Abstract
Background
Many sequenced bacterial genomes harbor phage-like elements or cryptic prophages. These elements have been implicated in pathogenesis, serotype conversion and phage immunity. The e14 element is a defective lambdoid prophage element present at 25 min in the E. coli K-12 genome. This prophage encodes important functional genes such as lit (T4 exclusion), mcrA (modified cytosine restriction activity) and pin (recombinase).
Results
Bioinformatic analysis of the e14 prophage sequence shows the modular nature of the e14 element which shares a large part of its sequence with the Shigella flexneri phage SfV. Based on this similarity, the regulatory region including the repressor and Cro proteins and their binding sites were identified. The protein product of b1149 was found to be a fusion of a replication protein and a terminase. The genes b1143, b1151 and b1152 were identified as putative pseudogenes. A number of duplications of the stfE tail fibre gene of the e14 are seen in plasmid p15B. A protein based comparative approach using the COG database as a starting point helped detect lambdoid prophage like elements in a representative set of completely sequenced genomes.
Conclusions
The e14 element was characterized for the function of its encoded genes, the regulatory regions, replication origin and homology with other phage and bacterial sequences. Comparative analysis at nucleotide and protein levels suggest that a number of important phage related functions are missing in the e14 genome including parts of the early left operon, early right operon and late operon. The loss of these genes is the result of at least three major deletions that have occurred on e14 since its integration. A comparative protein level approach using the COG database can be effectively used to detect defective lambdoid prophage like elements in bacterial genomes.
Keywords: e14, lambdoid phage, modular genome, decaying prophage
Background
Bacterial genomes harbor several types of mobile elements including transposons, insertion elements and temperate bacteriophages, both functional and defective. These elements can encode various important functions, including toxins, virulence factors, bacteriophage resistance, restriction modification systems and antibiotic resistance [1]. Prophages, both intact and defective, have a special role in this context as they are resident elements and play a special role in the physiology of the host bacteria. They have been implicated in serotype conversion, pathogenesis and phage immunity [reviewed by [2,3]].
The temperate lambda-like (lambdoid) phages have highly mosaic genomes with respect to each other. This forms the basis of the "modular genome hypothesis" proposed by Botstein in 1980 [4]. According to this hypothesis phages evolve by interchanging genetic elements (modules), each of which can be considered as a functional unit [5,6]. In spite of this diversity, E. coli and other enterobacterial genomes are recognized to contain a number of lambda-like cryptic prophages [reviewed by [7,8]]. For example the very well characterized E. coli K-12 genome carries eight convincingly identified prophages (λ itself and seven others; all of the latter are defective and six, DLP-12, e14, Rac, QIN, CPS-53, and Eut, are thought to be lambdoid in nature [reviewed by [7,9,10]]). The high rate of recombination, deletions and insertions present in such cryptic phage elements makes their unambiguous detection and determination of evolutionary linkages difficult (see below).
The e14 element, the subject of this report, is one such defective prophage element that is integrated into the E. coli K-12 genome at 25 min on the chromosome within the isocitrate dehydrogenase (icd) gene [11,12]. The sequence of the e14 element is available with the sequencing of the E. coli K-12 genome; it is 15.4 kbp long and lies between 1195432 bp and 1210646 bp on the K-12 chromosome [13]. The element has at one end 216 bp of homology with the C-terminal end of the host icd gene, and the actual crossover for integration (the attachment site) occurs between the first 11 bp at one end of the homology in e14 and an 11 bp sequence inside the host icd gene [12]. The integration event fused the e14 "icd replacement region" to the N-terminal portion of the host icd gene, causing only two amino acid changes in the isocitrate dehydrogenase protein [14]. The element is capable of excision if the host SOS response is triggered. Both excision and re-integration occur in a site-specific manner [11,15]. e14 shares its integration site with phage 21 and has a similar integration machinery to that of phage 21; both have slightly overlapping int and xis genes. These two genes are transcribed leftward and lie about 3 kb from the e14 att site [12,14]. However, e14 and phage 21 must have different specificities of site recognition since phage 21 Int and Xis cannot cure cells of the e14 element as demonstrated by Wang et al. [14].
Experimental data on e14 is scattered in the scientific literature. The e14 element was originally identified by Greener and Hill [16], and mapped on the E. coli K-12 chromosome and cloned by Plasterk et al. [17,18] and Maguin et al. [19]. A restriction map of the element was made which largely corresponds with the now available sequence [18]. Current E. coli genome databases attribute 20/21 ORFs to the e14 element [20-22]. Most of these are annotated as putative or hypothetical proteins and very few have a functional annotation. The element is known to encode several important functions including the lit gene involved in T4 exclusion [23,24], the rglA (mcrA) gene involved in restriction of hydroxymethylated non-glucosylated T4 phages [25,26], the pin gene involved in inversion of an adjacent 1794 bp segment within e14 [17,27]. In addition to these, it is also attributed to encode a kil function and a concomitant repressor protein [18], and an SOS induced cell division inhibition function attributed to the sfiC gene [19,28]. Defined regions of e14 encoding these latter functions have been implicated by mapping the kil, repressor and sfiC functions. However the actual genes corresponding to these functions have not been previously identified. Recent sequencing of numerous bacteriophage genomes now allows a much more sophisticated bioinformatic analysis of its genetic content and prediction of the function of many of the e14 genes.
E. coli is perhaps the best-understood cellular organism, and K-12 is the most highly studied E. coli strain. If this model genome is to be completely understood, and this goal now seems achievable, it is essential that we understand its prophage elements. Here, we use a sequence analysis approach to further understand the evolution, and phage- and host-related functions of the e14 element.
Results and Discussion
Overall genetic structure of e14
The e14 element is a relic lambdoid prophage element integrated into the icd gene of the E. coli K-12 genome. Little is known about where the e14 element came from or why it is maintained in the E. coli genome. We have attempted to characterize this prophage element keeping in mind that it is a lambdoid prophage that almost certainly was once functional. The element is 15204 bp in length and has an average G+C content of 0.45, while the E. coli chromosome has G+C content of 0.5. BLAST searches performed against the non-redundant database identified several particularly closely related sequences between e14. These regions of homology along with adjacent regions were then used in pair wise Blast and MegaBlast searches [29] in order to extend the similar regions. The important hits found after general and sequence specific BLAST are shown (Figure 1, Table 1). Important homologues of e14 include bacteriophages SfV, 21, ST64B, HK97, ΦP27 and prophage portions of several enterobacterial genomes and plasmid p15B (Figure 1 and Table 4 below). Figure 1 shows graphically that there are 24 open reading frames present in the e14 element and their relationships to a generic lambdoid phage genome. All but four e14 genes have convincing homology to genes present in the genomes of other lambdoid phages, and all the phage-encoded homologs of these twenty genes are present in similar locations on those phages. Seven of the e14 genes can be deduced to be functional and four appear to have obviously debilitating truncations. With its divergent early operons and late operon downstream of the early right operon, all of which contain genes that are homologous to other lambdoid phages, e14 is clearly derived from a lambdoid phage ancestor. It also seems clear that e14 no longer encodes a whole phage genome and that its ancestral prophage has suffered three major deletions, one in the early left operon, one that fused the early right and late operons and one within the late operon. In addition, there is a possible insertion near its left end since its original integration into the K-12 chromosome. We discuss these relationships in more detail below.
