Abstract
Background
The RNA-binding protein Sam68 has been implicated in a number of cellular processes, including transcription, RNA splicing and export, translation, signal transduction, cell cycle progression and replication of the human immunodeficiency virus and poliovirus. However, the precise impact it has on essential cellular functions remains largely obscure.
Results
In this report we show that conditional overexpression of Sam68 in fibroblasts results in both cell cycle arrest and apoptosis. Arrest in G1 phase of the cell cycle is associated with decreased levels of cyclins D1 and E RNA and protein, resulting in dramatically reduced Rb phosphorylation. Interestingly, cell cycle arrest does not require the specific RNA binding ability of Sam68. In marked contrast, induction of apoptosis by Sam68 absolutely requires a fully-functional RNA binding domain. Moreover, the anti-cancer agent trichostatin A potentiates Sam68-driven apoptosis.
Conclusions
For the first time we have shown that Sam68, an RNA binding protein with multiple apparent functions, exerts functionally separable effects on cell proliferation and survival, dependent on its ability to bind specifically to RNA. These findings shed new light on the ability of signal transducing RNA binding proteins to influence essential cell function. Moreover, the ability of a class of anti-cancer therapeutics to modulate its ability to promote apoptosis suggests that Sam68 status may impact some cancer treatments.
Background
Sam68 (Src associated in mitosis, 68 kDa) was first identified as a mitosis-specific substrate and binding partner of activated forms of the Src tyrosine kinase [1,2]. It has since been shown that it can bind to a variety of other signaling proteins through SH2, SH3 and WW domain-mediated interactions, suggesting a possible role as an adaptor protein [3-7]. It is a nuclear protein and contains a functional nucleic acid binding domain comprising a K homology (KH) module located within a larger "GSG" domain, which is present in Grp-33, Sam68 and Gld-1. This is required and sufficient for high-affinity binding to specific RNA sequences in vitro [8,9] and also mediates protein-protein interactions [10-12] and subnuclear localization [13,14]. Although Sam68 has so far only been shown to bind to RNA with sequence specificity [9], it remains possible that specific binding to single-stranded DNA may be biologically relevant.
Consistent with its in vitro binding of RNA, Sam68 has been implicated in processes linked to RNA usage and gene expression, including splicing [15-17] RNA export [18,19] and translation [20]. A role in transporting unspliced transcripts was suggested by finding that it could enhance function of the Rev protein of HIV, which promotes export of unspliced viral RNA through interaction with a Rev response element (RRE), and that a truncation mutant of Sam68 might act as a dominant inhibitor of Rev function [18,19]. Because of this, it has been suggested that Sam68 acts as a cellular analog of Rev. However, Sam68 can also drive expression from reporter plasmids in which the RRE has been deleted [21], and recent evidence suggests that Sam68 may stimulate protein expression without effect on RNA export [20]. In these studies Sam68 was shown to increase protein expression from RNA containing the constitutive transport element (CTE) of simpler retroviruses without affecting RNA transport. Instead, Sam68 appeared to stimulate RNA utilization in the cytoplasm.
Mutation of a conserved residue in the KH domain that renders Sam68 incapable of specific RNA binding in vitro abolished this effect, indicating a functional role of RNA binding. Moreover, its ability to promote cytoplasmic RNA utilization was inhibited by the Brk/Sik tyrosine kinase [20]. A recent study also suggests that Sam68 may play a role in transducing extracellular signals that regulate splicing [17]. It was shown that overexpression of Sam68 enhanced inclusion of exon v5 of CD44 in response to activation of the Ras-Erk pathway and that this was dependent on phosphorylation of Sam68 by activated Erk. These observations raise the possibility that Sam68 may integrate and transduce signals from cytoplasmic signaling pathways to control RNA utilization in a manner analogous to the role of p300/CBP in transcriptional regulation. Interestingly, Sam68 can physically interact with CBP [21].
Sam68 also has RNA binding-independent functions. It was recently shown that it can repress transcription from reporter constructs in a manner independent of RNA binding [21]. This, and other aspects of its control of gene expression, may in part reflect Sam68's ability to interact with proteins acting at different stages of gene expression, including the transcriptional co-activator CBP [21], the multi-functional DNA/RNA binding protein hnRNP K [22], and the splicing factor YT521-B [16]. It is not clear at which, and at how many, levels Sam68 exerts its effects on gene expression; the cellular consequences of these effects are also unknown. A role in control cell proliferation was predicted from its cell cycle dependent tyrosine phosphorylation [1,2]. Interestingly tyrosine phosphorylation of Sam68 by Src or Brk tyrosine kinases, or binding of Sam68 to the Src SH3 domain, decreases Sam68's ability to bind to homopolymeric or specific RNA in vitro [4,6,23], and downregulation of Sam68 may be important for signaling or transformation by these kinases. Consistent with this is the finding that antisense-mediated reduction of Sam68 expression transforms mouse fibroblasts [24]. On the other hand, deletion of sam68 in a chicken B cell line suppressed growth by elongating G2-M phase [25]. It has also been reported that expression of an RNA-binding-defective apparent splice variant of Sam68 lacking part of its KH domain can restrict G1/S progression, but that wild-type (wt) Sam68 does not, and that co-transfection of wt Sam68 eliminates the restriction induced by the splice-variant [26].
These reports of the biological effects of wt Sam68 seem somewhat contradictory and bear further examination. Because of the suggestion that Sam68 may exert a negative influence on growth (i.e., the anti-sense experiments), we studied the biological effects of wt and specific-RNA binding-defective Sam68 using an inducible expression system that allowed us to stably transfect cells with uninduced expression plasmids, thereby avoiding potential negative-selection effects. We found that expression of Sam68 conferred proliferative arrest and apoptosis, and that these effects were functionally separable based on the requirement for specific RNA binding by Sam68.
