Abstract
Mutations in components of the intraflagellar transport (IFT) machinery required for assembly and function of the primary cilium cause a subset of human ciliopathies characterized primarily by skeletal dysplasia. Recently, mutations in the IFT-A gene IFT144 have been described in patients with Sensenbrenner and Jeune syndromes, which are associated with short ribs and limbs, polydactyly and craniofacial defects. Here, we describe an N-ethyl-N-nitrosourea-derived mouse mutant with a hypomorphic missense mutation in the Ift144 gene. The mutant twinkle-toes (Ift144twt) phenocopies a number of the skeletal and craniofacial anomalies seen in patients with human skeletal ciliopathies. Like other IFT-A mouse mutants, Ift144 mutant embryos display a generalized ligand-independent expansion of hedgehog (Hh) signalling, in spite of defective ciliogenesis and an attenuation of the ability of mutant cells to respond to upstream stimulation of the pathway. This enhanced Hh signalling is consistent with cleft palate and polydactyly phenotypes in the Ift144twt mutant, although extensive rib branching, fusion and truncation phenotypes correlate with defects in early somite patterning and may reflect contributions from multiple signalling pathways. Analysis of embryos harbouring a second allele of Ift144 which represents a functional null, revealed a dose-dependent effect on limb outgrowth consistent with the short-limb phenotypes characteristic of these ciliopathies. This allelic series of mouse mutants provides a unique opportunity to uncover the underlying mechanistic basis of this intriguing subset of ciliopathies.
INTRODUCTION
The primary cilium, a microtubule-based organelle projecting from most quiescent vertebrate cells, plays a pivotal role in embryonic signalling and human disease (reviewed in 1). An expanding class of pleiotropic human diseases result from an underlying dysfunction of the primary cilium, with disease-causing mutations found in up to 40 genes that influence cilia function (2). These ciliopathies are commonly characterized by defects in the kidney, limb, eye and neural system, and in some instances different disorders are caused by mutations of varying severity in the same gene. There is also evidence that genetic modifiers and mutational load play a role in some ciliopathies, with variation at more than one locus thought to be responsible for the final phenotypic outcome (3).
Recently, a group of skeletal dysplasias have been classified as ciliopathies. These include short-rib polydactyly (SRP) syndrome, Jeune asphyxiating thoracic dystrophy and Sensenbrenner syndrome (or cranioectodermal dysplasia; 4–8). Patients with these disorders variably display limb truncation, short ribs, polydactyly and in some cases extra-skeletal features including renal defects. Sensenbrenner syndrome patients may also present with craniosynostosis, and dental, hair and retinal abnormalities. SRP comprises four distinct subtypes that are generally more severe than other skeletal ciliopathies, and may also present with cleft lip and/or palate (9). In each of these disorders, mutations have been identified in genes encoding intraflagellar transport (IFT) proteins, which drive the polarized trafficking system responsible for transporting proteins required for cilia assembly and function (reviewed in 10). IFT proteins organize into two complexes, with IFT-B proteins primarily mediating anterograde transport from the cell body to the cilium tip, and IFT-A proteins regulating the opposing retrograde trafficking. However, there is increasing evidence that IFT-A proteins also play a role in regulating anterograde IFT (11,12). Although mutations in the IFT-B gene IFT80 have been found in a subset of Jeune syndrome patients (4), IFT-A genes are more commonly altered in the skeletal ciliopathies. IFT139 is mutated in Jeune syndrome (13), IFT122 and IFT43 in Sensenbrenner syndrome (6,8), IFT121/WDR35 in both Sensenbrenner and SRP syndromes (5,7) and Ift144 in Sensenbrenner and Jeune syndromes (14). In addition, mutations affecting the retrograde IFT motor DYNC2H1 have been found in SRP and Jeune syndrome patients (15,16).
The past decade has seen accumulating evidence that the primary cilium mediates the activity of a number of developmental signalling pathways, including the hedgehog (Hh; 17), canonical Wnt and planar cell polarity (18), platelet-derived growth factor (19), fibroblast growth factor (FGF) (20), Notch (21) and Hippo cascades (22). Of these, Hh signalling in particular has been the major focus of studies to date. A number of components of the Hh pathway localize at or near the primary cilium, and are trafficked in and out in a highly regulated manner (23,24). Key to the regulation of Hh signalling by the cilium is the dynamic shuttling of the glioma-associated (GLI) transcriptional mediators between the cell body, cilia tip and the nucleus (24,25). In vertebrates, there are three GLI proteins, with full-length GLI1 and 2 (GLI-FL) primarily converted to transcriptional activators (GLI-A), whereas GLI3 is cleaved to a truncated repressor (GLI3-R; 26). There is still relatively little known about the role of the cilium in the post-translational modification of the GLI isoforms, and how the trafficking of these molecules is regulated. Analysis of a number of mouse models with mutations in genes encoding IFT-B proteins suggests that the formation of both GLI activators and repressors is reliant on IFT (27–29). These mutants show evidence of reduced Hh signalling in a number of contexts, particularly the limb and neural tube. However, mouse mutants for IFT-A genes such as Ift139 and Ift122 show increased Hh signalling (30,31), suggesting that complex A and B IFT proteins play differential roles in regulating Hh signalling.
Here, we describe a mouse with an N-ethyl-N-nitrosourea (ENU)-induced hypomorphic mutation in the gene encoding the IFT-A protein WDR19/IFT144. Phenotypically, this mutant, twinkle-toes (Ift144twt), displays a number of features reminiscent of the skeletal ciliopathies, including polydactyly, truncated ribs and craniofacial anomalies. Analysis of a more severe allele of Ift144 known as diamondhead (Ift144dmhd) further revealed a dosage-dependent effect on proximal–distal (PD) limb outgrowth that also mirrors the short limbs typical of these ciliopathies. Although cilia structure appears relatively normal in Ift144twt embryos, fewer cells possess a primary cilium in mutant mouse limbs and isolated fibroblasts. Furthermore, Ift144twt cells show an attenuated response to the upstream activation of Hh signalling. However, in keeping with other IFT-A mutants, enhanced downstream ligand-independent Hh signalling is observed in a number of contexts.
RESULTS
A mouse mutant with a recessive hypomorphic mutation in Wdr19/Ift144
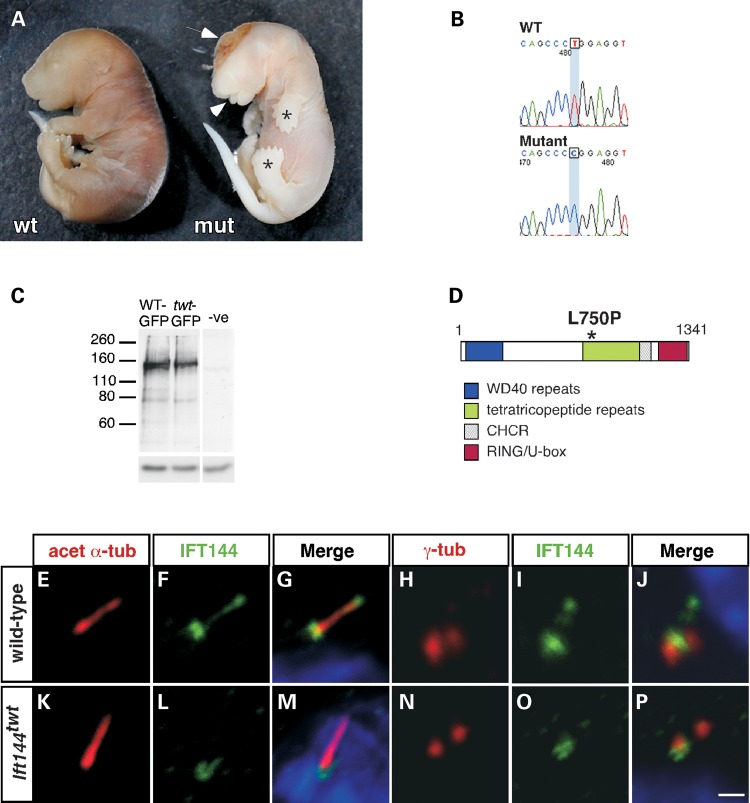
The twinkle-toes (twt) mouse was identified in an ENU mutagenesis screen for genes involved in epigenetic gene silencing (32), although this mutant is unrelated to the epigenetic phenotypes under study in that screen. In the homozygous state, twt embryos survive until close to birth and are characterized by almost completely penetrant exencephaly or exposed brain [Fig. 1A; n = 25/26 embryos analysed at 17.5–18.5 days post coitum (dpc)], in addition to a number of other dysmorphic features described in more detail below.
Figure 1.
The twt mouse has a causative mutation in the Ift144 gene. (A) Whole wild-type (left) and mutant (right) embryos at 17.5 dpc showing exencephaly (exposed brain, arrow), polydactyly (asterisks) and craniofacial abnormalities, including protruding tongue (arrowhead). (B) A homozygous T-to-C transition at position 2249 of the Ift144 gene causes a leucine-to-proline substitution at amino acid position 750 of the protein (D). (C) A mutant expression construct produces stable exogenous protein at a similar size to the wild-type construct. The negative lane represents protein from untransfected cells; the lower panel shows a tubulin loading control. (E–J) In most wild-type-ciliated MEFs (∼89%), endogenous IFT144 is enriched at the base and tip of the cilium, and appears in a punctate pattern along the axoneme, as shown by co-localization with acetylated α-tubulin at the axoneme (E–G) and staining close to γ-tubulin which marks the basal body (H–J). In Ift144twt MEFs, IFT144 appears preferentially restricted to the base of the cilium (∼78% of ciliated cells analysed; K–P), as confirmed by staining close to γ-tubulin (N–P). Scale bar: 1 µm.
Genetic mapping localized the twt mutation to a 2.4Mb region on mouse chromosome 5 between rs33231179 (64.35 Mb) and rs47670438 (66.72 Mb), a region predicted to contain 39 genes. Based on the similarity of the twt phenotype to mouse mutants with defective primary cilia, we prioritized the Wdr19 gene (RefSeq accession: NM_153391.2) from within this interval. Wdr19 is an orthologue of oseg6 in Drosophila, Dyf-2 in Caenorhabditis elegans and the IFT-A gene Ift144 in Chlamydomonas. In addition to a role in ciliopathies (14), human WDR19 has also been implicated in prostate cancer (33,34). Sequencing across the 37 exons of this gene revealed a T-to-C transition towards the end of exon 19 (2249T>C; Fig. 1B) in twt embryos and in obligate heterozygous carriers. We subsequently refer to Wdr19 as Ift144, and homozygotes for this recessive allele are termed Ift144twt.
