Abstract
Background
There is increasing evidence that gene order within the eukaryotic genome is not random. In yeast and worm, adjacent or neighboring genes tend to be co-expressed. Clustering of co-expressed genes has been found in humans, worm and fruit flies. However, in mice and rats, an effect of chromosomal distance (CD) on co-expression has not been investigated yet. Also, no cross-species comparison has been made so far. We analyzed the effect of CD as well as normalized distance (ND) using expression data in six eukaryotic species: yeast, fruit fly, worm, rat, mouse and human.
Results
We analyzed 24 sets of expression data from the six species. Highly co-expressed pairs were sorted into bins of equal sized intervals of CD, and a co-expression rate (CoER) in each bin was calculated. In all datasets, a higher CoER was obtained in a short CD range than a long distance range. These results show that across all studied species, there was a consistent effect of CD on co-expression. However, the results using the ND show more diversity. Intra- and inter-species comparisons of CoER reveal that there are significant differences in the co-expression rates of neighboring genes among the species. A pair-wise BLAST analysis finds 8 – 30 % of the highly co-expressed pairs are duplic ated genes.
Conclusion
We confirmed that in the six eukaryotic species, there was a consistent tendency that neighboring genes are likely to be co-expressed. Results of pair-wised BLAST indicate a significant effect of non-duplicated pairs on co-expression. A comparison of CD and ND suggests the dominant effect of CD.
Keywords: co-expression, inter-species comparison, chromosomal distance, eukaryotic genome
Background
As a consequence of DNA sequencing activities, whole-genome sequences for many microbial organisms as well as eukaryotic species are available in publicly accessible databases. DNA microarray technology makes it possible to simultaneously monitor expression patterns of thousand of genes. Expression profiles combined with whole-genome information, especially map information, enable us to investigate a relationship between co-expression of genes and a chromosomal distance (CD).
In the pioneering work in this field, Cohen et al. (2000) and Kruglyak and Tang (2000) independently showed that in yeast (Saccharomyces cerevisiae), adjacent pairs of genes show correlated expression [1,2]. In the nematode worm (Caenorhabditis elegans), a study of the relationship between physical distance and expression similarity found many co-expressed pairs of neighboring genes within a distance range of 20 kbp [3]. Clustering of co-expressed genes has been found in humans (Homo sapiens) [4], worm [5] and fruit flies (Drosophila melanogaster) [6,7]. However, in mice and rats, an effect of CD on co-expression has not been investigated thoroughly yet. Also, no cross-species comparison has been made so far. We analyzed the effect of distance using 24 expression datasets in six eukaryotic species: yeast, fruit fly, worm, rat (Rattus norvegicus), mouse (Mus musculus) and human and investigated inter-species differences.
Methods
Datasets
We used publicly available expression data in six eukaryotic species: yeast (two sets [8,9]), fruit fly (two sets [10-12]), worm (one set [13]), rat (three sets [14-16]), mouse (five sets [17-20]) and human (eleven sets [17], [21-28]) (for details see Table 1). Genes' chromosomal positions in yeast and worm were obtained from protein tables at GenBank ftp://ftp.ncbi.nih.gov/genbank/genomes/, and those in fruit fly and those in mouse, rat and human were obtained from FlyBase http://flybase.bio.indiana.edu/maps/lk/gnomap/ and from Resourcerer at TIGR http://pga.tigr.org/tigr-scripts/magic/r1.pl, respectively.
Table 1.
Dataset information
| species | label | genes | pairs | microarrays | weight |
| human | Su (a) [17] | 5510 | 770014 | 85 | 0.0482 |
| human | Armstrong [21] | 5492 | 765803 | 72 | 0.0406 |
| human | Pomeroy [22] | 3107 | 239951 | 266 | 0.0470 |
| human | Ramasawamy [23] | 6971 | 1217536 | 280 | 0.2508 |
| human | Yeoh [24] | 5503 | 768242 | 254 | 0.1436 |
| human | Houmard [25] | 15681 | 6344163 | 24 | 0.1120 |
| human | Diette [26] | 24810 | 15248313 | 29 | 0.3253 |
| human | Dyrskjot [27] | 3159 | 245900 | 40 | 0.0072 |
| human | Alizadeh (a) [28] | 2932 | 160672 | 31 | 0.0037 |
| human | Alizadeh (b) [28] | 2701 | 187917 | 35 | 0.0048 |
| human | Alizadeh (c) [28] | 3696 | 341437 | 67 | 0.0168 |
| mouse | Su (b) [17] | 7651 | 1616309 | 90 | 0.2437 |
| mouse | Schinke (a) [18] | 9124 | 2296857 | 30 | 0.1154 |
| mouse | Schinke (b) [18] | 7649 | 1615129 | 54 | 0.1461 |
| mouse | Neptune [19] | 13150 | 4744714 | 24 | 0.1908 |
| mouse | Scott [20] | 9124 | 2296857 | 79 | 0.3040 |
| rat | Faden [14] | 5519 | 884975 | 79 | 0.5741 |
| rat | Almon [15] | 4227 | 529217 | 47 | 0.2043 |
| rat | Almon [16] | 4227 | 529217 | 51 | 0.2216 |
| worm | Kim [13] | 15368 | 19987830 | 553 | 1 |
| fruit fly | Montalta-He and Egger [10,11] | 5534 | 3078761 | 31 | 0.8171 |
| fruit fly | Arbeitman [12] | 1138 | 133501 | 160 | 0.1829 |
| yeast | Eisen [8] | 2478 | 232362 | 79 | 0.4315 |
| yeast | Cho [9] | 6123 | 1422380 | 17 | 0.5685 |
Label provides the reference information. The genes, pairs and microarrays columns show the total number of the genes, pairs and microarrays in each dataset, respectively. Weight denotes the dataset's weight in each species.
