Abstract
The oncoprotein Bcr-Abl drives aberrant downstream activity through trans-autophosphorylation of homo-oligomers in chronic myelogenous leukemia (CML).1,2 The formation of Bcr-Abl oligomers is achieved through the coiled-coil domain at the N-terminus of Bcr.3, 4 We have previously reported a modified version of this coiled-coil domain, CCmut2, which exhibits disruption of Bcr-Abl oligomeric complexes and results in decreased proliferation of CML cells and induction of apoptosis.5 A major contributing factor to these enhanced capabilities is the destabilization of the CCmut2 homo-dimers, increasing the availability to interact with and inhibit Bcr-Abl. Here, we included an additional mutation (K39E) that could in turn further destabilize the mutant homo-dimer. Incorporation of this modification into CCmut2 (C38A, S41R, L45D, E48R, Q60E) generated what we termed CCmut3, and resulted in further improvements in the binding properties with the wild-type coiled-coil domain representative of Bcr-Abl. A separate construct containing one revert mutation, CCmut4, did not demonstrate improved oligomeric properties and indicated the importance of the L45D mutation. CCmut3 demonstrated improved oligomerization via a two-hybrid assay as well as through colocalization studies, in addition to showing similar biologic activity as CCmut2. The improved binding between CCmut3 and the Bcr-Abl coiled-coil may be used to redirect Bcr-Abl to alternative subcellular locations with interesting therapeutic implications.
Keywords: Chronic Myelogenous Leukemia (CML), Bcr-Abl, mutant coiled-coil, oligomerization disruption
Introduction
Chronic myelogenous leukemia (CML) has been transformed from a deadly cancer into a manageable disease through the development of tyrosine kinase inhibitors (TKIs).6, 7 Currently, there are three approved TKIs for the treatment of CML: imatinib, nilotinib, and dasatinib. All three are now FDA approved for newly diagnosed chronic phase CML, with the second-generation TKIs (nilotinib and dasatinib) more effective against mutant Bcr-Abl variants.8 While TKI therapy has proven effective for the management of CML, one particular resistant form, harboring an isoleucine mutation in the T315 “gatekeeper” residue, is uninhibited by all of the currently approved TKIs.9–12 Ponatinib 13, 14 and DCC-203615 are third generation TKIs currently in clinical trials that have shown to be effective against the T315I form. Nevertheless, TKIs are not without problems and are in fact not a cure for CML,8, 16–18 as patients will need to chronically manage the disease with TKIs. Thus, alternative approaches are still of scientific and therapeutic interest.
In the majority of CML cases, the Abl protein is fused onto the C-terminus of the breakpoint cluster region protein (Bcr), a fusion protein expressed from the Philadelphia chromosome.19–22 This expression leads to unregulated Y-kinase activity that phosphorylates and activates signaling pathways such as RAS, PI3K/AKT, and Src family kinases such as STAT5.23–25 The net result is inhibition of apoptosis, activation of cell proliferation, and an altered cell adhesion and motility, all engendering growth advantages and cancer. The trans-autophosphorylation of Bcr-Abl stems from the oligomeric state achieved through a coiled-coil (or oligomerization) domain at the N-terminus of the Bcr portion of the fusion protein.1,26,27 Bcr-Abl constructs lacking the coiled-coil domain have diminished transformation potential,1 validating oligomerization as a pivotal role in oncogenicity through Bcr-Abl.26, 28, 29 Formation of hetero-oligomeric structures between Bcr-Abl and an isolated coiled-coil domain prevents the trans-autophosphorylation necessary for constitutive activity leading to cancer, and is an interesting alternative to TKIs that bind at the Y-kinase domain.
The 72 amino acid Bcr-Abl coiled-coil domain forms two parallel α-helices connected through a short linker region.4 The dimerization interface is composed primarily from the second helix (α-2), residues 28–67. The first helix (α-1) functions as a swap domain and folds onto the backside of the opposing α-2 helix after dimerization. This dimer subsequently dimerizes to form a tetrameric dimer-of-dimers. While the tetrameric interface consists primarily of aromatic and hydrophobic interactions and is challenging for rational design, both hydrophobic and ionic interactions additionally contribute to formation of the dimer interface.3, 4 Given that formation of dimers is a preceding step to formation of the tetramer, coupled with the fact that modifications to charged interactions in α-helices are more readily identified, analysis of the dimer interface permits logical modifications for improved oligomerization.
While it is possible to use the wild-type coiled-coil domain to interfere with Bcr-Abl homo-oligomerization, problems may arise in that there is no preference for interacting with Bcr-Abl over another isolated coiled-coil domain. As Bcr-Abl in the cell will already be found in the tetrameric state, the feasibility of using the wild-type domain relies heavily on pushing the equilibrium towards formation of hetero-oligomers through increased concentration of the isolated coiled-coil domain. The ideal coiled-coil domain would exhibit minimal homo-oligomerization and form hetero-oligomers that are more stable than the Bcr-Abl oligomeric structures. In an attempt to generate a coiled-coil domain that more closely resembles the properties of the ideal coiled-coil, we have deliberately mutated residues in the Bcr-Abl coiled-coil domain. We previously demonstrated that modifications can be made to the Bcr-Abl coiled-coil domain to improve the oligomeric capabilities.5 The design approach was two-fold: decrease homo-dimer stability through incorporation of charge-charge repulsion, and increase hetero-dimer stability through formation of additional salt bridges. The resulting product, termed CCmut2, was found to not only have improved oligomeric properties, but further resulted in improved inhibition of Bcr-Abl and enhanced effects in preventing proliferation of CML cells. Regardless of this design approach, there is much evidence supporting that the improved oligomeric properties stem primarily from decreased stability of the mutant homo-dimer coupled with the retention of the ability to oligomerize with Bcr-Abl, not necessarily the formation of a more stable hetero-dimer. This indicates the importance of reducing the formation of mutant homo-dimers to allow for interaction with Bcr-Abl.
In an attempt to increase the hetero-dimer interaction, Q60 was mutated to glutamate in CCmut2 for the formation of an additional salt bridge with K39 of the Bcr-Abl coiled-coil domain. Notwithstanding, in the CCmut2 homo-dimer where both domains contain the Q60E mutation, an unintentional result is the possibility to form two sets of K39:E60 salt bridges (Figure 1B and C). In order to turn these undesired salt bridges (that may stabilize the mutant homo-dimer) into charge-charge repulsions (that can decrease the mutant homo-dimer stability), K39 was mutated to glutamate. This mutation, along with the other five mutations previously described for CCmut2, was termed CCmut3 (Figure 1D and E).