There are multiple predicted isoforms of mouse WDR19/IFT144 based on potential alternative splicing, but the reference protein (NP_700440.2; Q3UGF1) has a predicted size of ∼145 kDa (1341 amino acids). Like a number of other IFT proteins, IFT144 is characterized by regions of WD40 and tetratricopeptide (TPR) protein–protein interaction domains (12). Some prediction tools also suggest a C-terminal RING-like domain and a clathrin heavy chain repeat (Fig. 1D). The twt mutation introduces a leucine-to-proline substitution at amino acid position 750 (L750P), which lies within the first TPR repeat (asterisk, Fig. 1D). Several lines of evidence suggest that this is not a null mutation. First, although we were unable to optimize available antibodies to detect endogenous IFT144 by immunoblotting (IB), exogenous expression of an epitope-tagged construct encoding the mutant protein in mammalian cells produces a stable protein of equivalent size to a wild-type construct (Fig. 1C). Second, by immunofluorescence (IF) analysis, we confirmed the presence of endogenous protein in Ift144twt mouse embryonic fibroblasts (MEFs). In most cases, (∼78%) this was localized primarily at the base of the cilium (Fig. 1K–P), with the remainder of cells (∼22%) showing additional staining within the axoneme. In contrast, and generally consistent with previous studies in mammalian cells (12,14), most wild-type MEFs (∼89%) showed IFT144 enriched at the base and tip of the cilium, and in a punctate pattern along the axoneme (Fig. 1E–J). The remainder (∼11%) showed staining restricted to the base of the cilium. Given that the staining at the base of the cilium in both Ift144twt and wild-type MEFs does not completely co-localize with γ-tubulin, this is likely to represent the transition zone between the basal body and the axoneme (Fig. 1H–J, N–P). Finally, embryos homozygous for an alternative Ift144 allele dmhd have a splice-site mutation predicted to truncate the protein prior to the TPR domains, and die earlier (∼11.0 dpc) than Ift144twt embryos on the same FVB genetic background (Liem et al., submitted). Importantly, Ift144dmhd/twt compound heterozygous embryos die at ∼14.5 dpc with similar dysmorphic features to the Ift144twt and Ift144dmhd homozygotes (Fig. 2H–J), demonstrating that mutation of Ift144 is responsible for both the twt and dmhd phenotypes.
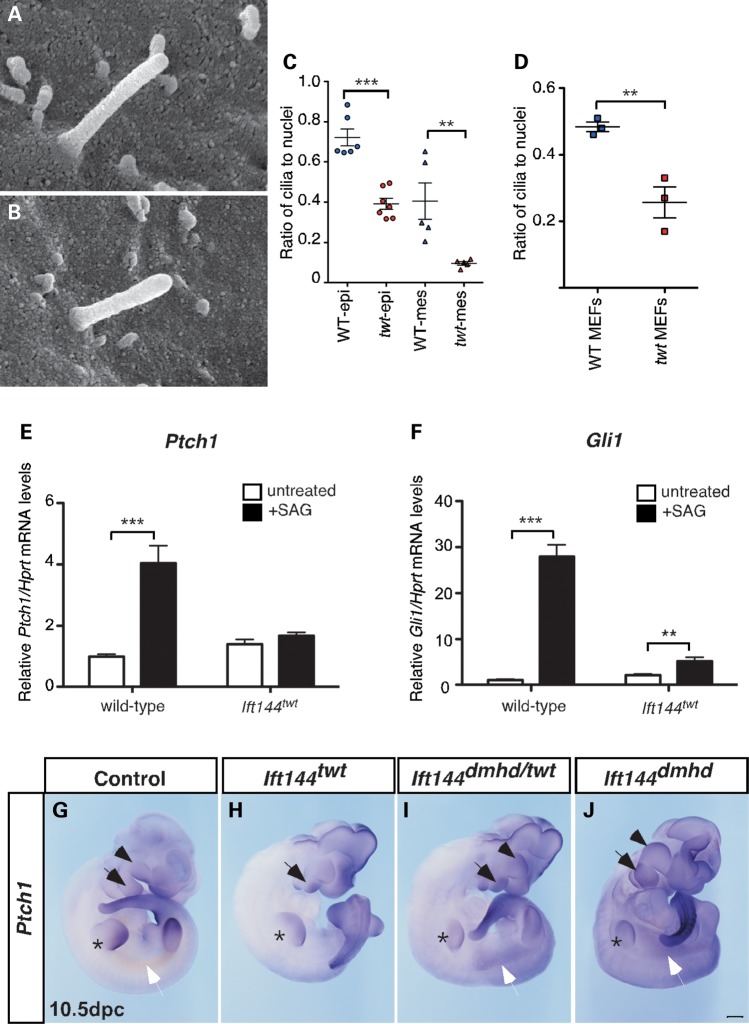
Figure 2.
Ift144twt embryos have fewer ciliated cells and altered Hh signalling. (A and B) SEM revealed no obvious consistent morphological differences in primary cilia projecting from the dorsal surface of the limb of wild-type (A) and Ift144twt mutant (B) embryos. (C and D) Whole-mount IF analysis for ARL13b to mark the axoneme revealed fewer ciliated epithelial and mesenchymal cells in Ift144twt mutant limbs (C) and MEFs (D) compared with wild-type. (E and F) MEFs from wild-type and Ift144twt mutant embryos were exposed to the SMO agonist SAG and the level of Hh signalling assessed by qRT-PCR analysis of Ptch1 (E) and Gli1 (F). The response of Ift144twt MEFs to SAG stimulation was reduced relative to wild-type MEFs. (G–J) WISH reveals enhanced expression of Ptch1 in 10.5 dpc Ift144 mutants. Arrowhead, maxilla; black arrow, mandible; asterisk, forelimb; white arrow, inter-limb flank mesenchyme Scale bar: 500 µm. P-values in (C)–(F) are based on Student's t-test (***P < 0.001;**P < 0.01). Error bars show standard error of the mean.
IFT144twt embryos show fewer ciliated cells but enhanced Hh signalling
In those mouse mutants described to date, perturbation of retrograde IFT results in cilia with a bulge either along the length of the cilium or at the distal tip (29–31). SEM analysis of the dorsal surface of 10.5 dpc limbs revealed no consistent bulging of the Ift144twt cilia, with overall morphology appearing relatively similar to wild-type (Fig. 2A and B). In a parallel study of the neural patterning and cilia structure in this allelic series of Ift144 mutants, Ift144dmhd mutant cilia appeared truncated and bulbous, suggesting a dose-dependent effect on cilia structure (Liem et al., submitted). In Ift144twt embryos, IF analysis, based on ARL13B staining to mark the axoneme, revealed a decrease in the percentage of cells with a cilium in both 10.5 dpc mouse limbs and serum-starved MEF cultures (Fig. 2C and D). A similar trend was found with anti-acetylated α-tubulin staining (not shown), suggesting that this effect was not due to a defect in localization of ARL13B to the axoneme in Ift144twt cells. In the limb, a significant decrease in the percentage of ciliated cells was observed in both the epithelium and mesenchyme of Ift144twt limbs (Fig. 2C), although the decrease in the mesenchyme (∼76%) was more marked than in the epithelium (∼46%). This suggests a cell-type-specific effect of IFT144 on cilia formation or maintenance.
Consistent with an effect on ciliogenesis, Ift144twt MEFs showed an attenuated response to the Hh agonist SAG which targets smoothened (SMO), a component of the Hh receptor complex. Pathway activity was measured by quantitative RT-PCR (qRT-PCR) for the Hh targets Ptch1 and Gli1. Levels of both transcripts were increased in response to SAG stimulation in wild-type MEFs (Fig. 2E and F). Ift144twt MEFs did not show a significant increase in Ptch1 levels in response to SAG (Fig. 2E), whereas Gli1 was upregulated to a much lesser degree than in wild-type MEFS (Fig. 2F). We also saw a similar decreased response to stimulation with SHH-conditioned media in Ift144twt MEFs (Supplementary Material, Fig. S1). These data suggest that compared with wild-type MEFs, Ift144twt cells respond weakly to stimulation with upstream Hh agonists.
Despite a decreased percentage of ciliated cells and an attenuated response to upstream stimulation of the Hh pathway in Ift144twt MEFs, whole-mount in situ-hybridization (WISH) analysis revealed a general increase or ectopic expansion of Ptch1 expression in Ift144twt embryos, particularly in the facial prominences and limbs (Fig. 2G and H, arrows mark the mandible, and asterisks the forelimbs). The increased expression in the facial prominences appeared more marked in Ift144dmhd and compound heterozygous Ift144dmhd/twt embryos (Fig. 2I and J, arrowheads mark the maxilla), which also showed evidence of upregulated Hh signalling in the inter-limb flank region (Fig. 2I and J, white arrows).
