Abstract
The assembly of proteins into defined complexes drives a plethora of cellular activities. These protein complexes often have a set of more stably interacting proteins as well as more unstable or transient interactions. Studying the in vivo components of these protein complexes is challenging as many of the techniques used for isolation result in the purification of only the most stable components and the transient interactions are lost. A technology called transient isotopic differentiation of interactions as random or targeted (transient I-DIRT) has been developed to identify these transiently interacting proteins as well as the stable interactions. Described here are the detailed methodological approaches used for a transient I-DIRT analysis of a multi-subunit complex, NuA3, that acetylates histone H3 and functions to activate gene transcription. Transcription is known to involve a concert of protein assemblies performing different activities on the chromatin/gene template, thus understanding the less stable or transient protein interactions with NuA3 will shed light onto the protein complexes that function synergistically, or antagonistically, to regulate gene transcription and chromatin remodeling.
Keywords: Chromatin, Protein–protein interactions, Mass spectrometry, Affinity purification, Isotope-labeling, I-DIRT
1. Introduction
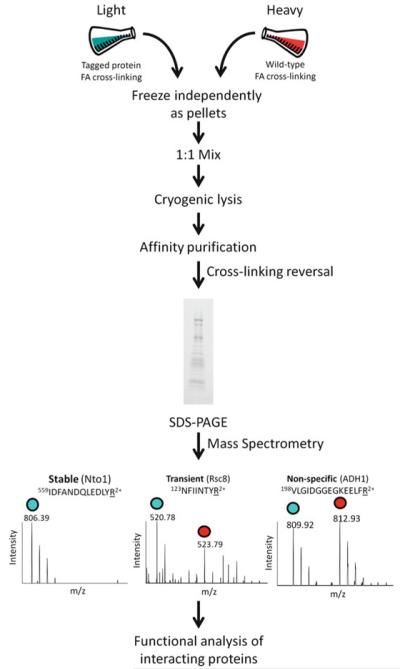
To identify networks of stable and transient protein–protein interactions, technology termed transient isotopic differentiation of interactions as random or targeted (transient I-DIRT) was developed (Fig. 1) (1) . This technique combines mild in vivo chemical cross-linking and isotopic-labeling with mass spectrometric readout to classify proteins co-purifying with an affinity-tagged “bait” protein as stable, transient, or nonspecific. There has been a multitude of protein–protein interaction studies using in vitro chemical cross-linking; however, there have only been a few using in vivo cross-linking with affinity purification followed by mass spectrometric analysis (1-3). Here, we describe the methodology for transient I-DIRT as it provides for a straightforward and quantifiable approach for defining the in vivo stable and less stable protein interactions. As an example of transient I-DIRT, we describe an analysis of the multi-subunit complex NuA3. NuA3 is a histone acetyltransferase in Saccharomyces cerevisiae that acetylates lysine 14 of histone H3 and activates gene transcription (4) . NuA3 is a five member protein complex composed of Sas3, Nto1, Yng1, Eaf6, and Taf30 (4) . Only Yng1 and Nto1 are found solely in the NuA3 complex. To identify the stable and transient protein–protein interactions with NuA3 specifically, the Yng1 protein was utilized as the purification “bait.” Specifically a TAP-tagged version of Yng1 was used in the methods described; however, other affinity tags can be utilized with an appropriate antibody for enriching.
Fig. 1.
Transient I-DIRT technology for identifying stable and less stable protein–protein interactions. One culture containing an affinity-tagged protein is grown in isotopically light media and cross-linked with formaldehyde (FA). A second culture of wild-type cells is grown in isotopically heavy media (e.g., 13C6-arginine) and also cross-linked with formaldehyde. Following independent harvesting and freezing, the cultures are mixed 1:1 and cryogenically lysed under liquid nitrogen temperature. The affinity-tagged protein is purified with associating proteins and resolved by SDS-PAGE. The gel lane is sliced into 2 mm sections and proteins are identified with mass spectrometry. Proteins interacting stably with the affinity-tagged protein complex have ~100% isotopically light tryptic peptides (i.e., these protein interactions have been preserved from the original isotopically light culture), while nonspecific proteins co-purifying with the affinity-tagged protein complex will have ~50% isotopically light tryptic peptides (i.e., contamination occurs during the purification procedure and these nonspecific protein associations have an equal probability to be isotopically light or heavy). Transiently interacting proteins will have an intermediate level of isotopically light tryptic peptides. Examples are shown of each interaction from a purification of the NuA3 histone acetyltrans-ferase. Following mass spectrometric analysis, the role of the specific protein interactions are explored with functional analyses.