Figure 1.
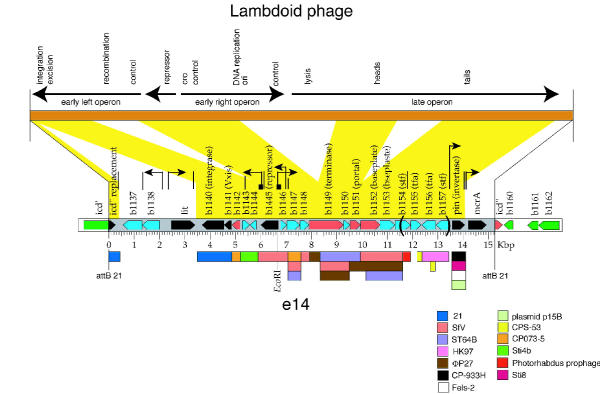
Overview of the e14 genome. The genetic functions of a generic lambdoid bacteriophage genome (brown rectangle) are shown above displayed with a transcriptional map (black arrows). In the middle, the section of the E. coli K-12 genome that contains e14 (gray rectangle) is shown with ORFs denoted by rectangular arrows oriented in the direction of transcription (green – host genes; red – e14 genes that are likely nonfunctional; black – e14 genes that are known to be functional; blue – e14 genes whose functionality cannot be assessed at present; parentheses indicate the boundaries of the P-invertable element). Small black arrows above the e14 map denote putative promoters, vertical lines denote putative terminators and small black squares putative operators. The yellow regions between the lambdoid and e14 maps indicate regions where e14 has homology to at least one known member of the lambdoid phage family (see text for details). Below, colored rectangles mark regions of highest homology between e14 and various known phages and prophages with regions of greater similarity closer to the e14 map (these are not meant to show all known homologies, only the closest ones); CPS-53 is a defective prophage in E. coli K-12, CP-933H is prophage in E. coli EDL933 and CP073-5, Sti4b, and Sti8 are provisional names for prophages in E. coli CFT073 and S. typhi CT18 (Supplementary Material of Ref. [39]).
Table 1.
Annotation of genes encoded by the e14 element. The functional annotation of the e14 genes along with the BLAST and FASTA hits, the closest structural homolog if any and the cluster to which the gene belongs are listed. TM indicates the transmembrane region, SS presence of signal sequence, COG, SM, PF, IPR, PS are prefixes to COG, SMART, PFAM, INTERPRO and PROSITE ids respectively. Genes for which direct or indirect evidence for transcriptional or translational expression is available have been indicated with a (+) sign and those genes which are inducible on SOS induction are marked with a (l+) in the last column of the table
| Name | Blattner number | Location | Protein length (orientation) | %G+C | Domain architecture and features | Function/similarity (Expression) |
| YmfD | b1137 | 659–1324 | 221 (-) | 0.35 | IPR001601 | Very weak match to Methyltranferase and tellurite resistance TehB (l+) |
| YmfE | b1138 | 1325–2029 | 234 (-) | 0.31 | TM(22–42, 59–79, 154–174, 186–206) | (l+) |
| Lit | b1139 | 2487–3380 | 297 (+) | 0.37 | PS00142 TM (61–82, 149–178) | T4 exclusion, Interacts with DNA, is a protease (l+) |
| IntE | b1140 | 3471–4598 | 375 (-) | 0.45 | IPR002104, PF00589 | phage integrase (l+) |
| Vxis | b1141 | 4579–4824 | 81 (-) | 0.44 | - | phage excisionase (l+) |
| YmfH | b1142 | 4861–5172 | 103 (-) | 0.54 | TM(42–62, 73–93) | similar to Q8FET3 of E. coli O6 (l+) |
| YmfI | b1143 | 5289–5630 | 113 (+) | 0.39 | - | (l+) |
| YmfJ | b1144 | 5568–5852 | 94 (-) | 0.47 | - | similar to Zinc finger protein Q8BGS3 (l+) |
| YmfK | b1145 | 6051–6725 | 224 (-) | 0.44 | IPR006198, PF00717 | cI/c2 repressor (l+) |
| b1146 | b1146 | 6513–7016 | 167 (+) | - | - | probable homolog of cro from Shigella flexneri (+) |
| YmfL | b1147 | 7048–7617 | 189 (+) | 0.49 | - | - |
| YmfM | b1148 | 7614–7952 | 112 (+) | 0.50 | - | - |
| YmfN | b1149 | 7962–9329 | 455(+) | 0.54 | IPR005021, PF03354, COG4626, SM00345 | Fusion of a replicase and a phage terminase |
| YmfR | b1150 | 9341–9523 | 60 (+) | 0.60 | TM(5–25, 26–46) | - |
| YmfO | b1151 | 9523–9934 | 137 (+) | 0.56 | IPR006944, PF04860 | Probable pseudogene, phage portal |
| YmfP | b1152 | 9935–10714 | 259 (+) | 0.58 | - | tail protein (baseplate?) |
| YmfQ | b1153 | 10705–11289 | 194 (+) | 0.57 | SS(1–32) | tail protein (baseplate?) |
| YcfK | b1154 | 11293–11922 | 209 (+) | 0.50 | COG3299 | tail fibre |
| YmfS | b1155 | 11924–12337 | 137 (+) | 0.42 | PF02413, IPR003458 | tail fibre assembly |
| TfaE | b1156 | 12309–12911 | 200 (-) | 0.48 | PF02413, IPR003458 | tail fibre assembly |
| StfE | b1157 | 12911–13411 | 166 (-) | 0.47 | - | side tail fibre |
| PinE | b1158 | 13477–14031 | 184 (+) | 0.49 | PF00239, PF02796, PS00397, PS00398 | DNA invertase – catalyses the inversion of 1800 bp P-region (+) |
| McrA | b1159 | 14138–14971 | 277 (+) | 0.38 | SM00507, IPR002711, IPR003615 | Modified cytosine restriction endonuclease A (+) |
Table 4.
