was performed using the ECL substrate (Pierce).
Electron microscopy
Tissue samples were fixed in 1% glutaraldehyde in 0.2 m HEPES buffer and post-fixed in OsO4 and uranyl acetate. After dehydration in graded series of ethanol, tissue samples were cleared in propylene oxide, embedded in Epoxy resin (Epon 812) and polymerized at 60°C for 24 h. For each sample, ultrathin sections (70 nm) were cut with a Leica EM UC6 ultramicrotome. Electron microscopy images were acquired using a FEI Tecnai-12 electron microscope equipped with an ULTRA VIEW CCD digital camera (FEI, Eindhoven, the Netherlands). Measurement of mitochondrial size (diameter) was performed using the AnalySIS software (Soft Imaging Systems GmbH, Munster, Germany).
Mitochondrial functional assays
Tissue mitochondria were isolated following the Mitochondria Isolation Kit protocol (MITOISO1; Sigma). Mitochondrial membrane potential was determined by incubation of isolated mitochondria with the JC-1 dye according to the manufacturer's instructions (Sigma). ATP content was measured using the ATP Determination Kit (A22066; Invitrogen, CA, USA).
Confocal microscopy
MEFs were transfected with GFP-LC3, DsREDmito and GFP-myc-Parkin. Twenty-four hours after transfection, cells were treated with CCCP (20 μm) for 20 h. After treatment, cells were gently washed with PBS, fixed in 4% paraformaldehyde (PFA) and mounted with vectashield (Vector Labs, CA, USA).
Images were taken using the 63×/1.4 Oil DiC M27 objective on a Zeiss LSM 710 confocal microscope. Laser lines at 488 nm (GFP-LC3, GFP-Parkin) and 561 nm (DsREDmito) were used.
Co-localization analysis was performed using the JaCoP plugin (just another co-localization plugin) for Image J. The results were expressed as the M1 coefficient (Mander's).
TUNEL assay
Paraffin-embedded tissues were sectioned (7 μm) in a Leica RM165 microtome and fixed in 4% PFA. Apoptotic nuclei were detected using the In Situ Cell Death Detection Kit (Roche) following the manufacturer's instructions.
Data analysis
All blots were quantified using the Image J software, and results were analyzed by Student's t-distribution. An analysis of variance (ANOVA) test (two-way ANOVA) was also used for mitochondrial morphological and functional assays, real-time PCRs and co-localization experiments. Results were expressed as the average of three or more independent experiments. A P-value <0.05 was considered statistically significant.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the Italian Telethon Foundation (R.dP., A.S., E.V.P., E.N., A.F. and A.B.); the European Research Council Advanced Investigator grant number 250154 (A.B.); the European Commission under the FP7 EUCLYD project (Grant No. HEALTH-2007-A-201678); the Beyond Batten Disease Foundation (A.B.); and the MPS Society (A.F. and A.B.). Funding to pay the Open Access publication charges for this article was provided by the Fondazione Telethon.
Supplementary Material
ACKNOWLEDGEMENTS
We thank D.C. Rubinsztein, G. Diez-Roux, A. Auricchio, A. De Matteis and C. Settembre for comments on the manuscript and A. Carissimo for statistical analysis. We thank L. Scorrano and W. Springer for material support (see Materials and Methods).
Conflict of Interest statement. The authors declare that they have no competing financial interests.