2. Materials
2.1. Cell Culture and Cryogenic Lysis
Synthetic complete media per liter: 6.7 g yeast nitrogen base without amino acids, 2 g synthetic drop-out media minus arginine, 900 mL dH2O. Media is sterilized by autoclaving. After cooling to room temperature, add 1 mL arginine (80 mg/mL in dH2O, sterile filtered) for isotopically light media, or add 1 mL of 13C6-arginine (80 mg/mL in dH2O, sterile filtered, Cambridge Isotope Laboratories CLM-2265) for isotopically heavy media. After adding light or heavy arginine, add 1 mL of ampicillin (100 mg/mL in 50% ethanol) and 100 mL of autoclaved 20% glucose (w/v).
Glycine is a 2.5 M stock, autoclaved.
37% formaldehyde w/v.
20 mM Hepes (pH 7.5)/1.2% polyvinylpyrrolidone.
Liquid nitrogen.
Retsch MM301 mixer mill with stainless steel cylinders (Retsch 25 mL screw top grinding jars) and ball bearings (Retsch 20 mm stainless steel).
2.2. Affinity Purification
Dynabeads M270-epoxy (Invitrogen 143-02D) are suspended in N,N-dimethylformamide at 30 mg/mL and stored at 4°C.
Purified rabbit IgG is resuspended at 17 mg/mL in dH2O and stored at −80°C in 100 μL aliquots (only thaw an aliquot once).
0.1 M sodium phosphate (pH 7.4) stock.
3 M ammonium sulfate stock.
Solutions for washing IgG-coated Dynabeads: 100 mM glycine pH 2.5, 10 mM Tris pH 8.8, PBS, PBS + 0.5% tritonX-100.
Affinity purification buffer: 20 mM Hepes pH 7.5, 300 mM NaCl, 2 mM MgCl2, 0.1% Tween-20, 1/100 protease inhibitor cocktail (stored at −20°C). Make buffer immediately prior to use and keep at 4°C.
Fisher Tissuemiser (or any available probe tissue homogenizer that can operate at 30,000 RPMs).
Branson Sonifier 450.
Rocking platform.
0.5 N ammonium hydroxide/0.5 mM EDTA. Make immediately prior to use.
2× SDS-PAGE loading buffer: 125 mM Tris–HCl pH 6.8, 20% glycerol, 4.1% SDS, 0.05% bromophenol blue, 4% 2-mercaptoethanol. Store at −20°C in 0.5 mL aliquots.
3. Methods
The methodological workflow for a transient I-DIRT analysis is shown in Fig. 1 . The key components of this methodology are (1) a strain with an affinity-tagged protein and (2) a strain without the tag on the protein that is grown isotopically heavy. The tagged and nontagged strains are grown to equivalent densities, subjected to a mild in vivo chemical cross-linking with formaldehyde, and frozen independently. The mild cross-linking helps to partially stabilize the protein interactions. Particularly for chromatin-associated protein complexes, the cross-linking has to be mild as extensive cross-linking precludes the efficient purification of protein complexes (1, 5) . Frozen strains are mixed 1:1 prior to cryolysis with a ball mill maintained under liquid nitrogen temperature, which provides a method for generating cell lysate without thawing. The ability to differentiate stable, transient, and nonspecific protein interactions with the affinity-tagged complex occurs at the point of thawing the cell lysate and extends for the duration of the purification procedure. During the course of the purification, the stable protein interactions (which are exclusively isotopically light) with the affinity-tagged complex are maintained, while the transient protein interactions (which are also isotopically light) are only somewhat maintained relative to their affinity for the tagged protein complex. Thus, for a transient protein interaction, some exchange of the isotopically light protein will occur with the isotopically heavy counterpart. For nonspecific protein associations with the affinity-tagged protein complex that occur during the purification procedure, there is an equal probability that the nonspecific protein will be isotopically light or isotopically heavy.
The ultimate readout for these stable, transient, and nonspecific protein interactions with the affinity-tagged protein complex is mass spectrometry. When peptides are assigned to a given protein co-purifying with the affinity-tagged complex, the type of protein interaction can be classified as one of the following: stable if the peptides are ~100% isotopically light, nonspecific if the peptides are ~50% isotopically light and ~50% isotopically heavy or transient if the peptides are in between 50 and 100% isotopically light. The following transient I-DIRT methodology is presented for an analysis of the NuA3 histone acetyltransferase, but the methodology can be extended to any affinity-tagged protein. Using the transient I-DIRT technique, we previously identified five stable (Sas3, Nto1, Yng1, Taf30, Eaf6), five transient (Pob3, Nap1, Spt16, Rsc8, Rsc7), and 278 nonspecific protein interactions with an NuA3 purification (1) . The analysis of NuA3 demonstrates the high level of nonspecific associations with affinity purifications, and the need for a technique like transient I-DIRT to identify the subpopulation of specific and transient interactions for subsequent functional studies.