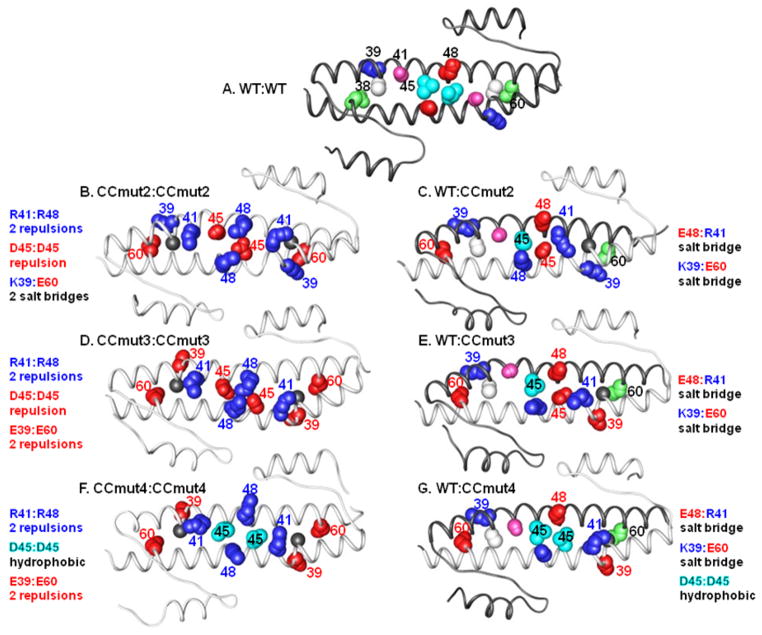
Figure 1.
Ribbon diagrams of the coiled-coil domains. White ribbons indicate wild type (WT) coiled-coil, grey ribbons indicate mutant coiled-coils, and each homo-dimer (A, B, D, and F) or hetero-dimer (C, E, and G) is labeled above. Blue numbering/spheres indicates positively charged amino acid residue; red numbering/spheres indicates negatively charged amino acid residue. For the WT, white = C38, blue (+ chg) = K39, purple = S41, cyan (hφ) = L45, red (− chg) = E48, green = Q60. Underlined residue is colored in the figure. For CC mutants, gray = C38A, red (− chg) = K39E, blue (+ chg) = S41R, red (− chg) = L45D, blue (+ chg) = E48R, red (− chg) = Q60E. Underlined residue is colored in the figure. A) WT:WT homo-dimer. Only the top strand is numbered. B) CCmut2:CCmut2 homo-dimer (CCmut2 contains C38A, S41R, L45D, E48R, Q60E mutations). The two R41:R48, and one D45:D45 charge-charge repulsion are shown, as well as the two sets of K39:E60 salt bridges. C) WT:CCmut2 hetero-dimer. The E48:R41 and K39:E60 salt bridges are indicated. D) CCmut3:CCmut3 homo-dimer (CCmut3 contains C38A, K39E, S41R, L45D, E48R, Q60E mutations). The two sets of K39:E60 salt bridges are now replaced with two sets of E39:E60 charge-charge repulsions. The two R41:R48 and one D45:D45 charge-charge repulsions are retained. E) WT:CCmut3 hetero-dimers. CCmut3 may form E48:R41 and K39:E60 salt bridges with WT as illustrated. F) CCmut4 homo-dimers (CCmut4 contains C38A, K39E, S41R, E48R, Q60E mutations). Similar to CCmut3 homo-dimer, the two sets of E39:E60, and two R41:R48 charge-charge repulsions are again illustrated. D45:D45 charge-charge repulsion is now replaced with a L45:L45 hydrophobic interaction in the middle. G) WT:CCmut4 hetero-dimers. The K39:E60 and E48:R41 salt bridges, and L45:L45 hydrophobic interactions are shown.
In analyzing possible mutations that led to CCmut2, it was discovered that incorporation of both L45D and V49D mutations, aimed at decreasing homo-dimer stability through charge-charge repulsions, introduced a kink in the α-2 helix and was unfavorable. However, making either L45D or V49D mutations alone contributed to specific interactions between the mutant and wild-type coiled-coils. Thus, L45D was chosen for incorporation into CCmut2. Residues 45 and 49 are unique among the set of possible mutations analyzed due to the fact that they are not located at “g” or “e” helical positions, but instead are located at the typical hydrophobic “d” and “a” positions. Although the L45D mutation was demonstrated to contribute to a decrease in homo-dimer stability (due to the D45:D45 charge-charge repulsion formed between the two mutant domains), it may have the drawback of decreased hydrophobic interactions in the hetero-dimer. The additional charge-charge repulsion designed through the K39E mutation may obfuscate the need for charge-charge repulsion through the L45D mutation, and reverting the L45D mutation back to the original leucine may provide the benefit of better hydrophobic interactions with the Bcr-Abl coiled-coil domain (Figure 1F and G). Thus, CCmut3 with the revert mutation D45L, was termed CCmut4.
For Figure 1, molecular graphics are shown displaying the average structures from 20–25 ns for the various “AB“ “std“ models of each class with the exception of CCmut3:WT model which was from 15–20 ns. In some of the mutated homodimers, such as CCmut2:CCmut2 and CCmut3:CCmut3, buckling at the center of the coiled coil is evidenced by bulging of the helices. MM-PBSA analysis30 proved inconclusive, even with inclusion of neutralizing salt and/or truncation of the non-coiled-coil regions. Further free energy investigation using thermodynamic integration, consistent with our previous work, is underway.
As a second iteration of a rationally designed coiled-coil domain for improved binding to Bcr-Abl, we demonstrate here that CCmut3, but not CCmut4, provides further enhancements of the oligomerization properties. CCmut3 produced similar effects as CCmut2 in inhibiting Bcr-Abl activity, decreasing CML cell proliferation, and inducing apoptosis in CML cells. In addition to an alternative inhibitor of Bcr-Abl, the enhanced binding may further prove beneficial for other interests currently underway in our laboratory involving the escorting of Bcr-Abl to other subcellular compartments.
Materials and Methods
Model Building from Molecular Dynamics Simulations
Snapshots were taken from molecular dynamics simulation using AMBER1131 initiated from crystal structure models from the 1K1F4 PDB structure for Figure 1. Model building: The four distinct coiled-coils in the unit cell were broken into four models of 67 residues each using chains A and B, C and D, E and F, and G and H. Missing terminal residues (chain D residue 67, chain E residues 1–2, chain G residues 66–67, and chain H residues 1–3 and 67) were grafted onto their respective structures based on the corresponding coordinates from model AB. Selenomethionine was altered back to methionine and residue 38 was reverted back to cysteine in the wild type structures. Mutated coiled coils (WT:mut2, WT:mut3, WT:mut4, mut2:mut2, mut3:mut3, and mut4:mut4) were built using the “swapaa“ utility in Chimera32 to swap amino acids with reasonable rotamers for the mutated residues. The models were built using the ff99SB force field33 and solvated with TIP3P34 solvent extending a minimum of 8 Å from the protein into a truncated octahedral box. Net-neutralizing Na+/Cl−35 was added followed by the addition of 45 extra Na+/Cl− ions for an excess salt concentration of ~200mM. The initial ion positions were based on favorable electrostatic fits as per AMBER’s LEaP adding single ions (for net-neutralization) or pairs of oppositely charged ions at a time (denoted “std“). Two additional simulations for each model separately randomized the ion positions by swapping with water molecules (denoted “rand“ and “rand2“). Equilibration: An initial minimization with electrostatics turned off was applied to all atoms to relax the added ions and remove large clashes, followed by 1000 steps of minimization (500 steepest descent and 500 conjugate gradient) and then 100 ps of heating (100K to 300K over the first 10 ps) with 25.0 kcal/mol-Å2 restraints on the protein atoms. Following the initial heating, five cycles of 1000 step minimization (as before) and 50 ps of MD were performed with restraint weights on the protein reduced sequentially from 5.0 to 1.0 kcal/mol-Å2 at each step in the cycle. Constant temperature and pressure were controlled by a Berendsen thermostat36 with a 0.2 coupling time. Production: Production simulations used a 5.0 coupling time without any positional restraints. All simulations, unless otherwise noted, applied a 9 Å cutoff to the direct space interactions with a homogeneous long range correction and automated 10 Å pairlist building if atoms moved more than 0.5 Å, particle mesh Ewald 37 with default parameters, SHAKE 38 on hydrogens, removal of center of mass motion every 5000 steps, and a 2 fs time step for molecular dynamics integration. During the 3 independent simulations for each pair on the ~25+ ns time scale, all of the coiled coil structures remained intact as judged by 1-D and 2-D RMSd measures and inspection of average structures.
