Abstract
Among US Latinas and Mexican women, those with higher European ancestry have increased risk of breast cancer. We combined an admixture mapping and genome-wide association mapping approach to search for genomic regions that may explain this observation. Latina women with breast cancer (n= 1497) and Latina controls (n= 1272) were genotyped using Affymetrix and Illumina arrays. We inferred locus-specific genetic ancestry and compared the ancestry between cases and controls. We also performed single nucleotide polymorphism (SNP) association analyses in regions of interest. Correction for multiple-hypothesis testing was conducted using permutations (Pcorrected). We identified one region where genetic ancestry was significantly associated with breast cancer risk: 6q25 [odds ratio (OR) per Indigenous American chromosome 0.75, 95% confidence interval (CI): 0.65–0.85, P= 1.1 × 10−5, Pcorrected= 0.02]. A second region on 11p15 showed a trend towards association (OR per Indigenous American chromosome 0.77, 95% CI: 0.68–0.87, P= 4.3 × 10−5, Pcorrected= 0.08). In both regions, breast cancer risk decreased with higher Indigenous American ancestry in concordance with observations made on global ancestry. The peak of the 6q25 signal includes the estrogen receptor 1 (ESR1) gene and 5′ region, a locus previously implicated in breast cancer. Genome-wide association analysis found that a multi-SNP model explained the admixture signal in both regions. Our results confirm that the association between genetic ancestry and breast cancer risk in US Latinas is partly due to genetic differences between populations of European and Indigenous Americans origin. Fine-mapping within the 6q25 and possibly the 11p15 loci will lead to the discovery of the biologically functional variant/s behind this association.
INTRODUCTION
Breast cancer incidence varies substantially across different racial and ethnic groups in the USA. The age-adjusted incidence of breast cancer from 2002 to 2006 in US non-Latino Whites and African Americans is 123.5 and 113.0 per 100 000 respectively, whereas US women of Latin American origin have an incidence of 90.2 (1). We have previously demonstrated that genetic ancestry is associated with breast cancer risk in US Latinas (2) and replicated these results in a sample of Mexican women (2,3). In both studies, higher European ancestry was associated with increased risk and higher Indigenous American ancestry was associated with decreased risk of breast cancer. We found that multiple non-genetic risk factors were associated with genetic ancestry and could confound the association between ancestry and breast cancer risk in both US Latinas and Mexican women. However, in both studies, genetic ancestry remained statistically significant after adjustment for known non-genetic risk factors, which suggests that genetic ancestry is a proxy for unmeasured risk factors and/or there is a genetic component to the difference in risk. Here, we explored the latter hypothesis using an admixture-based approach.
Admixture mapping leverages the demographic history of admixed populations to map susceptibility loci (4–12). A population is considered admixed if it results from the combination of two or more ancestral population groups (5). The principle of admixture mapping is to identify genomic regions in which cases share more of the same genetic ancestry compared with either population-based controls (case–control analysis) or compared with the average ancestry of the rest of the genome among cases (case-only analysis) (7). This approach has been used successfully to identify risk variants or risk regions for prostate cancer (9,11), obesity (10), white cell count (4,12), hypertension (8), interleukin 6 soluble receptor and interleukin 6 levels (6,9).
Genome-wide association studies (GWAS) in women of European and Asian origin have reported multiple risk variants for breast cancer (13–23). To date, there is no published breast cancer GWAS in Latinas. We have initiated a multi-stage GWAS of breast cancer in Latinas and in the present study we report results of an admixture mapping analysis in the stage 1 sample, which was motivated by our earlier findings of an ancestry association with breast cancer in Latinas. In this study, we have identified regions of the genome showing breast cancer associations with increased European ancestry that may be putative risk loci for breast cancer.
RESULTS
Participant characteristics
This study included a total of 1497 US Latina cases and 1160 controls from three studies [San Francisco Bay Area Breast Cancer Study (SFBCS), the Northern California Site of the Breast Cancer Family Registry (NC-BCFR) and the Multiethnic Cohort (MEC)] that are part of stage 1 in a GWAS of breast cancer (Table 1), as well as an additional 112 population controls from study of asthma in Latinos. Subjects from the NC-BCFR were younger and a higher proportion of cases had a family history of breast cancer, which reflects the oversampling of cases with indicators of increased genetic susceptibility. In contrast, women from the MEC were 50 years and older and were significantly older on average than women in either of the other studies. Results of univariate analyses comparing breast cancer cases and controls for the different characteristics are concordant with previously reported associations. Cases in all studies reported fewer full-term pregnancies and higher family history of breast cancer. In the SFBCS and MEC studies, the cases were slightly older than the controls. In each of the studies, the cases also had higher European ancestry (and lower Indigenous American ancestry) (Table 1).
Table 1.
Sample characteristics by study
| SFBCS |
NC-BCFR |
MEC |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Case, n= 345 | Control, n= 551 | P-valuea | Case, n= 625 | Control, n= 59 | P-valuea | Case, n= 546 | Control, n= 558 | P-valuea | |