IFT144twt embryos display craniofacial defects
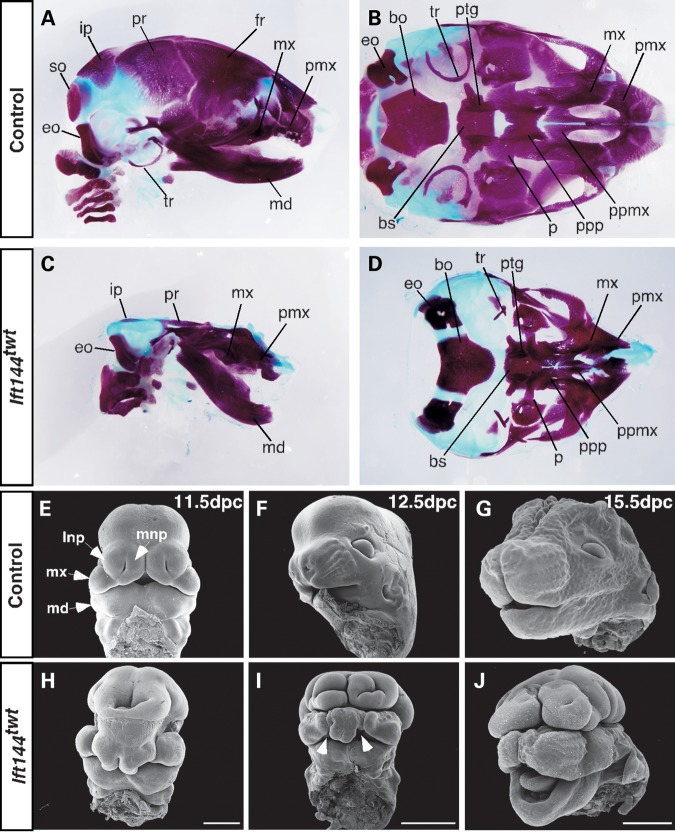
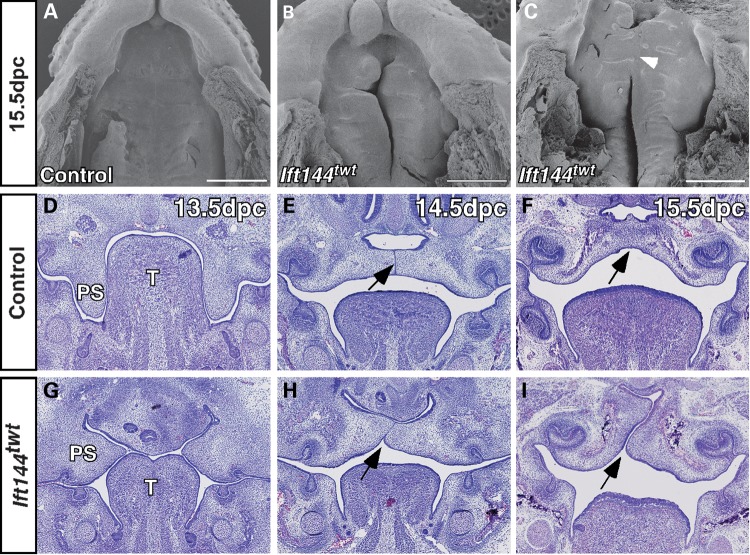
Skeletal preparations of 16.5 and 18.5 dpc embryos (n = 5) revealed that the craniofacial skeleton of Ift144twt embryos showed loss or truncation of several major bones of the skull, consistent with the exencephalic phenotype seen in these embryos (Fig. 3A–D). Most notably, the parietal (pr) and intraparietal (ip) bones are severely hypoplastic, and the frontal (fr) and supraoccipital (so) bones appear to be absent (Fig. 3A–D). The maxillary (mx) and premaxillary (pmx) bones are misshapen and truncated, and the palatine (p) bones and associated palatal processes (pp) are reduced (Fig. 3C and D), resulting in cleft palate (see below) and an overall truncation of the upper face in Ift144twt embryos. Most Ift144twt embryos display anopthalmia (lack of eyes; n = 22/26 embryos analysed at 17.5–18.5 dpc). The tympanic ring (tr), which supports the tympanic membrane or eardrum, also fails to form correctly, and this is consistent with the hearing defects characteristic of many ciliopathies. The mandible is truncated and rests at a wider angle in the mutant embryos (Fig. 3C), resulting in an open bite that is also clearly evident by scanning electron microscopy (SEM) analysis at 15.5 dpc (Fig. 3J). SEM analysis at earlier developmental stages revealed a failure in the medial (mnp) and lateral (lnp) nasal processes to meet and fuse with each other and with the maxilla (mx), resulting in severe bilateral cleft lip phenotype in mutant embryos (Fig. 3E, F, H and I). SEM analysis of 15.5 dpc embryonic heads with the lower jaw removed (n = 2) also revealed a cleft secondary palate in Ift144twt embryos (Fig. 4A–C), which was confirmed by histological analysis at a range of stages (n = 3 embryos per stage; Fig. 4G–I). The histological analysis suggests that mutant palatal shelves do not extend vertically to the tongue as in wild-type embryos at 13.5 dpc, but rather appear to crowd the oral cavity above the tongue (Fig. 4D and G). When wild-type palatal shelves have elevated at 14.5 dpc and fused by 15.5 dpc, mutant shelves abut each other but do not fuse correctly (Fig. 4E, F, H and I). In some cases, the shelves did appear to touch but the medial epithelial seam (MES; Fig. 4E, arrow) was retained. One of the 15.5 dpc mutants analysed by SEM showed some evidence of limited fusion of the anterior palate (Fig. 4C, arrowhead), but it is not possible to say whether the MES remains in this case.
Figure 3.
Craniofacial phenotype of Ift144twt embryos. (A–D) Skulls of 18.5 dpc control (A and B) and Ift144twt (C and D) embryos stained for bone (Alizarin red) and cartilage (Alcian blue). (E–J) SEM analysis of control (E–G) and Ift144twt mutant (H–J) heads at a range of gestational stages. White arrowheads in (I) mark the bilateral facial cleft in 12.5 dpc Ift144twt embryos. bo, basioccipital; bs, basisphenoid; eo, exoccipital; fr, frontal; ip, interparietal; lnp, lateral nasal process; md, mandible; mx, maxilla; mnp, medial nasal process; p, palatine; pmx, premaxilla; ppmx, palatal process maxilla; ppp, palatal process palatine; pr, parietal; ptg, pterygoid; so, supraoccipital; tr, tympanic ring. Scale bars: (E and H): 500 µm; (F, G, I and J): 1 mm.
Figure 4.
Ift144twt embryos display cleft palate. (A–C) SEM analysis of 15.5 dpc embryonic heads with the jaw removed revealed a cleft palate in Ift144twt embryos. One sample (C) showed some evidence of limited anterior fusion (arrowhead). (D–I) Histological analysis of frontal sections through the head of control (D–F), and Ift144twt mutant embryos (G–I) at a range of gestational stages. Mutant palatal shelves do not grow vertically alongside the tongue at 13.5 dpc (D and G), and do not fuse correctly despite often closely abutting one another (E, F, H and I). The arrow marks the MES in (E), its normal removal in (F) and the failure to form correctly in (H) and (I). ps, palatal shelf; t, tongue. Scale bar: 500 µm.
Rib defects in Ift144twt mutants reflect disrupted somitic patterning
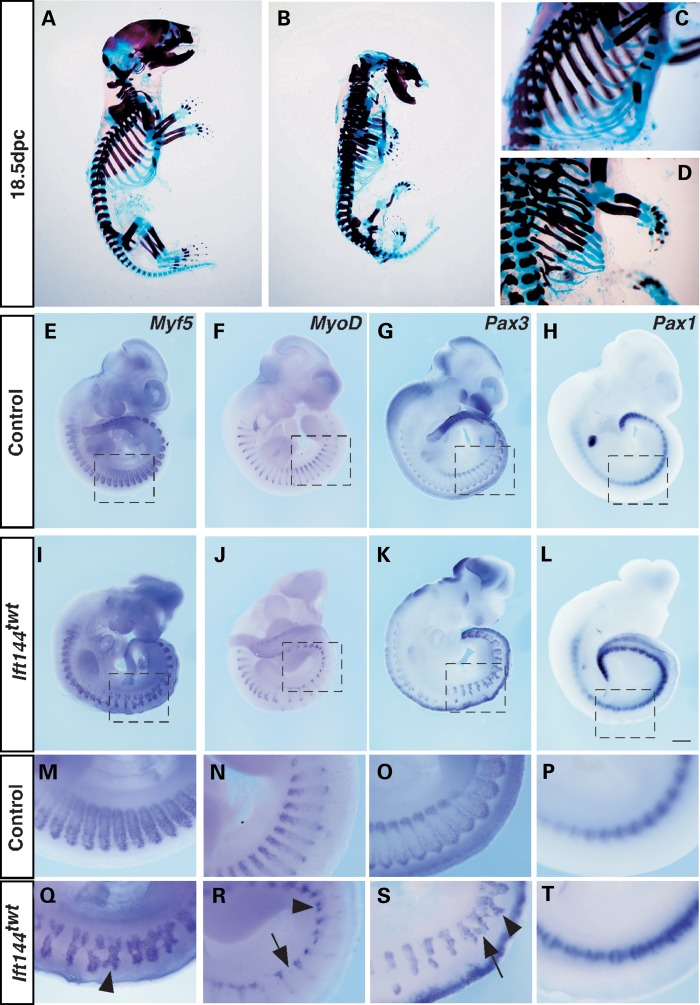
Skeletal preparations also revealed a short-rib phenotype in Ift144twt embryos, creating a small rib cage reminiscent of the phenotype in Jeune, SRP and Sensenbrenner syndromes (Fig. 5A–D). However, unlike these human phenotypes, Ift144twt embryos show marked fusion, branching and bifurcation of the ribs (Fig. 5D and Supplementary Material, Fig. S2), and the sternum is partially split and the ossification centres of the sternebrae are misaligned (Supplementary Material, Fig. S2). Defects in other somite-derived structures including a disordered arrangement of the vertebrae and a lack of curvature of the spine are also evident, but have not been analysed in detail (Fig. 5A and B and Supplementary Material, Fig. S2). Analysis of somitic markers at 10.5 and 11.5 dpc revealed major disruption to the organization of the medial and lateral myotome, as marked by Myf5 (Fig. 5E and I, M and Q) and MyoD (Fig. 5F, J, N and R and Supplementary Material, Fig. S2) respectively, and the dermomyotome, marked by Pax3 (Fig. 5G, K, O and S; Supplementary Material, Fig. S2). At these stages, the expression domains of these genes reveal bifurcation and incomplete separation of adjacent somite segments in Ift144twt embryos, in contrast to the highly ordered segmented pattern in wild-type embryos. The disruption is most marked for all genes in the interlimb region. By comparison, Pax1 expression in the medial sclerotome remains relatively unperturbed (Fig. 5H, L, P and T; Supplementary Material, Fig. S2), although expression of the lateral sclerotome marker scleraxis (Scx) is reduced, particularly at 11.5 dpc (Supplementary Material, Fig. S2).
Figure 5.
Ift144twt embryos display skeletal defects due to altered somite patterning. (A–D) 18.5 dpc control (A and C) and Ift144twt mutant (B and D) embryos stained for bone (Alizarin red) and cartilage (Alcian blue). Ift144twt embryos display truncated, fused and branched ribs. (E–T) WISH analysis of markers of somite compartments in control (E–H) and Ift144twt mutant (I–L) 10.5 dpc whole embryos. (M–T) Higher magnification of boxed regions in (E–L). Arrowheads show the regions' altered expression relative to controls. Scale bar: 500 µm.