Calculation of co-expression rate
We investigated only the genes of which the chromosomal position was known. In each dataset, the genes were divided into nchr subs ets, each of which consisted of genes/ORFs on the same chromosome (Here, nchr denotes the number of the chromosomes in that species). The Pearson correlation coefficient, r, and chromosomal distance, CD, measured in base pairs (bp) were calculated for every possible pair in a subset. After this calculation was repeated for all subsets, all results were merged and the total number of the pairs, N, was counted. The N pairs were sorted according to their values of r (regardless of CD), and then the top 20 % of the N pairs were selected from the sorted data as highly correlated (HC) pairs. The pairs were sorted into bins of equal sized intervals of CD. Because the six species have different genome compactness, different widths were used: 20 kbp for the mammals (human, rat and mouse), 2.5 kbp for the worm and fruit fly data and 1 kbp for yeast. To investigate an effect of CD on co-expression, we defined a co-expression rate (CoER) in each bin:
![]()
When the CoERs of the six species were plotted in a graph, the first bin of the mammals was further divided so that the starting point of the curves were almost the same.
As control groups, we selected two more groups of 0.2 N pairs from the sorted data: the lowest 20 % of N, negatively correlated (NC) pairs, and the 20 % of N centered at the median, zero-correlation (ZC) pairs. The CoER was calculated for these groups, and the results were compared with the HC group.
Weighted average
Because there were large differences in the number of the gene-pairs and microarrays in the datasets, we sought some measure of the reliability of each dataset. To do so, we used the product of two factors which inform regarding the breadth of the sampling of the organism's transcriptome: the numbers of the total pairs and microarrays in each dataset. The averaged CoER in each species was calculated using weights derived from the products. We considered the dataset having the largest weight in each species the most reliable one. It was expected that the CoER would scatter around 0.2 if there was no distance (or any other) effect on co-expression. To verify this, we also calculated the standard deviation of the CoER using the weights.
Normalized distance
When the six spec ies were compared a normalized distance (ND) was also used. The physical distance was normalized by the grand average of CD of all gene pairs next to each other in each species. The overall average was calculated as follows. First, the total length of a chromosome was divided by the number of genes on it. This was repeated for all chromosomes in a species and by averaging all values, we obtained the overall average in that species.
Gene ontology category
We also investigated Gene Ontology (GO) categories http://www.geneontology.org/ in the HC, NC and ZC groups in CD ranges between 0 and 20 kbp and between 980 and 1000 kbp. This analysis was carried out only for the most reliable dataset of yeast (Cho et al. 1998) and human (Diette, PGA Human CD4+Lymphocytes, http://microarray.cnmcresearch.org/pgadatatable.asp).
Pair-wise protein BLAST
Because duplicated genes are likely to be co-expressed due to their common history, we investigated how many pairs in the HC group were duplicated ones. In each of the yeast, worm and mouse datasets, HC pairs were sorted according to the CD (in ascending order) and the top 10,000 were selected for a pair-wise, stand-alone protein BLAST analysis. Of the 10,000, the pairs in which the protein sequences of both genes were known were subject to analyze. We employed a criterion previously proposed for the identification of duplicated genes in vertebrates [3,4]. Gene pairs having expected values (E) of less than 0.2 were deemed to be duplicated. Protein sequences were obtained from ExPASy Molecular Biology Server http://kr.expasy.org/ for mouse, from Wormpep.109 http://www.sanger.ac.uk/Projects/C_elegans/wormpep/ for worm and from Saccharomyces Genome Database http://www.yeastgenome.org/ for yeast.