| Mean age at diagnosis or recruitment (SD) | 56.0 (11.2) | 54.0 (11.0) | 0.006 | 47.7 (10.0) | 48.9 (11.4) | 0.366 | 65.9 (7.9) | 64.6 (7.7) | 0.005 |
| Mean European ancestry (SD)b | 0.55 (0.15) | 0.51 (0.15) | 0.002 | 0.54 (0.16) | 0.51 (0.15) | 0.218 | 0.55 (0.13) | 0.53 (0.14) | 0.021 |
| Mean Ind. American ancestry (SD)b | 0.37 (0.15) | 0.41 (0.15) | 0.003 | 0.38 (0.15) | 0.41 (0.13) | 0.159 | 0.38 (0.13) | 0.40 (0.13) | 0.007 |
| Mean African ancestry (SD)b | 0.08 (0.04) | 0.08 (0.07) | 0.582 | 0.08 (0.09) | 0.08 (0.05) | 0.753 | 0.06 (0.03) | 0.06 (0.03) | 0.199 |
| Premenopausal BMI | |||||||||
| <25 (%) | 38.7 | 21.3 | 0.008 | 38.3 | 24.1 | 0.264 | 43.1 | 40.9 | 0.755 |
| 25–29.9 (%) | 31.1 | 41.6 | 34.4 | 37.9 | 29.4 | 25.0 | |||
| 30+ (%) | 30.2 | 37.1 | 27.3 | 37.9 | 27.4 | 34.1 | |||
| Postmenopausal BMI | |||||||||
| <25 (%) | 24.3 | 16.0 | 0.063 | 32.6 | 50.0 | 0.331 | 27.5 | 29.7 | 0.913 |
| 25–29.9 (%) | 32.0 | 37.6 | 30.3 | 22.2 | 41.0 | 39.5 | |||
| 30+ (%) | 43.7 | 46.4 | 37.1 | 27.8 | 30.8 | 30.1 | |||
| Family history BC | 7.2 | 10.0 | 0.186 | 21.5 | 5.1 | 0.001 | 13.4 | 12.0 | 0.787 |
| Age at first FTP | |||||||||
| Nulliparous (%) | 13.0 | 6.3 | 0.003 | 17.2 | 13.5 | 0.302 | 10.1 | 7.3 | 0.006 |
| <20 years (%) | 20.0 | 26.3 | 21.1 | 32.2 | 31.9 | 39.4 | |||
| 20–30 years (%) | 55.1 | 57.0 | 48.5 | 44.1 | 48.5 | 48.2 | |||
| >30 years (%) | 11.6 | 10.3 | 13.1 | 10.2 | 6.8 | 3.6 | |||
| Age at menarche | |||||||||
| ≤12 years (%) | 54.8 | 45.2 | 0.02 | 48.8 | 28.8 | 0.005 | 48.3 | 48.2 | 0.120 |
| 13–14 years (%) | 32.1 | 39.0 | 39.6 | 49.2 | 36.8 | 40.7 | |||
| ≥15 years (%) | 13.1 | 15.8 | 11.6 | 22.0 | 12.8 | 10.4 | |||
| Number of FTP | |||||||||
| 0 (%) | 13.1 | 6.0 | <0.001 | 17.0 | 13.6 | 0.001 | 9.7 | 7.3 | 0.031 |
| 1–2 (%) | 35.5 | 28.7 | 43.2 | 35.6 | 25.6 | 19.2 | |||
| 3–5 (%) | 41.6 | 50.1 | 36.6 | 33.9 | 45.2 | 50.0 | |||
| 6+ (%) | 9.9 | 15.2 | 3.2 | 16.9 | 18.3 | 22.4 | |||
| Menopausal status | |||||||||
| Premenopausal (%) | 31.0 | 33.2 | 0.719 | 41.2 | 49.2 | 0.006 | 9.3 | 7.9 | 0.138 |
| Postmenopausal (%) | 60.3 | 59.2 | 50.2 | 32.2 | 65.9 | 62.7 | |||
| DK/Hyst (%) | 8.7 | 7.6 | 8.5 | 18.6 | 24.7 | 29.4 | |||
| HT use if postmenop. | |||||||||
| Never (%) | 38.0 | 44.8 | 0.057 | 57.8 | 42.1 | 0.276 | 39.7 | 44.1 | 0.126 |
| Ever (%) | 60.1 | 54.9 | 41.8 | 57.9 | 55.5 | 49.3 | |||
| DK (%) | 1.9 | 0.3 | 0.3 | 0.0 | 4.8 | 6.6 | |||
SD: standard deviation; Ind. American, Indigenous American; HT, hormone therapy; BC, breast cancer; BMI, body mass index; FTP, full-term pregnancies.
aP-values for two-way tables Fisher exact test of association for binary variables and two sample t test for continuous variables.
bThe difference in average global individual ancestry is not statistically significant between individuals with ER positive and negative tumors.
Association between locus-specific ancestry and breast cancer risk in US Latinas
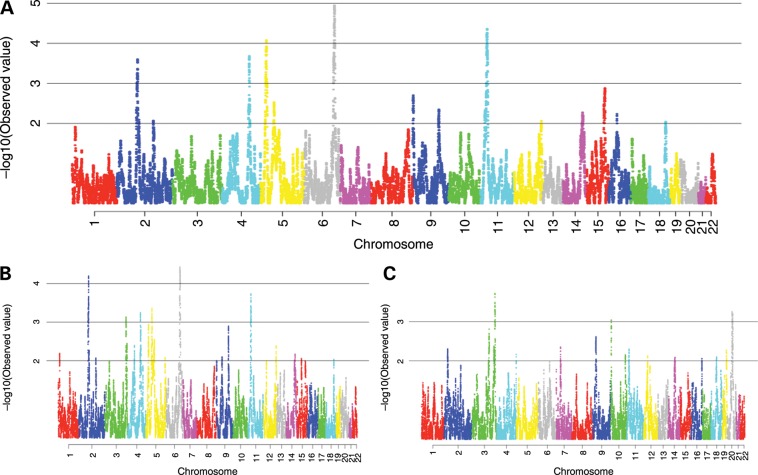
We found one region that showed a strong admixture mapping signal at 148–155Mb at 6q25 [odds ratio (OR) per Indigenous American chromosome 0.75, 95% confidence interval (CI): 0.65–0.85, P= 1.1 × 10−5]. A second region at 12–23 Mb at 11p15 showed a slightly weaker association (OR per Indigenous American chromosome 0.77, 95% CI: 0.68–0.87, P= 4.3 × 10−5). There were also regions on 5p15 (11–24 Mb), 4q28 (127–142 Mb) and 2p13 (68–76 Mb) showing suggestive associations (5p15 OR 1.31, 95% CI: 1.14–1.49, P= 8.3 × 10−5; 4q28 OR 1.29, 95% CI: 1.13–1.47, P= 2.0 × 10−4; 2p13 0.77, 95% CI: 0.68–0.89, P= 2.5 × 10−4) (Fig. 1). Previous GWAS have not reported risk variants within these last three regions (for a list of genes within each of these regions, see Supplementary Material, Table S1). For the two strongest signals at 6q25 and 11p15, increased Indigenous American ancestry was associated with reduced breast cancer risk. As expected, the results for the European component of ancestry were inversely correlated with the results of the Indigenous American component. We compared the results of the HAPMIX (24) -based admixture mapping analysis to results obtained using the LAMP 2.5 software for locus-specific ancestry estimation (25) and the signals were consistent between the two analyses.
Figure 1.
Results of breast cancer admixture mapping. On the X-axis are genomic positions by chromosome. On the Y-axis are the negative log10 P-values for the association between locus-specific ancestry and breast cancer risk. (A) Admixture mapping for Indigenous American component. (B) Admixture mapping for European component. (C) Admixture mapping for Africa component.
Genome-wide significance for admixture mapping has been empirically evaluated for African Americans (26), but has not yet been empirically evaluated for Latin American populations. Therefore, we evaluated the significance of the admixture mapping peaks by means of a permutation test (Supplementary Material, Table S3). Case/control status was permuted within five individual Indigenous American ancestry categories to reproduce the asymmetry of the global ancestry distribution between cases and controls at each permutation. A signal equal or stronger to the one at 6q25 occurred in 21 of 1000 permutations (Pcorrected = 0.022) and in 78 of 1000 (Pcorrected = 0.079) at 11p15. The signals at 5p15, 4q28 and 2p13 had a corrected P-value > 0.1 and are no longer discussed.
The 7 Mb region at 6q25 contains 74 genes, with the peak of the signal spanning the estrogen receptor 1 (ESR1) gene [MIM: 133430], which has been previously identified as a susceptibly locus for breast cancer reported in GWAS in Asian and European ancestry populations (18,27,28). The 11 Mb region at 11p15 contains 110 genes, with the peak of the signal spanning the neuron navigator 2 (NAV2) gene [MIM: 607026] (Supplementary Material, Table S1).