Construction of Plasmids and Mutagenesis
The plasmids pEGFP-CC, pEGFP-CCmut2, pM1-CC, pM1-CCmut2, pEFVP16-CC, and pEFVP16-CCmut2 were constructed as previously described.5 pM1-CCmut3 and pEFVP16-CCmut3 were created through site-directed mutagenesis using pM1-CCmut2 and pEFVP16-CCmut2 as the templates, respectively. The mutagenic primers were 5′-GCTGGAGCGCGCCGAGGCCCGCATTCGG-3′ and 5′-CCGAATGCGGGCCTCGGCGCGCTCCAGC-3′. pM1-CCmut4 and pEFVP16-CCmut4 were created through site-directed mutagenesis using pM1-CCmut3 and pEFVP16-CCmut3 as the templates, respectively. The mutagenic primers were 5′-CCGCATTCGGCGCCTGGAGCAGCGGGTGAAC-3′ and 5′-GTTCACCCGCTGCTCCAGGCGCCGAATGCGG-3′. The genes encoding CCmut3 and CCmut4 were then amplified through PCR using pM1-CCmut3 and pM1-CCmut4 as the templates, respectively, and the primers 5′-TGTAACTCGAGTTATGGTGGACCCGGTG-3′ and 5′-ATGCTCTCGAGACCGGTCATAGCTCTTC-3′. The PCR products were then inserted into pEGFP-C1 (Clontech, Mountain View, CA, USA) at the XhoI restriction site. To create pmCherry-Bcr-Abl, the gene encoding Bcr-Abl was digested out of pEGFP-Bcr-Abl39 with EcoRI (New England Biolabs, Ipswich, MA, USA), and inserted into pmCherry-C1 (Clontech).
Cell Lines and Transient Transfection
Cells were maintained at 37°C in a 5% CO2 incubator. K562, Ph+ leukemia cells (gifted from Kojo Elenitoba-Johnson, Univ. of Michigan), and Cos-7 cells, African monkey kidney cells (ATCC), were grown in RPMI (Invitrogen, Carlsbad, CA, USA) media with 10% FBS (Hyclone Laboratories, Logan, UT, USA), 1% penicillin-streptomycin-glutamine (Invitrogen), and 0.1% gentamicin (Hyclone). Cells were passaged every two to three days at a density of 5.0 × 104 cells/mL. Two days after passaging, 2.0 × 106 cells per treatment group were transfected with 5–8 μg DNA according to the Cell Line Nucleofector® Kit V protocol (program T-013) using the Amaxa Nucleofector II (Lonza Group, Basel, Switzerland). Cos-7 cells were passaged every 2–3 days and transfected 24 hrs after seeding the cells using Lipofectamine LTX (Invitrogen) as recommended by the supplier. Both K562 and Cos-7 cells were transfected consistently between cell passages 3 and 10, as the most optimal transfection efficiencies are seen in that range.
Mammalian Two-Hybrid Assay
An in depth description of how the mammalian two-hybrid assay was carried out is described elsewhere.5 In short, pM1-CC (or mutant), pEFVP16-CC (or mutant), pG5-Fluc (Promega, Madison, WI, USA), and pRL-CMV (Promega) plasmids were co-transfected into Cos-7 cells in a 10:10:10:1 ratio. pAD-SV40 and pBD-p53 (Stratagene, Agilent Technologies Inc., Santa Clara, CA, USA) plasmids were used for the positive control, and pM1 lacking the coiled-coil gene was used as the negative control. 48 hrs after transfection both firefly and renilla luminescence were measured using the Dual-Glo Luciferase Assay (Promega) reagents per the manufacturer’s recommendations. The mean from duplicate transfections were taken from 5 separate experiments. A relative response ratio was calculated using the following equation on the firefly values normalized to the renilla values: (Experiment − Ctrl−)/(Ctrl+ − Ctrl−). For ease of comparing to the wild-type coiled-coil interaction, the results were then normalized to the wild-type interaction (n=4 or 5).
Confocal Microscopy and Colocalization
K562 cells were transfected with Lipofectamine LTX (Invitrogen) 24 hrs after seeding into 4-well live-cell chambers (Lab-Tek chamber slide system, Nalge NUNC International, Naperville, IL, USA). At least 24 hrs after transfection, the cells were imaged. All images of cells were acquired on an Olympus IX81 FV1000-XY confocal microscope equipped with 405 diode, 488 argon, and 543 HeNE lasers using a 60X PlanApo oil immersion objective (NA 1.45) using Olympus FluoView software. Excitation and emission filters were as follows: EGFP, 488 nm excitation, emission filter 500–530 nm; mCherry, 543 nm excitation, emission filter 555–655 nm. Images were collected in sequential line mode. The exposure settings and gain of laser were kept constant and below detected pixel saturation for each group of cells. No crosstalk was observed between channels as determined by excitation with either the 488 nm or 543 nm laser lines, independently, while collecting fluorescence in both channels. Pixel resolution was kept at 1024 × 1024 with maximum of 2.5X digital zoom. Prior to statistical colocalization analysis, all images were corrected for background noise (i.e., mean background intensity outside of cells). All experiments were completed in triplicate (n≥3). Region of interests (ROIs) were created around whole cells. Image and statistical analysis was performed with JACoP in ImageJ (http://rsb.info.nih.gov/ij).40 Costes’ automatic threshold was used to generate the quantitative colocalization coefficient. 41
Flow Cytometry
48 hrs following transfection of K562 cells with pEGFP-C1, pEGFP-CC, pEGFP-CCmut2, or pEGFP-CCmut3, 5 mL of cells were pelleted and resuspended in 0.5 mL of 1X annexin binding buffer (Invitrogen). Immediately before flow cytometry analysis, 0.5 μL of 7-aminoactinomycin D (7AAD, Invitrogen, 1 mg/mL) and 5 μL annexin V conjugated with allophycocyanin (annexin-APC, Invitrogen) were added to the cells. Flow cytometric analysis was performed on a FACSCantoII analyzer (Becton Dickinson, Franklin Lakes, NJ, USA) using BD FACSDiva v6.1.3 (BD) software. Both EGFP and 7AAD were excited with a blue laser with 488 nm wavelength, while APC was excited with a red laser with 635 nm wavelength. The fluorescence detector used for EGFP was 530/30 nm, the detector for 7AAD was 660/20 nm, and the detector for APC was 660/20 nm. The untransfected cells were excluded from analysis through gating on the EGFP fluorescence, and the percentage of transfected, apoptotic cells was quantified through combining the number of cells that stained positively for annexin-APC only (Q2) with the number of cells that stained positively for annexin-APC and 7AAD (Q4). All four plasmids were transfected 3 separate times for separate analysis (n=3).