Statistical test
To investigate intra- and inter-species differences, a multiple comparison of CoER was carried out using the Ryan procedure [29] at the significance level of α = 0.01. The underlying concept of this procedure is the incremental adjustment of the significance thresholds. Except for the significance thresholds, each of the tests was made in exactly the same manner as for a single pair. First, the highest and lowest CoERs were compared at the significance level of 2α / n(n-1), where n was the number of samples to be compared (in the inter-species comparison, n = 6). If the extremes differed significantly, we tested each of them against the CoER next to the other extreme at the significance level of 2α / n(n-2). If we found a significant difference in the previous step, we continued to test the highest CoER vs. the third lowest, the lowest vs. the third highest and the second highest vs. the second lowest using 2α / n(n-3). These steps were repeated until 2α / n(n-5). Because different bin widths were used in yeast, worm and fruit fly, the CoER was recalculated for the range between 0 and 20 kbp when the six species were compared. This comparison method was also employed to investigate intra-species differences in the eleven human datasets.
Results and Discussion
Co-expression rate
In all 24 datasets the distribution of Pearson correlation coefficient appeared to be a bell-shaped curve centered at approximately zero. In what follows, we will describe the results of the HC pairs unless otherwise noted. A higher CoER was obtained in the first bin than a long CD range between 980 k – 1000 kbp in all datasets.
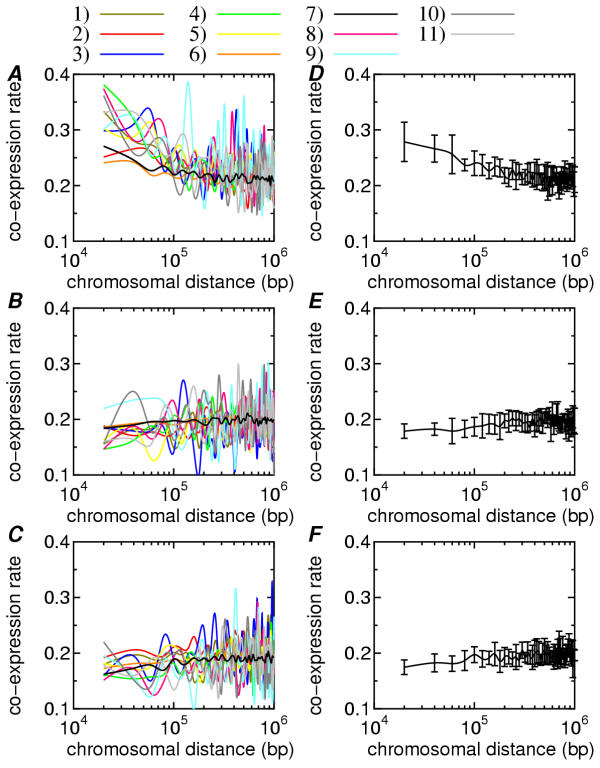
In Fig. 1, the CoERs obtained from the eleven human datasets are plotted as a function of CD: Fig. 1(A),1(B) and 1(C) show the results of the HC, ZC and NC pairs, respectively. The CoERs from the HC pairs decreased as the CD increased although there were large swings in CoER in a CD range above 100 kbp. These swings were larger in smaller data sets because of stochastic effects and undersampling of the transcriptome. In contrast, in the CD range below 100 kbp, the CoERs from the ZC and NC groups were relatively flat (compare Fig. 1(A) with 1(B) and 1(C)).
Figure 1.
The co-expression rates (CoERs) in the eleven human datasets are plotted as a function of chromosomal distance (CD): the results from the highly-correlated (HC) pairs (A), the zero-correlation (ZC) pairs (B) and the negatively-correlated (NC) pairs (C). The reference information is as follows: 1) Armstrong [21], 2) Ramasawamy [23], 3) Pomeroy [22], 4) Su (a) [17], 5) Yeoh [24], 6) Houmard [25], 7) Diette [26], 8) Dyrskjot [27], 9) Alizadeh (a) [28], 10) Alizadeh (b) [28], 11) Alizadeh (c) [28]. The weighted average and standard deviation of the CoER over the eleven datasets: the results from the HC pairs (D), the ZC pairs (E) and the NC pairs (F).