REFERENCES
- 1.Klionsky D.J., Emr S.D. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–1721. doi: 10.1126/science.290.5497.1717. doi:10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eskelinen E.L., Saftig P. Autophagy: a lysosomal degradation pathway with a central role in health and disease. Biochim. Biophys. Acta. 2009;1793:664–673. doi: 10.1016/j.bbamcr.2008.07.014. doi:10.1016/j.bbamcr.2008.07.014. [DOI] [PubMed] [Google Scholar]
- 3.Brunk U.T., Terman A. The mitochondrial–lysosomal axis theory of aging: accumulation of damaged mitochondria as a result of imperfect autophagocytosis. Eur. J. Biochem. 2002;269:1996–2002. doi: 10.1046/j.1432-1033.2002.02869.x. doi:10.1046/j.1432-1033.2002.02869.x. [DOI] [PubMed] [Google Scholar]
- 4.Tatsuta T., Langer T. Quality control of mitochondria: protection against neurodegeneration and ageing. EMBO J. 2008;27:306–314. doi: 10.1038/sj.emboj.7601972. doi:10.1038/sj.emboj.7601972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim I., Rodriguez-Enriquez S., Lemasters J.J. Selective degradation of mitochondria by mitophagy. Arch. Biochem. Biophys. 2007;462:245–253. doi: 10.1016/j.abb.2007.03.034. doi:10.1016/j.abb.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kissova I., Deffieu M., Manon S., Camougrand N. Uth1p is involved in the autophagic degradation of mitochondria. J. Biol. Chem. 2004;279:39068–39074. doi: 10.1074/jbc.M406960200. doi:10.1074/jbc.M406960200. [DOI] [PubMed] [Google Scholar]
- 7.Kanki T., Wang K., Cao Y., Baba M., Klionsky D.J. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev. Cell. 2009;17:98–109. doi: 10.1016/j.devcel.2009.06.014. doi:10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okamoto K., Kondo-Okamoto N., Ohsumi Y. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev. Cell. 2009;17:87–97. doi: 10.1016/j.devcel.2009.06.013. doi:10.1016/j.devcel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 9.Novak I., Kirkin V., McEwan D.G., Zhang J., Wild P., Rozenknop A., Rogov V., Lohr F., Popovic D., Occhipinti A., et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11:45–51. doi: 10.1038/embor.2009.256. doi:10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keating D.J. Mitochondrial dysfunction, oxidative stress, regulation of exocytosis and their relevance to neurodegenerative diseases. J. Neurochem. 2008;104:298–305. doi: 10.1111/j.1471-4159.2007.04997.x. [DOI] [PubMed] [Google Scholar]
- 11.Jiang X., Wang X. Cytochrome c-mediated apoptosis. Annu. Rev. Biochem. 2004;73:87–106. doi: 10.1146/annurev.biochem.73.011303.073706. doi:10.1146/annurev.biochem.73.011303.073706. [DOI] [PubMed] [Google Scholar]
- 12.Seo A.Y., Joseph A.M., Dutta D., Hwang J.C., Aris J.P., Leeuwenburgh C. New insights into the role of mitochondria in aging: mitochondrial dynamics and more. J. Cell. Sci. 2010;123:2533–2542. doi: 10.1242/jcs.070490. doi:10.1242/jcs.070490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karbowski M., Youle R.J. Dynamics of mitochondrial morphology in healthy cells and during apoptosis. Cell Death Differ. 2003;10:870–880. doi: 10.1038/sj.cdd.4401260. doi:10.1038/sj.cdd.4401260. [DOI] [PubMed] [Google Scholar]
- 14.Lee Y.J., Jeong S.Y., Karbowski M., Smith C.L., Youle R.J. Roles of the mammalian mitochondrial fission and fusion mediators Fis1, Drp1, and Opa1 in apoptosis. Mol. Biol. Cell. 2004;15:5001–5011. doi: 10.1091/mbc.E04-04-0294. doi:10.1091/mbc.E04-04-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Twig G., Elorza A., Molina A.J., Mohamed H., Wikstrom J.D., Walzer G., Stiles L., Haigh S.E., Katz S., Las G., et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. doi:10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vives-Bauza C., Zhou C., Huang Y., Cui M., de Vries R.L., Kim J., May J., Tocilescu M.A., Liu W., Ko H.S., et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc. Natl Acad. Sci. USA. 2010;107:378–383. doi: 10.1073/pnas.0911187107. doi:10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsuda N., Sato S., Shiba K., Okatsu K., Saisho K., Gautier C.A., Sou Y.S., Saiki S., Kawajiri S., Sato F., et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J. Cell Biol. 2010;189:211–221. doi: 10.1083/jcb.200910140. doi:10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Narendra D.P., Jin S.M., Tanaka A., Suen D.F., Gautier C.A., Shen J., Cookson M.R., Youle R.J. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. doi:10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanaka A. Parkin-mediated selective mitochondrial autophagy, mitophagy: Parkin purges damaged organelles from the vital mitochondrial network. FEBS Lett. 2010;584:1386–1392. doi: 10.1016/j.febslet.2010.02.060. doi:10.1016/j.febslet.2010.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geisler S., Holmstrom K.M., Skujat D., Fiesel F.C., Rothfuss O.C., Kahle P.J., Springer W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. doi:10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 21.Ziviani E., Tao R.N., Whitworth A.J. Drosophila parkin requires PINK1 for mitochondrial translocation and ubiquitinates mitofusin. Proc. Natl Acad. Sci. USA. 2010;107:5018–5023. doi: 10.1073/pnas.0913485107. doi:10.1073/pnas.0913485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kirkin V., McEwan D.G., Novak I., Dikic I. A role for ubiquitin in selective autophagy. Mol. Cell. 2009;34:259–269. doi: 10.1016/j.molcel.2009.04.026. doi:10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 23.Okatsu K., Saisho K., Shimanuki M., Nakada K., Shitara H., Sou Y.S., Kimura M., Sato S., Hattori N., Komatsu M., et al. p62/SQSTM1 cooperates with Parkin for perinuclear clustering of depolarized mitochondria. Genes Cells. 2010;15:887–900. doi: 10.1111/j.1365-2443.2010.01426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Narendra D., Kane L.A., Hauser D.N., Fearnley I.M., Youle R.J. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy: VDAC1 is dispensable for both. Autophagy. 2010;6:1090–1106. doi: 10.4161/auto.6.8.13426. doi:10.4161/auto.6.8.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Laar V.S., Berman S.B. Mitochondrial dynamics in Parkinson's disease. Exp. Neurol. 2009;218:247–256. doi: 10.1016/j.expneurol.2009.03.019. doi:10.1016/j.expneurol.2009.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martinez-Vicente M., Talloczy Z., Wong E., Tang G., Koga H., Kaushik S., de Vries R., Arias E., Harris S., Sulzer D., et al. Cargo recognition failure is responsible for inefficient autophagy in Huntington's disease. Nat. Neurosci. 2010;13:567–576. doi: 10.1038/nn.2528. doi:10.1038/nn.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Futerman A.H., van Meer G. The cell biology of lysosomal storage disorders. Nat. Rev. Mol. Cell Biol. 2004;5:554–565. doi: 10.1038/nrm1423. doi:10.1038/nrm1423. [DOI] [PubMed] [Google Scholar]
- 28.Neufeld E.F. Lysosomal storage diseases. Annu. Rev. Biochem. 1991;60:257–280. doi: 10.1146/annurev.bi.60.070191.001353. doi:10.1146/annurev.bi.60.070191.001353. [DOI] [PubMed] [Google Scholar]
- 29.Ballabio A., Gieselmann V. Lysosomal disorders: from storage to cellular damage. Biochim. Biophys. Acta. 2009;1793:684–696. doi: 10.1016/j.bbamcr.2008.12.001. doi:10.1016/j.bbamcr.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 30.Settembre C., Fraldi A., Jahreiss L., Spampanato C., Venturi C., Medina D., de Pablo-Latorre R., Tacchetti C., Rubinsztein D.C., Ballabio A. A block of autophagy in lysosomal storage disorders. Hum. Mol. Genet. 2008;17:119–129. doi: 10.1093/hmg/ddm289. doi:10.1093/hmg/ddm289. [DOI] [PubMed] [Google Scholar]
- 31.Cosma M.P., Pepe S., Annunziata I., Newbold R.F., Grompe M., Parenti G., Ballabio A. The multiple sulfatase deficiency gene encodes an essential and limiting factor for the activity of sulfatases. Cell. 2003;113:445–456. doi: 10.1016/s0092-8674(03)00348-9. doi:10.1016/S0092-8674(03)00348-9. [DOI] [PubMed] [Google Scholar]
- 32.Dierks T., Schmidt B., Borissenko L.V., Peng J., Preusser A., Mariappan M., von Figura K. Multiple sulfatase deficiency is caused by mutations in the gene encoding the human C(alpha)-formylglycine generating enzyme. Cell. 2003;113:435–444. doi: 10.1016/s0092-8674(03)00347-7. doi:10.1016/S0092-8674(03)00347-7. [DOI] [PubMed] [Google Scholar]