Increasing severity of limb outgrowth defects across the Ift144 allelic series
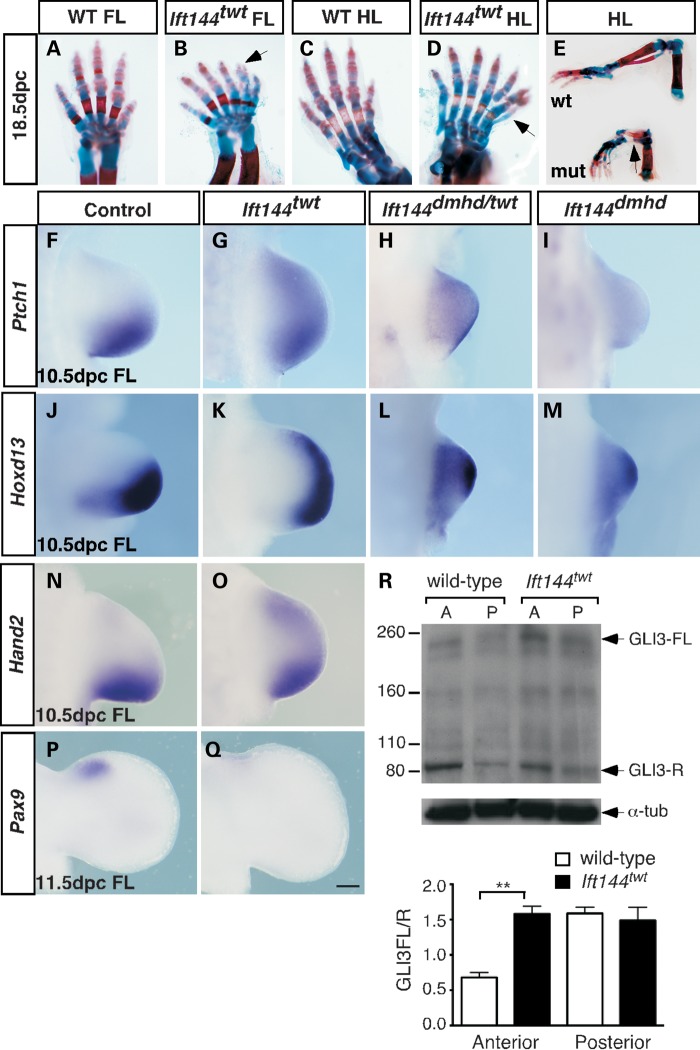
In keeping with many human ciliopathies, homozygous Ift144twt embryos exhibit forelimb and hindlimb polydactyly (Fig. 6A–D). The normal biphalangeal digit 1 is triphalangeal in mutant embryos, and an ectopic digit was often observed anterior to digit 1 (Fig. 6D, arrow). Ectopic digit tips generated by branching at the joints were sometimes evident (Fig. 6B, arrow), and tibial agenesis was consistently observed (Fig. 6E, arrow). Interestingly, in contrast to the slightly wider limb paddles characteristic of Ift144twt embryos at 10.5 dpc, Ift144dmhd and Ift144dmhd/twt compound heterozygous embryos display a smaller forelimb paddle, with truncation in the PD and to a lesser extent the anterior–posterior (AP) axes (Fig. 6H, I, L and M). The hindlimbs of these embryos do not display such an obvious truncation and appear much more consistent in morphology across all three genotypes (Supplementary Material, Figs. S3 and S4). The earlier embryonic lethality of Ift144dmhd and Ift144dmhd/twt embryos precludes analysis of limb skeletal structure. These data suggest there is a forelimb-specific outgrowth defect that increases in severity across the Ift144 allelic series.
Figure 6.
The forelimbs of Ift144 mutants display a dose-dependent effect on patterning. (A–E) Limbs of wild-type (A, C and E—top) and Ift144twt mutant (B, D and E—bottom) 18.5 dpc embryos stained for bone (Alizarin red) and cartilage (Alcian blue). Ift144twt mutant limbs display preaxial polydactyly (arrow in D) with branching at the digit tips (Arrow in C), and tibial agenesis (arrow in E). (F–Q) WISH analysis of a range of markers in the forelimb. At 10.5 dpc, Ptch1 expression is expanded into the anterior of Ift144twt (G), Ift144dmhd/twt (H) and Ift144dmhd (I) mutant forelimbs (FL) compared with control (F). Hoxd13 expression is expanded anteriorly in Ift144twt (K), Ift144dmhd/twt (L) and Ift144dmhd (M) 10.5 dpc mutant forelimbs (FL) compared with control (J). Hand2 expression is upregulated in the anterior of the 10.5 dpc Ift144twt (O) forelimb compared with control (N). The control Pax9 anterior expression domain (P) is undetectable in Ift144twt (Q) mutant limbs at 11.5 dpc (R) IB revealed a subtle increase in the GLI3FL/R ratio specifically in the anterior (A) but not the posterior (P) 11.5 dpc Ift144twt limb. Quantification is based on three independent blots using the same protein samples from pooled limbs. P-value is based on Student's t-test (**P = 0.0022). Error bars show standard error of the mean. Limbs in (A)–(D) are oriented anterior to the right; (F)–(Q), anterior to the top. Scale bar: 200 µm.
Hh signalling is ectopically expanded into the anterior of both the forelimb and hindlimb buds of embryos from all three genotypes, as determined by the expression of Ptch1 and Gli1 by WISH (Fig. 6F–I, and Supplementary Material, Fig. S3). In general, this expansion is accompanied by a decrease in the normal posterior expression of these genes, particularly in Ift144dmhd/twt and Ift144dmhd limbs. Hoxd13 and Hand2 expression, which is normally restricted to the posterior limb by GLI3-R function, both show ectopic expansion into the anterior of Ift144twt limbs at 10.5 dpc (Fig. 6J, K, N and O). Similarly, expression of Pax9 is normally maintained in the anterior limb by GLI3-R from 11.5 dpc (35), and this is virtually undetectable in Ift144twt limb buds (Fig. 6P and Q). Expression of Hoxd13 in Ift144dmhd/twt and Ift144dmhd limbs at 10.5 dpc shows an anterior shift accompanied by a general upregulation across the limb paddle (Fig. 6L, M, and Supplementary Material, Fig. S4). These data suggest that GLI3-R function is reduced in the Ift144 mutant limbs, or that the ratio of GLI3-FL:R is altered. To assess this, IB for GLI3 was performed in protein samples from pooled anterior and posterior Ift144twt and wild-type limb buds. This revealed an increased ratio of the GLI3-FL:R isoforms specifically in the anterior of Ift144twt limb buds (Fig. 6R). The data presented graphically are from three blots based on the same sample from pooled limbs, but a similar trend was observed using an independent pool and using a different GLI3 antibody (not shown). Although this alteration is relatively subtle, based on analysis of a range of other polydactylous mouse mutants, we believe this is responsible for the polydactyly seen in these limbs.
SHH–GREM1–FGF interactions are disturbed in a dose-dependent manner in Ift144 mutant forelimbs
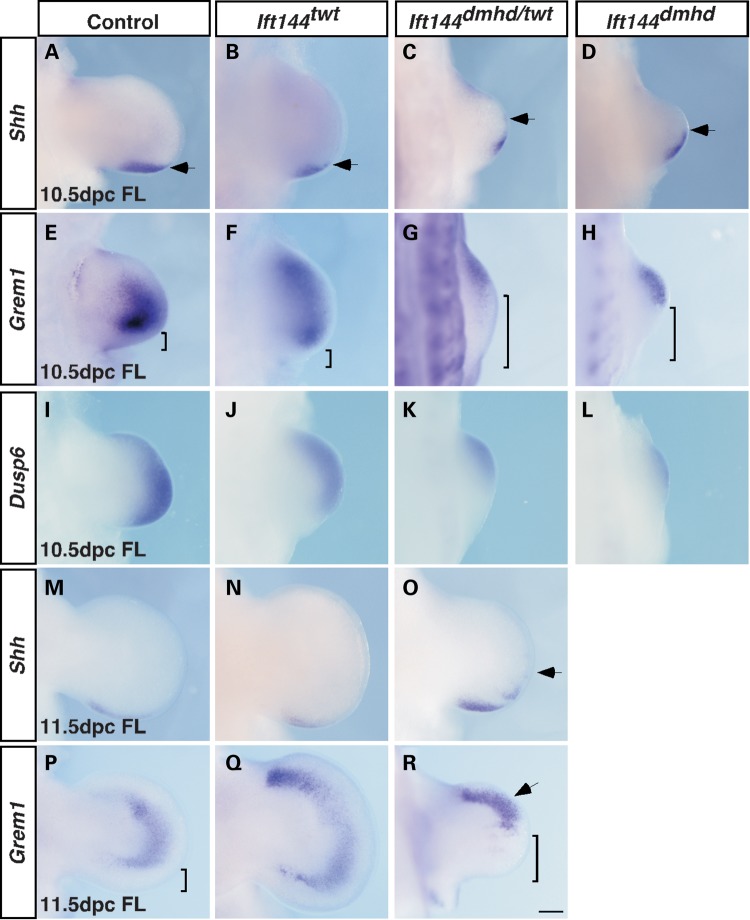
To investigate the forelimb outgrowth defect in the Ift144 embryos, we analysed key markers of the SHH-GREM1-FGF signalling loop responsible for regulating outgrowth and patterning of the early vertebrate limb bud (36). Expression of Shh itself is not obviously altered in Ift144twt 10.5 or 11.5 dpc limbs, with no anterior ectopic domain of expression as seen in a number of other polydactylous mouse mutants, including Gli3extra-toes (Fig. 7B and N; 37). In both IFT144dmhd/twt and IFT144dmhd limbs, expression of Shh extends anteriorly around the distal margin of the forelimb, but not the hindlimb bud at 10.5 dpc, with a more discontinuous expanded domain of expression in IFT144dmhd/twt limbs at 11.5 dpc (Fig. 7C and D, O; arrows mark anterior limit of expression).
Figure 7.
The SHH–GREM1–FGF signalling loop is prematurely disrupted in Ift144 mutant forelimbs. (A–D, M–O) Shh expression is unaltered in Ift144twt forelimbs at 10.5 dpc (B) and 11.5 dpc (N) relative to controls (A and M). Shh expression is expanded along the distal periphery in Ift144dmhd/twt (C) and Ift144dmhd (D) forelimbs at 10.5 dpc, and in Ift144dmhd/twt at 11.5 dpc (O). Arrows in (A)–(D) and (O) mark the anterior limit of Shh expression. (E–H, P–R) Grem1 expression is expanded anteriorly in 10.5 dpc (F) and 11.5 dpc (Q) Ift144twt forelimbs compared with controls (E, P). In both Ift144dmhd/twt (G) and Ift144dmhd (H) 10.5 dpc forelimbs, Grem1 expression is expanded anteriorly but reduced in the posterior relative to control (E). A similar pattern is seen in Ift144dmhd/twt forelimbs at 11.5 dpc (R). Square brackets in (E)–(H), (P), (R) denote the region of the posterior limb devoid of Grem1 expression. (I–L) Dusp6 expression suggests attenuated FGF signalling in the posterior of Ift144dmhd/twt (K) and Ift144dmhd (L) and, to a lesser extent, Ift144twt (J) forelimbs at 10.5 dpc relative to control (I). Limbs are oriented anterior to the top. Scale bar: 200 µm.
Expression of Grem1, a key intermediate linking Hh signalling from the zone of polarizing activity (ZPA) with FGF signalling from the apical ectodermal ridge (AER), is expanded anteriorly in the 10.5 dpc Ift144twt forelimb relative to control (Fig. 7E and F). The expression of Grem1 in 10.5 dpc forelimbs of Ift144dmhd/twt and Ift144dmhd mutants is also expanded into the anterior, but there is an obvious decrease in posterior expression, resulting in an expanded posterior domain devoid of Grem1 expression (Fig. 7E and G, H, brackets). At 11.5 dpc, Grem1 expression is similarly expanded into the anterior of both Ift144twt and Ift144dmhd/twt forelimbs, but reduced posterior expression is specific to Ift144dmhd/twt forelimbs, with evidence of patchy expression (Fig. 7R, bracket). These data suggest an expansion of the posterior region refractory to Grem1 expression in forelimbs of mutants carrying the more severe Ift144 alleles. This domain is thought to arise because, although SHH from the ZPA induces Grem1 expression, Shh-expressing cells and their descendants are unable to express Grem1, and increasing distance between the SHH source and Grem1 is thought to play a role in termination of limb outgrowth (38). An alternative model for termination of limb outgrowth involves the FGF–GREM1 inhibitory loop, whereby FGF levels in the AER eventually reach a threshold which inhibits Grem1 expression, which in turn leads to downregulated FGF signalling (39). This normally results in a domain devoid of Grem1 expression immediately under the AER. In the small 11.5 dpc Ift144dmhd/twt forelimbs, anterior Grem1 expression extends to the distal edge of the limb (Fig. 7R, arrow), suggesting premature termination of the circuitry governing forelimb outgrowth in the more severe Ift144 alleles.