Homologous regions of e14 with other phage and bacterial genomes. Regions of similarity of e14 with other genomes. All the regions indicated show greater than 85% identity in the region of the match. The matching regions in e14 are ordered based on position in the e14 genome. Figure 1 provides a schematic representation of this table.
| Organism | Region | e14 Region | Correspond e14 protein |
| S. flexneri phage V | 24627–25019 | 4859–5249 | ymfI |
| 25540–27256 | 5903–9926 | ymfK, ymfL, ymfM | |
| 1156–2211 | 8192–9247 | ymfN | |
| 15305–16983 | 9926–11604 | ymfO, ycfK | |
| S. typhimurium phage ST64B | 27440–27600 | 5031–5190 | ymfI |
| 29587–29625 | 7544–7582 | ymfL | |
| 1140–1405 | 8228–8493 | ymfN | |
| 1608–2840 | 8696–9928 | ymfN | |
| E. coli CFT073 | 38985–38623 | 4859–5220 | ymfI |
| 37962–37900 | 5900–5963 | - | |
| 36862–36316 | 7035–7581 | ymfL | |
| 36307–35709 | 7596–8194 | ymfM | |
| E. coli O157:H7 | 153052–153211 | 6–165 | icd |
| 25330–25963 | 11395–14028 | pin | |
| 23421–23500 | 11517–11596 | ycfK | |
| 24611–24263 | 12232–12580 | ymfS, tfaA | |
| S. flexneri 2a str. 301 | 1197399–1197608 | 7–216 | icd |
| 325210–325530 | 11604–11284 | ycfk | |
| 921146–921257 | 13459–13570 | pin | |
| 2684817–2685390 | 14032–13459 | pin | |
| Bacteriophage HK97 | 20864–20264 | 12256–12856 | ymfS, tfaA |
| 19918–19709 | 13205–13414 | pin | |
| Bacteriophage p27 | 22957–23180 | 8225–8448 | ymfN |
| 23410–23498 | 8678–8766 | ymfN | |
| 23764–23883 | 9035–9154 | ymfN | |
| 23735–23883 | 9006–9154 | ymfN | |
| 23941–24443 | 9212–9714 | ymfR, ymfO | |
| Plasmid p15B (X62121) | 3853–3216 | 12324–12961 | tfaA, stfE (-) |
| 3890–4390 | 12918–13415 | stfE | |
| 6180–6377 | 13218–13415 | stfE | |
| 4700–4880 | 13211–13415 | stfE | |
| 2998–2803 | 13215–13410 | stfE | |
| 5203–5418 | 13200–13415 | stfE | |
| 5701–5905 | 13211–13415 | stfE | |
| 6438–6879 | 13476–13917 | Pin | |
| S. enterica serovar Typhi Ty2 | 3525667–3526015 | 12581–12233 | tfaA, stfE |
| 3526698–3527201 | 13396–13898 | stfE, pin | |
| 3524880–3525072 | 13410–13218 | stfE | |
| 2754201–2754696 | 13898–13404 | stfE, pin | |
| 1410749–1410959 | 13205–13415 | stfE | |
| 1411306–1411507 | 13218–13419 | stfE | |
| 1408359–1408791 | 13908–13476 | Pin | |
| 1409336–1409551 | 13415–13200 | stfE | |
| 1408852–1409071 | 13415–13196 | stfE | |
The regulatory switch in the e14 genome
The regulatory switch that determines whether a lambdoid phage will follow a lytic or lysogenic life cycle includes an operator/promoter sequence and two major regulatory proteins similar to the Cro and CI repressor proteins of phage lambda [30]. Analysis of the e14 sequence suggests that its regulatory mechanism is similar to other functional lambdoid phages, especially Shigella flexneri phage SfV. This similarity has been reported previously by Allison et al. [31]. The e14 regulatory switch region has 96% identity to that of SfV. This region encodes the b1145 and b1146 proteins of e14 and the homologous P34 and P35 of SfV. (We will use mostly the "b" gene nomenclature system of Blattner et al. [13] for E. coli K-12 genes, because the genes are numerically named in their order on the chromosome, which makes their relative locations obvious; Table 1 gives both names for each open reading frame). The b1145 (e14) and P34 (SfV) proteins are identical except for a single amino acid difference – V4 of P34 is I4 of b1145 protein. A domain search with b1145 protein shows that it belongs to the LexA group of SOS-response transcriptional repressors similar to the lambda repressor (Interpro id: IPR006198, PFAM id: PF00717). In addition, b1145 protein is similar in sequence to the "CI repressors" of several other bacteriophages including Bordetella phage BPP-1 (accession no. AAK40284), and Salmonella phages P22 [32] and ST64B [33]. The experimental results of Plasterk and van de Putte [18] and Maguin et al. [19] suggest that disruption of the single e14 EcoRI site (Figure 1) is sufficient to negate repression of the kil function (see below). Since this site lies within the b1145 gene, one can conclude with reasonable certainty that b1145 encodes a functional prophage repressor that is responsive to an SOS signal. Its location between the early left and early right operons and leftward orientation is identical to prophage repressor genes in all known lambdoid phages. The second protein player in this regulation is the Cro protein, which is structurally similar to CI repressor and binds to the same operators, but performs the opposite function of facilitating lytic rather than a lysogenic life cycle. The putative b1146 protein matches the SfV P35 protein with 99% identity over 66 amino acid residues with a single residue change: C49 of P35 is Y48 of the b1146 protein. We note that the original annotation of b1146 is about 100 residues longer than the SfV P35 protein and other known Cro protein homologs. It is very likely that the first 101 residue section was wrongly predicted as part of this ORF. The smaller 66 codon ORF has a plausible RBS (Figure 2) and is identified by the gene prediction program GeneMark [34] (using E. coli as a typical model) as a separate ORF.
Figure 2.

The regulatory region of the e14 element. The possible ORFs for the cro (b1146) and cI repressors (b1145) are indicated in blue and orientation indicated by the arrow. The inverted repeats as detected by Allison et al. [31] for SfV are boxed. The palindromic regions are underlined. The ribosome binding sites (RBS) and putative -10 and -35 for the early right operon are indicated by different color letters within the box.
The b1145 protein is deduced to be functional, since the early left operon functions IntE and Vxis and early right operon kil function are normally off in K-12 (see below), and b1146 also seems likely to be functional by virtue of its near identity to its SfV homologue. In the well-characterized lambdoid phages the CI and Cro repressors bind to the same operators which overlap the promoters for the two divergent early operons. SfV and e14 are 95.2% identical in nucleotide sequence over a 1643 bp region that includes b1145, b1146 and the two potential operator regions. Allison et al. [31] predicted three inverted repeats (we note that they are all closely related to the consensus palindrome TTGTACCTNNNAGGTACAA) in SfV in the cI-cro intergenic region that might act as OR of lambda phage. These sites are maintained in the e14 sequence with two differences in the first repeat, one in the second repeat and two in the third repeat (Figure 2). Since it has been experimentally shown that LexA repression controls expression of functions in both the e14 early left and early right operons [18,19], and transcription of the majority of genes tested in these operons have been found to increase following UV irradiation [35], the promoters and operators on both sides of b1145 appear to have remained intact. We note that there are also two putative operator sequences (above) between b1144 and b1145 centered on e14 bp 5976 and 5996 (and similar sequences in SfV), and that plausible, correctly oriented promoters (see below) overlap both the left and right putative early operator regions.