Cell Proliferation and Western Blotting
Cell proliferation analysis and western blotting were carried out as previously described.5 Briefly, for proliferation analysis, 48 hrs following transfection into 2.5 × 106 K562 cells, trypan blue exclusion was used to determine the number of viable cells. For western blots, cell pellets were resuspended in 200 μL lysis buffer/106 cells and freeze thawed at −80°C. After protein gel electrophoresis and transfer to a PVDF membrane, the membrane was probed with primary antibodies (anti-pAbl(Y245): 73E5, Cell Signaling Technology; anti-pSTAT5(Y694): E208, Abcam; anti-pCrkl(Y207): #3181, Cell Signaling Technology; anti-actin: mAbcam 8226, Abcam), followed by HRP-conjugated secondary antibodies (Ab6814, Abcam) or anti-rabbit (#7074, Cell Signaling Technology) before the addition of ChemiGlo (Alpha Innotech, Cell Biosciences, Santa Clara, CA, USA) chemiluminescent substrate and detection with a FluorChem FC2 imager (AlphaInnotech). Plasmids were transfected three separate times, and a western was performed on each lysate for analysis (n=3).
Colony Forming Assay
The colony forming assay was carried out as previously described.5 In short, 24 hrs following transfection into K562 cells, 1000 cells in Iscove’s modified Dulbecco’s media (Stem Cell Technologies, Vancouver, BC, Canada) with 2% FBS were seeded in Methocult H4230 methylcellulose medium (Stem Cell Technologies) in the absence of cytokines. Each group of treated cells was seeded in duplicate, and colony formation was assessed seven days after seeding cells by counting colonies in two 200 μm2 areas of the plate. Experiments were repeated at least 3 times and compared to control (cells transfected with pEGFP-C1) (n=3).
Caspase-3/7 Assay
Caspase-3/7 assays were performed as previously described.5 Briefly, 48 hrs following transfection into K562 cells, cell counts were performed and 3 × 106 cells were resuspended in 50 μL EnzChek Caspase-3/7 lysis buffer (kit #2, Invitrogen). Cells were frozen at −80 °C, thawed, and the cell debris removed by centrifuged at 5,000 × g for 5 min. Lysates were mixed with 50 μL 2x AMC-DEVD substrate in a black 96-well plate (Cellstar, Greiner Bio-One, Monroe, NC, USA), and incubated at room temperature for 30 min before measuring the fluorescence with excitation 342 nm and emission 441 nm on a SpectraMax M2 plate reader (Molecular Devices, Sunnyvale, CA, USA). Lysates from three separate transfections for each plasmid were assayed for caspase-3/7 activity (n=3).
Fluorescence Microscopy & DNA Segmentation
A detailed description of the fluorescence microscopy and DNA segmentation can be found elsewhere.5 In short, 48 hrs following transfection K562 cells were transferred to 2-well live cell chambers. 0.3 μL H33342 (nuclear dye, Invitrogen) was added, and the cells were incubated at 37°C for 15 min. Cells were then analyzed with an inverted fluorescence microscope (Olympus IX701F, Scientific Instrument Co., Sunnyvale, CA, USA) with high-quality narrow band GFP filter (excitation HQ480/20 nm, emission HQ510/20 nm, beam splitter Q4951p, Chroma Technology Corp., Brattleboro, VT, USA) equipped with an F-view Monochrome CCD camera. Fields of view for imaging were selected based on EGFP fluorescence (blinded to nuclei), and were imaged with a 40X oil immersion objective. The nuclei of at least 50 transfected cells (EGFP fluorescence) per group were classified as either healthy (round or kidney shaped) or segmented (punctate) 42, 43 and the percentage of cells with segmented DNA was calculated (n=3).
Statistical Analysis
All experiments were analyzed using one-way ANOVA with Tukey’s post test (as previously).5
Results
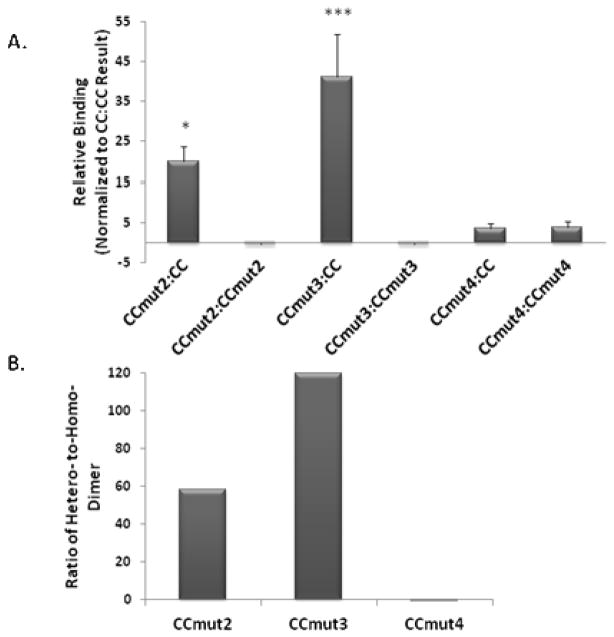
Mammalian two-hybrid assays, to determine interactions between CC and coiled-coil mutants were carried out in Cos-7 cells. For the hetero-dimer interactions, both possible fusion constructs were assayed (i.e. pM1-CC/pEFVP16-CCmutX and pM1-CCmutX/pEFVP16-CC). While the same trend was observed regardless of which fusion construct was the wild-type, slightly higher values were obtained when the wild-type was fused to the DNA binding domain (pM1). As illustrated in Figure 2A, the greatest interaction resulted from the hetero-dimer between CCmut3 and the wild-type coiled-coil domain (CC) (Figure 2A, third column). Equally important, the CCmut3:CCmut3 interaction (homo-dimer) resulted in negligible binding (Figure 2A, fourth column). Although it is difficult, if not impossible, in this assay to separate out the binding contributions from increased availability versus increased affinity, the design to decrease the stability of the mutant homo-dimer did correlate to an increased ability to bind wild-type as predicted. The result from the CCmut3 hetero-dimer and homo-dimer together indicate the improved interaction exhibited by CCmut3.
Figure 2.
Mammalian two-hybrid assay. A) Relative response ratios determined for each interaction and normalized to the wild-type homo-oligomerization (CC:CC) to indicate the relative binding efficiency. Values indicated are means ± S.D. Statistics were performed on the values prior to normalizing to CC so as to include the wild-type interaction in the statistics (n=4 or 5). *p<0.05, ***p<0.01 versus CC:CC interaction; one-way ANOVA, Tukey’s post test. B) The quotient of the hetero-dimer and homo-dimer inte raction (absolute value) is graphed for all three mutants, indicating the preferential binding to CC over the formation of a homo-dimer.
Although the CCmut4 hetero-dimer did result in a slightly greater interaction than wild-type (all results are normalized to the wild-type CC:CC interaction, Figure 2A, fifth column), the CCmut4 homo-dimer produced an almost equivalent result (Figure 2A, sixth column). This finding indicates the importance of the L45D mutation in generating a preferential binding towards wild-type. Whereas the other mutations designed for charge-charge repulsion are found on the helix at “g” and “e” postions and may more readily adopt rotamer conformations that position the like charges away from each other, the L45D mutation is in the hydrophobic core where such rotamer flexibility may be more limited. As demonstrated from the dramatic differences between CCmut3 and CCmut4, the L45D mutation is critical.