To confirm early disruption of SHH–GREM1–FGF interactions, we analysed expression of Dusp6 (or Mkp3) as a downstream marker of FGF signalling. As expected, FGF signalling appears downregulated in Ift144dmhd/twt and Ift144dmhd forelimbs at 10.5 dpc, with the domain of expression shifted anteriorly and reduced in the posterior region (Fig. 7K and L). This posterior reduction in expression was much less obvious in the Ift144twt forelimb (Fig. 7J), and is likely to correspond to reduced Grem1 expression since GREM1 maintains FGF expression in the AER (40). Consistent with the lack of an obvious outgrowth phenotype in the early hindlimb of this allelic series, Grem1 and Dusp6 expression is similar in the hindlimb of all three mutant genotypes (Supplementary Material, Fig. S4).
DISCUSSION
To date, two siblings with Sensenbrenner syndrome have been shown to carry compound heterozygous mutations in IFT144, and one Jeune syndrome patient is homozygous for a missense mutation (14). This adds to a host of other IFT-A gene mutations responsible for the skeletal ciliopathies (5–8,13). Mouse models of cilia dysfunction have been instrumental in revealing the importance of the primary cilium in both cellular signalling and human disease, and provide the opportunity to explore the genetic and mechanistic basis of ciliopathies. Analysis of mice with mutations in genes encoding anterograde IFT-B proteins, as well as both the anterograde and retrograde motors, initially led to the conclusion that IFT and primary cilia are required for optimal Hh signalling (27–29). Here, we describe an allelic series of mouse mutants for the Ift144 gene, which encodes a component of the IFT-A complex. The Ift144twt mouse carries a hypomorphic allele of Ift144. Homozygous embryos survive until late in gestation and phenocopy aspects of the skeletal defects seen in human skeletal ciliopathies. Like a number of other recently analysed IFT-A mutants, Ift144twt and the Ift144dmhd null allele show expanded rather than reduced Hh signalling in a number of contexts, including the limb and facial prominences. Enhanced Hh signalling likely results from a combination of ligand-independent activation and de-repression of the pathway, and in fact cells derived from Ift144twt mutant embryos show an attenuated response to upstream activation of signalling.
Ift144twt embryos display facial and palatal clefting
Sensenbrenner patients with compound heterozygous mutations in IFT144 display dental and retinal defects, and minor palatal anomalies consisting of a bony protrusion (14). In contrast, the craniofacial defects in Ift144twt embryos are extremely severe and some, such as the loss of the frontal bone and anopthalmia, may be secondary to the exencephaly phenotype. However, K14-Shh transgenic mice ectopically expressing Shh in the facial epithelia display a similar bone truncation phenotype with no overt exencephaly (41). Other aspects of the IFT144twt phenotype also mirror the K14-Shh phenotype, including the open bite and cleft palate, suggesting that increased Hh signalling contributes to this phenotype in the Ift144twt mouse. Upregulated Hh signalling has also been implicated in both syndromic and sporadic forms of cleft palate in humans through variation in the Hh receptor PTCH1 (42). Several cilia-related mouse models also display cleft palate (43–45), and this is also a feature of a number of human ciliopathies, including Meckel–Gruber syndrome (2), hydrolethalus and acrocallosal syndromes (46) and oral–facial–digital syndrome (47). Notably, SRP syndrome is also characterized by cleft lip and palate (9). To date, no IFT144 mutations have been reported in SRP patients, but in the one published study only two patients were screened (14). Our data suggest that more extensive screening may uncover Ift144 mutations in SRP syndrome patients, and that genes involved in cilia function are promising candidates for variation in sporadic complex forms of clefting.
The role of Hh signalling in craniofacial development is complex and involves signalling from a number of domains in both the forebrain and frontonasal ectoderm (48). In general, increased Hh signalling is thought to lead to hypertelorism or wide-spacing of the eyes, and decreased signalling to collapse of the midline or holoprosencephaly. This has been extended to cilia models, with analysis of a neural crest cell-specific conditional deletion of the Kif3a gene revealing upregulated Hh signalling in the facial prominences, an expanded midline and cleft palate (45). An expanded midline is often indicated by an increased distance between the medial nasal process at 11.5 dpc. In this sense, the Ift144twt mutant does not show overt midline expansion, since the nasal processes do not form correctly and the medial processes fail to separate. However, at later stages, the facial clefting phenotype could be considered a midline expansion. Although the Ift144 mutant craniofacial phenotypes are consistent with altered Hh signalling, a number of other signalling pathways linked to cilia function are also implicated in similar phenotypes. Most notably, defective Wnt (49) and PDGF signalling (50) both cause cleft palate in mice, and Pdgfc−/−Pdgfa−/− mice also display midline expansion (50). The interaction between these developmental signalling pathways and their convergence at the primary cilium makes it difficult to unequivocally dissect their individual roles in the Ift144 mutant craniofacial phenotypes.
Aspects of the rib phenotype in Ift144twt embryos mimic human ciliopathies
Jeune, Sensenbrenner and SRP syndromes are all characterized by shortened ribs and a narrow chest, among other phenotypic features (4). Late-stage Ift144twt embryos show a small rib cage reminiscent of the human phenotypes but, unlike human patients reported to date, display marked branching and bifurcation of the ribs. A similar phenotype is seen in the alien mutant mouse due to a mutation in the IFT-A encoding Ift139/Ttc21b gene, further linking these disruptions to IFT-A function (51). It has been proposed that the shortened ribs in the human ciliopathies result from aberrant Hh signalling since Indian hedgehog (Ihh) null mice show a similar phenotype (52). However, the branching and fusion of the ribs seen in the Ift144twt embryos is likely to result from perturbed somite patterning early in development before IHH plays a role in the chondrogenic and osteogenic programmes, although we cannot rule out that ectopic early Hh signalling is responsible for these defects. Analysis of markers of somitic compartments revealed considerable disruption to the myotome and dermomyotome compartments as revealed by the altered expression domains of Myf5 and Pax3. Reduced Scx expression also revealed a defect in the lateral sclerotome, although altered Pax1 expression in the medial sclerotome was less obvious. These somitic defects are likely to underlie the rib defects seen in Ift144twt mutant embryos and are consistent with the notion that the rib precursors arise in the dermomyotome (53), and that rib patterning is co-ordinated by signalling interactions between the dermomyotome, myotome and sclerotome (54). Several pathways linked to the primary cilium could potentially mediate this interaction, with rib branching and fusion defects similar to those in Ift144twt embryos also seen in mouse mutants affecting the canonical Wnt and PCP (55), and PDGF (50) signalling pathways. Interestingly, the open-brain (opb) mouse with a mutation in the gene encoding RAB23, a negative regulator of Hh signalling, also displays similar rib defects (56). This may indicate a role for Hh signalling in this phenotype, especially given that SHH from the notochord and floorplate is involved in specifying epaxial or dorsal myotome (57) and sclerotome (58). However, RAB23 is also thought to function at the primary cilium (59,60), raising the possibility that the defects in these mice are secondary to the effects of cilia dysfunction on other signalling pathways. The association of branched and fused ribs with defective IFT-A in mice suggests that this phenotype may occur in human ciliopathies yet to be described, and may in fact prove valuable as a predictive marker for classification of such ciliopathies.
Analysis of an Ift144 allelic series reveals dose-dependent effects on limb patterning
The polydactyly phenotype in the Ift144twt mutant embryos is consistent with that seen in most cilia-related mouse models and human ciliopathies, and likely results from defective cleavage of GLI3, leading to altered ratios of GLI3-FL:R across the limb. Similarly, the polydactyly in anterograde IFT-B mutants is thought to result from reduced levels of GLI3-R, but these mutants also show a reduction in GLI-A function, as indicated by Ptch1 and Gli1 levels (27). In contrast, marker analysis in the Ift144 mutants analysed in this study suggests enhanced GLI-A function as evidenced by the anterior expansion of Ptch1 and Gli1 expression. There is still some debate as to whether GLI3-FL can act as a transcriptional activator in the limb (61,62), suggesting that upregulated Ptch1 and Gli1 expression may be mediated by ectopic GLI2-A in this context. However, this is difficult to determine unequivocally, as it is now widely accepted that GLI-FL isoforms undergo an additional step to convert them to transcriptional activators, meaning that levels alone are only partially informative.
Although the enhanced GLI-A function is unlikely to be a major contributor to the polydactyly phenotype, it may in part cause the small limb paddle seen in the more severe Ift144dmhd and Ift144dmhd/twt embryos. The smaller limb in this case is unlikely to be due to loss of GLI3R alone because the null Gli3 extra-toes mice display a broadened limb paddle. Although Ift144dmhd embryos do not survive long enough to assess digit number, Ift144dmhd/twt forelimbs ultimately display at least five digits, although the number of digits in these embryos is statistically fewer than Ift144twt forelimbs (data not shown). The morphological and molecular phenotype of Ift144dmhd and Ift144dmhd/twt forelimbs displays some similarity to the forelimbs of embryos conditionally deleted for Ptch1 in the limb mesenchyme, resulting in high-level constitutive ligand-independent Hh signalling across the limb paddle (63). These mutants show hindlimb polydactyly and variable forelimb oligodactyly. We previously proposed that the forelimb phenotype in the conditional Ptch1 mutants resulted from the very early perturbation of the SHH–GREM1–FGF loop that controls limb growth in the AP and PD axes. We see similar forelimb-specific effects on this signalling loop in the Ift144dmhd and Ift144dmhd/twt embryos, and further suggest that the FGF–GREM1 inhibitory loop controlling termination of limb growth is activated prematurely, resulting in the severe outgrowth defects seen in these forelimbs. Although these are germline mutations not subject to differences in timing of Cre-mediated deletion, we suggest that the forelimb is subject to a lower threshold level above which early widespread ligand-independent signalling perturbs the signalling networks essential for the correct outgrowth and patterning of the limb. The absence of such marked effects in the early Ift144twt forelimb suggests a dosage-dependent effect on Hh signalling in these mutants. A similar small forelimb is also observed in mouse embryos with a mutation in the Ift122 and Ift121/Wdr35 IFT-A genes (5,64). This correlates with the shortened limbs seen in Jeune, SRP, Sensenbrenner and Ellis van Creveld syndromes, another ciliopathy similar in presentation to SRP syndrome (2,5,8), and provides a molecular explanation for these defects. The fact that the forelimb defects vary in a dose-dependent manner across the Ift144 allelic series has similarities with the mutational spectrum in certain ciliopathies whereby mutations of differing severity in the same gene can lead to distinct disorders (65).