DNA replication functions
Where it has been studied, canonical lambdoid phage DNA replication starts from a single origin (ori) in a bidirectional (θ) mode and later by a rolling circle (σ) mode. The origin of replication typically lies within the early right operon and is characterized by the presence of several repeats and palindromic sequences. Cumulative GC skew plots can provide a way of identifying origins of replication. The lowest point in such a plot usually corresponds to the origin of replication and the highest point to the replication termination point [36,37]. A cumulative GC skew plot was made for the e14 element. As is evident from this plot (Figure 3), two significant minima are present. The first of these minima occurs about 2000 bp from the left end of the element and the second is about 6000 residues from the left end. Neither of these regions show associated repeat regions. By analogy with the various canonical lambdoid phages, the replication origin should be in or near the replication genes that are located in the middle of the early right operon. The N-terminal portion of b1149 appears to be a replication-related protein, suggesting the original replication origin was near b1149. This region is close to the second minima seen in Figure 3. e14 is a decaying prophage element, and it is likely that the origin of replication has been deleted in the course of evolution; Figure 1 shows that it was likely removed by the middle proposed major deletion that has affected e14 (see below).
Figure 3.
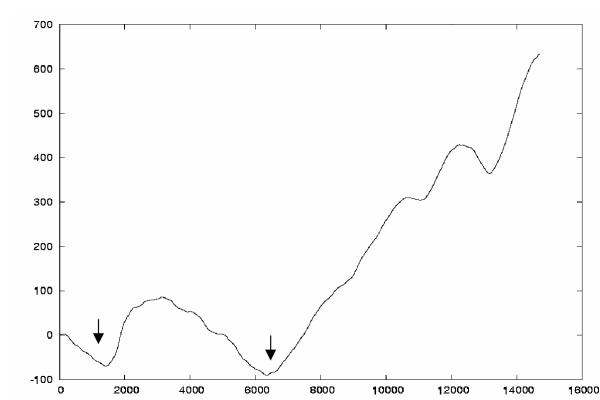
Cumulative GC-plot of e14. Cumulative GC-plot of e14 using a window size of 500 showing the regions of minima, which were analyzed for possible origins of replication. The y-axis represents ∑ (G-C)/(G+C) multiplied by 1000. The x-axis gives the base positions in e14.
Modular genome organization
The presently annotated e14 genome contains 20/21 predictable ORFs, which are available from the public databases including the Ecogene database [20], Genobase [21] and the Swissprot database [22]. Most of these encode putative proteins with no current functional annotation. Based on available data, sequence similarity, domain and motif searches an attempt was made to provide functional annotation for all the ORFs (Table 1). In the following paragraphs we will comment on the e14 genes from left to right across the element.
Most lambdoid bacteriophages do not have any complete genes between the att site and the int gene. However, e14 genes b1137, b1138 and lit lie between the att site and integrase gene. This has resulted in speculations regarding the origin of these genes. The three genes also show a significantly lower G+C content (Table 1) than the remainder of e14. All the three genes show LexA-dependent transcriptional induction on UV irradiation [35], but this could be an indirect result of e14 induction. Interestingly, the intergenic region between b1138 and lit harbors a region with eight bp multiple exact repeats which are highly AT rich. b1137 was previously annotated as a putative methyltransferase and involved in tellurite resistance, but these matches are very weak; it also shows four possible transmembrane segments and low similarity to certain eukaryotic proteins. The next ORF encodes the lit function. Expression of this protein inhibits protein expression late in phage T4 development. The protein interacts with a short sequence, the gol region within gene 23 that is the major head protein gene of phage T4 [38]. Lit is a protease known to cleave EF-Tu resulting in global inhibition of translation and death of E. coli cells infected with T4 phage [39]. These three e14 genes are unique in that none have convincing homologs in the current database that are phage bacteria encoded. Therefore the origins of this region are difficult to establish. It could have been picked up "recently" by the original functional phage ancestor of e14 through a specialized transduction (imprecise excision) mechanism before its integration here, or it could have been inserted here by some other process after e14's integration; its location next to att makes the former path more attractive.
By sequence homology with phage 21 and other phages, the integrase and excisionase function are encoded by intE and vxis, respectively, which form overlapping ORFs that are almost certainly functional as e14 is capable of SOS induced excision from the chromosome. Both IntE (b1140) and Vxis (b1141) show LexA-dependent transcriptional induction on UV-irradiation [35].
The small hypothetical b1142 protein is about 11 kDa in size and is similar to the N-terminal region of gene c3200 of E. coli O6:H1 CFT073 (87 % identity over 54 residues) [40]. The latter protein is much larger than its e14 homolog, and is encoded in a similar position in a lambdoid prophage in that genome. The C-terminal region of this CFT073 protein shows close sequence similarity to hypothetical proteins in SfV, ST64B, CPS-53, and Xylella fastidiosa prophage XfP4 [41], and each of these homologs lies in the early left operon in these lambdoid elements. It is possible that b1142 is a remnant of a larger gene and the deletion event that truncated it could be the left major e14 deletion in Figure 1. Gene b1143 encodes a protein with weak similarity to the putative protein encoded by gene STY2069 of Salmonella enterica CT18 [42] which lies in the early left operon of a prophage there. b1144 encodes a 94 amino acid protein which matches prophage-encoded hypothetical proteins early left operon from S. flexneri and S. enterica. b1144 also shows high transcriptional induction upon UV-irradiation [35].
The next two ORFs, b1145 and b1146, correspond to the CI repressor and Cro proteins as discussed in the previous section.
The b1147 and b1148 genes have no known function, but both show convincing similarity to hypothetical proteins of lambdoid phage origin. For example, phages SfV and ST64B carry homologs of b1147 and b1148 in similar locations as in e14. The roles of these homologs have not been studied, however a lethal (kil) function that kills the host bacterium was mapped by Plasterk and van de Putte [18] to what we now can deduce is the b1146–b1149 interval. Since b1146 and b1149 are homologs of non-lethal genes, it seems most likely that b1147 and/or b1148 encode this lethal function. We also note that a lethal sfiC function was mapped to the e14 element by Maguin et al. [19]. Their data are consistent with sfiC being a CI repressor-controlled gene, but its location was not accurately mapped. It is not known whether kil and sfiC are the same or different functions. Experimental evidence suggests that the sfiC gene product interacts with the FtsZ cell division protein and is responsible for an irreversible blockage of cell division [19], unlike the reversible inhibition brought about by SulA [28]. The protein product is highly stable even in lon+ strains and does not show significant similarity to any non-phage protein. It is interesting that other lambdoid phages are known to encode FtsZ inhibitors in their early left operons [43-47].