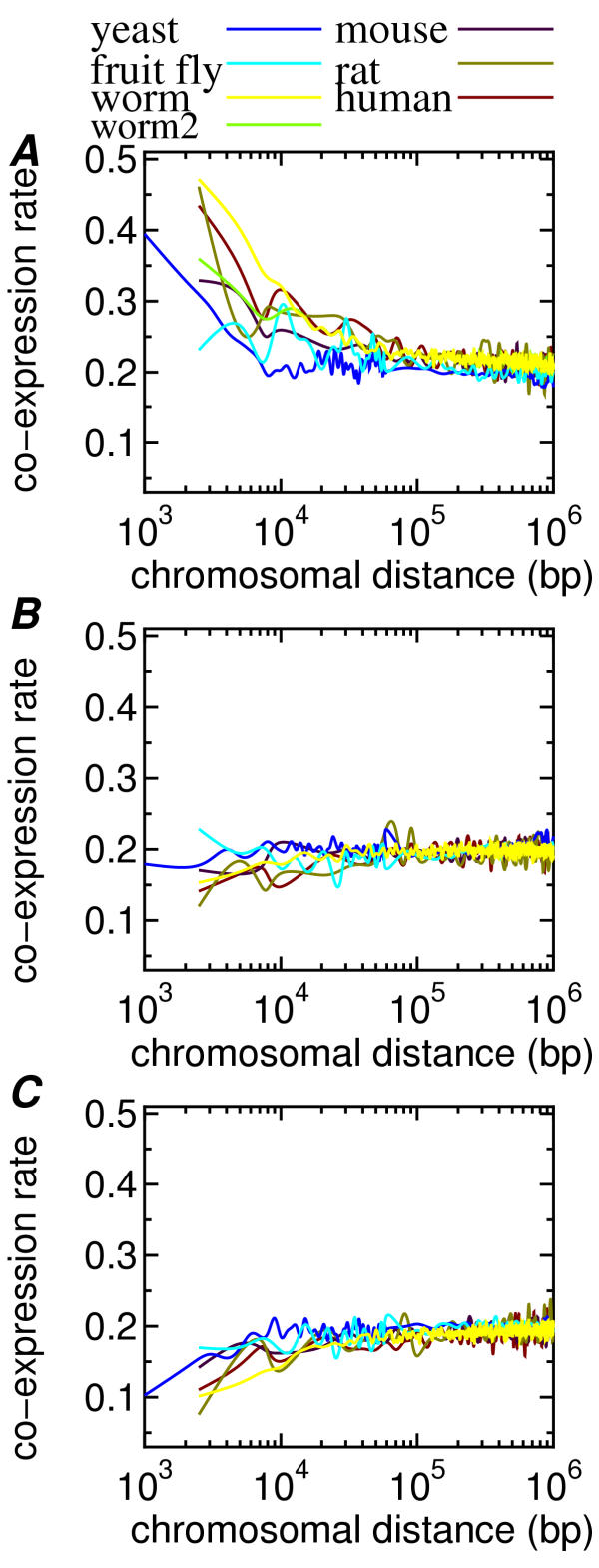
The weighted average and standard deviation over the eleven curves in (A), (B) and (C) are shown in (D), (E) and (F), respectively. The average in (D) showed the same tendency of gradual decrease to 0.2, whereas the averages in (E) and (F) showed no distance effect. The weighted averages for all six species are given in Fig. 2. The phenomena shown in Fig. 1 were consistently observed in all species. These results strongly suggest that co-expression of neighboring genes is common in many eukaryotic species and also that the CD, especially a short CD below 10 kbp, is associated with increased frequency of co-expression.
Figure 2.
A comparison of the six species using the weighted average of each species. (A) the highly correlated pairs, (B) the zero correlation pairs and (C) the negatively correlated pairs. Note that worm2 represents the CoERs of the worm dataset without the pairs in operons and duplicates.
GO category
The results of pair-wise analysis of GO category appeared to be different in the yeast and human datasets (Table 2). In yeast, only the HC pairs shared the same category and most of the pairs were not duplicates. Four pairs were found in a long CD range (980 – 1000 kbp). In the human data, more variety was obtained and pairs having the same category were found even in the NC group. A category, GO:5887 (integral to plasma membrane), was seen in the three groups while GO:6954 (inflammatory response) was seen in the HC and ZC. In the long CD range, eleven pairs were found. The results suggest the human genome involves more complicated functional relationships over a substantial CD.
Table 2.