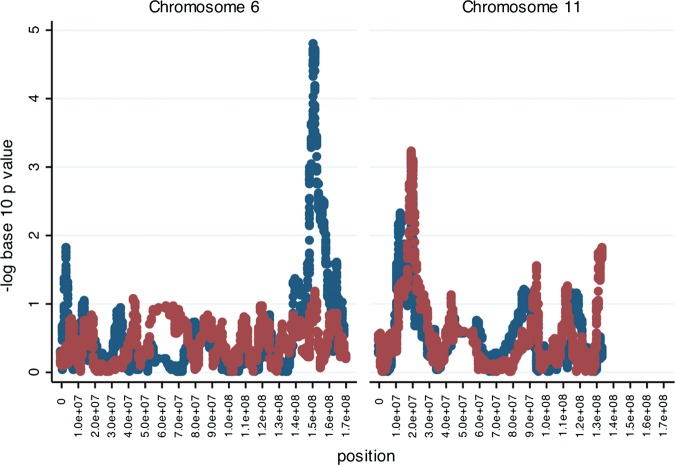
There is increasing evidence of etiologic heterogeneity due to both genetic and non-genetic risk factors for breast cancer subtypes (19,20,29–31). Therefore, we performed hypothesis-generating analyses by subgroups defined by ER status (Fig. 2, Supplementary Material, Fig. S2) for the two regions with significant admixture mapping signals. The signal at 6q25 was stronger for ER-positive breast cancer (n= 827 cases) as opposed to the association with ER-negative breast cancer (n= 297 cases). There was no apparent difference at 11p15 by ER status. In examining the signals for ER-positive breast cancer, we observed no other significant or near-significant peaks besides the 6q25 locus. The difference in the strength of the ER-positive signal could be due to the smaller sample size for the ER-negative tumors. We further tested for heterogeneity by evaluating the effect of locus-specific ancestry at these two regions by ER-positive versus ER-negative disease. We did not find statistically significant associations. For the 6q25 region, the OR for the association of Indigenous American locus-specific ancestry and risk of ER-positive disease (versus ER-negative disease) was 0.76 (95%CI: 0.47–1.23, P= 0.27) and, for the 11p15 region, it was 1.25 (95%CI: 0.78–2.01, P= 0.36).
Figure 2.
Results of ER subtype-specific breast cancer admixture mapping for the Indigenous American component. On the X-axis are genomic positions within each chromosome. On the Y-axis are the negative log10 P-values for the association between locus-specific ancestry and ER-specific breast cancer subtype. Results for ER-positive analyses are in blue and for ER-negative analyses are in red.
Body mass index is a strong predictor of post-menopausal breast cancer risk (32–35) and also varies with genetic ancestry (36). Therefore, we re-tested the models for locus-specific ancestry after adjustment for age and body mass index. We found that the results were unchanged for both 6q25 (OR per Indigenous = 0.74, P= 1.4 × 10−5) and 11p15 (OR per Indigenous chromosome = 0.76, P= 5.6 × 10−5).
Attempt to fine map the admixture signals at 6q25 and 11p15
6q25: multiple single nucleotide polymorphisms (SNPs) at 6q25 in the 5′ of ESR1 have been associated with breast cancer in previous studies (18,22,27,28). In Latinas, we found no statistically significant associations between these previously reported variants and breast cancer risk [rs3757318 (22) (G/A) OR = 1.18, P= 0.15, risk allele frequency (RAF): 0.07 (A allele); rs2046210 (18) (G/A) OR = 1.07, P= 0.23, RAF: 0.28 (A allele); rs9397435 (28) (A/G) OR = 1.15, P= 0.22, RAF: 0.06 (G allele); rs6913578 (27) (A/C) OR = 1.09, P= 0.17, RAF: 0.25 (C allele)], nor evidence that these variants account for the admixture signal in the region (OR per Indigenous American chromosome 0.75, 95%CI: 0.69–0.81, P= 3.0 × 10−5 adjusting for known risk SNPs). We then investigated the possibility that a single variant not previously reported may explain the association between locus-specific ancestry and breast cancer risk. We tested genotyped and imputed SNPs (imputed from the 1000 genome project, http://www.1000genomes.org) that were nominally significant for association with breast cancer risk in analyses adjusted for global and locus-specific ancestry; within the 7 Mb admixture signal at 6q25, there were 206 SNPs (out of 16 690 with minor allele frequency (MAF) > 5%) with a P< 0.001 (Supplementary Material, Table S2). We found that no individual SNP could completely attenuate the locus-specific ancestry signal (Fig. 3). The strongest single association was with rs79692348 which attenuated the association with locus-specific ancestry but did not completely eliminate it (OR = 0.82 per Indigenous chromosome, P= 0.0099).
Figure 3.
Admixture mapping and SNP association results. The solid line represents the admixture mapping signal. The blue dots represent the negative log10 P-values of association between single SNPs and breast cancer risk (all SNPs that showed an association with breast cancer risk at a P-value ≤ 0.05 were included). The red dots represent the locus-specific ancestry association after adjustment for selected individual SNPs (SNPs that showed an association with breast cancer at a P-value ≤ 0.001). The blue dots with black hollow squares are the SNPs that modified the locus-specific ancestry association signal represented by the red dots. (A) 6q25 region. (B) 11p15 region.
Based on this observation, we investigated models with more than one SNP. We used a step-forward approach and successively included the SNPs with the lowest P-values within the region that were weakly correlated with each other (r2≤ 0.2). We stopped adding SNPs at the point in which the locus-specific ancestry association had a P-value > 0.05. At 6q25, a multi-SNP model could explain the locus-specific ancestry signal with four SNPs (rs79692348, rs4304175, rs9383928, rs7747474). Three of these SNPs are within the 5′ region of the ESR1 locus (within the C6orf97 gene). In addition, none of these SNPs is correlated with those previously reported to be associated with breast cancer at the ESR1 locus (r2 ≤ 0.1). The rs79692348 marker, which shows the strongest association with risk in the SNP association analysis, has an allele frequency of <1% in African and European populations (Supplementary Material, Table S4).
11p15: there were 128 SNPs (out of 26 051 with MAF > 5%) that were nominally significant (P< 0.001) in the GWAS case–control analysis in the 11 Mb region (Supplementary Material, Table S2). In conditional analyses with locus-specific ancestry, no individual SNP could fully account for the local ancestry signal (Fig. 3). However, a two-SNP model attenuated the locus-specific ancestry completely (rs10444227, rs4757841) (Table 2). These two SNPs are within the NAV2 gene. Allele frequencies for these markers differ slightly between Asians/Indigenous American and European populations and are less common in Africans (Supplementary Material, Table S4).
Table 2.