The b1149 protein appears to be a unique fusion between a replication protein and phage terminase. While the first 78 residues are quite similar to the N-termini of putative replication proteins from E. coli O157:H7 prophage CP-933P [10] (sprot id: Q8XAD8) and phages ΦP27 [48], ST64T [49] and SfV. The rest of the b1149 protein is extremely similar to the C-termini of terminase proteins of ST64B (98% identical) and SfV (96%) and other phages. The deletion that caused this gene fusion is the middle major e14 deletion in Figure 1, and it seems unlikely that the b1149 protein product is now functional. b1150 is a very small protein that is highly similar to proteins encoded by genes in the same location by phages ST64B, SfV and ΦP27. b1151 closely resembles portal proteins involved in head assembly from phages ST64B and ΦP27 over the N-terminal 135 amino acid residues. In bacteriophage ST64B the portal protein is 414 residues and ΦP27 protein is 413 residues. b1151 is almost certainly a C-terminally truncated pseudogene derived from a homolog of these larger proteins. This truncation and the N-terminal truncation relative to its homologs of the next gene, b1152, represent the boundaries of the right major e14 deletion in Figure 1. b1152 and b1153 are tail protein homologs of gene 47 and 48 proteins of phage Mu, which has a contractile tail. SfV phage tail proteins are their closest homologs and occur in similar relative positions. The N-terminal 106 residues of b1154 are similar to a 22 kDa protein from SfV (85% identity over 100 residues) and show similarity to side tail fiber proteins in other phages. The remaining C-terminal 103 residues are weakly related to the predicted protein of gene plu2959 of Photorhabdus luminescens TT01 [50], which lies in the tail region of a prophage in that genome. The left boundary of the Pin-invertible element which starts 11582 bp from the left attachment site of e14, lies within b1154, 96 codons from the 5'-terminus. b1155 shows close resemblance in its C-terminal region to genes in prophages CPS-53 of E. coli K-12, CP933H of E. coli EDL933, and Sti8 of S. enterica CT18. TfaE (b1156) shows 90% identity to the tail fibre assembly protein of bacteriophage HK97. b1154 and b1155 proteins are members of the large tail fibre assembly (Tfa) protein family that includes phage T4 gene 38 and Mu gene 50 proteins.
The pin (b1158) protein is a site-specific DNA invertase like the Min invertase of p15B, Gin of phage Mu, Hin of S. enterica, and Cin of phages P1 and P7, as well as putative invertases on a number of prophages in the sequenced bacterial genomes such as Sp1 of E. coli Sakai [51], Sti3 and Sti7 of S. enterica CT18, and Fels-2 of S. enterica LT2 [52]. These invertases in turn belong to a larger family of site-specific resolvase and recombinase proteins. The Pin protein catalyses the inversion of a 1794 bp long fragment referred to as the P-element [18]. This invertible element lies between 11582–13405 bases from the left att site and encompasses the four ORFs b1154, b1155, b1156 and stfE (b1157). When the early right/late operon fusion, in which these genes lie, is transcribed, genes b1155 and b1156 are not expected to be expressed in the shown (Figure 1) orientation of the P-element (and b1157 is not expected to have any bone fide translation start), but after inversion, the b1157 open reading frame would be fused to the N-terminal 96 codons of b1154 and b1156 would be placed in the correct orientation for expression. StfE (b1157) and b1154 appear to encode the C-termini of alternate side tail fibre proteins, and b1155 and b1156 appear to encode alternate as tail fibre assembly proteins.
The last gene of the e14 element, mcrA, encodes a methylation-dependent restriction endonuclease belonging to the HNH family of proteins found in several bacterial and bacteriophage systems [25,53]. In vivo studies on McrA suggest that it restricts T-even phage DNA that is hydroxymethylated and non-glucosylated (RglA activity) and also cleaves HpaII and SssI methylated DNA [25]. No close homologs of mcrA are known on other phage or prophage genomes, but many temperate phages carry genes that protect the host bacterium from attack by other phages.
Operons in the e14 element were predicted based on the co-occurrence of genes in the same order in different genomes [54] (Figure 1), but putative promoters and terminators that seem reasonable for expression of the various genes in light of their analogy to lambdoid phage genes could be identified (Tables 2 &3) based on the known coding regions and operons. These need to be experimentally verified.
Table 2.
Predicted promoters for the e14 element. Putative promoters predicted using BPROM available at the website http://www.softberry.com. Scores are as given by BPROM. Promoters with a score above 3 were considered for the study. Only those promoters which could be associated with some gene are listed. Promoter for the shorter ORF of b1146 was predicted based on Allison et al. [31] and GeneMark program and hence is omitted from the table.
| Position | Strand | -10 box | -35 box | Score | Gene | ||
| -10 | -35 | Overall | |||||
| 2070 | - | gtatataat | ttgtaa | 72 | 47 | 9.24 | ymfE, ymfD |
| 2461 | + | gtatatact | ctgaag | 62 | 19 | 6.95 | lit |
| 6003 | - | ttttatact | tttatg | 76 | 33 | 8.09 | ymfJ |
| 6919 | - | cacaaaact | ttgctc | 17 | 31 | 3.24 | ymfK |
| 6800 | + | atgtaatat | ttgaag | 61 | 54 | 3.5 | ymfL |
| 13200 | + | acttaaaat | ttgcat | 67 | 50 | 4.68 | pinE |
| 14113 | + | aagtagtat | ttgcaa | 44 | 55 | 5.55 | mcrA |
Table 3.
Predicted terminators for the e14 element. 'rho' independent terminators in the e14 genome as predicted by the GCG terminator program. Only terminators, which could be associated with genes are listed here.
| Start | Strand | Sequence |
| 3379 | + | gatatggctgtccgccgctcgcttaaagtggactttttagtttttatcatg |
| 5769 | + | tgctaacaaaatgcgggcctcagtgcctgcatttggctctatctgctgcaa |
| 7072 | + | cactggaaaatagaaaaacagcctgagtggtacgtgaaagctgtcagaaaa |
| 11921 | + | aagatgaaaatatactgttgcttaaataccgttggtttttttatggatggc |
| 14045 | + | ttgtgtacaaaagaaagtaaaacaacagcaacttgttgcaattttatcaat |
| 15382 | + | ttaaatattgaaacgggcgtataacacgcccgttgttttatttatgtggat |
| 622 | - | ctaaagatgtatgtgaaggggccgcgctcgcggccttttttacattccgca |
| 1385 | - | agtcggaaaaatcccggacgataaaataaaagaatttttcactaaaaataa |
| 6057 | - | agcctaatcaatgtttatgaacctgcttcggcaggtttttttatacttgac |
The mosaic nature of the e14 genome is evident from the various similarity searches conducted using this sequence as query (Figure 1). A large section of the e14 genome is very similar to the SfV phage genome. However, unlike the SfV phage, which is a functional temperate bacteriophage and encodes 53 proteins, the e14 element encodes only about 23 proteins. As has been discussed above, this suggests that large deletions must have occurred during the course of evolution of the e14 element. These deletions have removed important genes like the major head coat protein and major tail shaft protein genes, lysis genes and replication genes, and we have made suggestions as to where each of the three major deletions might have occurred. This is not the first case of such deletions in defective prophage elements, for example the K-12 Rac prophage also appears to have suffered at least one large deletion [7]. An interesting observation is the presence of a number of paralogs of StfE protein in different orientations in p15B and Salmonella enterica. p15B is a plasmid in E. coli 15T- which shows 81% homology to bacteriophage P1 [55]. The site-specific DNA inversion system of the plasmid however is very similar to the pin recombinase gene and to part of the invertible DNA of the Pin system as shown in Figure 1. The p15B recombinase system is known to be more complex compared to other recombinase systems including the Cin recombinase system in P1 and is capable of alternately assembling one out of six different ORFs [55,56]. This suggests that such recombinase systems which interchange alternate virion host specificity proteins (tail fibres) could potentially also be of greater complexity, however, there is no experimental evidence for this idea.