- 33.Diez-Roux G., Ballabio A. Sulfatases and human disease. Annu. Rev. Genomics Hum. Genet. 2005;6:355–379. doi: 10.1146/annurev.genom.6.080604.162334. doi:10.1146/annurev.genom.6.080604.162334. [DOI] [PubMed] [Google Scholar]
- 34.Settembre C., Annunziata I., Spampanato C., Zarcone D., Cobellis G., Nusco E., Zito E., Tacchetti C., Cosma M.P., Ballabio A. Systemic inflammation and neurodegeneration in a mouse model of multiple sulfatase deficiency. Proc. Natl Acad. Sci. USA. 2007;104:4506–4511. doi: 10.1073/pnas.0700382104. doi:10.1073/pnas.0700382104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fraldi A., Annunziata F., Lombardi A., Kaiser H.J., Medina D.L., Spampanato C., Fedele A.O., Polishchuk R., Sorrentino N.C., Simons K., et al. Lysosomal fusion and SNARE function are impaired by cholesterol accumulation in lysosomal storage disorders. EMBO J. 2010;29:3607–3620. doi: 10.1038/emboj.2010.237. doi:10.1038/emboj.2010.237. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Otomo T., Higaki K., Nanba E., Ozono K., Sakai N. Inhibition of autophagosome formation restores mitochondrial function in mucolipidosis II and III skin fibroblasts. Mol. Genet. Metab. 2009;98:393–399. doi: 10.1016/j.ymgme.2009.07.002. doi:10.1016/j.ymgme.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 37.Jennings J.J., Jr, Zhu J.H., Rbaibi Y., Luo X., Chu C.T., Kiselyov K. Mitochondrial aberrations in mucolipidosis Type IV. J. Biol. Chem. 2006;281:39041–39050. doi: 10.1074/jbc.M607982200. doi:10.1074/jbc.M607982200. [DOI] [PubMed] [Google Scholar]
- 38.Takamura A., Higaki K., Kajimaki K., Otsuka S., Ninomiya H., Matsuda J., Ohno K., Suzuki Y., Nanba E. Enhanced autophagy and mitochondrial aberrations in murine G(M1)-gangliosidosis. Biochem. Biophys. Res. Commun. 2008;367:616–622. doi: 10.1016/j.bbrc.2007.12.187. doi:10.1016/j.bbrc.2007.12.187. [DOI] [PubMed] [Google Scholar]
- 39.Sano R., Annunziata I., Patterson A., Moshiach S., Gomero E., Opferman J., Forte M., d'Azzo A. GM1-ganglioside accumulation at the mitochondria-associated ER membranes links ER stress to Ca2+-dependent mitochondrial apoptosis. Mol. Cell. 2009;36:500–511. doi: 10.1016/j.molcel.2009.10.021. doi:10.1016/j.molcel.2009.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cao Y., Espinola J.A., Fossale E., Massey A.C., Cuervo A.M., MacDonald M.E., Cotman S.L. Autophagy is disrupted in a knock-in mouse model of juvenile neuronal ceroid lipofuscinosis. J. Biol. Chem. 2006;281:20483–20493. doi: 10.1074/jbc.M602180200. doi:10.1074/jbc.M602180200. [DOI] [PubMed] [Google Scholar]
- 41.Spampanato C., De Leonibus E., Dama P., Gargiulo A., Fraldi A., Sorrentino N.C., Russo F., Nusco E., Auricchio A., Surace E.M., Ballabio A. Efficacy of a combined intracerebral and systemic gene delivery approach for the treatment of a severe lysosomal storage disorder. Mol. Ther. 2011;19:860–869. doi: 10.1038/mt.2010.299. doi:10.1038/mt.2010.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raben N., Shea L., Hill V., Plotz P. Monitoring autophagy in lysosomal storage disorders. Methods Enzymol. 2009;453:417–449. doi: 10.1016/S0076-6879(08)04021-4. doi:10.1016/S0076-6879(08)04021-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Settembre C., Fraldi A., Rubinsztein D.C., Ballabio A. Lysosomal storage diseases as disorders of autophagy. Autophagy. 2008;4:113–114. doi: 10.4161/auto.5227. [DOI] [PubMed] [Google Scholar]
- 44.Zhong L., Tan Y., Zhou A., Yu Q., Zhou J. RING finger ubiquitin–protein isopeptide ligase Nrdp1/FLRF regulates parkin stability and activity. J. Biol. Chem. 2005;280:9425–9430. doi: 10.1074/jbc.M408955200. doi:10.1074/jbc.M408955200. [DOI] [PubMed] [Google Scholar]
- 45.Luo S., Rubinsztein D.C. Apoptosis blocks Beclin 1-dependent autophagosome synthesis—an effect rescued by Bcl-xL. Cell Death Differ. 2010;17:268–277. doi: 10.1038/cdd.2009.121. doi:10.1038/cdd.2009.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Navratil M., Terman A., Arriaga E.A. Giant mitochondria do not fuse and exchange their contents with normal mitochondria. Exp. Cell Res. 2007;314:164–172. doi: 10.1016/j.yexcr.2007.09.013. doi:10.1016/j.yexcr.2007.09.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.