Locus-specific ancestry (LSA) association adjusting for multiple SNPs
| Multi-SNP model |
Single-SNP association |
||||||
|---|---|---|---|---|---|---|---|
| SNP id | Position | ORa | P-valueb | Allelesc | Allele freqd | ORe | P-value |
| LSA 6q25 | 0.74 | 9.42×106 | |||||
| s79692348 | 151937679 | 0.82 | 0.0099 | C/T | 0.87 (0.99) | 1.52 | 2.10×106 |
| rs4304175 | 151923982 | 0.83 | 0.0134 | T/C | 0.41 (0.30) | 0.79 | 2.60×105 |
| rs9383928 | 151896937 | 0.84 | 0.0178 | A/G | 0.72 (0.79) | 1.30 | 2.90×105 |
| rs7747474 | 149565986 | 0.88 | 0.0955 | T/C | 0.12 (0.25) | 1.44 | 3.00×105 |
| LSA 11p15 | 0.77 | 3.81×105 | |||||
| rs10444227 | 19945852 | 0.83 | 0.0051 | A/C | 0.70 (0.52) | 0.76 | 6.40×106 |
| rs4757841 | 19767614 | 0.89 | 0.0915 | A/G | 0.39 (0.49) | 1.26 | 4.20×105 |
aOR shows effect of LSA per chromosome.
bThese are the P-values for the effect of LSA in the model with no adjustment and the models adjusting for multiple SNPs.
cTested allele/reference allele.
dAllele frequency for the tested allele in US Latinas (in parenthesis is the frequency in Europeans, Supplementary Material, Table S4).
eOR per allele for each SNP (per allele) and breast cancer risk.
The 11p15 signal is 18 Mb from the LSP1 gene [MIM: 153432] found to be associated with breast cancer risk in previous GWAS (17,22), and therefore does not overlap with the LSP1 region association previously reported. The published associated SNPs within the LSP1 region did not show a statistically significant association in the Latinas GWAS analysis [rs909116 (C/T), P= 0.14 and rs3817198 (C/T), P= 0.45]. The allele frequencies for the two published SNPs are similar between Europeans and the US Latinas (0.53/0.57 and 0.30/0.22). Therefore, it is unlikely that an association with the SNPs within the LSP1 gene is driving the admixture mapping results on 11p15.
DISCUSSION
In the first breast cancer admixture mapping scan in US Latinas, we found a genome-wide statistically significant signal of increased European ancestry among cases compared with controls at 6q25. A second region at 11p15 also demonstrated a trend towards increased European ancestry among cases. At both regions, higher Indigenous American ancestry was associated with reduced breast cancer risk, whereas higher European ancestry was associated with increased risk. The direction of the locus-specific ancestry associations is consistent with our previous result showing that global individual ancestry in US Latina and Mexican women is associated with breast cancer risk, with higher Indigenous American ancestry being associated with reduced risk (2,3). The associations with locus-specific ancestry could not be explained by any single common SNP from the GWAS data. Thus, these signals are likely to be reflecting associations with variants in these regions that are biologically functional and that are not well captured by the SNPs we tested.
The peak of the signal at 6q25 is located at the ESR1 locus, which has been associated with breast cancer in previous GWAS in Asians and Europeans (18,22). Even though different studies have reported associations near the ESR1 gene, results are not consistent as different SNPs seem to be responsible for the association in different populations (22,28). The original GWAS study that reported an association between variant rs2046210 and breast cancer was conducted in a sample of Chinese women from Shanghai (18). Turnbull et al. (22) evaluated the 6q25.1 region in breast cancer cases and controls of European ancestry and reported that another SNP (rs3757318) showed the strongest association (r2 with rs2046210 of 0.48 and 0.09 in Asian and European populations from HapMap, respectively). A multiethnic fine-mapping study in samples of European, Asian and African origin reported a third marker (rs9397435) to be consistently associated with risk across populations (r2 > 0.65 with rs2046210 and rs3757318 in Asians) (28). A later study involving Chinese, Japanese, European and African American samples reported that the effect of the rs2046210 variant might be due to its association (r2 = 0.91 in Chinese and 0.83 in individuals of European ancestry) with another putatively functional variant identified in the region, rs6913578 (27).
None of these SNPs was significantly associated with breast cancer risk in our analyses, although statistical power to detect the reported OR's was limited (we had 25–80% power to detect the reported effect sizes for these risk variants). The ORs for these previously published SNPs in our study were in the same direction as those reported.
Our results showed that there were no individual SNPs within the admixture mapping signal that accounted for the locus-specific ancestry association. Instead, we found that a multi-SNP model, comprised of SNPs that were associated with risk (P< 0.001) and that were weakly correlated, could attenuate the admixture mapping signal. Our results suggest that the locus-specific ancestry signal as well as the multi-SNP model may be capturing the presence of a separate single or multiple risk variants for breast cancer at this locus. Three of the SNPs in the multi-SNP model are located within the C6orf97 gene. A recent study showed three open reading frames located upstream 50–250 Kb of ESR1 (C6orf97, C6orf96 and C6orf211) that are co-expressed with ESR1 (37). Therefore, the biologically functional variant(s) at this region may either directly affect ESR1 expression and activity or affect the expression and activity of one or more of these other genes.
In the 11p15 region, several SNPs were independently associated with breast cancer risk (the lowest P-value was 6.4 × 10−6) (Supplementary Material, Table S2); however, none of these SNPs alone could account for the strong association with local ancestry. All of the variants are located within the NAV2 gene also known as helicase, APC down-regulated 1 (HELAD1) and retinoic acid inducible in neuroblastoma cells (RAINB1). NAV2 spans 400 Kb and has been reported to be up-regulated in colorectal carcinomas (38) and to be involved in neuronal development (39). Like at 6q25, multiple SNPs define the admixture signal; however, their independent associations were not strong which suggests that local ancestry is the best proxy of any biologically functional variant(s) in the region.
We were not able to find any individual SNP that could explain the association between breast cancer risk and local ancestry in these regions. Prior studies utilizing admixture mapping have revealed a single common variant that underlies the association between IL6 soluble receptor level (6) and white blood cell count (12). However, admixture mapping for prostate cancer in African Americans revealed several different independent SNPs that were responsible for the association with African ancestry at 8q24 (40). In addition, the association of end-stage renal disease and ancestry at the 22q12 locus, initially identified by admixture mapping, was thought to be in MYH9 [MIM: 160775] (41,42). However, fine-mapping and sequencing at these region revealed that the association was actually best explained, and more likely due to two variants in the APOL1 [MIM: 603743] gene which is nearby (43,44). Therefore, care should be taken with interpreting SNP associations in an admixture mapping locus until detailed fine-mapping data are available.
Recently, simulation studies have demonstrated that rare variants may underlie some of the associations with common variants detected by GWAS (45). Since no single common variant in our data explains the signal, but multiple common variants attenuate the signal, one plausible explanation may be that multiple rare variants at the 6q25 locus may be accounting for the admixture mapping result that we observed. This would explain why unlinked common variants are explaining the admixture mapping signal. The ideal way to resolve this question would be through sequencing studies at both loci, followed by functional work.
Our results demonstrate that there are genomic regions that may harbor risk variants that vary in frequency between ancestral populations and influence differences in breast cancer incidence among US racial/ethnic groups. In previous studies, we demonstrated that at least part of the association between breast cancer and genetic ancestry among US Latinas and Mexican women was due to confounding between genetic ancestry and non-genetic risk factors (2,3,36). In addition, there is evidence showing that breast cancer risk increases for second and third generation US Latinas compared with recent immigrants (46) which supports the hypothesis that reproductive, lifestyle and demographic factors strongly contribute to breast cancer risk. Therefore, differences in both genetic and non-genetic factors are likely to contribute to differences in breast cancer risk between racial/ethnic groups.