Gene Ontology categories shared by the two genes in pairs with a CD below 20 kbp
| (A) highly correlated pairs | ||||
| category | description | # pairs | ||
| h | molecular functions | GO:0005132 | interferon-alpha/beta receptor ligand | 2 |
| h | GO:0005194 | cell adhesion | 2 | |
| h | GO:0008089 | anterograde axon cargo transport | 4 | |
| h | biological process | GO:0006954 | inflammatory response | 6 |
| h | GO:0006955 | immune response | 3 | |
| h | GO:0007165 | signal transduction | 5 | |
| h | cellular component | GO:0005634 | nucleus | 5 |
| h | GO:0005886 | plasma membrane | 5 | |
| h | GO:0005887 | integral to plasma membrane | 2 | |
| y | molecular functions | GO:0003735 | structural cons tituent of ribosome | 12 (1) |
| y | biological process | GO:0006412 | protein biosynthesis | 13 (1) |
| y | cellular component | GO:0005737 | cytoplasm | 12 (2) |
| (B) zero-correlated pairs | ||||
| h | molecular functions | GO:0005200 | cytoskeletal structural protein | 3 |
| h | GO:0008009 | chemokine | 2 | |
| h | GO:0008417 | fucosyltransferase | 2 | |
| h | GO:0015464 | acetylcholine receptor | 2 | |
| h | biological process | GO:0006954 | inflammatory response | 3 |
| h | GO:0007048 | oncogenesis | 4 | |
| h | GO:0007165 | signal transduction | 8 | |
| h | cellular component | GO:0005882 | intermediate filament | 3 |
| h | GO:0005886 | plasma membrane | 4 | |
| h | GO:0005887 | integral to plasma membrane | 5 | |
| (C) negatively correlated pairs | ||||
| h | molecular functions | GO:0005624 | membrane fraction | 3 |
| h | biological process | GO:0006508 | proteolysis and peptidolysis | 2 |
| h | GO:0007165 | signal transduction | 2 | |
| h | cellular component | GO:0005624 | membrane fraction | 3 |
| h | GO:0005887 | integral to plasma membrane | 2 | |
Pair-wise protein BLAST
Table 3 summarizes the BLAST results. In yeast, about 780 out of the 10,00 pairs were deemed to be duplicated pairs (E < 0.2). In the worm dataset, 8,370 pairs were available for the analysis and 2,658 (31.8 %) were regarded as duplicates. In the mouse datasets, out of about 3,500 analyzed pairs, 11.2 % were putative duplicates. However, in the three species, most pairs had expected values larger than 1. These results indicate that there are many non-duplicate pairs in the HC group, suggesting that the high CoERs in the HC group were due to not only effects of duplicated pairs but also non-duplicate effects. The distributions of the expected values in the five mouse datasets were almost the same (Table 3), suggesting that differences in microarrays were not significant.
Table 3.
Distribution of expected values obtained in pair-wise protein BLAST
| Expected value | ||||||||
| species | label | # of analyzed pairs | 0 – 0.2 | 0.2 – 0.4 | 0.4 – 0.6 | 0.6 – 0.8 | 0.8 – 1 | > 1 |
| yeast | Eisen [8] | 10000 | 800 | 515 | 402 | 316 | 369 | 7598 |
| yeast | Cho [9] | 9980 | 768 | 422 | 377 | 325 | 322 | 7766 |
| worm | Kim [13] | 8370 | 2658 | 267 | 206 | 182 | 210 | 4847 |
| mouse | Su (b) [17] | 3631 | 437 | 151 | 111 | 116 | 101 | 2715 |
| mouse | Schinke (a) [18] | 3554 | 340 | 149 | 131 | 107 | 99 | 2728 |
| mouse | Schinke (b) [18] | 3599 | 396 | 139 | 110 | 109 | 129 | 2716 |
| mouse | Neptune [19] | 3122 | 344 | 118 | 115 | 77 | 101 | 2367 |
| mouse | Scott [20] | 3659 | 408 | 117 | 110 | 104 | 111 | 2809 |
Intra- and inter-species comparisons
In Fig. 1(D), the standard deviations for the first three bins are relatively large. For example, the CoERs in the first bin in the eleven datasets were in a range between 0.24 and 0.38. To investigate intra-species differences in humans, a multiple comparison with the Ryan procedure was carried out (α = 0.01). Forty-nine out of the 55 possible combinations were not significantly different. In the five mouse datasets, 12,778 HC pairs out of approximately 20,000 used for the BLAST analysis were commonly seen in two or more datasets. The distributions of the expected values in the five mouse datasets were almost the same (Table 3). This suggests that there was no significant intra-species difference. Accordingly, the noise in the microarray data and the differences in microarray design appear to have minor influences on our results.