3.1. Cell Culture and Cryogenic Lysis
S. cerevisiae BY4741 YNG1::TAP-HIS is grown to midlog phase (~2.5 × 107 cells/mL) at 30°C with shaking at 200 RPM in isotopically light synthetic complete media. The arginine auxotroph strain BY4741 arg4::KAN is grown to ~2.5 × 107 cells/mL in isotopically heavy synthetic complete media with 13C6-arginine (see Note 1). Four liters of each strain are typically grown to give ~5 g of wet cell pellet. Large-scale growths are inoculated from 3 mL stationary phase cultures (in the respective synthetic media) with an estimated doubling time of 2.5 h.
Protein–protein interactions are partially trapped with mild in vivo chemical cross-linking. When the cultures reach ~2.5 × 107 cells/mL, remove the flasks from the incubator and add formaldehyde (37% stock concentration) to a final amount of 0.05%. Swirl the flask to mix and leave at room temperature for 5 min. Quench the cross-linking by adding 2.5 M glycine to a final concentration of 125 mM. Swirl the flask to mix and leave at room temperature for 5 min.
Cells are collected by centrifugation (2,500 × g) at 4°C for 30 min in 1 L bottles, washed with 100 mL of ice cold dH2O, and re-collected by centrifugation. Add 20 mM Hepes (pH 7.5)/1.2% polyvinylpyrrolidone to the wet cell pellet (1 mL buffer/10 g of wet cells) and mix by pipetting.
Cells are frozen as a suspension in liquid nitrogen. Use scissors to cut ~1 cm off the end of a 1 mL pipette tip – increasing the size of the opening will provide for easier pipetting. Next, fill a 50 mL polypropylene conical tube with liquid nitrogen. Slowly pipette the cell suspension drop-wise into the liquid nitrogen. Liquid nitrogen should be added at intervals during the freezing procedure to keep the tube full. Once the cells are frozen as pellets, pour off the excess liquid nitrogen. Poke three holes in the cap of the conical tube with a needle and place it on the tube. The frozen cells are now stored at −80°C.
For cryolysis, the isotopically light YNG1-TAP and isotopically heavy arg4::KAN cells are mixed 1:1 by cell weight. Cells are kept at liquid nitrogen temperature as much as possible during this process to avoid thawing. Weigh each set of pellets, mix 1:1 by weight, and add to a 50 mL polypropylene conical tube. Shake this tube thoroughly to ensure mixing of the pellets.
Cryogenic lysis is performed with a Retsch MM301 mixer mill using stainless steel cylinders (Retsch 25 mL screw top grinding jars) and ball bearings (Retsch 20 mm stainless steel). Cell pellets and cell powder are kept at liquid nitrogen temperature as much as possible during this process to avoid thawing. Stainless steel cylinders and ball bearings are precooled in liquid nitrogen (the cylinders and ball bearings are cooled once the nitrogen stops “boiling”). Tongs should be used to retrieve the cylinders from the liquid nitrogen. Approximately 3 g of mixed cell pellets are added to a cylinder with a ball bearing and then placed into liquid nitrogen. Once the cylinder with ball bearing and yeast pellets is cooled, it is attached to the mixer mill, processed for 3 min at 30 Hz, and then returned to the liquid nitrogen. The cycle of cryolysing is repeated five times. Following the final cycle, the cylinder is opened and the cell powder is scooped out into a 50 mL polypropylene conical tube that is in a bath of liquid nitrogen (no liquid nitrogen in the tube). The tube is sealed with a cap containing three holes made with a needle and placed at −80°C for storage.
Cryogenic lysis with a mixer mill is the preferred method for lysing and blending the cells. One should avoid methods, such as lysis, with glass beads as the samples will thaw during the procedure, which precludes uniform blending of the samples prior to thawing. If a mixer mill is not readily available, a reasonable alternative for cryogenic lysis is manual grinding of the cells in the presence of liquid nitrogen with a mortar and pestle. When manually grinding, the cells should be covered with liquid nitrogen during the lysis process. The cells should be ground into a fine powder. Grinding should continue until >75% lysis is visually observed with a light microscope. After the cells are ground, the cells are stored at −80°C as described above immediately after allowing the liquid nitrogen to evaporate.