Search for prophage elements other genomes
With the recent surge in the availability of genomic DNA sequences it has been found that many microbial genomes harbor prophage elements. These elements can encode key functions including virulence factors, toxins and phage immunity proteins. Thus, detection of such elements in bacterial genome sequences becomes very important. Given the low sequence similarity between parts of the phage elements and their modular nature (nonhomologous alternatives are known for many of the modules [5]), the search for such elements is no simple task. Even if the search is restricted to the tailed temperate phages (there are other kinds of temperate DNA phages [57,58]) or even to the lambdoid phages, none of the phage genes are sufficiently conserved to serve as a single marker for all prophages, and to make it more difficult, in any given case any particular gene could have been deleted from a defective prophage [7,8]. We attempted to search for e14-like modules in the sequenced prokaryotic genomes keeping the mosaic nature of these elements in mind. This approach is different from other approaches in that it does not rely on a single gene like integrase or terminase for phage detection but has the potential to use the entire known pool of temperate tailed phage-encoded genes for detection. The initial search involved looking at the COG [59] database in the eight bacterial genomes listed in Table 5 (these eight include representatives from the rather distantly related Proteobacteria and Firmicutes). COG entries on these genomes for each of the e14 proteins were analyzed. In cases where the COG entries were not available, the COGNITOR program was used to obtain hits. The COG hits were sorted by organism and on the locus of occurrence in the organism. Genes encoding the COG hits for the different e14 proteins, which were within 30,000 base pairs of each other, were then grouped together. Any region with greater than two genes in this cluster was considered to be a putative prophage element and further analyzed. Twenty-six phage-related regions were identified by this analysis of which 23 are already known phage-like areas in the bacterial genomes. Two (labeled P2 and P3 in Table 5) of the remaining three regions are probably non-phage areas. The region identified in Bacillus Subtilis and marked as P1 could be a decaying prophage region as it has at least 4 genes in this area which perform phage related functions apart from the two used in the detection. This includes yneA (which contains a lysin motif), ynzC (a site specific recombinase which shows similarity to integrase of phage phi-FC1, Lactococcus lactis phage TP901-1 and Listeria innocua phage A118), yndG (virulence factor and is a KicB toxin homolog), yndB (related to a prophageXfP2 protein in Xylella fastidiosa XF2524).
Table 5.
Predicted phage like elements using a comparative protein based approach. Phage elements detected in other genomes using orthology to e14 proteins as a criterion. Clustering of orthologous proteins (COG hits) for the e14 proteins in different organisms was examined. Only those organisms with two or more COG hits in the e14 element are listed. Estimates of the boundaries of the phage element are provided. 26 phage related regions could be identified by this analysis out of which 23 are already known phage areas in the bacterial genomes. Two (labeled P2 and P3) of the remaining three regions are probably non-phage areas. *The regions which have not been previously identified as prophage element have been marked as P1, P2 and P3. $Denotes approximate boundaries.
| Organism | Proteins in e14 | Related COG member | locus (approx.) | prophage name |
| E. coli O157:H7 EDL933 | b1149, b1151, b1159 | z1359, z1362, z1356 | 1250302–1295563 | CP-933M |
| b1149, b1151, b1158, b1159 | z1803, z1806, z1817, z1800 | 1626570–1674696 | CP-933N | |
| b1149, b1149, b1151, b1159 | z6045, z6070, z6042, z6047 | 2271618–2331237 | CP-933P | |
| b1145, b1154, b1155, b1157, b1158 | z0309, z0314, z0315, z0317, zpinH | 300060–310646 | CP-933H | |
| b1140, b1141, b1145, b1155, b1157 | z1866, z1867, zumuD, z1920, z1918 | 1701990–1749459 | CP-933X | |
| b1140, b1143, b1155 | zintT, z2978, z2983 | 2668339–2689384 | CP-933T | |
| b1149, b1151 | z1854, z1849 | 1678701–1693737 | CP-933C | |
| b1140, b1145 | zintU, z3126 | 2743223–2788401 | CP-933U | |
| b1140, b1145 | zintO, z2090 | 1849324–1929903 | CP-933O | |
| b1145, b1149, b1151 | z3358, z3332, z3328 | 2966157–3015089 | CP-933V | |
| E. coli K-12 | b1156, b1158 | ybcx, ybck | 564025–585326 | DLP12 |
| b1156, b1157, b1158 | ynac, stfr, pinr | 1409966–1433025 | rac | |
| b1156, b1157, b1158 | ydfm, ydfn, pinq | 1630450–1646830 | qin | |
| b1154, b1156 | yfdk, tfaS | 2464404–2474619 | KpLE1, CPS-53 | |
| B. subtilis 168 | b1149, b1158 | BS_lexA, BS_yneB | $1902658–1919056 | *P1 |
| b1151, b1152 | BS_ykeA, BS_xkdT | 1316849–1347491 | PBSX | |
| b1152, b1158 | BS_yqbT, BS_spoIVCA | $2652219–2700977 | SKIN | |
| M. loti MAFF303099 | b1149, b1151 | Mlr8521, Mlr8522 | $6974260–7021772 | Meso2 |
| b1143, b1149 | Mlr4761, Mlr4759 | $3776480–3781495 | *P2 | |
| C. crescentus | b1150, b1154, b1157 | CC1890, CC1902, CC1904 | $2096699–2098302 | *P3 |
| X. fastidiosa 9a5c | b1140, b1149 | XF1642, XF1645 | 1595657–1629967 | XfP4 |
| b1140, b1152 | XF1555, XF1598 | 1519081–1532748 | XfP3 | |
| N. meningitidis Z2491 (serogroup A) | b1152, b1153, b1157 | NMA1323, NMA1324, NMA1325 | 1207176–1236496 | Pnm2 |
| b1152, b1153 | NMA1826, NMA1825 | 1768530 to 1807766 | Pnm1 | |
| S. pyogenens SF370 (serotype M1) | b1140, b1159 | spy1488, spy1468 | 1189125–1222634 | 370.2 |
| b1157, b1158 | spy0671, spy0655, spy1468 | 529591–570493 | 370.1 | |
Further, this region is also identified as a possible phage element "5" and shows compositional variation compared to the rest of the Bacillus subtilis genome [60]. We could thus identify several lambdoid prophage elements in a representative set of bacterial genomes using such a protein level approach. This approach takes into consideration the modular nature of phage genomes and looks for orthologs of the genes of the defective prophage e14 that exist in proximity of each other. We hasten to mention that a number of putative prophages were not found by this analysis. But this exercise was knowingly severely limited by only taking orthologs of e14 genes into consideration, and a similar approach using the entire pool of known lambdoid phage (or even all temperate phage) genes should make a much more sensitive and robust technique for detecting phage elements, and, importantly, it can be automated.