In summary, we used an admixture mapping approach to identify two regions where genetic ancestry is associated with breast cancer susceptibility among Latina women. One of them, 6q25, is genome-wide statistically significant and has been previously associated with breast cancer risk, but the specific variants reported to affect risk do not explain the association in our data. The other signal, 11p15, is novel but is marginally significant at the genome-wide level and will require further replication to be confirmed. We were not able to find any one variant that explained the ancestry associations at either locus. Fine-mapping within these regions will lead to the discovery of the variant or variants that contribute to the admixture mapping associations and will improve our understanding of the architecture of breast cancer risk predisposition in the Latina population.
MATERIALS AND METHODS
All participants provided written informed consent as approved by local Human Subjects Committees.
Samples
The SFBCS is a population-based case–control study of breast cancer, which includes 821 Latina breast cancer cases and 916 Latina controls (47,48). Cases aged 35–79 years and diagnosed with invasive breast cancer from 1995 to 2002 were identified through the Greater Bay Area Cancer Registry. Controls were identified by random-digit dialing in the same geographic region and were frequency matched by 5-year age intervals. Blood specimen collection was initiated in 1999. The present analysis includes 351 cases and 579 controls from this study who self-identified their ethnic background as Latina or Hispanic.
The BCFR is an international, NCI-funded resource that has recruited and followed over 13 000 breast cancer families (49). The present study includes BCFR samples from the population-based Northern California site of the BCFR (NC-BCFR). Cases aged 18–64 years and diagnosed from 1995 to 2007 were ascertained through the Greater Bay Area Cancer Registry. Cases with indicators of increased genetic susceptibility (diagnosis at age <35 years, bilateral breast cancer with the first diagnosis at age <50 years, a personal history of ovarian or childhood cancer, a family history of breast or ovarian cancer in first-degree relatives) were over-sampled. Cases not meeting these criteria were randomly sampled (50). Population controls were identified through random-digit dialing and frequency matched on 5-year age group to cases diagnosed from 1995 to 1998. The present study includes 641 cases and 61 controls from the NC-BCFR.
The MEC is a large prospective cohort study in California (mainly Los Angeles County) and Hawaii (51). The breast cancer study is a nested case–control study, including women with invasive breast cancer age >50 and controls matched by age and self-identified ethnicity (51). For the current study, we used data and DNA samples from 546 Latina women with breast cancer and 558 matched controls.
Genetics of Asthma in Latino Americans (GALA1): the GALA1 study is a family-based study (including children with asthma and their parents) of pediatric asthma in Latino Americans (52). The sample includes 294 individuals of Mexican origin and 365 individuals from Puerto Rico. We included 112 females of Mexican origin from the GALA1 study to our set of population controls. The individuals are between 11 and 42 years of age (85% are older than 20).
Genotyping and quality-control procedures
The SFBCS, NC-BCFR and GALA samples were genotyped with the Affymetrix 6.0 array according to the manufacturer's instructions (https://www.affymetrix.com) in the laboratory of Esteban Gonzalez Burchard at UCSF. The MEC samples were genotyped with the Illumina Infinitum 660W array (http://www.illumina.com) in the Epigenome Data Production Center at USC.
We excluded 15 cases and 30 controls from the SFBCS/NC-BCFR/GALA set that had a genotyping call rate <95% or showed either known or cryptic relatedness. We excluded 48 samples from the MEC that had a genotyping call rate of <95% and 34 that showed either known or cryptic relatedness. The final sample included 1699 individuals from the SFBCS/NC-BCFR/GALA set (977 cases and 722 controls) and 1070 from the MEC (520 cases and 550 controls).
We excluded all SNPs with minor allele frequency of <1% and call rate of <99%. In order to reduce the noise of our locus-specific ancestry estimates as a result of stranding issues and genotyping error, we filtered G/C and A/T SNPs (since for those changes it is more difficult to detect stranding issues) and SNPs that had large differences between expected and observed frequencies in the admixed individuals based on the allele frequencies of the ancestral populations [Europeans from the Human Genome Diversity Project (HGDP) and Behar et al. (53), HapMap Yorubans (http://hapmap.ncbi.nlm.nih.gov) and Indigenous Americans from Reich et al., in submission]. The final number of SNPs included in the admixture mapping meta-analysis after filtering and matching with ancestral genotypes was 59 211.
Locus-specific ancestry estimation
There are various available methods for locus-specific ancestry estimation using genomewide data (24,25,54) that have relatively low error rates when the admixed populations are composed of well-differentiated ancestral groups (e.g. African Americans, Latinos) (24,55). In our analysis, we used the HAPMIX software to estimate locus-specific ancestry, which is a model-based method that uses detailed haplotype information (24). We developed an add-on extension to the HAPMIX method that supports local ancestry inference from three source populations. By default, HAPMIX supports ancestry inference from two source populations, but Latinos are three-way mixed. Briefly, this method works by first running HAPMIX three different ways, assigning reference panel 1 with haplotypes from one of Indigenous American, European or African samples and reference panel 2 with the combined haplotypes from the other two ancestries. The output from each of these three runs provides probabilistic estimates for the number of copies (0, 1 or 2) of ancestry from panel 1. To determine the ancestral state at each locus, the method uses ordinary least squares to solve a system of nine equations equivalent to these output probabilities, determining ancestry probabilities for the six unknown possible diploid ancestral types (Indigenous American/Indigenous American, Indigenous American/European, Indigenous American/African, European/European, European/African, African/African). In simulations, this method has between 93.2 and 97.8% accuracy. We also used the program LAMP (version 2.5) (25) to obtain locus-specific ancestry and compared the results with those obtained using HAPMIX. We estimated global individual ancestry as the average locus-specific ancestry across all loci for each individual.
Statistical analysis
The significance of the difference in mean values for age at diagnosis and overall genetic ancestry between cases and controls was assessed using t-tests. We used Fisher's exact test for evaluating the difference in the distribution of categorical variables between cases and controls (number of full-term pregnancies, family history of breast cancer, age at menarche, age at first full-term pregnancy, menopausal status and hormone therapy and body mass index). The program Stata 11 was used for these analyses (56).
We applied logistic regression models to test the association between locus-specific ancestry and breast cancer risk, including Indigenous American and African individual ancestry as covariates for the SFBCS/NC-BCFR/GALA samples and the MEC samples separately. We combined the results of the two analyses using the inverse variance method. These analyses were conducted using R (57).
To test if a particular SNP would account for the locus-specific ancestry association in any of the two regions with strong admixture mapping signals, we conducted a logistic regression for each of the SNPs within the signal that was associated with breast cancer risk at a 5% level of significance (Supplementary Material, Table S2). The SNP association analysis included genotyped as well as imputed SNPs for better coverage. Sample data were phased and missing markers were imputed using the software Beagle 3.3 (58). Phased data of 1094 samples from the 1000 Genomes Project (www.1000genomes.org) were used as the reference data set. These samples are from African, African American, Asian, Caucasian and Native American populations. The correlation between the allele dosage with highest posterior probability and the true allele dosage for the marker was evaluated by Beagle 3.3. Markers with the square of correlation coefficient >0.8 were used in further analyses. The logistic regression included the locus-specific ancestry at the peak of the signal, the individual Indigenous American and African ancestries and the genotypes for the particular SNP being tested. The data from the different studies were combined and a study variable was included in the analysis as a covariate.