3.2. Affinity Purification
For affinity purification via the TAP-tag on Yng1, 40 mg of M270-epoxy Dynabeads are coated with 3 mg of rabbit IgG (see Note 2). The following mixture is incubated overnight at 30°C with rocking: 40 mg of M270-epoxy Dynabeads, 3 mg of rabbit IgG, 1 M ammonium sulfate, 60 mM sodium phosphate (pH 7.4). Following the overnight coupling, the beads are collected with a magnet and washed with: 1 mL of 100 mM glycine pH 2.5, 1 mL of 10 mM Tris pH 8.8, 4 times with 1 mL of PBS, 1 mL of PBS/0.5% tritonX-100, 1 mL of PBS. All washes are done quickly except for the PBS/tritionX-100 wash that should be done for 15 min. For each purification, the beads should be coupled fresh.
For a typical purification of a chromatin-associated protein complex like NuA3, we use 10 g of cryogenically lysed cell powder. All steps are performed at 4°C. To 10 g of cell powder containing isotopically light YNG1-TAP and isotopically heavy arg4::KAN cell lysates, add 50 mL of affinity purification buffer (see Note 3). The cell lysate is suspended by gentle inversion. Cell lysate is thoroughly blended with a handheld Fisher Tissuemiser (or any available probe tissue homogenizer) for 20 s at 30,000 RPMs. Chromatin is sheared to ~500 bp with a Branson Sonifier 450 with five 10-second cycles (optimal shearing occurs with a maximum of 3 mL of lysate per tube and at least 1 min of incubation on ice following each cycle). The number of cycles and duration of sonication will vary between sonicators, thus parameters for sonication of chromatin to ~500 bp should be empirically determined. The supernatant is collected by centrifugation at 2,500 × g for 10 min. The 40 mg of IgG-coated Dynabeads are added for ~16 h with gentle inversion on a rocking platform (see Note 4). Beads are collected with a magnet and washed five times with 1 mL of affinity purification buffer. Proteins are eluted from the beads with 0.5 mL of 0.5 N ammonium hydroxide/0.5 mM EDTA for 5 min at room temperature and the eluant is lyophilized. This elution procedure minimizes the amount of heavy and light chain antibody that is released from the resin. The lyophilized proteins are resuspended in 20 μL SDS-PAGE loading buffer and heated to 95°C for 20 min.
Proteins collected from the affinity purification are resolved by 4–20% SDS-PAGE. In our laboratory, SDS-PAGE is performed with the pre-cast Invitrogen Tris–glycine gel system, and the gel is stained with the Thermo GelCode Blue Stain (a colloidal Coomassie stain) prior to imaging for documentation. A colloidal Coomassie stain is recommended, as this stain is sensitive for imagine purposes and easily removed for mass spectrometry. All handling of the gel should be done with powder-free gloves and clean labware. Cleaning items with a commercial glass cleaner such as Windex is good for minimizing keratin contamination, which is the major source of contamination during processing. The gel lane is sliced into 2 mm bands in preparation for mass spectrometry. The easiest way to slice the gel is to place it on a clean glass plate with a ruler underneath and use a clean razor to excise 2 mm sections. The excised gel bands are placed into 1.7 mL microcentrifuge tubes and stored at −20°C.
Gel bands are submitted to a proteomics facility for protein identification and high resolution analysis of tryptic peptides (see Note 5). The proteomics facility performs the following: destaining of the gel bands, in-gel trypsin digestion and tandem mass spectrometric identification of the tryptic peptides. A high resolution mass analyzer should be used to collect the mass spectra. Tandem mass spectra do not necessarily need to be collected with a high resolution mass analyzer. Proteins will be identified with database searching of the tandem mass spectrometric data. Peak areas of peptides corresponding to the assigned proteins will be extracted from the mass spectra. Note that peak areas only need to be extracted for peptides that will contain the heavy amino acid(s). The percent isotopically light peptide is calculated: (light area/(heavy area + light area)) × 100. Percent light peptide values are averaged together for a given protein and the standard deviation is calculated. A typical representation of this data is a bar graph with percent light on the y-axis and the given protein on the x-axis (1) . Stable protein interactions are seen as ~100% isotopically light (e.g., Fig. 1 shows Nto1 as a stable component of NuA3). Nonspecific protein associations are seen as ~50% isotopically light (e.g., Fig. 1 shows Adh1 as a contamination during the purification of NuA3). Transient protein interactions are observed as an intermediate between the stable and nonspecific levels (e.g., Fig. 1 shows Rsc8 as a transiently interacting protein with NuA3). Proteins qualify as transient interactions if the percent isotopically light is >2 standard deviations above the contaminant threshold and less than ~100% isotopically light (1) .