Results
Sam68 confers proliferative arrest in the absence of specific RNA binding
To study the effect of overexpression of Sam68 on cell function we attempted to generate NIH 3T3 cell lines stably overexpressing Sam68. However, we were unable to obtain any overexpressor clones, suggesting that Sam68 overexpression suppressed growth. We therefore established "tet-off" NIH 3T3-derived cell lines in which expression of wt or specific-RNA binding-defective Sam68 could be repressively controlled by doxycycline (Fig. 1A). We chose to study a mutant of Sam68 in which Gly178 in the KH domain is substituted by Glu (G178E) as a potential loss of function mutant, rather than deletion mutants, for the following reasons: 1) The G178E substitution mimics a mutation in the C. elegans gld-1 tumor suppressor gene which is sufficient for loss of Gld-1 function and tumor formation [28]. 2) We previously showed that Sam68G178E did not bind in vitro-selected, high affinity Sam68 RNA ligands, but bound to homopolymeric RNA, whereas KH domain deletion mutants did not bind to either ligand [9]. These two observations indicate that loss of high affinity, specific RNA binding is sufficient for loss of protein function. 3) KH domain deletion, but not the G178E substitution, compromises Sam68 self-interaction [8] and results in mislocalization of Sam68 within the nucleus [13,14], which complicates the interpretation of data obtained with such proteins.
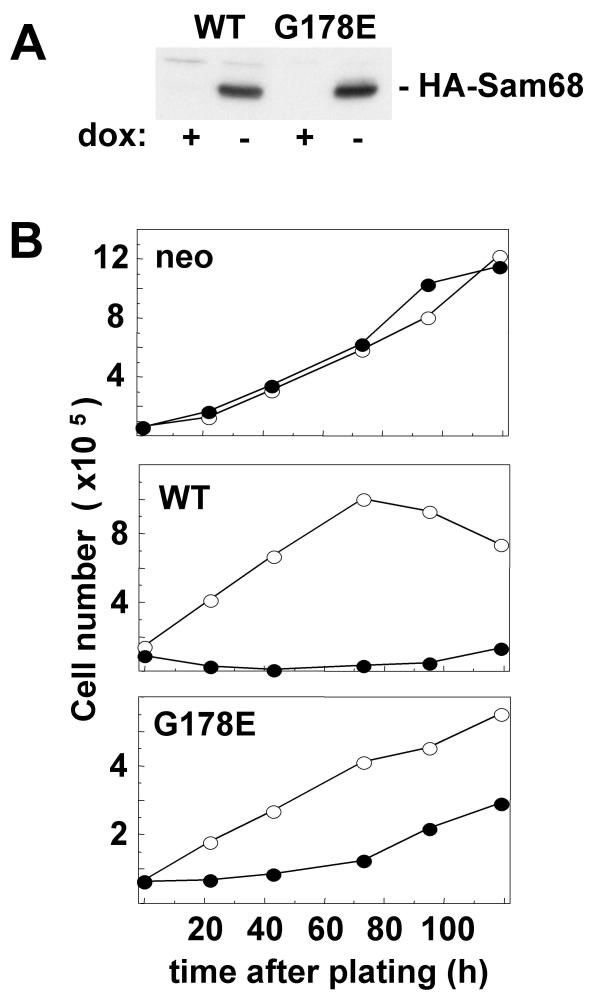
Figure 1.
Sam68 overexpression inhibits cell proliferation independently of RNA binding. A. Tet-off cells conditionally expressing wild-type (WT) or RNA binding-defective (G178E) HA-epitope tagged Sam68 were grown in the presence or absence of 100 ng/ml doxycycline for 24 h. HA-Sam68 expression was assessed by immunoblotting with anti-HA antibody. B. Cells conditionally expressing wt or G178E HA-Sam68 or no cDNA (neo) were plated at low density and cultured in the presence or absence of doxycycline. Adherent cells were collected by trypsinization and counted at the indicated times after plating. Results are representative of three separate experiments.
We first assessed the effect of Sam68 overexpression on cell proliferation: The cell lines were cultured at sub-confluence in the presence or absence of doxycycline and adherent cells were counted over 7 days (Fig 1B). The rate of proliferation of the tet-off wt or G178E Sam68-expressing cells was dramatically decreased in the absence of doxycycline. The number of cells overexpressing wt Sam68 actually decreased over 4 d, indicating Sam68-induced cell death. Since wt, but not G178E Sam68, provoked cell death, it was not possible to compare the relative effectiveness of the two proteins in retarding proliferation. Removal of doxycycline had no effect on the rate of proliferation of control cells that had been selected with an empty vector (neo; top panel).
It was previously reported that an RNA binding-defective, apparent, splice variant of Sam68 lacking part of the KH domain was able to compromise G1 phase progression, but that wt Sam68 could not [26]. Our results with wt Sam68 seemed at variance with those studies, so we examined the effect of overexpression of wt Sam68 or specific-RNA binding-defective Sam68G178E on the G1 to S phase transition. Following brief induction, or not, of Sam68 expression, cells were synchronized in mitosis by treatment with nocodazole and then released into G1 by drug washout. DNA synthesis was measured by BrdU incorporation (Fig. 2A). Twenty-four hours after release from mitosis most uninduced cells had incorporated BrdU and were in the subsequent G2/M or G1 phases, as determined by propidium iodide staining (data not shown). Overexpression of wt Sam68 reduced the number of BrdU positive cells from 89% to 24%, indicating G1 delay or arrest. Moreover, most of the BrdU positive cells were still in S phase, suggesting that the preceding G1 phase had been extended.