Correlation between defective IFT-A proteins, enhanced Hh signalling and skeletal dysplasia
Our data support an increasing body of evidence suggesting that defects in IFT-A protein function contribute to a specific subset of ciliopathies characterized by short ribs and limbs, polydactyly and craniofacial dysmorphology. In general, the defects seen in the hypomorphic Ift144twt embryos are far more severe than in human patients with IFT144 mutations, despite the fact that at least one of these patients is thought to possess no functional IFT144 protein (14). This may reflect species-specific differences, but equally may be due to genetic background effects, especially given the high propensity for genetic modification in ciliopathies. It is likely that these phenotypes are related in part to expanded ligand-independent Hh signalling, but the potential effect of cilia structure on multiple signalling cascades makes it difficult to dissect out primary causative signalling events.
Why IFT-A deficiency can lead to enhanced ligand-independent Hh signalling in some contexts is not yet known. The swollen cilia normally characteristic of defects in retrograde IFT are thought to result from the build up of proteins within the axoneme. Recent studies showing that reduced dosage of Ift122 rescues the swollen cilia phenotype in Dync2h1 retrograde motor mutants suggest that IFT-A proteins are also involved in mediating anterograde trafficking of cargo into the cilia (11). This is also seen in Ift144twt and Dync2h1 double mutants (Liem et al., submitted), and early studies in C. elegans suggested that DYF-2, the orthologue of WDR19/IFT144, affects both anterograde and retrograde trafficking (12). The precise role of IFT-A proteins in mediating trafficking, ciliogenesis and consequent effects on signalling remains to be determined. In the case of Hh signalling, the answer is likely to lie in the trafficking of GLI isoforms between the cell body, cilium and the nucleus, and the role of the cilium in conversion of these molecules to transcriptional regulators. How the balance between GLI-A and GLI-R function is affected in a context-specific manner by IFT-A mutations, and whether this is due to the role of IFT-A proteins in anterograde or retrograde IFT or is related to the effects on cilia structure, awaits more detailed analyses.
MATERIALS AND METHODS
Mouse strains and mutation detection
The twinkletoes and diamondhead lines were maintained on an FVB/NJ inbred background. Genotyping details are available on request. Linkage analysis was carried out using 227 F2 mice produced from either an FVB/C57BL6 F1 twt+/− intercross or an FVB/C57BL6 F1 twt+/−× FVB twt+/− backcross. A panel of microsatellite markers was used to localize the mutation to a chromosomal region, and recombination events and analysis of polymorphic SNPs refined the interval. Based on the annotated sequence for mouse Wdr19 (Accession no. NM_153391), all exons and exon/intron junctions were amplified by PCR, and fragments sequenced using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems). All animal experimentation was performed under approval from QIMR and UQ animal ethics committees and conformed to ethical guidelines.
Skeletal preparations and in situ hybridization
Skeletal preparation (66), WISH techniques (67) and probes (63) have been described previously. In situ hybridization images are representative of a minimum of three independent embryos, with each experiment conducted with a somite-matched control embryo (wild-type or heterozygote, which are phenotypically and molecularly indistinguishable).
Scanning electron microscopy
Limbs and heads were fixed at room temperature or on ice for 1–2h in 2.5% glutaraldehyde in PBS, washed two to three times in PBS, transferred to a 1% aqueous solution of osmium tetroxide for 1h and washed three times in distilled water. Samples were immediately dehydrated in an ethanol series (30, 50, 70, 90, 100%). Dehydration was conducted in a Pelco Biowave system (Pelco International, CA, USA) set at 250 W, and each ethanol change given a 40s excitation. Critical point drying was carried out in a Balzers11 120 or Autosamdri-815 CPD, samples were mounted on metal stubs, sputter coated with platinum or gold and viewed in a Jeol 3600 SEM (limbs) or JCM-500 Benchtop Scanning Electron Neoscope (heads).
Antibodies
Primary antibodies were anti-ARL13b (1:1000 for IF analysis) (68); rabbit-anti-γ-tubulin (1:500 for IF; Sigma-Aldrich T5192); rabbit-anti-acetylated α-tubulin (1:1000 for IF; Cell Signalling Technology D20G3); mouse-anti-WDR19 (1:500 for IF; Abnova H00057728-B01); rabbit-anti-GLI3 (1:300 for IB; Santa Cruz Biotechnology H-280:sc-2068); mouse-anti-α-tubulin (1:10 000 for IB; Sigma T9026, DMIA); rabbit-anti-GFP (1:2000 for IB; Invitrogen A-6455). Secondary antibodies were Alexa 488 goat anti-mouse (1:500 for IF; Invitrogen A-11001); Alexa 594 goat anti-rabbit (1:500 for IF; Invitrogen A-11037); HRP-conjugated goat anti-rabbit (1:3000 for IB; Invitrogen 62–6120); HRP-conjugated goat anti-mouse (1:5000 for IB; Zymed/Invitrogen 62-6520).
Isolation of MEFs and IF analysis
MEFs were isolated from trunks of 13.5 dpc embryos by standard methods (69), and cultured for up to five passages in Dulbecco's modified Eagle's medium (DMEM, Invitrogen) containing 10% FCS. For IF analysis, MEFs were grown on glass cover slips to ∼80% confluency in DMEM+10% FCS, at which time the medium was replaced with serum-free DMEM or DMEM containing 0.5% FCS for 24 h. For the WDR19/IFT144 antibody only, cells were washed briefly in PBS and treated with saponin (0.05% in PBS) for 1min prior to fixation in 4% paraformaldehyde (PFA) for 10min. After washing in PBS, cells were permeabilized in PBTX (PBS, 0.1% Triton X-100) for 10min, washed in PBS and incubated with 5% normal goat serum in PBS for 1h, followed by 1h with primary antibody. After brief washes in PBS, cells were incubated with secondary antibody for 1h, mounted on glass slides with ProLong Gold antifade reagent with DAPI (Invitrogen) and imaged on a Zeiss LSM-710 upright laser scanning confocal microscope with a 63× objective. For quantification of cilia, ARL13b-stained cilia were counted manually in three independent experiments using two different wild-type and Ift144twt MEF cultures (100 cells counted per experiment). Statistical analyses used the Prism 5 (Graphpad) software. P-values are based on Student's t-test.
Whole-mount IF and quantification of cilia
For whole-mount IF analysis, 10.5 dpc embryos were fixed in 4%PFA/PBS overnight at 4°C and dehydrated using a methanol/PBTX series. Tissue autoflourescence was reduced using Dent's bleach (four parts MeOH:one part DMSO:one part 30% H2O2) for 2h at room temperature, and the limbs subsequently washed overnight in methanol. Samples were rehydrated using a methanol/PBTX series, blocked for 4 h in 10% horse serum/PBTX and incubated with primary antibody in PBTX overnight at 4°C. Following washing in PBTX, secondary antibody was applied overnight at 4°C. Samples were incubated with DAPI (50 nm) for 10 min, washed for 2 h with PBTX, rinsed in PBS and mounted in Prolong Gold on concave microscope slides.
For epithelial cilia counts, Z-stack imaging was conducted perpendicular to the dorsal surface of the limbs and those images containing the first cell layer, encompassing only epithelial cells, were used for quantitation. Both forelimbs from two independent embryos of each genotype were imaged, with a total of six data points (each consisting of approximately 200 nuclei) included in the statistical analysis. For mesenchymal cilia counts, whole-mount-stained limbs were cryo-sectioned in the transverse plane prior to confocal imaging. Sections selected for cilia counts lay between 80 and 150 µm from the distal tip of the limb. The ImageJ software was used in blind experiments to count ARL13B-positive axonemes and DAPI-stained nuclei. Again, both forelimbs from two embryos were used, with each data point (five in total) consisting of one whole section incorporating approximately 1000–1500 nuclei. Statistical analyses used the Prism 5 (Graphpad) software. P-values are based on Student's t-test.
Histology
Embryonic heads were fixed in 4% PFA in PBS overnight, washed in PBS, followed by EtOH/PBS (1:1), and 70% EtOH for 10 min. Samples were stored in 70% EtOH and when required processed through xylene and into paraffin wax using a Leica tissue processor. Samples were embedded using a Tissue-Tek Embedding Station (Miles Scientific) and sectioned at 8 μm in the frontal plane. Sections were stained with haematoxylin and eosin (H&E) using standard procedures.
Immunoblotting
For the detection of GLI3, pooled anterior and posterior 11.5 dpc autopod halves were lysed in RIPA buffer plus complete protease inhibitor cocktail (Roche). Samples in 1× Laemmlie sample buffer with 10% β-mercaptoethanol were boiled for 5min at 100°C and sonicated prior to electrophoresis through an 8% Tris–glycine gel and transferred onto PDVF membrane (Millipore) overnight at 30 V. Following antibody incubation, protein was detected with HRP-conjugated secondary antibodies and ECL detection reagents (Amersham). Three immunoblots prepared from the same pools of control- and mutant-bisected limb buds were quantified by densitometry using a GS-800 Calibrated Densitometer and the Quantity ONE software (Bio-Rad Laboratories). Student's t-test compared the ratios of the total intensity for GLI3FL and GLI3R bands in each lane.
For detection of GFP-tagged IFT144, NIH-3T3 cells at 80% confluency were transiently transfected with the open-reading frame of wild-type or twt mutant Ift144 in pEGFP-N1 (Clontech), using Lipofectamine 2000 (Invitrogen). Cells were recovered in DMEM (Invitrogen) with 10% FCS overnight before culturing in DMEM supplemented with 0.5% FCS for 24 h. IB was essentially as described above excluding the sonication step.
Quantitative real-time RT-PCR
MEFs were cultured in DMEM containing 10% FCS to 80% confluency, before replacing with DMEM+0.5% FCS supplemented with SAG (100 nm, Alexis Biochemicals) or diluent alone (H2O). Twenty-four hours after SAG administration, MEFs were collected in TriReagent (Sigma-Aldrich) for RNA extraction. First-strand cDNA synthesis was performed using the SuperScript III First-Strand Synthesis system (Invitrogen) on total RNA using oligo dT primers. The cDNA was amplified with ABI SYBR Green PCR master mix using transcript sequence-specific oligonucleotide primer pairs (sequence available on request). Temperature cycling and real-time detection of SYBR Green fluorescence were performed in an Applied Biosystems 7900HT Fast Real-Time PCR system. For each gene of interest, as well as for the normalizing gene Hprt1, triplicate PCRs were performed for each sample (n = 6, comprised of three samples per treatment, for two independent cell lines of each genotype). Relative transcript levels were determined using the delta-delta ct method, and normalized relative to wild-type-untreated samples which were set to 1.0. Data were statistically analysed using Student's t-test.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the National Health and Medical Research Council (NHMRC) of Australia (project grant number 631493 to C.W.) and National Institutes of Health (R01 NS044385 to K.V.A.). C.W. is an NHMRC Senior Research Fellow, and E.W. and R.G.P. are NHMRC Australia Fellows.