We evaluated the effect of multiple SNPs on the admixture mapping signal by means of a logistic regression model that included the locus-specific ancestry at the peak of the signal, individual Indigenous American and African ancestries, and we added one SNP at a time, starting with the SNP that showed the strongest association with breast cancer risk. The final model included the minimum number of SNPs that contributed to the attenuation of the locus-specific ancestry signal. We stopped adding SNPs to the model once the locus-specific ancestry signal had an associated P-value of more than 0.05. We ran one model in which the successively added SNPs could not be highly correlated (r2 ≤ 0.2) and another model in which we allowed for correlation up to an r2 of 0.4. Results were similar. Samples were pooled for these analyses.
Permutation procedure
A permutation procedure was conducted to obtain an empirical distribution of Z statistics that would allow us to evaluate the significance of the admixture mapping signals. Case/control status was permuted for the SFBCS/NC-BCFR/GALA and MEC samples separately within five individual Indigenous American ancestry categories (0–20, 20–40, 40–60, 60–80 and 80–100) and at each permutation we calculated the association between locus-specific ancestry and breast cancer risk controlling for global Indigenous American and African ancestry. By permuting within the five ancestry categories, we were able to reproduce the asymmetry of the global ancestry distribution between cases and controls at each permutation. We then combined the results of each permutation for the two sets of samples using the inverse variance method. The significance of the admixture mapping signals was evaluated by comparing the value of the Z statistics of the original results with the maximum Z statistic for a thousand permutations (Supplementary Material, Table S3).
Adding to the ancestry-based permutation procedure, we evaluated how well controlling for global Indigenous American and African ancestry in the admixture mapping analysis addressed the issue of the asymmetry in the distribution of global ancestry between cases and controls. We compared the distribution of the (Z)2 statistics from our analysis to the expected χ2 distribution under the null and we observed that the inflation of high Z statistics in our study was due to the statistics obtained within the regions that show a strong locus-specific ancestry association. Once those regions were removed, the QQ plot showed that there was no inflation in our statistics compared with the expectation (Supplementary Materials, Fig. S3).
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the National Cancer Institute [R01 CA120120 (E.Z.); R25 CA112355 (L.F.), R01 CA132839 (C.A.H.)]. L.F. was partially supported by a BIRCWH K12 training grant sponsored by NIH/NICHHD (5K12HD052163) and a K01 award by NIH/NCI (CA160607). Additional support came from the National Institutes of Health [R01 HG006399 (B.P.)], the NIH Ruth Kirschstein F32 award (A.W.) and the Beatriu de Pinos Program [2006 BP-A 10144 and 2009 BP-B 00274 (MV)]. Each of the participating studies was supported by the following grants: SFBCS (National Institutes of Health grant R01-63446 and R01-CA77305, California Breast Cancer Research Program grant 7PB-0068); Cancer Prevention Institute of California (U01 CA69417); University of California, Irvine Informatics Support Center (U01 CA078296); MEC (National Institutes of Health grants R01 CA63464 and R37 CA54281); GALA1 (The National Institutes of Health (HL078885, AI077439, HL088133), the Flight Attendant Medical Research Institute (FAMRI), American Asthma Foundation and the Sandler Foundation). The Breast Cancer Family Registry (BCFR) was supported by the National Cancer Institute, National Institutes of Health under RFA CA-06-503 and through cooperative agreements with members of the BCFR and Principal Investigators. The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in the BCFR, nor does mention of trade names, commercial products or organizations imply endorsement by the US Government or the BCFR. The funders of this research had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Supplementary Material
ACKNOWLEDGEMENTS
We want to thank the study participants.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Jemal A., Siegel R., Xu J., Ward E. Cancer statistics, 2010. CA Cancer J. Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Fejerman L., John E.M., Huntsman S., Beckman K., Choudhry S., Perez-Stable E., Burchard E.G., Ziv E. Genetic ancestry and risk of breast cancer among U.S. Latinas. Cancer Res. 2008;68:9723–9728. doi: 10.1158/0008-5472.CAN-08-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fejerman L., Romieu I., John E.M., Lazcano-Ponce E., Huntsman S., Beckman K.B., Perez-Stable E.J., Gonzalez Burchard E., Ziv E., Torres-Mejia G. European ancestry is positively associated with breast cancer risk in Mexican women. Cancer Epidemiol. Biomarkers Prev. 2010;19:1074–1082. doi: 10.1158/1055-9965.EPI-09-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nalls M.A., Wilson J.G., Patterson N.J., Tandon A., Zmuda J.M., Huntsman S., Garcia M., Hu D., Li R., Beamer B.A., et al. Admixture mapping of white cell count: genetic locus responsible for lower white blood cell count in the Health ABC and Jackson Heart studies. Am. J. Hum. Genet. 2008;82:81–87. doi: 10.1016/j.ajhg.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pritchard J.K., Stephens M., Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reich D., Patterson N., Ramesh V., De Jager P.L., McDonald G.J., Tandon A., Choy E., Hu D., Tamraz B., Pawlikowska L., et al. Admixture mapping of an allele affecting interleukin 6 soluble receptor and interleukin 6 levels. Am. J. Hum. Genet. 2007;80:716–726. doi: 10.1086/513206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Winkler C.A., Nelson G.W., Smith M.W. Admixture mapping comes of age. Annu. Rev. Genomics Hum. Genet. 2010;11:65–89. doi: 10.1146/annurev-genom-082509-141523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deo R.C., Patterson N., Tandon A., McDonald G.J., Haiman C.A., Ardlie K., Henderson B.E., Henderson S.O., Reich D. A high-density admixture scan in 1,670 African Americans with hypertension. PLoS Genet. 2007;3:e196. doi: 10.1371/journal.pgen.0030196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freedman M.L., Haiman C.A., Patterson N., McDonald G.J., Tandon A., Waliszewska A., Penney K., Steen R.G., Ardlie K., John E.M., et al. Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-American men. Proc. Natl Acad. Sci. USA. 2006;103:14068–14073. doi: 10.1073/pnas.0605832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng C.Y., Kao W.H., Patterson N., Tandon A., Haiman C.A., Harris T.B., Xing C., John E.M., Ambrosone C.B., Brancati F.L., et al. Admixture mapping of 15,280 African Americans identifies obesity susceptibility loci on chromosomes 5 and X. PLoS Genet. 