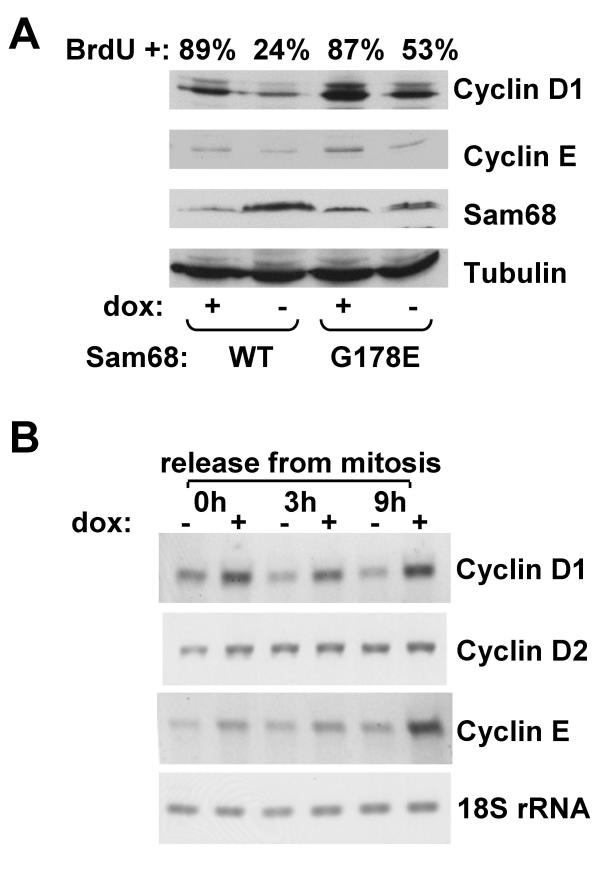
Figure 2.
Sam68 overexpression inhibits G1 to S phase progression and decreases G1 cyclin levels. A. Cells in the presence or absence of doxycycline were synchronized in mitosis by nocodazole treatment and released into G1 phase by drug washout. Cells were lysed in SDS-PAGE sample buffer 8 h after release and lysates were immunoblotted with the indicated antibodies. BrdU incorporation was assessed 24 h after release and quantitated by flow cytometry. B. Cells were released from mitosis and total RNA was prepared at the indicated times. Levels of the indicated transcripts were determined by RT-PCR with specific primers. Results are representative of three separate experiments.
Overexpression of Sam68G178E also decreased the number of BrdU positive cells; from 87% to 53%. The lesser effect of this mutant may reflect an additional specific-RNA binding-dependent effect on G1 progression. Nonetheless, it is clear that this mutant severely inhibits proliferation and G1 passage. Immunoblotting 8 h after mitotic release showed that expression of wt or mutant Sam68 reduced the levels of cyclins D1 and E (Fig. 2A, top three panels, dox-lanes). [The clone expressing Sam68G178E appeared to express a higher level of cyclin D1 in the uninduced state (Fig. 2A, top panel, dox+ lanes), which may further explain the weaker ability of this mutant to inhibit G1 progression.]
To determine if the G1 cyclins were down-regulated by control of their RNA levels, we measured these levels by RT-PCR (Fig. 2B). Cyclin D1 transcript levels were reduced in cells overexpressing Sam68, most notably at 9 h after mitosis (late G1). However, levels of the related cyclin D2 RNA were unchanged by Sam68 overexpression. The normal increase in Cyclin E RNA level noted 9 h after release from mitosis in uninduced cells was abolished by induction of Sam68. These Sam68-induced changes in cyclin RNA levels are sufficient to explain the observed changes in cyclin protein levels.
To follow the consequences of cyclin D1 and E downregulation we compared the kinetics of the cyclin level changes with the activities of G1 cyclin-dependent kinases and the phosphorylation of Rb following release from mitosis or G0 into G1. Cells were synchronized in mitosis with nocodazole or in G0 by serum withdrawal and released by drug removal or serum replacement, respectively. Cyclin expression levels were determined by immunoblotting (Fig. 3A). In agreement with the RNA level changes (Fig. 2B), cyclin D1 levels were high in mitotic cells and stayed at the same levels in uninduced cells during G1 progression. Also in agreement, cyclin D1 expression was lower in mitotic cells overexpressing Sam68 and was barely detectable 5 and 10 h after release from mitosis. As expected, cyclin D1 levels were low in serum-starved cells and increased after 10 and 15 h of serum treatment of uninduced cells. This increase was reduced in cells overexpressing Sam68. Cyclin E was not detectably expressed in mitotic cells but expression in uninduced cells increased 5 and 10 h after release. This increase was substantially blocked by overexpressed Sam68. Sam68 overexpression had no noticeable effect on cyclin E levels in the serum-treated cells; this may be due to the much lower levels of Sam68 in the serum-depleted cells compared to the cells released from mitosis (Fig. 3A, top panel).
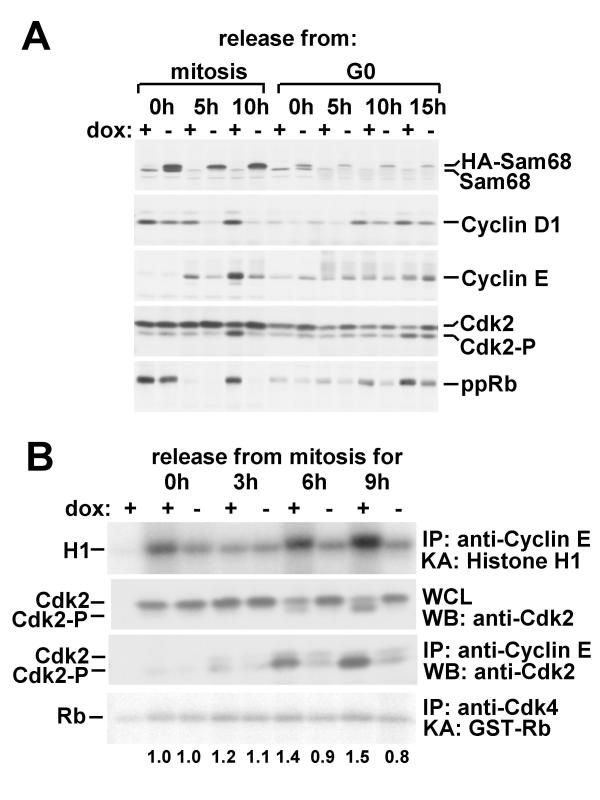
Figure 3.