As both the hetero-dimer and homo-dimer formations are critical in evaluating the improvements made through the mutations, the quotient of the hetero-dimer and homo-dimer can be used to more easily compare the overall effect. Figure 2B reports these quotients for CCmut2, CCmut3, and CCmut4. Although the quotient for CCmut2 indicates it is a favorable domain for interacting with Bcr-Abl (at least the wild-type coiled-coil domain of Bcr-Abl), the best set of mutations are those comprising CCmut3. While CCmut4 may interact favorably with wild-type (Figure 2A, fifth column), it does not exhibit any preferential binding (Figure 2B, third column) and did not result in any improvement as compared to the wild-type domain. Following these results, CCmut4 was dropped from further analysis due to the lack of improvement in oligomerization properties.
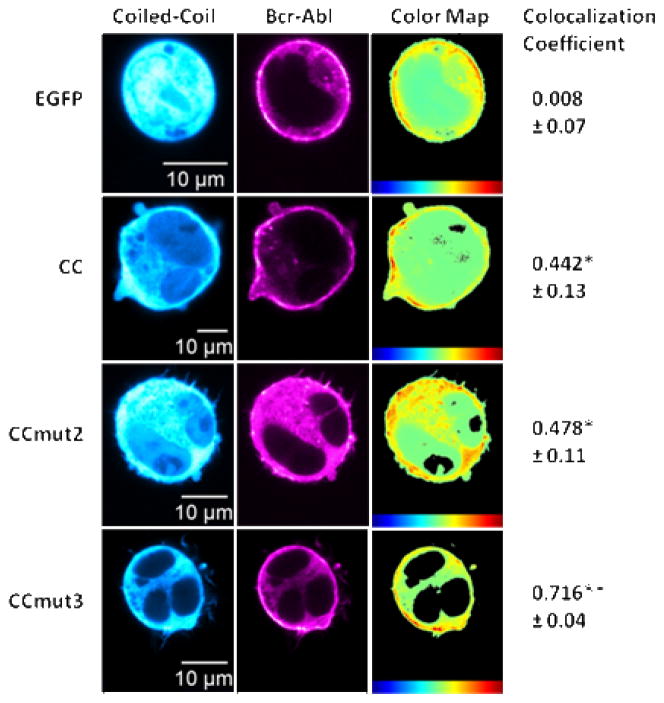
While the mammalian two-hybrid assays are a quantitative method of studying the effects of the mutations, these assays were carried out with the isolated coiled-coil domains and not full-length Bcr-Abl. To validate the modifications corresponding to the binding to Bcr-Abl, colocalization experiments were performed in K562 cells. A rigorous analysis for determining colocalization, one utilizing the Costes’ colocalization coefficient, was employed.41 Given that CC, CCmut2, and CCmut3 should all bind Bcr-Abl, the colocalization was aimed at simply confirming the interaction with the full-length protein, and not necessarily at distinguishing their binding affinities. Nevertheless, Figure 3 shows that CCmut3 was found to exhibit a much higher degree of colocalization with Bcr-Abl (colocalization coefficient of 0.716 ± 0.04) than CC (colocalization coefficient of 0.442 ± 0.13) or CCmut2 (colocalization coefficient of 0.478 ± 0.11), and further validated the improved oligomeric properties. Not entirely surprising, there was no difference between CC and CCmut2 (Figure 3). These colocalization results confirm the CCmut3 set of mutations have improved the ability to bind Bcr-Abl inside a live cell.
Figure 3.
Colocalization with Bcr-Abl. Representative images of either EGFP or coiled-coil domain seen in left column (false colored cyan for visualization) with representative images of Bcr-Abl distribution in middle column (false colored magenta for visualization). Heat maps indicating the colocalization of the two fluorophores seen in the right column with colocalization scale at the bottom (red highest) followed by the colocalization coefficient. The mean colocalization coefficient was determined from at least three cells per transfection (n=4), with values reported as means ± S.E.M. *p<0.05, **p<0.01 control; one-way ANOVA, Tukey’s post test.
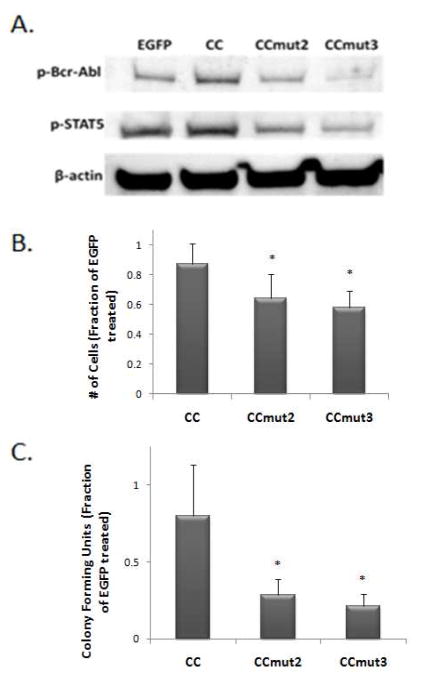
Analysis of the oligomeric disruption of Bcr-Abl was further explored to confirm the interaction between it and CCmut3. As Bcr-Abl trans-autophosphorylation is dependent on the coiled-coil domain, the formation of hetero-oligomers between a coiled-coil domain and full-length Bcr-Abl should reduce the phosphorylation of Bcr-Abl. As a measure of the phosphorylation state of Bcr-Abl, western blotting was carried out probing K562 lysates with an antibody that recognizes the phosphorylated form of Bcr-Abl. As seen in Figure 4A, the phosphorylation of Bcr-Abl was the least after transfection of CCmut3. A secondary measure of the phosphorylation of Bcr-Abl, and its corresponding kinase activity, is the phosphorylation of Bcr-Abl substrates such as STAT5. The phosphorylation of STAT5 was also the least after transfection of CCmut3 (Figure 4A). The western blot results corroborate the mammalian two-hybrid and colocalization data.
Figure 4.
Effects of CCs on Bcr-Abl and proliferation of K562 cells. A) Western blot indicating the phosphorylation of Bcr-Abl (top, p-Bcr-Abl) and STAT5 (middle, p-STAT5). B) Colony forming units counted seven days after seeding 103 transfected K562 cells into methylcellulose medium, normalized to colony growth from EGFP control (n=3). Values reported as means ± S.D. *p<0.05 control; one-way ANOVA, Tukey’s post test. C) Viable K562 cells were counted 48 hrs following transfection, with the number of proliferating cells normalized to the number resulting from the EGFP transfection (n=4). Values reported as means ± S.D. *p<0.05 control; one-way ANOVA, Tukey’s post test.
Next, the improved Bcr-Abl binding of CCmut3 was analyzed for its effect on CML cells. As CML cells are dependent on Bcr-Abl signaling for growth, inhibition of Bcr-Abl will affect the proliferation rate of the cells, which can be monitored simply by performing cell counts after treatment. 48 hrs following transfections of either EGFP, CC, CCmut2, or CCmut3 into a CML model cell line (K562), cell counts were performed using trypan blue exclusion. As seen in Figure 4B, the proliferation of the cells decreased in the order of CC, CCmut2, and CCmut3. Although indirect, this provides evidence of the disruption of Bcr-Abl oligomerization resulting in inhibition of its signaling.