Supplementary Material
ACKNOWLEDGEMENTS
The authors acknowledge the staff from the Queensland Biosciences Precinct animal house for help with animal care, and Vicki Metzis for helpful discussion. Confocal microscopy was performed at the ACRF Cancer Biology Imaging Facility at the IMB, established with the generous support of the Australian Cancer Research Foundation. The authors acknowledge the facilities as well as scientific and technical assistance from the staff in the Australian Microscopy & Microanalysis Facility at the Centre for Microscopy and Microanalysis at The University of Queensland.
Conflict of Interest statement. None of the authors of this manuscript have any conflicts of interest to declare.
REFERENCES
- 1.Goetz S.C., Anderson K.V. The primary cilium: a signalling centre during vertebrate development. Nat. Rev. Genet. 2010;11:331–344. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Waters A.M., Beales P.L. Ciliopathies: an expanding disease spectrum. Pediatr. Nephrol. 2011;26:1039–1056. doi: 10.1007/s00467-010-1731-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Novarino G., Akizu N., Gleeson J.G. Modeling human disease in humans: the ciliopathies. Cell. 2011;147:70–79. doi: 10.1016/j.cell.2011.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beales P.L., Bland E., Tobin J.L., Bacchelli C., Tuysuz B., Hill J., Rix S., Pearson C.G., Kai M., Hartley J., et al. IFT80, which encodes a conserved intraflagellar transport protein, is mutated in Jeune asphyxiating thoracic dystrophy. Nat. Genet. 2007;39:727–729. doi: 10.1038/ng2038. [DOI] [PubMed] [Google Scholar]
- 5.Mill P., Lockhart P.J., Fitzpatrick E., Mountford H.S., Hall E.A., Reijns M.A., Keighren M., Bahlo M., Bromhead C.J., Budd P., et al. Human and mouse mutations in WDR35 cause short-rib polydactyly syndromes due to abnormal ciliogenesis. Am. J. Hum. Genet. 2011;88:508–515. doi: 10.1016/j.ajhg.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walczak-Sztulpa J., Eggenschwiler J., Osborn D., Brown D.A., Emma F., Klingenberg C., Hennekam R.C., Torre G., Garshasbi M., Tzschach A., et al. Cranioectodermal dysplasia, Sensenbrenner syndrome, is a ciliopathy caused by mutations in the IFT122 gene. Am. J. Hum. Genet. 2010;86:949–956. doi: 10.1016/j.ajhg.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gilissen C., Arts H.H., Hoischen A., Spruijt L., Mans D.A., Arts P., van Lier B., Steehouwer M., van Reeuwijk J., Kant S.G., et al. Exome sequencing identifies WDR35 variants involved in Sensenbrenner syndrome. Am. J. Hum. Genet. 2010;87:418–423. doi: 10.1016/j.ajhg.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arts H.H., Bongers E.M., Mans D.A., van Beersum S.E., Oud M.M., Bolat E., Spruijt L., Cornelissen E.A., Schuurs-Hoeijmakers J.H., de Leeuw N., et al. C14ORF179 encoding IFT43 is mutated in Sensenbrenner syndrome. J. Med. Genet. 2011;48:390–395. doi: 10.1136/jmg.2011.088864. [DOI] [PubMed] [Google Scholar]
- 9.Turkmen M., Temocin K., Acar C., Levi E., Karaman C., Inan G., Elcioglu N. Short rib-polydactyly syndrome: a case report. Turk. J. Pediatr. 2003;45:359–362. [PubMed] [Google Scholar]
- 10.Pedersen L.B., Veland I.R., Schroder J.M., Christensen S.T. Assembly of primary cilia. Dev. Dyn. 2008;237:1993–2006. doi: 10.1002/dvdy.21521. [DOI] [PubMed] [Google Scholar]
- 11.Ocbina P.J., Eggenschwiler J.T., Moskowitz I., Anderson K.V. Complex interactions between genes controlling trafficking in primary cilia. Nat. Genet. 2011;43:547–553. doi: 10.1038/ng.832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Efimenko E., Blacque O.E., Ou G., Haycraft C.J., Yoder B.K., Scholey J.M., Leroux M.R., Swoboda P. Caenorhabditis elegans DYF-2, an orthologue of human WDR19, is a component of the intraflagellar transport machinery in sensory cilia. Mol. Biol. Cell. 2006;17:4801–4811. doi: 10.1091/mbc.E06-04-0260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis E.E., Zhang Q., Liu Q., Diplas B.H., Davey L.M., Hartley J., Stoetzel C., Szymanska K., Ramaswami G., Logan C.V., et al. TTC21B contributes both causal and modifying alleles across the ciliopathy spectrum. Nat. Genet. 2011;43:189–196. doi: 10.1038/ng.756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bredrup C., Saunier S., Oud M.M., Fiskerstrand T., Hoischen A., Brackman D., Leh S.M., Midtbo M., Filhol E., Bole-Feysot C., et al. Ciliopathies with skeletal anomalies and renal insufficiency due to mutations in the IFT-A gene WDR19. Am. J. Hum. Genet. 2011;89:634–643. doi: 10.1016/j.ajhg.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Merrill A.E., Merriman B., Farrington-Rock C., Camacho N., Sebald E.T., Funari V.A., Schibler M.J., Firestein M.H., Cohn Z.A., Priore M.A., et al. Ciliary abnormalities due to defects in the retrograde transport protein DYNC2H1 in short-rib polydactyly syndrome. Am. J. Hum. Genet. 2009;84:542–549. doi: 10.1016/j.ajhg.2009.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dagoneau N., Goulet M., Genevieve D., Sznajer Y., Martinovic J., Smithson S., Huber C., Baujat G., Flori E., Tecco L., et al. DYNC2H1 mutations cause asphyxiating thoracic dystrophy and short rib-polydactyly syndrome, type III. Am. J. Hum. Genet. 2009;84:706–711. doi: 10.1016/j.ajhg.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huangfu D., Liu A., Rakeman A.S., Murcia N.S., Niswander L., Anderson K.V. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature. 2003;426:83–87. doi: 10.1038/nature02061. [DOI] [PubMed] [Google Scholar]
- 18.Wallingford J.B., Mitchell B. Strange as it may seem: the many links between Wnt signaling, planar cell polarity, and cilia. Genes Dev. 2011;25:201–213. doi: 10.1101/gad.2008011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schneider L., Stock C.M., Dieterich P., Jensen B.H., Pedersen L.B., Satir P., Schwab A., Christensen S.T., Pedersen S.F. The Na+/H+ exchanger NHE1 is required for directional migration stimulated via PDGFR-alpha in the primary cilium. J. Cell Biol. 2009;185:163–176. doi: 10.1083/jcb.200806019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neugebauer J.M., Amack J.D., Peterson A.G., Bisgrove B.W., Yost H.J. FGF signalling during embryo development regulates cilia length in diverse epithelia. Nature. 2009;458:651–654. doi: 10.1038/nature07753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ezratty E.J., Stokes N., Chai S., Shah A.S., Williams S.E., Fuchs E. A role for the primary cilium in notch signaling and epidermal differentiation during skin development. Cell. 2011;145:1129–1141. doi: 10.1016/j.cell.2011.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Habbig S., Bartram M.P., Muller R.U., Schwarz R., Andriopoulos N., Chen S., Sagmuller J.G., Hoehne M., Burst V., Liebau M.C., et al. NPHP4, a cilia-associated protein, negatively regulates the Hippo pathway. J. Cell Biol. 2011;193:633–642. doi: 10.1083/jcb.201009069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rohatgi R., Milenkovic L., Scott M.P. Patched1 regulates hedgehog signaling at the primary cilium. Science. 2007;317:372–376. doi: 10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]
- 24.Kim J., Kato M., Beachy P.A. Gli2 trafficking links Hedgehog-dependent activation of Smoothened in the primary cilium to transcriptional activation in the nucleus. Proc. Natl Acad. Sci. USA. 2009;106:21666–21671. doi: 10.1073/pnas.0912180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen M.H., Wilson C.W., Li Y.J., Law K.K., Lu C.S., Gacayan R., Zhang X., Hui C.C., Chuang P.T. Cilium-independent regulation of Gli protein function by Sufu in Hedgehog signaling is evolutionarily conserved. Genes Dev. 2009;23:1910–1928. doi: 10.1101/gad.1794109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang B., Fallon J.F., Beachy P.A. Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell. 2000;100:423–434. doi: 10.1016/s0092-8674(00)80678-9. [DOI] [PubMed] [Google Scholar]
- 27.Liu A., Wang B., Niswander L.A. Mouse intraflagellar transport proteins regulate both the activator and repressor functions of Gli transcription factors. Development. 2005;132:3103–3111. doi: 10.1242/dev.01894. [DOI] [PubMed] [Google Scholar]
- 28.May S.R., Ashique A.M., Karlen M., Wang B., Shen Y., Zarbalis K., Reiter J., Ericson J., Peterson A.S. Loss of the retrograde motor for IFT disrupts localization of Smo to cilia and prevents the expression of both activator and repressor functions of Gli. Dev. Biol. 2005;287:378–389. doi: 10.1016/j.ydbio.2005.08.050. [DOI] [PubMed] [Google Scholar]
- 29.Huangfu D., Anderson K.V. Cilia and Hedgehog responsiveness in the mouse. Proc. Natl Acad. Sci. USA. 2005;102:11325–11330. doi: 10.1073/pnas.0505328102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tran P.V., Haycraft C.J., Besschetnova T.Y., Turbe-Doan A., Stottmann R.W., Herron B.J., Chesebro A.L., Qiu H., Scherz P.J., Shah J.V., et al. THM1 negatively modulates mouse sonic hedgehog signal transduction and affects retrograde intraflagellar transport in cilia. Nat. Genet. 2008;40:403–410. doi: 10.1038/ng.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qin J., Lin Y., Norman R.X., Ko H.W., Eggenschwiler J.T. Intraflagellar transport protein 122 antagonizes Sonic Hedgehog signaling and controls ciliary localization of pathway components. Proc. Natl. Acad. Sci. USA. 2011;108:1456–1461. doi: 10.1073/pnas.1011410108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blewitt M.E., Vickaryous N.K., Hemley S.J., Ashe A., Bruxner T.J., Preis J.I., Arkell R., Whitelaw E. An N-ethyl-N-nitrosourea screen for genes involved in variegation in the mouse. Proc. Natl Acad. Sci. USA. 2005;102:7629–7634. doi: 10.1073/pnas.0409375102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin B., White J.T., Utleg A.G., Wang S., Ferguson C., True L.D., Vessella R., Hood L., Nelson P.S. Isolation and characterization of human and mouse WDR19, a novel WD-repeat protein exhibiting androgen-regulated expression in prostate epithelium. Genomics. 2003;82:331–342. doi: 10.1016/s0888-7543(03)00151-4. [DOI] [PubMed] [Google Scholar]