Sam68 overexpression decreases G1 Cdk activities and Rb phosphorylation. A. Cells were synchronized in mitosis by nocodazole treatment or in G0 by serum deprivation and released into G1 by drug removal or serum replacement, respectively. At the indicated times after release cells were lysed in 3T3 lysis buffer and immunoblotted with antibodies against Sam68, cyclin D1, cyclin E, Cdk2 or phospho-Rb. B. Lysates from cells released from mitosis for the indicated times were subjected to immunoprecipitation with anti-cyclin E or -Cdk4 antibodies. Immunoprecipitates were assayed for in vitro kinase activity (KA) with histone H1 or GST-Rb as substrates, respectively. Indicated below the Cdk4 assay gel are relative 32P incorporations into Rb per lane, as determined by phosphorimager analysis. Whole cell lysates (WCL) or immunoprecipitates (IP) were also Western blotted (WB) with anti-Cdk2 antibodies. Results are representative of three separate experiments.
Cdk2 activity was indirectly determined by immunoblotting and examining the downward gel shift that accompanies its activation. Whereas there was a substantial increase in the level of downshifted Cdk2 10 h after release from mitosis in uninduced cells, this was abolished in the induced cells (Fig. 3A). There was also a small but noticeable decrease in Cdk2 activation in late G1 after serum stimulation, in spite of the unchanged cyclin E levels in these cells.
As expected from its effect on cyclin levels, Sam68 overexpression reduced phosphorylation of Rb (determined with an anti-phospho-Rb antibody) in cells treated with serum for 10 or 15 h, and completely abolished it in cells released from mitosis for 10 h (Fig. 3A, bottom panel). To confirm that the reduced Rb phosphorylation was due to decreased G1 Cdk activation, we measured the activities of Cdk2 and Cdk4 in immune complex kinase assays (Fig. 3B). Cyclin E-associated Cdk2 activity increased 6 and 9 h after exiting mitosis, and this was completely blocked by Sam68 overexpression (Fig. 3B, top panel). Immunoblotting of the anti-cyclin E immunoprecipitates with anti-Cdk2 confirmed that only the downshifted, activated form of Cdk2 was co-immunoprecipitated and that this form was greatly diminished in cells overexpressing Sam68 (Fig. 3B, third panel). Overexpression of Sam68 caused a less striking, but reproducible, decrease in Cdk4 activity after release from mitosis (53% activity at 9 h after mitosis in anti-Cdk4 immunoprecipitates from cells overexpressing Sam68 compared to uninduced cells; Fig. 3B, bottom panel). In summary, overexpression of Sam68 arrests or delays progression through G1. This is associated with reduced G1 cyclin levels and Rb phosphorylation and does not require specific RNA binding. The kinetics of cyclin reduction, which precedes S phase, are consistent with this being responsible for the decrease in Rb phosphorylation and consequent G1 arrest.
Sam68 induces apoptosis by a mechanism requiring specific RNA binding
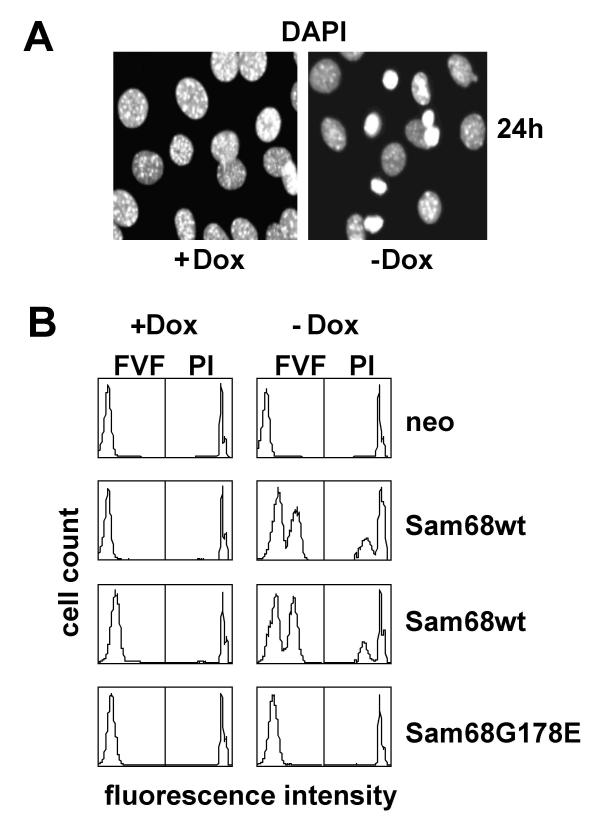
As shown in Fig. 1B, overexpression of wt, but not specific-RNA binding-defective Sam68 caused cell death as well as proliferative arrest. In the absence of doxycycline a substantial number of Sam68 tet-off cells underwent nuclear condensation within 24 h, revealed by DAPI staining, indicative of apoptosis (Fig. 4A). As an alternative, more quantitative measure of apoptosis we used a cell-based fluorescence assay of caspase activation. Induction of Sam68 overexpression induced significant apoptosis (i.e., a large fraction of caspase-positive cells) 72 h after doxycycline removal (Fig. 4B). No caspase positive cells were detectable in cells expressing the RNA binding-defective G178E mutant. The increase in caspase-positive cells was matched by an increase in cells with sub-G1 DNA content, revealed by propidium iodide staining. These results show that wt, but not specific-RNA binding-defective, Sam68 strongly promotes apoptosis.