As an alternative method of measuring the proliferation of the cells, a colony forming assay was performed. 24 hrs following transfection of the plasmids into K562 cells, 103 cells were seeded into methylcellulose medium and incubated at 37 °C for seven days before counting the colony forming units in a 200 μm2 area. Both CCmut2 and CCmut3 were found to produce the fewest number of colonies, indicative of the decrease in cell proliferation (figure 4C). Notwithstanding, there was no difference in the number of colonies after transfection of CCmut2 or CCmut3. The results achieved by CCmut2 and CCmut3 were comparable to the result after treating K562 cells with 0.5 μM imatinib,5 demonstrating the therapeutic potential.
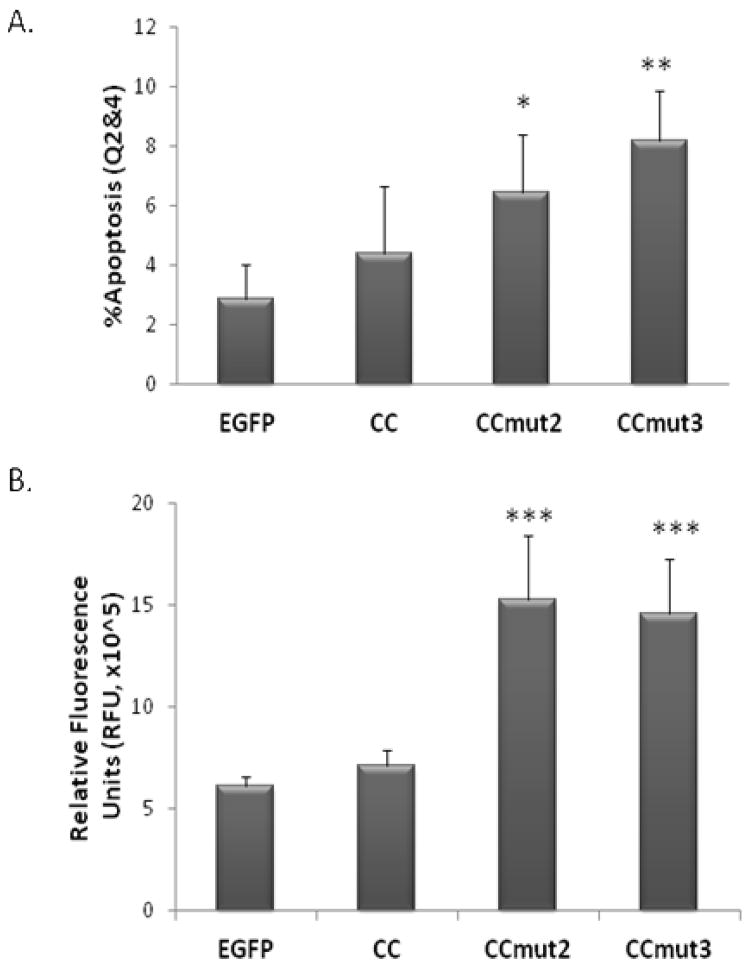
Further exploration of the effects on CML cells was carried out though analysis of the induction of apoptosis. First, flow cytometry was used to analyze the number of cells with externalized phosphatidylserine, an established marker of apoptosis. As seen in Figure 5A, the level of cell death incrementally increased in the order of CC, CCmut2, and CCmut3. Although the statistical significance compared to the EGFP control was greater for CCmut3 than for CCmut2, there was no statistically significant difference between CCmut2 and CCmut3. To further validate the induction of apoptosis, the activity of caspase-3/7 was also measured. A substantial increase in caspase-3/7 activity was observed after transfection of either CCmut2 or CCmut3 compared to both EGFP and CC (Figure 5B), but no statistical difference was observed between CCmut2 and CCmut3.
Figure 5.
Apoptosis assays. A) 48 hrs following transfection into K562 cells phosphatidylserine externalization was assessed through flow cytometry. After gating on EGFP fluorescence to select only the transfected cells, the number of 7AAD−/Annexin+ cells (Q2, early apoptosis) was combined with the 7AAD+/Annexin+ cells (Q4, late apoptosis) and used to determine the percentage of apoptotic cells (n=3). Values reported as means ± S.D. *p<0.05, **p<0.01 control; one-way ANOVA, Tukey’s post test. B) 48 hrs following transfection into K562 cells the activation of caspase-3/7 was analyzed in the cell lysates through a fluorescence-based assay (n=3). Values reported as means ± S.D. ***p<0.01; one-way ANOVA, Tukey’s post test.
Analysis of the cell morphology is an alternative method of studying the induction of apoptosis. Healthy K562 cells are relatively large, round cells with kidney shaped (or round) nuclei (Figure 6A, top row). Apoptotic cells, however, exhibit very distinct morphological signs readily detectable through microscopy. Some morphological signs of apoptosis include cell shrinkage (as opposed to necrotic cells that may expand), cytoplasmic blebbing, echnoid spikes, and nuclear segmentation.42, 43 Of these signs, nuclear segmentation is one indication that can be quantified (each individual cell determined to have either a normal or segmented nucleus) as a measurement of the induction of apoptosis. 48 hrs after the transfection of the plasmids into K562 cells, the nuclei were stained with H33342 and photographed on a fluorescence microscope. The fields of view photographed were selected based on the EGFP fluorescence, while being blinded to the nuclear stain, which reduces biased selection of nuclei. Representative images are illustrated in Figure 6A. The percentage of transfected cells with segmented nuclei is graphed in Figure 6B. Similar to the cell proliferation, and flow cytometry, an incremental effect was observed on nuclear segmentation with CC, CCmut2, and CCmut3. Nevertheless, CCmut2 and CCmut3 produced statistically significant differences versus control, but were indistinguishable from each other. Thus, the flow cytometry, caspase assay, and nuclear segmentation together demonstrate the ability of the modified coiled-coils (both CCmut2 and CCmut3) to inhibit Bcr-Abl, decrease cell proliferation, and induce apoptosis in CML cells.
Figure 6.
Nuclear segmentation of K562 cells. A) Representative images of K562 cells 48 hrs after transfection of EGFP (top row) or CCmut3 (bottom row). Left column = phase contrast images with black arrow indicating example of cell shrinkage; middle column = fluorescence from EGFP; right column = stained nuclei with white arrow indicating example of nuclear segmentation. B) Quantitative results of nuclear segmentation. Percentage of transfected cells with segmented nuclei was determined from three or four fields of view, and repeated with three separate transfections (n=3). Values reported as means ± S.D. *p<0.05; one-way ANOVA, Tukey’s post test.