After identification of stable, transient, and nonspecific interactions, the investigator must select an appropriate system to explore the functional significance of the identified associating proteins. Systems for knocking out or knocking down a particular protein provide a good method for studying the significance of the protein interaction.
Acknowledgments
Funding was provided by NIH grants P20RR015569, P20RR016460, and R01DA025755.
Footnotes
The methodology presented is for incorporation of isotopically heavy arginine using the S. cerevisiae BY4741 arg4::KAN strain. If one is using a different auxotroph, heavy amino acid(s) or organism, then the incorporation efficiency of the heavy amino acid must be measured. For the transient I-DIRT procedure, one needs to approach complete incorporation of the heavy amino acid. To measure incorporation of heavy amino acid, the strain under study should be grown to mid-log phase and the proteins isolated. Proteins should be subjected to high resolution mass spectrometric analysis of tryptic peptides using a proteomics facility. Tryptic peptides containing the heavy amino acid should be completely labeled. It is always a good idea to label arginine or lysine as these are represented in tryptic peptides (since trypsin cuts after arginines and lysines). Additionally, it is a good idea to use isotopically heavy versions of carbon that are at least six Daltons heavy because (1) carbon isotopes (relative to deuterated) of amino acids do not affect peptide elution from reverse phase columns that are a key component of most mass spectrometric setups and (2) peptides with an amino acid that is less than six Daltons heavy result in isotopic overlap when compared to their isotopically light counterparts.
The procedure outlined for affinity purification was optimized for NuA3. NuA3 is a relatively low abundance chromatin-associated protein complex. The amount of cell lysate necessary for other protein complexes must be determined empirically. The amount of cell lysate used in this section is a good starting point for most protein complexes that have been studied with the transient I-DIRT technique. Note that all steps in this section are scalable to the amount of starting lysate (e.g., 4 mg of coupled Dynabeads are used for each gram of cell powder).
The affinity purification buffer described in this section is an empirically determined buffer that provides good yields for chromatin-associated protein complexes. The components of the buffer can be varied in accordance to the protein complex under study. Some of the components typically varied include: NaCl (200–400 mM) and TritonX-100 (0–1% v/v).
The time of incubation with coated Dynabeads is empirically determined. Shorter affinity isolation times (e.g., 1–4 h) can be explored and may result in fewer nonspecific interactions (6).
The mass spectrometric processing and data analysis are standard procedures for a proteomics facility. Proteomics facilities are available at many universities and are also available commercially.
References
- 1.Smart SK, Mackintosh SG, Edmondson RD, Taverna SD, Tackett AJ. Mapping the local protein interactome of the NuA3 histone acetyltransferase. Protein Sci. 2009;18:1987–1997. doi: 10.1002/pro.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gingras AC, Gstaiger M, Raught B, Aebersold R. Analysis of protein complexes using mass spectrometry. Nat Rev Mol Cell Biol. 2007;8:645–654. doi: 10.1038/nrm2208. [DOI] [PubMed] [Google Scholar]
- 3.Guerrero C, Tagwerker C, Kaiser P, Huang L. An integrated mass spectrometry-based proteomic approach: quantitative analysis of tandem affinity-purified in vivo cross-linked protein complexes (QTAX) to decipher the 26 S proteasome-interacting network. Mol Cell Proteomics. 2006;5:366–378. doi: 10.1074/mcp.M500303-MCP200. [DOI] [PubMed] [Google Scholar]
- 4.Taverna SD, Ilin S, Rogers RS, Tanny JC, Lavender H, Li H, Baker L, Boyle J, Blair LP, Chait BT, Patel DJ, Aitchison JD, Tackett AJ, Allis CD. Yng1 PHD finger binding to histone H3 trimethylated at lysine 4 promotes NuA3 HAT activity at lysine 14 of H3 and transcription at a subset of targeted ORFs. Mol. Cell. 2006;24:785–796. doi: 10.1016/j.molcel.2006.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Byrum S, Mackintosh SG, Edmondson RD, Cheung WL, Taverna SD, Tackett AJ. Analysis of histone exchange during chromatin purification. JIOMICS. 2010 doi: 10.5584/jiomics.v1i1.26. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cristea IM, Williams R, Chait BT, Rout MP. Fluorescent proteins as proteomic probes. Mol Cell Proteomics. 2005;4:1933–1941. doi: 10.1074/mcp.M500227-MCP200. [DOI] [PubMed] [Google Scholar]