2009;5:e1000490. doi: 10.1371/journal.pgen.1000490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jia L., Landan G., Pomerantz M., Jaschek R., Herman P., Reich D., Yan C., Khalid O., Kantoff P., Oh W., et al. Functional enhancers at the gene-poor 8q24 cancer-linked locus. PLoS Genet. 2009;5:e1000597. doi: 10.1371/journal.pgen.1000597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reich D., Nalls M.A., Kao W.H., Akylbekova E.L., Tandon A., Patterson N., Mullikin J., Hsueh W.C., Cheng C.Y., Coresh J., et al. Reduced neutrophil count in people of African descent is due to a regulatory variant in the Duffy antigen receptor for chemokines gene. PLoS Genet. 2009;5:e1000360. doi: 10.1371/journal.pgen.1000360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahmed S., Thomas G., Ghoussaini M., Healey C.S., Humphreys M.K., Platte R., Morrison J., Maranian M., Pooley K.A., Luben R., et al. Newly discovered breast cancer susceptibility loci on 3p24 and 17q23.2. Nat. Genet. 2009;41:585–590. doi: 10.1038/ng.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gold B., Kirchhoff T., Stefanov S., Lautenberger J., Viale A., Garber J., Friedman E., Narod S., Olshen A.B., Gregersen P., et al. Genome-wide association study provides evidence for a breast cancer risk locus at 6q22.33. Proc. Natl Acad. Sci. USA. 2008;105:4340–4345. doi: 10.1073/pnas.0800441105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hunter D.J., Kraft P., Jacobs K.B., Cox D.G., Yeager M., Hankinson S.E., Wacholder S., Wang Z., Welch R., Hutchinson A., et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat. Genet. 2007;39:870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomas G., Jacobs K.B., Kraft P., Yeager M., Wacholder S., Cox D.G., Hankinson S.E., Hutchinson A., Wang Z., Yu K., et al. A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1) Nat. Genet. 2009;41:579–584. doi: 10.1038/ng.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Easton D.F., Pooley K.A., Dunning A.M., Pharoah P.D., Thompson D., Ballinger D.G., Struewing J.P., Morrison J., Field H., Luben R., et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng W., Long J., Gao Y.T., Li C., Zheng Y., Xiang Y.B., Wen W., Levy S., Deming S.L., Haines J.L., et al. Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nat. Genet. 2009;41:324–328. doi: 10.1038/ng.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stacey S.N., Manolescu A., Sulem P., Thorlacius S., Gudjonsson S.A., Jonsson G.F., Jakobsdottir M., Bergthorsson J.T., Gudmundsson J., Aben K.K., et al. Common variants on chromosome 5p12 confer susceptibility to estrogen receptor-positive breast cancer. Nat. Genet. 2008;40:703–706. doi: 10.1038/ng.131. [DOI] [PubMed] [Google Scholar]
- 20.Stacey S.N., Manolescu A., Sulem P., Rafnar T., Gudmundsson J., Gudjonsson S.A., Masson G., Jakobsdottir M., Thorlacius S., Helgason A., et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat. Genet. 2007;39:865–869. doi: 10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- 21.Long J., Cai Q., Shu X.O., Qu S., Li C., Zheng Y., Gu K., Wang W., Xiang Y.B., Cheng J., et al. Identification of a functional genetic variant at 16q12.1 for breast cancer risk: results from the Asia Breast Cancer Consortium. PLoS Genet. 2010;6:e1001002. doi: 10.1371/journal.pgen.1001002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Turnbull C., Ahmed S., Morrison J., Pernet D., Renwick A., Maranian M., Seal S., Ghoussaini M., Hines S., Healey C.S., et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat. Genet. 2010;42:504–507. doi: 10.1038/ng.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fletcher O., Johnson N., Orr N., Hosking F.J., Gibson L.J., Walker K., Zelenika D., Gut I., Heath S., Palles C., et al. Novel breast cancer susceptibility locus at 9q31.2: results of a genome-wide association study. J. Natl Cancer Inst. 2011;103:425–435. doi: 10.1093/jnci/djq563. [DOI] [PubMed] [Google Scholar]
- 24.Price A.L., Tandon A., Patterson N., Barnes K.C., Rafaels N., Ruczinski I., Beaty T.H., Mathias R., Reich D., Myers S. Sensitive detection of chromosomal segments of distinct ancestry in admixed populations. PLoS Genet. 2009;5:e1000519. doi: 10.1371/journal.pgen.1000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pasaniuc B., Sankararaman S., Kimmel G., Halperin E. Inference of locus-specific ancestry in closely related populations. Bioinformatics. 2009;25:i213–i221. doi: 10.1093/bioinformatics/btp197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Montana G., Pritchard J.K. Statistical tests for admixture mapping with case-control and cases-only data. Am. J. Hum. Genet. 2004;75:771–789. doi: 10.1086/425281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cai Q., Wen W., Qu S., Li G., Egan K.M., Chen K., Deming S.L., Shen H., Shen C.Y., Gammon M.D., et al. Replication and functional genomic analyses of the breast cancer susceptibility locus at 6q25.1 generalize its importance in women of Chinese, Japanese, and European ancestry. Cancer Res. 2011;71:1344–1355. doi: 10.1158/0008-5472.CAN-10-2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stacey S.N., Sulem P., Zanon C., Gudjonsson S.A., Thorleifsson G., Helgason A., Jonasdottir A., Besenbacher S., Kostic J.P., Fackenthal J.D., et al. Ancestry-shift refinement mapping of the C6orf97-ESR1 breast cancer susceptibility locus. PLoS Genet. 2010;6:e1001029. doi: 10.1371/journal.pgen.1001029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bauer K.R., Brown M., Cress R.D., Parise C.A., Caggiano V. Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype: a population-based study from the California cancer Registry. Cancer. 2007;109:1721–1728. doi: 10.1002/cncr.22618. [DOI] [PubMed] [Google Scholar]
- 30.Millikan R.C., Newman B., Tse C.K., Moorman P.G., Conway K., Dressler L.G., Smith L.V., Labbok M.H., Geradts J., Bensen J.T., et al. Epidemiology of basal-like breast cancer. Breast Cancer Res. Treat. 2008;109:123–139. doi: 10.1007/s10549-007-9632-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carey L.A., Perou C.M., Livasy C.A., Dressler L.G., Cowan D., Conway K., Karaca G., Troester M.A., Tse C.K., Edmiston S., et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 2006;295:2492–2502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 32.Schapira D.V., Kumar N.B., Lyman G.H., Cox C.E. Abdominal obesity and breast cancer risk. Ann. Intern. Med. 1990;112:182–186. doi: 10.7326/0003-4819-112-3-182. [DOI] [PubMed] [Google Scholar]
- 33.Morimoto L.M., White E., Chen Z., Chlebowski R.T., Hays J., Kuller L., Lopez A.M., Manson J., Margolis K.L., Muti P.C., et al. Obesity, body size, and risk of postmenopausal breast cancer: the Women's Health Initiative (United States) Cancer Causes Control. 2002;13:741–751. doi: 10.1023/a:1020239211145. [DOI] [PubMed] [Google Scholar]