Methods
Sequence manipulation and analysis was done using the EMBOSS [61] and GCG [62] suite of sequence analysis tools. Perl scripts were used for calculating cumulative GC plots and facilitating other searches. The cumulative GC was calculated as ∑(G-C)/(G+C) using a window size of 500. Domain searches were done using the NCBI CDD server [63], Interpro [64], PFAM [65] and SMART [66] databases were used where ever necessary. Promoter sequences in the e14 genome were detected using the BPROM [67] utility. The predicted promoters were then analyzed for the presence of ORFs in close vicinity. The promoters for which an ORF could not be assigned are not listed in this work. Rho-independent terminators were detected using the terminator program available with the GCG package. The program is an adaptation of the terminator program by Brendel and Trifonov 1984 [68]. Promoters and terminators that could not be explained functionally were ignored though the prediction servers identified several with high scores. Information on operons within the e14 genome was obtained with TIGROperons [54]. The COG database [59] was used to find orthologs of proteins encoded by the e14 element. For the proteins, which are not known to belong to any of the COGs listed, the COGNITOR application was used to identify orthologs.
Authors contribution
PM was responsible for data collection and analysis. SK conceived of the study, and participated in its design and analysis. SC helped to bring this information into the biological context of past and current prophage research.
Acknowledgments
Acknowledgements
PM and SK thank Sri Vidhya for help during manuscript preparation. We acknowledge the use of the Bioinformatics Centre facility funded by DBT, Govt of India, and CSIR for fellowship to PM, DBT Indo-Israel project to SK, and NIH grant AI49003 to SC.
Contributor Information
Preeti Mehta, Email: mehta_p74@yahoo.com.
Sherwood Casjens, Email: sherwood.casjens@path.utah.edu.
Sankaran Krishnaswamy, Email: krishna@mrna.tn.nic.in.
References
- Craig NL, Craigie R, Gellert M, Lambowitz A. Mobile DNA II Amer Society for Microbiology. 2002.
- Banks DJ, Beres SB, Musser JM. The fundamental contribution to GAS evolution, genome diversification and strain emergence. Trends in Microbiol. 2002;10:515–521. doi: 10.1016/S0966-842X(02)02461-7. [DOI] [PubMed] [Google Scholar]
- Boyd EF, Brussow H. Common themes among bacteriophage-encoded virulence factors and diversity among the bacteriophages involved. Trends in Microbiol. 2002;10:521–529. doi: 10.1016/S0966-842X(02)02459-9. [DOI] [PubMed] [Google Scholar]
- Botstein D. A theory of modular evolution for bacteriophages. Ann N Y Acad Sci. 1980;354:484–490. doi: 10.1111/j.1749-6632.1980.tb27987.x. [DOI] [PubMed] [Google Scholar]
- Casjens S, Hatfull G, Hendrix R. Evolution of dsDNA tailed bacteriophage genomes. Semin Virology. 1992;3:383–397. [Google Scholar]
- Hendrix RW, Smith MC, Burns RN, Ford ME, Hatfull GF. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc Natl Acad Sci USA. 1999;96:2192–2197. doi: 10.1073/pnas.96.5.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens S. Prophages and bacterial genomics: what have we learned so far? Mol Microbiol. 2003;49:277–300. doi: 10.1046/j.1365-2958.2003.03580.x. [DOI] [PubMed] [Google Scholar]
- Canchaya C, Proux C, Fournous G, Bruttin A, Brussow H. Prophage Genomics. Microbiol Mol Biol Rev. 2003;67:238–276. doi: 10.1128/MMBR.67.2.238-276.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd KE. Novel intergenic repeats of Escherichia coli K-12. Res Microbiol. 1999;150:653–664. doi: 10.1016/S0923-2508(99)00126-6. [DOI] [PubMed] [Google Scholar]
- Perna NT, Plunkett G, 3rd, Burland V, Mau B, Glasner JD, Rose DJ, Mayhew GF, Evans PS, Gregor J, Kirkpatrick HA, Posfai G, Hackett J, Klink S, Boutin A, Shao Y, Miller L, Grotbeck EJ, Davis NW, Lim A, Dimalanta ET, Potamousis KD, Apodaca J, Anantharaman TS, Lin J, Yen G, Schwartz DC, Welch RA, Blattner FR. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001;409:529–533. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- Brody H, Hill CW. Attachment site of the genetic element e14. J Bacteriol. 1988;170:2040–2044. doi: 10.1128/jb.170.5.2040-2044.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill CW, Gray JA, Brody H. Use of the Isocitrate Dehydrogenase Structural Gene for attachment of e14 in Escherichia coli K-12. J Bacteriol. 1989;171:4083–4084. doi: 10.1128/jb.171.7.4083-4084.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner FR, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- Wang H, Yang C, Lee G, Chang F, Wilson H, Campillo-Campbell A, Campbell A. Integration specificities of two lambdoid phages (21 and e14) that insert at the same attB site. J Bacteriol. 1997;179:5705–5711. doi: 10.1128/jb.179.18.5705-5711.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody H, Greener A, Hill CW. Excision and reintegration of the Escherichia coli K-12 chromosomal element e14. J Bacteriol. 1985;161:1112–1117. doi: 10.1128/jb.161.3.1112-1117.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greener A, Hill CW. Identification of a novel genetic element in Escherichia coli K-12. J Bacteriol. 1980;144:312–321. doi: 10.1128/jb.144.1.312-321.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Putte P, Plasterk RHA, Kuypers A. A Mu gin complementing function and an invertible DNA region in Escherichia coli K-12 are situated on the genetic element e14. J Bacteriol. 1984;158:517–522. doi: 10.1128/jb.158.2.517-522.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasterk RHA, Van de Putte P. The invertible P-DNA segment in the chromosome of Escherichia coli. EMBO J. 1985;4:237–242. doi: 10.1002/j.1460-2075.1985.tb02341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguin E, Brody H, Hill CW, D'Ari R. SOS-associated division inhibition gene sfiC is part of excisable element e14 in Escherichia coli. J Bacteriol. 1986;168:464–466. doi: 10.1128/jb.168.1.464-466.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecogene http://bmb.med.miami.edu/EcoGene/EcoWeb/index.html
- Genobase http://ecoli.aist-nara.ac.jp
- Swissprot http://www.expasy.ch/sprot
- Kao C, Snyder L. The lit gene product which blocks bacteriophage T4 late gene expression is a membrane protein encoded by a cryptic DNA element, e14. J Bacteriol. 1988;170:2056–2062. doi: 10.1128/jb.170.5.2056-2062.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao C, Gumbs E, Snyder L. Cloning and characterization of the Escherichia coli lit gene, which blocks bacteriophage T4 late gene expression. J Bacteriol. 1987;169:1232–1238. doi: 10.1128/jb.169.3.1232-1238.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravi RS, Sozhamannan S, Dharmalingam K. Transposon mutagenesis and genetic mapping of the rglA and rglB loci of Escherichia coli. Mol Gen Genet. 1985;198:390–392. doi: 10.1007/BF00332928. [DOI] [PubMed] [Google Scholar]