Figure 4.
Sam68 overexpression induces apoptosis dependent on RNA binding. A. Tet-off cells conditionally expressing wild-type HA-Sam68 were cultured on coverslips in 10% serum with or without doxycycline for 24 h, fixed and stained with DAPI. B. Tet-off cell lines expressing the indicated Sam68 proteins were cultured in the presence or absence of doxycycline for 72 h and then assayed for caspase activation with a cell-permeable fluorescent caspase inhibitor, FAM-VAD-FMK (FVF). Fixed cells were counterstained prior to flow cytometry with propidium iodide (PI). Results with two different clones of cells expressing wt Sam68 are shown.
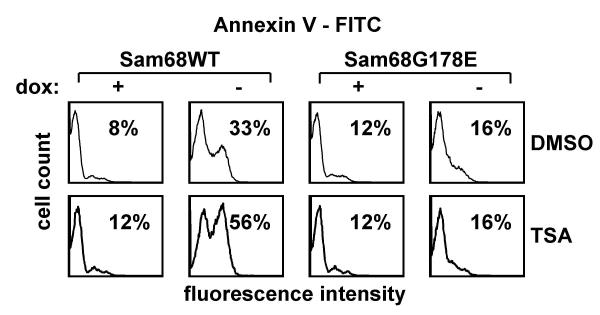
We were curious to see if any anti-cancer agents were able to enhance Sam68-induced apoptosis. Of the drugs screened, only trichostatin A (TSA) enhanced Sam68-driven apoptosis: In the absence of Sam68 overexpression, TSA (200 nM for 16 h) did not cause apoptosis. However, in cells overexpressing Sam68, TSA treatment increased the fraction of apoptotic cells, as determined by Annexin V-FITC binding, from 33% to 56% (Fig. 5). As expected, Sam68G178E did not induce apoptosis, and this was unaltered by TSA treatment.
Figure 5.
Trichostatin A potentiates Sam68-induced apoptosis. Tet-off cells conditionally expressing wt or G178E HA-Sam68 were cultured with or without doxycycline and then with 200 nM TSA or DMSO. Apoptotic cells were detected by annexin V-FITC binding and flow cytometry.
Discussion
Despite abundant circumstantial clues to its cellular function, an understanding of the precise cellular role of Sam68 and the significance of its RNA binding ability has proved elusive. Among the processes that it appears to regulate are transcription, splicing, RNA export and translation. The possibility that Sam68 might play multiple roles in gene expression is supported by the present study in which we show that its effects on cell proliferation and death are functionally separable.
We found that Sam68 compromises cell proliferation without the need for specific RNA binding. At least part of its proliferative block resulted from cell cycle arrest in G1 phase. This involved downregulation of cyclin D1 and E transcripts, suggesting that transcription of these genes was repressed. This effect was recapitulated by an specific-RNA binding-defective mutant of Sam68, and is reminiscent of the findings of Hong et al. [21], who showed that Sam68 expression can repress transcription of reporter constructs in an RNA-binding-independent manner. It is of course possible that cyclin RNA diminution is attributable to post-transcriptional intervention by Sam68 since modulation of RNA export, stability or translatability could result in alteration of total RNA levels. However, the effect of RNA-binding-defective mutants favors a transcriptional repression interpretation. If so, it will be important to assess whether Sam68 directly affects cyclin transcription or acts upstream of transcription. A direct effect on cyclin transcription could involve Sam68 interaction with the transcriptional co-activator CBP [21] or with hnRNP K [22]. So far we have not observed any effects of Sam68 overexpression on potential upstream regulators of cyclin transcription such as Ras/Erk signaling or AP-1 expression (our unpublished results).
It was previously reported that an apparent splice variant of Sam68, in which part of the KH domain was deleted, could block G1 progression and cyclin D1 accumulation in NIH 3T3 cells, whereas wt Sam68 did not [26]. Clearly this contrasts with our findings in which both wt and mutant Sam68 block progression and cyclin D1 accumulation. Also we have been unable to detect the reported splice variant in NIH 3T3 cells. It is not clear why our results differ.
In contrast to its effect on proliferation, induction of apoptosis by Sam68 requires its RNA-binding ability. This ability is required for other Sam68 functions as well: For example, its ability to enhance the cytoplasmic utilization of reporter constructs containing retroviral RRE or CTE elements requires RNA-binding [20]. This enhancement does not result from an effect on RNA export (as previously surmised) since it occurs without appreciable effect on nuclear export or global transcript level. It has been hypothesized that Sam68 "marks" the transcripts in a way that directs their cytoplasmic fate [20]. It has also recently been reported that Sam68 can regulate splicing in response to phosphorylation by activated Erk [17]. Clearly the identification of relevant RNA targets of Sam68 will be important to elucidate its precise functions. A recent report identified RNAs capable of binding to Sam68 [27]. Many of these possessed a motif identified by in vitro selection procedures [9]. This lends some credence to their authenticity, but also opens the question of whether such strategies uncover true in vivo targets or reflect in vitro binding.