- 34.Petrelli J.M., Calle E.E., Rodriguez C., Thun M.J. Body mass index, height, and postmenopausal breast cancer mortality in a prospective cohort of US women. Cancer Causes Control. 2002;13:325–332. doi: 10.1023/a:1015288615472. [DOI] [PubMed] [Google Scholar]
- 35.Harvie M., Hooper L., Howell A.H. Central obesity and breast cancer risk: a systematic review. Obes. Rev. 2003;4:157–173. doi: 10.1046/j.1467-789x.2003.00108.x. [DOI] [PubMed] [Google Scholar]
- 36.Ziv E., John E.M., Choudhry S., Kho J., Lorizio W., Perez-Stable E.J., Burchard E.G. Genetic ancestry and risk factors for breast cancer among Latinas in the San Francisco Bay Area. Cancer Epidemiol. Biomarkers Prev. 2006;15:1878–1885. doi: 10.1158/1055-9965.EPI-06-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dunbier A.K., Anderson H., Ghazoui Z., Lopez-Knowles E., Pancholi S., Ribas R., Drury S., Sidhu K., Leary A., Martin L.A., et al. ESR1 is co-expressed with closely adjacent uncharacterised genes spanning a breast cancer susceptibility locus at 6q25.1. PLoS Genet. 2011;7:e1001382. doi: 10.1371/journal.pgen.1001382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ishiguro H., Shimokawa T., Tsunoda T., Tanaka T., Fujii Y., Nakamura Y., Furukawa Y. Isolation of HELAD1, a novel human helicase gene up-regulated in colorectal carcinomas. Oncogene. 2002;21:6387–6394. doi: 10.1038/sj.onc.1205751. [DOI] [PubMed] [Google Scholar]
- 39.Merrill R.A., Plum L.A., Kaiser M.E., Clagett-Dame M. A mammalian homolog of unc-53 is regulated by all-trans retinoic acid in neuroblastoma cells and embryos. Proc. Natl Acad. Sci. USA. 2002;99:3422–3427. doi: 10.1073/pnas.052017399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haiman C.A., Patterson N., Freedman M.L., Myers S.R., Pike M.C., Waliszewska A., Neubauer J., Tandon A., Schirmer C., McDonald G.J., et al. Multiple regions within 8q24 independently affect risk for prostate cancer. Nat. Genet. 2007;39:638–644. doi: 10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kao W.H., Klag M.J., Meoni L.A., Reich D., Berthier-Schaad Y., Li M., Coresh J., Patterson N., Tandon A., Powe N.R., et al. MYH9 is associated with nondiabetic end-stage renal disease in African Americans. Nat. Genet. 2008;40:1185–1192. doi: 10.1038/ng.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kopp J.B., Smith M.W., Nelson G.W., Johnson R.C., Freedman B.I., Bowden D.W., Oleksyk T., McKenzie L.M., Kajiyama H., Ahuja T.S., et al. MYH9 is a major-effect risk gene for focal segmental glomerulosclerosis. Nat. Genet. 2008;40:1175–1184. doi: 10.1038/ng.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Genovese G., Friedman D.J., Ross M.D., Lecordier L., Uzureau P., Freedman B.I., Bowden D.W., Langefeld C.D., Oleksyk T.K., Uscinski Knob A.L., et al. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science. 2010;329:841–845. doi: 10.1126/science.1193032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tzur S., Rosset S., Shemer R., Yudkovsky G., Selig S., Tarekegn A., Bekele E., Bradman N., Wasser W.G., Behar D.M., et al. Missense mutations in the APOL1 gene are highly associated with end stage kidney disease risk previously attributed to the MYH9 gene. Hum. Genet. 2010;128:345–350. doi: 10.1007/s00439-010-0861-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dickson S.P., Wang K., Krantz I., Hakonarson H., Goldstein D.B. Rare variants create synthetic genome-wide associations. PLoS Biol. 2010;8:e1000294. doi: 10.1371/journal.pbio.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.John E.M., Phipps A.I., Davis A., Koo J. Migration history, acculturation, and breast cancer risk in Hispanic women. Cancer Epidemiol. Biomarkers Prev. 2005;14:2905–2913. doi: 10.1158/1055-9965.EPI-05-0483. [DOI] [PubMed] [Google Scholar]
- 47.John E.M., Horn-Ross P.L., Koo J. Lifetime physical activity and breast cancer risk in a multiethnic population: the San Francisco Bay area breast cancer study. Cancer Epidemiol. Biomarkers Prev. 2003;12:1143–1152. [PubMed] [Google Scholar]
- 48.John E.M., Schwartz G.G., Koo J., Wang W., Ingles S.A. Sun exposure, vitamin D receptor gene polymorphisms, and breast cancer risk in a multiethnic population. Am. J. Epidemiol. 2007;166:1409–1419. doi: 10.1093/aje/kwm259. [DOI] [PubMed] [Google Scholar]
- 49.John E.M., Hopper J.L., Beck J.C., Knight J.A., Neuhausen S.L., Senie R.T., Ziogas A., Andrulis I.L., Anton-Culver H., Boyd N., et al. The Breast Cancer Family Registry: an infrastructure for cooperative multinational, interdisciplinary and translational studies of the genetic epidemiology of breast cancer. Breast Cancer Res. 2004;6:R375–R389. doi: 10.1186/bcr801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.John E.M., Miron A., Gong G., Phipps A.I., Felberg A., Li F.P., West D.W., Whittemore A.S. Prevalence of pathogenic BRCA1 mutation carriers in 5 US racial/ethnic groups. JAMA. 2007;298:2869–2876. doi: 10.1001/jama.298.24.2869. [DOI] [PubMed] [Google Scholar]
- 51.Kolonel L.N., Henderson B.E., Hankin J.H., Nomura A.M., Wilkens L.R., Pike M.C., Stram D.O., Monroe K.R., Earle M.E., Nagamine F.S. A multiethnic cohort in Hawaii and Los Angeles: baseline characteristics. Am. J. Epidemiol. 2000;151:346–357. doi: 10.1093/oxfordjournals.aje.a010213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Burchard E.G., Avila P.C., Nazario S., Casal J., Torres A., Rodriguez-Santana J.R., Toscano M., Sylvia J.S., Alioto M., Salazar M., et al. Lower bronchodilator responsiveness in Puerto Rican than in Mexican subjects with asthma. Am. J. Respir. Crit. Care Med. 2004;169:386–392. doi: 10.1164/rccm.200309-1293OC. [DOI] [PubMed] [Google Scholar]
- 53.Behar D.M., Yunusbayev B., Metspalu M., Metspalu E., Rosset S., Parik J., Rootsi S., Chaubey G., Kutuev I., Yudkovsky G., et al. The genome-wide structure of the Jewish people. Nature. 2010;466:238–242. doi: 10.1038/nature09103. [DOI] [PubMed] [Google Scholar]
- 54.Tang H., Coram M., Wang P., Zhu X., Risch N. Reconstructing genetic ancestry blocks in admixed individuals. Am. J. Hum. Genet. 2006;79:1–12. doi: 10.1086/504302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sankararaman S., Kimmel G., Halperin E., Jordan M.I. On the inference of ancestries in admixed populations. Genome Res. 2008;18:668–675. doi: 10.1101/gr.072751.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.StataCorp. College Station, TX: 2009. StataCorp LP. [Google Scholar]
- 57.R_Development_Core_Team. R Foundation for Statistical Computing. Vienna, Austria: 2010. [Google Scholar]
- 58.Browning B.L., Browning S.R. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am. J. Hum. Genet. 2009;84:210–223. doi: 10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.