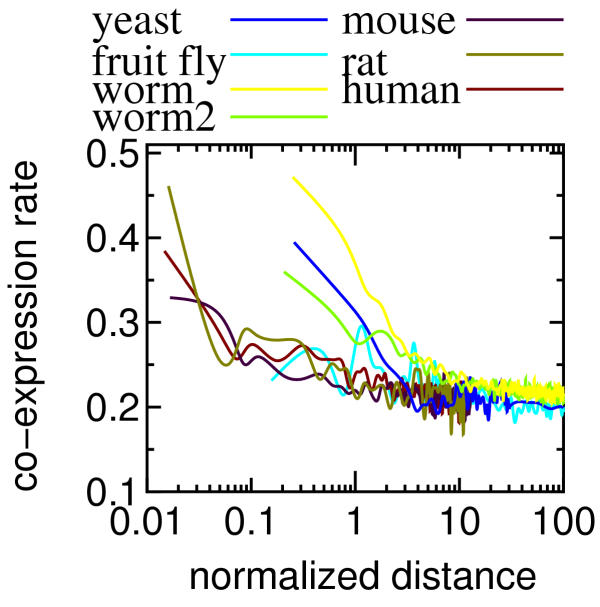
The CoERs in the weighted averages of the six species were compared. The results of the intra- and inter-species comparisons indicate that there are significant differences (p < 0.01) in the CoER in a short CD range (0 – 20 kbp) between any pair among worm, mammal (human, rat and mouse), fruit fly and yeast except two pairs of (worm and rat), and (mouse and fruit fly) (Table 4). Although the rat CoERs in the first three bins are almost the same as those of the worm (Fig. 2(A)), the results using the normalized distance (Fig. 3) strongly suggest that the multicellular organisms except worm show similar CoERs. The CoERs of worm and yeast were much larger than the others for a ND range between 0.3 and 1. In yeast, which is a unic ellular eukaryote with a compact genome, the organization of coordinated gene regulation is probably different from the other species with more dispersed genomes. As previously reported [3], the worm genome involved much more duplicated pairs than the yeast and mouse (Table 3). According to Blumenthal et al. [30], the worm genome involves at least 1,000 operons, which correspond about 15 % of all C. elegans genes. After excluding the pairs in the duplicates and operons, the worm CoERs are similar to those of the other multicellular organisms (see worm2 in Fig. 2(A)), indicating that the duplicates and operons are the main reason for the larger CoERs in the worm. However, when the ND was used, the worm curves (both with and without duplicates and operons) were similar to the yeast data.
Table 4.
The results of a multiple comparison with the Ryan procedure
| worm | rat | human | mouse | fly | yeast | |
| worm | * | * | * | * | ||
| rat | p = 0.703 | * | * | |||
| human | p < 0.001 | p = 0.094 | * | * | ||
| mouse | p < 0.001 | p = 0.017 | p = 0.250 | * | ||
| fly | p < 0.001 | p < 0.001 | p < 0.001 | p = 0.077 | * | |
| yeast | p < 0.001 | p < 0.001 | p < 0.001 | p < 0.001 | p = 0.001 |
A * denotes significant difference (p < 0.01).
Figure 3.
The co-expression rates shown in Figure 2(A) were re-plotted against the normalized distance. A value on the horizontal axis can be smaller than 1 because all possible pairs next to each other were involved in the calculation (i.e. we did not exclude a non-coding region, cetromere, etc from the calculation). Note that worm2 represents the CoERs of the worm dataset without the pairs in operons and duplicates.
A comparison of Figs. 2(A) and 3 provides some information on mechanisms behind the distance effect. If the physical distance has the dominant effect (chromatin remodeling is a possible cause), the CoERs in Fig. 2(A) should be similar across the species. On the other hand, if the effect of ND is major, Fig. 3 should show similar curves. The actual results seem to be the former and suggest that the CD plays the dominant role. In the multicellular organisms, the CoERs were higher than 0.2 up to about 50 kbp whereas the yeast curve was flat above 10 kbp. The mechanism behind this difference is currently not clear, but there are some clues. For example, several factors have been identified as controlling localized gene transcription [31]. These include the size of euchromatic chromosome territories and the spacing of chromatin "insulators" which provide impedance to non-specific enhancer activity upon neighboring genes [32]. Variation across species of these factors could explain our findings although this has not been systematically studied to date. Further analysis is required to advance our understanding of the mechanisms.
Conclusions
In this study, the effect of chromosomal as well as normalized distance on co-expression was analyzed using expression data from six eukaryotic species. We confirmed that in the six species, there was an effect of distance on CoER and a consistent tendency that neighboring genes are likely to be co-expressed. The results of intra- and inter-species comparisons of CoER show that there are significant differences in the co-expression rates of neighboring genes among yeast, worm and the other multicellular organisms. The pair-wise protein BLAST analysis indicates that effects of non-duplicates are not negligible. A comparison between the effects of chromosomal and normalized distance revealed that the physical distance has the dominant effect.