Our findings suggest that Sam68 can act as a tumor suppressor, and are consistent with a previous study showing that antisense-treatment to reduce Sam68 levels transforms 3T3 fibroblasts [24]. Interestingly we found that at least one anti-cancer agent, TSA, potentiated the ability of Sam68 to induce apoptosis. At the concentrations used, TSA alone did not induce apoptosis, but it increased the level of apoptosis in cells overexpressing Sam68. TSA inhibits protein deacetylases and is generally assumed to exert its effects by enhancing histone acetylation, thereby relaxing transcriptional constraints. Sam68 overexpression did not have any detectable effect on the extent of histone hyperacetylation induced by TSA (data not shown), so it seems unlikely that Sam68 modulates TSA-induced transcription. Rather, pro-apoptotic signals generated by Sam68 and TSA may synergize to enforce apoptosis. Although we have not detected reactivity of Sam68 with anti-acetyl lysine antibodies, since Sam68 associates with the acetyltransferase CBP, the potential role of acetylation in regulating Sam68 function bears closer examination.
We and others have not observed cell status-dependent modulation of Sam68 expression levels; e.g., in response to cell cycle progression, growth factors or stress. We have also not detected marked differences in its expression levels in several human cancer cell lines. Thus, it would seem most likely that Sam68 activity is regulated post-translationally. This regulation could be conferred by covalent modification, allosteric modulation or changes in subnuclear localization. Sam68 is phosphorylated at C-terminal tyrosine residues by Src family kinases and the nuclear kinase Brk/Sik. These phosphorylations reduce its ability to bind to RNA in vitro, so we would predict that tyrosine phosphorylation by these kinases would decrease its ability to induce apoptosis. Indeed we have found that co-expressing activated forms of Src or Brk does suppress apoptosis in cells overexpressing Sam68. However, until we are able to generate non-phosphorylatable, yet functional, mutants of Sam68, it will not be possible to determine whether this effect is attributable to Sam68 phosphorylation.
Serine/threonine phosphorylation may also be involved in regulation of Sam68's pro-apoptotic function: it has recently been shown that phosphorylation of Sam68 by Erk enables Sam68 to regulate splice site selection in CD44 pre-RNA [17]. Our results with the deacetylase inhibitor also raise the possibility of regulation by acetylation. Sam68 is also dimethylated on arginine residues, and this can influence its ability to interact with other proteins [3].
Sam68 might also be allosterically regulated by its interactions with multiple nuclear proteins. For example, it has been reported that binding of hnRNP K inhibits Sam68's ability to enhance export of RRE-containing messages (Yang, et al. 2002). Finally, it is known that Sam68 assumes a distinctive, punctate subnuclear distribution in certain cancer cell lines (Chen et al. 1999), and this might influence its ability to execute its normal function. In light of our results showing enhancement of Sam68 pro-apoptotic ability by the anti-cancer agent TSA it will be important to find out whether, and how, the functional status of Sam68 is altered in cellular transformation.
Conclusion
Overexpression of Sam68 results in either cell cycle arrest or apoptosis in mouse fibroblasts. Induction of apoptosis, but not proliferative arrest, requires an intact specific-RNA binding function of Sam68. The separable effects of Sam68 on these key cellular events may in part explain the many roles previously attributed to Sam68 in cellular and viral function. The ability of Sam68 to provoke apoptosis is enhanced by TSA, suggesting that Sam68 status may influence the effectiveness of some anti-cancer drugs.
Methods
Cell lines and plasmids
cDNAs encoding HA-epitope-tagged Sam68 (wild-type or G178E; [9] were cloned into the tet-inducible vector pUHD-10-3. Conditional "tet-off" cell lines were generated by co-transfection of NIH 3T3 cells with pTetTak and the drug-resistance plasmid pSV2neo. Stable cell lines were selected in the presence of 400 μg/ml G418 and maintained in the presence of 100 ng/ml doxycycline to repress expression from the CMV promoter. Single colonies were selected for further study. For the key experiments presented at least two different clones were tested to exclude clonal variation influences (in Fig. 4C results obtained using two different clones expressing wt Sam68 are shown). The particular clones of wt and RNA binding defective Sam68 expressing cells used in the figures shown were chosen based on approximately equal levels of epitope-tagged protein expression. Expression levels of wt and mutant HA-Sam68 varied in parallel, relative to endogenous Sam68, depending on time of induction and growth conditions. For instance, inducible expression was lower in serum-starved cells (see Fig. 3A). Plasmids encoding YFP-tagged Sam68 were constructed by cloning sam68 cDNA into pEYFP-C1 (Clontech).
Apoptosis and proliferation assays
Cells were synchronized in mitosis by nocodazole treatment. Tet-off cells were trypsinized and plated in medium without or with 0.1 ng/ml doxycycline for 6–8 h. (We found that maintaining a low level of doxycycline was important for efficient collection of mitotic cells, presumably by preventing high levels of Sam68 expression and consequent cell cycle arrest prior to the mitotic block.) Nocodazole (0.1 μg/ml) was added and mitotic cells were collected by shake-off 13–15 h later. To release cells from the mitotic block, cells were washed by centrifugation three times with medium with or without doxycycline and then plated in medium containing 10% serum with or without 100 ng/ml doxycycline. Cells were either: (1) lysed in 3T3 lysis buffer [1], without Na deoxycholate, or SDS-PAGE sample buffer at the indicated times after release or (2) BrdU was added 7 h after release and BrdU incorporation was assessed 24 h after release as described by the manufacturer (Roche) and quantitated by flow cytometry.
Trichostatin A (TSA) treatment of cells was as follows: Sam68 wt or G178E tet-off cells were incubated with or without doxycycline for 48 h and then in medium containing doxycycline with 200 nM TSA or dimethyl sulfoxide (DMSO) for 16 h. It was necessary to switch off HA-Sam68 expression by including doxycycline in all samples since we found that TSA treatment increased expression from the regulatable CMV promoter, presumably by increasing histone acetylation at the promoter.