Conclusions
Analysis of the structure of the Bcr-Abl coiled-coil domain, and the previously designed CCmut2, guided the identification of residues 39 and 45 as interesting locations for possible mutations. Incorporation of a K39E mutation into CCmut2 produced CCmut3. Subsequently, reverting the CCmut3 L45D mutation back to leucine produced CCmut4. The effects of these modifications on the oligomerization properties were studied through a mammalian two-hybrid assay in Cos-7 cells and compared to the effects observed from CCmut2 as well as the wild-type coiled-coil domain. While CCmut3 yielded impressive improvements in the binding capabilities, CCmut4 was found to bind almost equivalently to another CCmut4 as it binds CC. The binding results from CCmut4 indicate the importance of the L45D mutation in preventing homo-oligomerization. These binding results, including both CCmut3 and CCmut4, further support the idea that decreased homo-dimer stability is a major driving force in the improved oligomerization capability with Bcr-Abl. The mammalian two-hybrid assay using isolated coiled-coil domains was then followed by colocalization with full-length Bcr-Abl. CC, CCmut2, and CCmut3 were all found to colocalize with Bcr-Abl, with CCmut3 resulting in the greatest degree of colocalization. The improved binding of CCmut3 to Bcr-Abl also decreased the phosphorylation of Bcr-Abl, a parameter indicative of the oligomeric state and activity of Bcr-Abl. These binding studies validated the additional mutation used in CCmut3 led to enhanced oligomeric properties and demonstrated its usefulness as a Bcr-Abl binding domain.
The improved binding by CCmut3 was then tested for its effect on CML cells. Cell proliferation was first measured through cell counts and colony forming assays following transfection of the coiled-coil domains. Both CCmut2 and CCmut3 decreased the cell proliferation, but there was no statistical difference between the two compounds.
CCmut3 was also demonstrated to induce apoptosis in K562 cells as measured through 7AAD staining, activation of caspase-3/7, and hallmark morphological signs of apoptosis. Both the decreased cell proliferation and the induction of apoptosis are indirect evidence of the ability of CCmut3 to interfere with the homo-oligomerization of Bcr-Abl, as oligomerization is essential for Bcr-Abl activity. Due to the fact that CCmut3 does not contain the Y-kinase domain necessary for activation through trans-autophosphorylation, the binding between CCmut3 and Bcr-Abl results in inactivation of Bcr-Abl and induction of apoptosis in CML cells.
These results can be divided into two distinct categories of experiments: testing the binding, and testing the effect the compound has on CML cells. The analysis of the binding revealed a marked improvement in CCmut3, even over the proven CCmut2. Nevertheless, this improved binding did not correlate to enhancements over CCmut2 in effects on CML cells. This may be explained by insufficient assay sensitivity to distinguish a modest difference, indicating the improvement is only marginal. An alternative explanation lies in the idea that the disruption of Bcr-Abl oligomerization is already maximized with the binding capabilities of CCmut2. If this is the case, any improvements in binding with an alternative coiled-coil domain would not instigate enhancements in the effect on CML cells. This theory is supported by the inability to increase the effect of CCmut2 on CML cells through combinatorial treatment with CCmut2 and a TKI (unpublished data).
Although use of CCmut3 for interfering with the oligomeric state of Bcr-Abl is an interesting alternative to the standard TKIs, a further application currently in pursuit in our laboratory is the use as a capture motif for escort of Bcr-Abl to alternative subcellular locations. As opposed to a passive induction of apoptosis when Bcr-Abl is inhibited, we are seeking an active induction of apoptosis that may lead to a more potent therapeutic strategy. With this goal in mind, the improved binding properties are of utmost interest. This work establishes CCmut3 as an efficient binding partner to Bcr-Abl with implications in inhibiting Bcr-Abl as well as other interesting therapeutic strategies that require tight binding to Bcr-Abl.
Acknowledgments
We acknowledge the use of DNA/Peptide Core (NCI Cancer Center Support Grant P30 CA042014, Huntsman Cancer Institute). This work was funded by NIH R01-CA129528. We would like to thank Rian Davis, Mohanad Mossalam, Abdul Okal, and Karina Matissek for scientific discussions.
References
- 1.McWhirter JR, Galasso DL, Wang JY. A coiled-coil oligomerization domain of Bcr is essential for the transforming function of Bcr-Abl oncoproteins. Mol Cell Biol. 1993;13(12):7587–95. doi: 10.1128/mcb.13.12.7587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guo XY, Cuillerot JM, Wang T, Wu Y, Arlinghaus R, Claxton D, Bachier C, Greenberger J, Colombowala I, Deisseroth AB. Peptide containing the BCR oligomerization domain (AA 1-160) reverses the transformed phenotype of p210bcr-abl positive 32D myeloid leukemia cells. Oncogene. 1998;17(7):825–33. doi: 10.1038/sj.onc.1201999. [DOI] [PubMed] [Google Scholar]
- 3.Taylor CM, Keating AE. Orientation and oligomerization specificity of the Bcr coiled-coil oligomerization domain. Biochemisty. 2005;44(49):16246–56. doi: 10.1021/bi051493t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao X, Ghaffari S, Lodish H, Malashkevich VN, Kim PS. Structure of the Bcr-Abl oncoprotein oligomerization domain. Nat Struct Biol. 2002;9(2):117–20. doi: 10.1038/nsb747. [DOI] [PubMed] [Google Scholar]
- 5.Dixon AS, Pendley SS, Bruno BJ, Woessner DW, Shimpi AA, Cheatham TE, 3rd, Lim CS. Disruption of BCR-ABL coiled coil oligomerization by design. J Biol Chem. 2011;286(31):27751–60. doi: 10.1074/jbc.M111.264903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Okimoto RA, Van Etten RA. Navigating the road toward optimal initial therapy for chronic myeloid leukemia. Curr Opin Hematol. 18(2):89–97. doi: 10.1097/MOH.0b013e32834399a5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldman JM. Chronic myeloid leukemia: a historical perspective. Semin Hematol. 47(4):302–11. doi: 10.1053/j.seminhematol.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 8.Wei G, Rafiyath S, Liu D. First-line treatment for chronic myeloid leukemia: dasatinib, nilotinib, or imatinib. J Hematol Oncol. 3:47. doi: 10.1186/1756-8722-3-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jabbour E, Cortes JE, Kantarjian H. Second-line therapy and beyond resistance for the treatment of patients with chronic myeloid leukemia post imatinib failure. Clin Lymphoma Myeloma. 2009;9 (Suppl 3):S272–9. doi: 10.3816/CLM.2009.s.023. [DOI] [PubMed] [Google Scholar]
- 10.Quintas-Cardama A, Kantarjian HM, Cortes JE. Mechanisms of primary and secondary resistance to imatinib in chronic myeloid leukemia. Cancer Control. 2009;16(2):122–31. doi: 10.1177/107327480901600204. [DOI] [PubMed] [Google Scholar]