Authors' contributions
YF conceived the methodology, performed the statistical tests and drafted the manuscript. HI developed all software and performed the bioinformatical analysis. ISK guided the study and coordinated the project. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
We would like to thank M. Kimura for comments on an earlier version of the manuscript. This research was supported in part by the National Institute of Health through grants HL66805-01 and NS40828-01A1.
Contributor Information
Yutaka Fukuoka, Email: fukuoka.bmi@tmd.ac.jp.
Hidenori Inaoka, Email: inaoka.bmi@tmd.ac.jp.
Isaac S Kohane, Email: isaac_kohane@harvard.edu.
References
- Cohen BA, Mitra RD, Hughes JD, Church GM. A computational analysis of whole-genome expression data reveals chromosomal domains of gene expression. Nat Genet. 2000;26:183–186. doi: 10.1038/79896. [DOI] [PubMed] [Google Scholar]
- Kruglyak S, Tang H. Regulation of adjacent yeast genes. Trends in Genetics. 2000;16:109–111. doi: 10.1016/S0168-9525(99)01941-1. [DOI] [PubMed] [Google Scholar]
- Lercher MJ, Blumenthal T, Hurst LD. Coexpression of neighboring genes in Caenorhabditis elegans is mostly due to operons and duplicate genes. Genome Res. 2003;13:238–243. doi: 10.1101/gr.553803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lercher MJ, Urrutia AO, Hurst LD. Clustering of housekeeping genes provides a unified model of gene order in human genome. Nat Genet. 2002;31:180–183. doi: 10.1038/ng887. [DOI] [PubMed] [Google Scholar]
- Roy PJ, Stuart JM, Lund J, Kim SK. Chromosomal clustering of muscle-expressed genes in Canenorhabditis elegans. Nature. 2002;418:975–979. doi: 10.1038/nature01012. [DOI] [PubMed] [Google Scholar]
- Spellman PT, Rubin GM. Evidence for large domains of similarly expressed genes in Drosophila genome. J Biol. 2002;1:5. doi: 10.1186/1475-4924-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutanaev AM, Kalmykova AI, Shevelyov YY, Nurminsky DI. Large clusters of co-expressed genes in the Drosophila genome. Nature. 2002;420:666–669. doi: 10.1038/nature01216. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho RJ, Fromont-Racine M, Wodicka L, Feierbach B, Stearns T, Legrain P, Lockhart DJ, Davis RW. Parallel analysis of genetic selections using whole genome oligonucleotide arrays. Proc Natl Acad Sci. 1998;95:3752–3757. doi: 10.1073/pnas.95.7.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montalta-He H, Leemans R, Loop T, Strahm M, Certa U, Primig M, Acampora D, Simeone A, Reichert H. Evolutionary conservation of otd/Otx2 transcription factor action: a genome-wide microarray analysis in Drosophila. Genome Biol. 2002;3:RESEARCH0015. doi: 10.1186/gb-2002-3-4-research0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger B, Leemans R, Loop T, Kammermeier L, Fan Y, Radimerski T, Strahm MC, Certa U, Reichert H. Gliogenesis in Drosophila : genome-wide analysis of downstream genes of glial cells missing in the embryonic nervous system. Development. 2002;129:3295–3309. doi: 10.1242/dev.129.14.3295. [DOI] [PubMed] [Google Scholar]
- Arbeitman MN, Furlong EE, Imam F, Johnson E, Null BH, Baker BS, Krasnow MA, Scott MP, Davis RW, White KP. Gene expression during life cycle of Drosophila melanogaster. Science. 2002;297:2270–2275. doi: 10.1126/science.1072152. [DOI] [PubMed] [Google Scholar]
- Kim SK, Lund J, Kiraly M, Duke K, Jiang M, Stuart JM, Eizinger A, Wylie BN, Davidson GS. A gene expression map for Caenorhabditis elegans. Science. 2001;293:2087–2092. doi: 10.1126/science.1061603. [DOI] [PubMed] [Google Scholar]
- Faden A. CNS Regeneration http://microarray.cnmcresearch.org/pgadatatable.asp
- Almon R. PGA Rat Liver Methylprednisolone http://microarray.cnmcresearch.org/pgadatatable.asp
- Almon R. PGA Rat Muscle Methylprednisolone http://microarray.cnmcresearch.org/pgadatatable.asp