Three methods were used to assess apoptosis: (1) Cells were grown on coverslips, fixed in formaldehyde and stained with 4',6-diamidino-2-phenylindole (DAPI) to score condensed nuclei. (2) Caspase activity in whole cells was determined using a fluorescent caspase inhibitor (FAM-VAD-FMK) as described by the manufacturer (Intergen). (3) Cells were fixed in 70% ethanol and stained with propidium iodide prior to flow cytometry analysis. Annexin V-FITC or -PE labeling of live cells was performed according to manufacturer's protocols (Pharmingen) and cells were analyzed by flow cytometry. Flow cytometry data were analyzed and quantitated using FlowJo software (TreeStar).
Protein, RNA and protein kinase assays
Protein expression levels were determined by Western blotting with specific antibodies. RNA levels were determined by RT-PCR. Reverse transcription was primed with a mixture of oligo dT and random primers. The number of PCR cycles required for quantitative assessment of the amount of each transcript was determined experimentally. 18S ribosomal RNA was used as a loading control. PCR products were analyzed on agarose gels and stained with ethidium bromide. Cyclin E-associated Cdk2 activity and Cdk4 activity were assayed by immunoprecipitation of cell lysates, prepared in 3T3 lysis buffer [1]; without Na deoxycholate, containing equal amounts of total protein with anti-cyclin E (Santa Cruz Biotechnology; sc-481) or anti-Cdk4 antibodies (Santa Cruz Biotechnology; sc-260). Immune complexes were incubated with substrate and [γ-32P]ATP. Histone H1 (Sigma) and GST-Rb (Santa Cruz Biotechnology) were used as substrates for Cdk2 and Cdk4 kinase assays, respectively. The assays were stopped by adding SDS-PAGE sample buffer, and radioactive products were resolved by SDS-PAGE.
At least three similar experiments were performed to confirm each finding. All results shown are representative of these multiple experiments.
Author's Contributions
SJT conceived the study, designed, and performed several of the experiments (cell cycle analyses, RNA analysis, BRDU incorporation, annexin staining, and flow cytometry), analyzed the data, and drafted the manuscript. RJR contributed to the design and performance of several of the experiments (growth curves, caspase activation, immunofluorescence, and kinase assays) and participated in writing the manuscript. DS contributed to the analysis of the data and participated in writing the manuscript. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
We thank Gerd Blobel, Lou Hammarskjold and Quan Lu for valuable discussions. We thank Hector Nolla for expert assistance with flow cytometry. S.T. wishes to express his gratitude to G. Steve Martin for his generous support, G.S.M. and the members of the Martin lab for providing a stimulating and enlightening environment, and M.J. for inspiration. This work was supported by NIH/NCI grant CA32317.
Contributor Information
Stephen J Taylor, Email: suburbantrain@yahoo.com.
Ross J Resnick, Email: rjr9@cornell.edu.
David Shalloway, Email: dis2@cornell.edu.
References
- Taylor SJ, Shalloway D. An RNA-binding protein associated with Src through its SH2 and SH3 domains in mitosis. Nature. 1994;368:867–871. doi: 10.1038/368867a0. [DOI] [PubMed] [Google Scholar]
- Fumagalli S, Totty NF, Hsuan JJ, Courtneidge SA. A target for Src in mitosis. Nature. 1994;368:871–874. doi: 10.1038/368871a0. [DOI] [PubMed] [Google Scholar]
- Bedford MT, Frankel A, Yaffe MB, Clarke S, Leder P, Richard S. Arginine methylation inhibits the binding of proline-rich ligands to Src homology 3, but not WW, domains. J Biol Chem. 2000;275:16030–16036. doi: 10.1074/jbc.M909368199. [DOI] [PubMed] [Google Scholar]
- Derry JJ, Richard S, Valderrama Carvajal H, Ye X, Vasioukhin V, Cochrane AW, Chen T, Tyner AL. Sik (Brk) phosphorylates Sam68 in the nucleus and negatively regulates its RNA binding ability. Mol Cell Biol. 2000;20:6114–6126. doi: 10.1128/MCB.20.16.6114-6126.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard S, Yu D, Blumer KJ, Hausladen D, Olszowy MW, Connelly PA, Shaw AS. Association of p62, a multifunctional SH2- and SH3-domain-binding protein, with src family kinases, Grb2 and phospholipase C gamma-1. Mol Cell Biol. 1995;15:186–197. doi: 10.1128/mcb.15.1.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SJ, Anafi M, Pawson T, Shalloway D. Functional interaction between c-Src and its mitotic target, Sam 68. J Biol Chem. 1995;270:10120–10124. doi: 10.1074/jbc.270.17.10120. [DOI] [PubMed] [Google Scholar]
- Weng Z, Thomas SM, Rickles RJ, Taylor JA, Brauer AW, Seidel-Dugan C, Michael WM, Dreyfuss G, Brugge JS. Identification of Src, Fyn and Lyn SH3-binding proteins; implications for a function of SH3 domains. Mol Cell Biol. 1994;14:4509–4521. doi: 10.1128/mcb.14.7.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Damaj BB, Herrera C, Lasko P, Richard S. Self association of the single-KH-domain family members Sam68, GRP33, GLD-1, and Qk1: role of the KH domain. Mol Cell Biol. 1997;17:5707–5718. doi: 10.1128/mcb.17.10.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q, Taylor SJ, Shalloway D. Specificity and determinants of Sam68 RNA binding. Implications for the biological function of K homology domains. J Biol Chem. 1997;272:27274–27280. doi: 10.1074/jbc.272.43.27274. [DOI] [PubMed] [Google Scholar]