- 34.Lin B., Utleg A.G., Gravdal K., White J.T., Halvorsen O.J., Lu W., True L.D., Vessella R., Lange P.H., Nelson P.S., et al. WDR19 expression is increased in prostate cancer compared with normal cells, but low-intensity expression in cancers is associated with shorter time to biochemical failures and local recurrence. Clin. Cancer Res. 2008;14:1397–1406. doi: 10.1158/1078-0432.CCR-07-1535. [DOI] [PubMed] [Google Scholar]
- 35.McGlinn E., van Bueren K.L., Fiorenza S., Mo R., Poh A.M., Forrest A., Soares M.B., Bonaldo Mde F., Grimmond S., Hui C.C., et al. Pax9 and Jagged1 act downstream of Gli3 in vertebrate limb development. Mech. Dev. 2005;122:1218–1233. doi: 10.1016/j.mod.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 36.Butterfield N.C., McGlinn E., Wicking C. The molecular regulation of vertebrate limb patterning. Curr. Top. Dev. Biol. 2010;90:319–341. doi: 10.1016/S0070-2153(10)90009-4. [DOI] [PubMed] [Google Scholar]
- 37.Buscher D., Bosse B., Heymer J., Ruther U. Evidence for genetic control of Sonic hedgehog by Gli3 in mouse limb development. Mech. Dev. 1997;62:175–182. doi: 10.1016/s0925-4773(97)00656-4. [DOI] [PubMed] [Google Scholar]
- 38.Scherz P.J., Harfe B.D., McMahon A.P., Tabin C.J. The limb bud Shh-Fgf feedback loop is terminated by expansion of former ZPA cells. Science. 2004;305:396–399. doi: 10.1126/science.1096966. [DOI] [PubMed] [Google Scholar]
- 39.Verheyden J.M., Sun X. An Fgf/Gremlin inhibitory feedback loop triggers termination of limb bud outgrowth. Nature. 2008;454:638–641. doi: 10.1038/nature07085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zuniga A., Haramis A.P., McMahon A.P., Zeller R. Signal relay by BMP antagonism controls the SHH/FGF4 feedback loop in vertebrate limb buds. Nature. 1999;401:598–602. doi: 10.1038/44157. [DOI] [PubMed] [Google Scholar]
- 41.Cobourne M.T., Xavier G.M., Depew M., Hagan L., Sealby J., Webster Z., Sharpe P.T. Sonic hedgehog signalling inhibits palatogenesis and arrests tooth development in a mouse model of the nevoid basal cell carcinoma syndrome. Dev. Biol. 2009;331:38–49. doi: 10.1016/j.ydbio.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mansilla M.A., Cooper M.E., Goldstein T., Castilla E.E., Lopez Camelo J.S., Marazita M.L., Murray J.C. Contributions of PTCH gene variants to isolated cleft lip and palate. Cleft Palate Craniofac. J. 2006;43:21–29. doi: 10.1597/04-169R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Friedland-Little J.M., Hoffmann A.D., Ocbina P.J., Peterson M.A., Bosman J.D., Chen Y., Cheng S.Y., Anderson K.V., Moskowitz I.P. A novel murine allele of Intraflagellar Transport Protein 172 causes a syndrome including VACTERL-like features with hydrocephalus. Hum. Mol. Genet. 2011;20:3725–3737. doi: 10.1093/hmg/ddr241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weatherbee S.D., Niswander L.A., Anderson K.V. A mouse model for Meckel syndrome reveals Mks1 is required for ciliogenesis and Hedgehog signaling. Hum. Mol. Genet. 2009;18:4565–4575. doi: 10.1093/hmg/ddp422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brugmann S.A., Allen N.C., James A.W., Mekonnen Z., Madan E., Helms J.A. A primary cilia-dependent etiology for midline facial disorders. Hum. Mol. Genet. 2010;19:1577–1592. doi: 10.1093/hmg/ddq030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Putoux A., Thomas S., Coene K.L., Davis E.E., Alanay Y., Ogur G., Uz E., Buzas D., Gomes C., Patrier S., et al. KIF7 mutations cause fetal hydrolethalus and acrocallosal syndromes. Nat. Genet. 2011;43:601–606. doi: 10.1038/ng.826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ferrero G.B., Valenzise M., Franco B., Defilippi C., Gregato G., Corsello G., Pepe E., Silengo M. Oral, facial, digital, vertebral anomalies with psychomotor delay: a mild form of OFD type Gabrielli? Am. J. Med. Genet. 2002;113:291–294. doi: 10.1002/ajmg.10815. [DOI] [PubMed] [Google Scholar]
- 48.Hu D., Marcucio R.S. A SHH-responsive signaling center in the forebrain regulates craniofacial morphogenesis via the facial ectoderm. Development. 2009;136:107–116. doi: 10.1242/dev.026583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.He F., Xiong W., Wang Y., Li L., Liu C., Yamagami T., Taketo M.M., Zhou C., Chen Y. Epithelial Wnt/beta-catenin signaling regulates palatal shelf fusion through regulation of Tgfbeta3 expression. Dev. Biol. 2011;350:511–519. doi: 10.1016/j.ydbio.2010.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ding H., Wu X., Bostrom H., Kim I., Wong N., Tsoi B., O'Rourke M., Koh G.Y., Soriano P., Betsholtz C., et al. A specific requirement for PDGF-C in palate formation and PDGFR-alpha signaling. Nat. Genet. 2004;36:1111–1116. doi: 10.1038/ng1415. [DOI] [PubMed] [Google Scholar]
- 51.Herron B.J., Lu W., Rao C., Liu S., Peters H., Bronson R.T., Justice M.J., McDonald J.D., Beier D.R. Efficient generation and mapping of recessive developmental mutations using ENU mutagenesis. Nat. Genet. 2002;30:185–189. doi: 10.1038/ng812. [DOI] [PubMed] [Google Scholar]
- 52.St-Jacques B., Hammerschmidt M., McMahon A.P. Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes Dev. 1999;13:2072–2086. doi: 10.1101/gad.13.16.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kato N., Aoyama H. Dermomyotomal origin of the ribs as revealed by extirpation and transplantation experiments in chick and quail embryos. Development. 1998;125:3437–3443. doi: 10.1242/dev.125.17.3437. [DOI] [PubMed] [Google Scholar]
- 54.Seo K.W., Wang Y., Kokubo H., Kettlewell J.R., Zarkower D.A., Johnson R.L. Targeted disruption of the DM domain containing transcription factor Dmrt2 reveals an essential role in somite patterning. Dev. Biol. 2006;290:200–210. doi: 10.1016/j.ydbio.2005.11.027. [DOI] [PubMed] [Google Scholar]
- 55.Hamblet N.S., Lijam N., Ruiz-Lozano P., Wang J., Yang Y., Luo Z., Mei L., Chien K.R., Sussman D.J., Wynshaw-Boris A. Dishevelled 2 is essential for cardiac outflow tract development, somite segmentation and neural tube closure. Development. 2002;129:5827–5838. doi: 10.1242/dev.00164. [DOI] [PubMed] [Google Scholar]
- 56.Sporle R., Schughart K. Paradox segmentation along inter- and intrasomitic borderlines is followed by dysmorphology of the axial skeleton in the open brain (opb) mouse mutant. Dev. Genet. 1998;22:359–373. doi: 10.1002/(SICI)1520-6408(1998)22:4<359::AID-DVG6>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 57.Borycki A.G., Brunk B., Tajbakhsh S., Buckingham M., Chiang C., Emerson C.P., Jr Sonic hedgehog controls epaxial muscle determination through Myf5 activation. Development. 1999;126:4053–4063. doi: 10.1242/dev.126.18.4053. [DOI] [PubMed] [Google Scholar]
- 58.Bumcrot D.A., McMahon A.P. Somite differentiation. Sonic signals somites. Curr. Biol. 1995;5:612–614. doi: 10.1016/s0960-9822(95)00123-0. [DOI] [PubMed] [Google Scholar]
- 59.Yoshimura S., Egerer J., Fuchs E., Haas A.K., Barr F.A. Functional dissection of Rab GTPases involved in primary cilium formation. J. Cell Biol. 2007;178:363–369. doi: 10.1083/jcb.200703047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boehlke C., Bashkurov M., Buescher A., Krick T., John A.K., Nitschke R., Walz G., Kuehn E.W. Differential role of Rab proteins in ciliary trafficking: Rab23 regulates smoothened levels. J. Cell Sci. 2010;123:1460–1467. doi: 10.1242/jcs.058883. [DOI] [PubMed] [Google Scholar]
- 61.Hill P., Gotz K., Ruther U. A SHH-independent regulation of Gli3 is a significant determinant of anteroposterior patterning of the limb bud. Dev. Biol. 2009;328:506–516. doi: 10.1016/j.ydbio.2009.02.017. [DOI] [PubMed] [Google Scholar]
- 62.Wang C., Ruther U., Wang B. The Shh-independent activator function of the full-length Gli3 protein and its role in vertebrate limb digit patterning. Dev. Biol. 2007;305:460–469. doi: 10.1016/j.ydbio.2007.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Butterfield N.C., Metzis V., McGlinn E., Bruce S.J., Wainwright B.J., Wicking C. Patched 1 is a crucial determinant of asymmetry and digit number in the vertebrate limb. Development. 2009;136:3515–3524. doi: 10.1242/dev.037507. [DOI] [PubMed] [Google Scholar]
- 64.Cortellino S., Wang C., Wang B., Bassi M.R., Caretti E., Champeval D., Calmont A., Jarnik M., Burch J., Zaret K.S., et al. Defective ciliogenesis, embryonic lethality and severe impairment of the Sonic Hedgehog pathway caused by inactivation of the mouse complex A intraflagellar transport gene Ift122/Wdr10, partially overlapping with the DNA repair gene Med1/Mbd4. Dev. Biol. 2009;325:225–237. doi: 10.1016/j.ydbio.2008.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hildebrandt F., Benzing T., Katsanis N. Ciliopathies. N. Engl. J. Med. 2011;364:1533–1543. doi: 10.1056/NEJMra1010172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bruce S.J., Butterfield N.C., Metzis V., Town L., McGlinn E., Wicking C. Inactivation of Patched1 in the mouse limb has novel inhibitory effects on the chondrogenic program. J. Biol. Chem. 2010;285:27967–27981. doi: 10.1074/jbc.M109.091785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fowles L.F., Bennetts J.S., Berkman J.L., Williams E., Koopman P., Teasdale R.D., Wicking C. Genomic screen for genes involved in mammalian craniofacial development. Genesis. 2003;35:73–87. doi: 10.1002/gene.10165. [DOI] [PubMed] [Google Scholar]
- 68.Caspary T., Larkins C.E., Anderson K.V. The graded response to Sonic Hedgehog depends on cilia architecture. Dev. Cell. 2007;12:767–778. doi: 10.1016/j.devcel.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 69.Todaro G.J., Green H. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J. Cell Biol. 1963;17:299–313. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.