- Su AI, Cooke MP, Ching KA, Hakak Y, Walker JR, Wiltshire T, Orth AP, Vega RG, Sapinoso LM, Moqrich A, Patapoutian A, Hampton GM, Schultz PG, Hogenesch JB. Large-scale analysis of the human and mouse transcriptomes. Proc Natl Acad Sci. 2002;99:4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schinke M, Riggi LE, Chen I, Izumo S. Exercise induced hypertrophy http://cardiogenomics.med.harvard.edu/groups/proj1/pages/swim_home.html
- Neptune ER. PGA Murine Fibrillin-1 Deficient http://microarray.cnmcresearch.org/pgadatatable.asp
- Scott A. PGA Alternatively Activated Macrophages, Massaro: PGA Murine Calories Restriction, O'Donnell C: PGA Murine Glucose Metabolism, Rose M: PGA Murine Goblet Cells, Kleeberger: PGA Murine Air Hyperpermability, Clerch: PGA Murine Lung Septation, Moller and Chen: PGA Murine Pulumonary Fibrosis, all at http://microarray.cnmcresearch.org/pgadatatable.asp
- Armstrong SA, Staunton JE, Silverman LB, Pieters R, den Boer ML, Minden MD, Sallan SE, Lander ES, Golub TR, Korsmeyer SJ. MLL translocations specify a distinct gene expression profile that distinguishers a unique leukemia. Nat Genet. 2002;30:41–47. doi: 10.1038/ng765. [DOI] [PubMed] [Google Scholar]
- Pomeroy SL, Tamayo P, Gaasenbeek M, Sturla LM, Angelo M, McLaughlin ME, Kim JY, Goumnerova LC, Black PM, Lau C, Allen JC, Zagzag D, Olson JM, Curran T, Wetmore C, Biegel JA, Poggio T, Mukherjee S, Rifkin R, Califano A, Stolovitzky G, Louis DN, Mesirov JP, Lander ES, Golub TR. Prediction of central nervous system embryonal tumor outcome based on gene expression. Nature. 2002;415:436–442. doi: 10.1038/415436a. [DOI] [PubMed] [Google Scholar]
- Ramaswamy S, Tamayo P, Rifkin R, Mukherjee S, Yeang CH, Angelo M, Ladd C, Reich M, Latulippe E, Mesirov JP, Poggio T, Gerald W, Loda M, Lander ES, Golub TR. Multiclass cancer diagnosis using tumor gene expression signatures. Proc Natl Acad Sci. 2001;98:15149–15154. doi: 10.1073/pnas.211566398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeoh EJ, Ross ME, Shurtleff SA, Williams WK, Patel D, Mahfouz R, Behm FG, Raimondi SC, Relling MV, Patel A, Cheng C, Campana D, Wilkins D, Zhou X, Li J, Liu H, Pui CH, Evans WE, Naeve C, Wong L, Downing JR. Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell. 2002;1:133–143. doi: 10.1016/S1535-6108(02)00032-6. [DOI] [PubMed] [Google Scholar]
- Houmard JA. PGA Human Muscle Obese http://microarray.cnmcresearch.org/pgadatatable.asp
- Diette G. PGA Human CD4+Lymphocytes http://microarray.cnmcresearch.org/pgadatatable.asp
- Dyrskjot L, Thykjaer T, Kruhoffer M, Jensen JL, Marcussen N, Hamilton-Dutoit S, Wolf H, Orntoft TF. Identifying distinct classes of bladder carcinoma using microarrays. Nat Genet. 2003;33:90–96. doi: 10.1038/ng1061. [DOI] [PubMed] [Google Scholar]
- Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, Powell JI, Yang L, Marti GE, Moore T, Hudson J, Jr, Lu L, Lewis DB, Tibshirani R, Sherlock G, Chan WC, Greiner TC, Weisenburger DD, Armitage JO, Warnke R, Levy R, Wilson W, Grever MR, Byrd JC, Botstein D, Brown PO, Staudt LM. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- Ryan TA. Significance tests for multiple comparison of proportions, variances, and other statistics. Psychological Bull. 1960;57:318–328. doi: 10.1037/h0044320. [DOI] [PubMed] [Google Scholar]
- Blumenthal T, Evans D, Link CD, Guffanti A, Lawson D, Thierry-Mieg J, Thierry-Mieg D, Chiu WL, Duke K, Kiraly M, Kim SK. A global analysis of Caenorhabditis elegans operons. Nature. 2002;417:851–854. doi: 10.1038/nature00831. [DOI] [PubMed] [Google Scholar]
- Alvarez M, Rhodes SJ, Bidwell JP. Context-dependent transcription: all politics is local. Gene. 2003;313:43–57. doi: 10.1016/S0378-1119(03)00627-9. [DOI] [PubMed] [Google Scholar]
- Bell AC, West AG, Felsenfeld G. Insulators and boundaries: versatile regulatory elements in the eukaryotic genome. Science. 2001;291:447–450. doi: 10.1126/science.291.5503.447. [DOI] [PubMed] [Google Scholar]