- Chen T, Richard S. Structure-function analysis of Qk1: a lethal point mutation in mouse quaking prevents homodimerization. Mol Cell Biol. 1998;18:4863–4871. doi: 10.1128/mcb.18.8.4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Fruscio M, Chen T, Bonyadi S, Lasko P, Richard S. The identification of two Drosophila K homology domain proteins. Kep1 and SAM are members of the Sam68 family of GSG domain proteins. J Biol Chem. 1998;273:30122–30130. doi: 10.1074/jbc.273.46.30122. [DOI] [PubMed] [Google Scholar]
- Di Fruscio M, Chen T, Richard S. Characterization of Sam68-like mammalian proteins SLM-1 and SLM-2: SLM-1 is a Src substrate during mitosis. Proc Natl Acad Sci USA. 1999;96:2710–2715. doi: 10.1073/pnas.96.6.2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Boisvert F, Bazett-Jones D, Richard S. A role for the GSG domain in localizing Sam68 to novel nuclear structures in cancer cells. Mol Biol Cell. 1999;10:3015–3033. doi: 10.1091/mbc.10.9.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride AE, Taylor SJ, Shalloway D, Kirkegaard K. KH domain integrity is required for wild-type localization of Sam68. Exp Cell Res. 1998;241:84–95. doi: 10.1006/excr.1998.4047. [DOI] [PubMed] [Google Scholar]
- Grossman JS, Meyer MI, Wang YC, Mulligan GJ, Kobayashi R, Helfman DM. The use of antibodies to the polypyrimidine tract binding protein (PTB) to analyze the protein components that assemble on alternatively spliced pre-mRNAs that use distant branch points. RNA. 1998;4:613–625. doi: 10.1017/S1355838298971448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann AM, Nayler O, Schwaiger FW, Obermeier A, Stamm S. The interaction and colocalization of Sam68 with the splicing-associated factor YT521-B in nuclear dots is regulated by the Src family kinase p59(fyn) Mol Biol Cell. 1999;10:3909–3926. doi: 10.1091/mbc.10.11.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matter N, Herrlich P, Konig H. Signal dependent regulation of splicing via phosphorylation of Sam68. Nature. 2002;420:691–695. doi: 10.1038/nature01153. [DOI] [PubMed] [Google Scholar]
- Li J, Liu Y, Kim BO, He JH. Direct participation of Sam68, the 68-Kilodalton Src-associated protein in mitosis, in the CRM1-mediated Rev nuclear export pathway. J Virol. 2002;76:8374–8382. doi: 10.1128/JVI.76.16.8374-8382.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy TR, Xu W, Mau JK, Goodwin CD, Suhasini M, Tang H, Frimpong K, Rose DW, Wong-Staal F. Inhibition of HIV replication by dominant negative mutants of Sam68, a functional homolog of HIV-1 Rev. Nat Med. 1999;5:635–642. doi: 10.1038/9479. [DOI] [PubMed] [Google Scholar]
- Coyle JH, Guzik BW, Bor Y-C, Jin L, Eisner-Smerage L, Taylor SJ, Rekosh D, Hammarskjold M-L. Sam68 enhances the cytoplasmic utilization of intron-containing RNA and is functionally regulated by the nuclear kinase Sik/Brk. Mol Cell Biol. 2003;23:92–103. doi: 10.1128/MCB.23.1.92-103.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong W, Resnick RJ, Rakowski C, Shalloway D, Taylor SJ, Blobel GA. Physical and functional interaction between the transcriptional cofactor CBP and the KH domain protein Sam68. Mol Cancer Res. 2002;1:48–55. [PubMed] [Google Scholar]
- Yang JP, Reddy TR, Truong KT, Suhasini M, Wong-Staal F. Functional interaction of Sam68 and heterogeneous nuclear ribonucleoprotein K. Oncogene. 2002;21:7187–7194. doi: 10.1038/sj.onc.1205759. [DOI] [PubMed] [Google Scholar]
- Wang LL, Richard S, Shaw AS. p62 association with RNA is regulated by tyrosine phosphorylation. J Biol Chem. 1995;270:2010–2013. doi: 10.1074/jbc.270.5.2010. [DOI] [PubMed] [Google Scholar]
- Liu K, Li L, Nisson PE, Gruber C, Jessee J, Cohen SN. Neoplastic transformation and tumorigenesis associated with Sam68 protein deficiency in cultured murine fibroblasts. J Biol Chem. 2000;275:40195–40201. doi: 10.1074/jbc.M006194200. [DOI] [PubMed] [Google Scholar]
- Li QH, Haga I, Shimizu T, Itoh M, Kurosaki T, Fujisawa J. Retardation of the G2-M phase progression on gene disruption of RNA binding protein Sam68 in the DT40 cell line. FEBS Lett. 2002;525:145–150. doi: 10.1016/S0014-5793(02)03103-4. [DOI] [PubMed] [Google Scholar]
- Barlat I, Maurier F, Duchesne M, Guitard E, Tocque B, Schweighoffer F. A role for Sam68 in cell cycle progression antagonized by a spliced variant within the KH domain. J Biol Chem. 1997;272:3129–3132. doi: 10.1074/jbc.272.6.3129. [DOI] [PubMed] [Google Scholar]
- Itoh M, Haga I, Li QH, Fujisawa J. Identification of cellular mRNA targets for RNA binding protein Sam68. Nuc Acids Res. 2002;30:5452–5464. doi: 10.1093/nar/gkf673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AR, Schedl T. Mutations in gld-1, a female germ cell-specific tumor suppressor gene in C. elegans, affect a conserved domain also found in Src-associated Sam68. Genes Dev. 1995;9:1491–504. doi: 10.1101/gad.9.12.1491. [DOI] [PubMed] [Google Scholar]