- 11.Deininger M. Resistance and relapse with imatinib in CML: causes and consequences. J Natl Compr Canc Netw. 2008;6 (Suppl 2):S11–S21. [PubMed] [Google Scholar]
- 12.Hochhaus A, Erben P, Ernst T, Mueller MC. Resistance to targeted therapy in chronic myelogenous leukemia. Semin Hematol. 2007;44(1 Suppl 1):S15–24. doi: 10.1053/j.seminhematol.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 13.Thomas X. Is AP24534 (Ponatinib) the next treatment of Philadelphia chromosome-positive acute lymphoblastic leukemia? Bull Cancer. doi: 10.1684/bdc.2011.1390. [DOI] [PubMed] [Google Scholar]
- 14.Zhou T, Commodore L, Huang WS, Wang Y, Thomas M, Keats J, Xu Q, Rivera VM, Shakespeare WC, Clackson T, Dalgarno DC, Zhu X. Structural mechanism of the Pan-BCR-ABL inhibitor ponatinib (AP24534): lessons for overcoming kinase inhibitor resistance. Chem Biol Drug Des. 77(1):1–11. doi: 10.1111/j.1747-0285.2010.01054.x. [DOI] [PubMed] [Google Scholar]
- 15.Eide CA, Adrian LT, Tyner JW, Mac Partlin M, Anderson DJ, Wise SC, Smith BD, Petillo PA, Flynn DL, Deininger MW, O’Hare T, Druker BJ. The ABL switch control inhibitor DCC-2036 is active against the chronic myeloid leukemia mutant BCR-ABLT315I and exhibits a narrow resistance profile. Cancer Res. 71(9):3189–95. doi: 10.1158/0008-5472.CAN-10-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fausel C. Targeted chronic myeloid leukemia therapy: seeking a cure. J Manag Care Pharm. 2007;13(8 Suppl A):8–12. doi: 10.18553/jmcp.2007.13.s8-a.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Radich JP. Chronic myeloid leukemia 2010: where are we now and where can we go? Hematology Am Soc Hematol Educ Program. 2010:122–8. doi: 10.1182/asheducation-2010.1.122. [DOI] [PubMed] [Google Scholar]
- 18.Pavlovsky C, Kantarjian H, Cortes JE. First-line therapy for chronic myeloid leukemia: Past, present, and future. Am J Hematol. 2009;84(5):287–93. doi: 10.1002/ajh.21380. [DOI] [PubMed] [Google Scholar]
- 19.Rowley JD. Letter: A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243(5405):290–3. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- 20.Groffen J, Stephenson JR, Heisterkamp N, Bartram C, de Klein A, Grosveld G. The human c-abl oncogene in the Philadelphia translocation. J Cell Physiol Suppl. 1984;3:179–91. doi: 10.1002/jcp.1041210421. [DOI] [PubMed] [Google Scholar]
- 21.Groffen J, Stephenson JR, Heisterkamp N, de Klein A, Bartram CR, Grosveld G. Philadelphia chromosomal breakpoints are clustered within a limited region, bcr, on chromosome 22. Cell. 1984;36(1):93–9. doi: 10.1016/0092-8674(84)90077-1. [DOI] [PubMed] [Google Scholar]
- 22.Kurzrock R, Kantarjian HM, Druker BJ, Talpaz M. Philadelphia chromosome-positive leukemias: from basic mechanisms to molecular therapeutics. Ann Intern Med. 2003;138(10):819–30. doi: 10.7326/0003-4819-138-10-200305200-00010. [DOI] [PubMed] [Google Scholar]
- 23.Ren R. The molecular mechanism of chronic myelogenous leukemia and its therapeutic implications: studies in a murine model. Oncogene. 2002;21(56):8629–42. doi: 10.1038/sj.onc.1206090. [DOI] [PubMed] [Google Scholar]
- 24.Sattler M, Griffin JD. Molecular mechanisms of transformation by the BCR-ABL oncogene. Semin Hematol. 2003;40(2 Suppl 2):4–10. doi: 10.1053/shem.2003.50034. [DOI] [PubMed] [Google Scholar]
- 25.Hazlehurst LA, Bewry NN, Nair RR, Pinilla-Ibarz J. Signaling networks associated with BCR-ABL-dependent transformation. Cancer Control. 2009;16(2):100–7. doi: 10.1177/107327480901600202. [DOI] [PubMed] [Google Scholar]
- 26.Beissert T, Hundertmark A, Kaburova V, Travaglini L, Mian AA, Nervi C, Ruthardt M. Targeting of the N-terminal coiled coil oligomerization interface by a helix-2 peptide inhibits unmutated and imatinib-resistant BCR/ABL. Int J Cancer. 2008;122(12):2744–52. doi: 10.1002/ijc.23467. [DOI] [PubMed] [Google Scholar]
- 27.McWhirter JR, Wang JY. Effect of Bcr sequences on the cellular function of the Bcr-Abl oncoprotein. Oncogene. 1997;15(14):1625–34. doi: 10.1038/sj.onc.1201342. [DOI] [PubMed] [Google Scholar]
- 28.Beissert T, Puccetti E, Bianchini A, Guller S, Boehrer S, Hoelzer D, Ottmann OG, Nervi C, Ruthardt M. Targeting of the N-terminal coiled coil oligomerization interface of BCR interferes with the transformation potential of BCR-ABL and increases sensitivity to STI571. Blood. 2003;102(8):2985–93. doi: 10.1182/blood-2003-03-0811. [DOI] [PubMed] [Google Scholar]
- 29.Mian AA, Oancea C, Zhao Z, Ottmann OG, Ruthardt M. Oligomerization inhibition, combined with allosteric inhibition, abrogates the transformation potential of T315I-positive BCR/ABL. Leukemia. 2009;23(12):2242–7. doi: 10.1038/leu.2009.194. [DOI] [PubMed] [Google Scholar]
- 30.Srinivasan J, Cheatham TE, III, Cieplak P, Kollman PA, Case DA. Continuum solvent studies of the stability of DNA, RNA and phosphoramidate helices. J Amer Chem Soc. 1998;120:9401–9409. [Google Scholar]
- 31.Case DA, Cheatham TE, III, Darden TA, Gohlker H, Luo R, Merz KM, Jr, Onufriev AV, Simmerling C, Wang B, Woods R. The AMBER biomolecular simulation programs. J Comp Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comp Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 33.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 35.Joung IS, Cheatham TE., III Determination of alkali halide monovalent ion parameters for use in explicitly solvated biomolecular simulations. J Phys Chem B. 2008;112:9020–9041. doi: 10.1021/jp8001614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR. Molecular dynamics with coupling to an external bath. J Comp Phys. 1984;81:3684–3690. [Google Scholar]
- 37.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. A Smooth Particle Mesh Ewald Method. J Chem Phys. 1995;103(19):8577–8593. [Google Scholar]
- 38.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J Comp Phys. 1977;23:327–341. [Google Scholar]
- 39.Dixon AS, Kakar M, Schneider KM, Constance JE, Paullin BC, Lim CS. Controlling subcellular localization to alter function: Sending oncogenic Bcr-Abl to the nucleus causes apoptosis. J Control Release. 2009 doi: 10.1016/j.jconrel.2009.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bolte S, Cordelieres FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc. 2006;224(Pt 3):213–32. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- 41.Costes SV, Daelemans D, Cho EH, Dobbin Z, Pavlakis G, Lockett S. Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys J. 2004;86(6):3993–4003. doi: 10.1529/biophysj.103.038422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barrett KL, Willingham JM, Garvin AJ, Willingham MC. Advances in cytochemical methods for detection of apoptosis. J Histochem Cytochem. 2001;49(7):821–32. doi: 10.1177/002215540104900703. [DOI] [PubMed] [Google Scholar]
- 43.Willingham MC. Cytochemical methods for the detection of apoptosis. J Histochem Cytochem. 1999;47(9):1101–10. doi: 10.1177/002215549904700901. [DOI] [PubMed] [Google Scholar]