- Kelleher JE, Raleigh EA. Response to UV damage by four Escherichia coli K-12 Restriction Systems. J Bacteriol. 1994;176:5888–5896. doi: 10.1128/jb.176.19.5888-5896.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto M, Oosawa K, Momota H. Mapping of the pin locus coding for a site-specific recombinase that causes flagellar-phase variation in Escherichia coli K-12. J Bacteriol. 1983;156:663–668. doi: 10.1128/jb.156.2.663-668.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguin E, Lutkenhaus J, D'Ari R. Reversibility of SOS-associated division inhibition in Escherichia coli. J Bacteriol. 1986;166:733–738. doi: 10.1128/jb.166.3.733-738.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MegaBLAST http://www.ncbi.nlm.nih.gov/BLAST
- Ptashne MA. Genetic Switch: Phage Lambda and Higher Organisms. Cell Press and Blackwell Scientific Publications, Cambridge MA; 1986. [Google Scholar]
- Allison GE, Angeles D, Tran-Dinh N, Verma NK. Complete genomic sequence of SfV, a serotype-converting temperate bacteriophage of Shigella flexneri. J Bacteriol. 2002;184:1974–1987. doi: 10.1128/JB.184.7.1974-1987.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedulla ML, Ford ME, Karthikeyan T, Houtz JM, Hendrix RW, Hatfull GF, Poteete AR, Gilcrease EB, Winn-Stapley DA, Casjens SR. Corrected sequence of the bacteriophage P22 genome. J Bacteriol. 2003;185:1475–1477. doi: 10.1128/JB.185.4.1475-1477.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mmolawa PT, Schmieger H, Heuzenroeder MW. Bacteriophage ST64B, a genetic mosaic of genes from diverse sources isolated from Salmonella enterica serovar typhimurium DT 64. J Bacteriol. 2003;185:6481–6485. doi: 10.1128/JB.185.21.6481-6485.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borodovsky M, McIninch J. GeneMark: parallel gene recognition for both DNA strands. Computers & Chemistry. 1993;17:123–133. doi: 10.1016/0097-8485(93)85004-V. [DOI] [Google Scholar]
- Courcelle J, Khodursky A, Peter B, Brown PO, Hanawalt PC. Comparative gene expression profiles following UV exposure in wild-type and SOS deficient Escherichia coli. Genetics. 2001;158:41–64. doi: 10.1093/genetics/158.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoriev A. Analyzing genomes with cumulative skew diagrams. Nucl Acids Res. 1998;26:2286–2290. doi: 10.1093/nar/26.10.2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S, Nuttall S, Ngui K, Fisher C, Lopez P, Dyall-Smith M. HF2: a double stranded DNA tailed haloarcheal virus with a mosaic genome. Mol Microbiol. 2002;44:283–296. doi: 10.1046/j.1365-2958.2002.02890.x. [DOI] [PubMed] [Google Scholar]
- Bergsland KJ, Kao C, Yu YN, Gulati R, Snyder L. A site in the T4 bacteriophage major head protein gene that can promote the inhibition of all translation in Escherichia coli. J Mol Biol. 1990;213:477–494. doi: 10.1016/S0022-2836(05)80209-8. [DOI] [PubMed] [Google Scholar]
- Bingham R, Ekunwe SI, Falk S, Snyder L, Kleanthous C. The Major Head Protein of Bacteriophage T4 Binds Specifically to Elongation Factor Tu. J Biol Chem. 2000;275:23219–23226. doi: 10.1074/jbc.M002546200. [DOI] [PubMed] [Google Scholar]
- Welch RA, Burland V, Plunkett G, 3rd, Redford P, Roesch P, Rasko D, Buckles EL, Liou SR, Boutin A, Hackett J, Stroud D, Mayhew GF, Rose DJ, Zhou S, Schwartz DC, Perna NT, Mobley HL, Donnenberg MS, Blattner FR. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc Natl Acad Sci U S A. 2002;99:17020–17024. doi: 10.1073/pnas.252529799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson AJ, Reinach FC, Arruda P, Abreu FA, Acencio M, Alvarenga R, Alves LM, Araya JE, Baia GS, Baptista CS, et al. The genome sequence of the plant pathogen Xylella fastidiosa The Xylella fastidiosa Consortium of the Organization for Nucleotide Sequencing and Analysis. Nature. 2000;406:151–157. doi: 10.1038/35018003. [DOI] [PubMed] [Google Scholar]
- Parkhill J, Dougan G, James KD, Thomson NR, Pickard D, Wain J, Churcher C, Mungall KL, Bentley SD, Holden MT, et al. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature. 2001;413:848–852. doi: 10.1038/35101607. [DOI] [PubMed] [Google Scholar]
- Greer H. The kil gene of bacteriophage lambda. Virology. 1975;66:589–604. doi: 10.1016/0042-6822(75)90231-7. [DOI] [PubMed] [Google Scholar]
- Sergueev K, Yu D, Austin S, Court D. Cell toxicity caused by products of the p(L) operon of bacteriophage lambda. Gene. 2001;272:227–235. doi: 10.1016/S0378-1119(01)00535-2. [DOI] [PubMed] [Google Scholar]
- Conter A, Bouche JP, Dassain M. Identification of a new inhibitor of essential division gene ftsZ as the kil gene of defective prophage Rac. J Bacteriol. 1996;178:5100–5104. doi: 10.1128/jb.178.17.5100-5104.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisinger GR, Rietsch A, Lubitz W, Blasi U. Lambda kil-mediated lysis requires the phage context. Virology. 1993;193:1033–1036. doi: 10.1006/viro.1993.1222. [DOI] [PubMed] [Google Scholar]
- Semerjian AV, Malloy DC, Poteete AR. Genetic structure of the bacteriophage P22 PL operon. J Mol Biol. 1989;207:1–13. doi: 10.1016/0022-2836(89)90437-3. [DOI] [PubMed] [Google Scholar]
- Recktenwald J, Schmidt H. The nucleotide sequence of Shiga toxin (Stx) 2e-encoding phage ΦP27 is not related to other Stx phage genomes, but the modular genetic structure is conserved. Infect Immun. 2002;70:1896–1908. doi: 10.1128/IAI.70.4.1896-1908.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mmolawa PT, Schmieger H, Tucker CP, Heuzenroeder MW. Genomic structure of the Salmonella enterica serovar Typhimurium DT 64 bacteriophage ST64T: evidence for modular genetic architecture. J Bacteriol. 2003;185:3473–3475. doi: 10.1128/JB.185.11.3473-3475.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchaud EC, Rusniok L, Frangeul C, Buchrieser A, Givaudan S, Taourit S, Bocs C, Boursaux-Eude M, Chandler JF, Charles , et al. The genome sequence of the entomopathogenic bacterium Photorhabdus luminescens. Nat Biotechnol. 2003;21:1307–1313. doi: 10.1038/nbt886. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Makino K, Ohnishi M, Kurokawa K, Ishii K, Yokoyama K, Han CG, Ohtsubo E, Nakayama K, Murata T. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 2001;8:11–22. doi: 10.1093/dnares/8.1.11. [DOI] [PubMed] [Google Scholar]
- McClelland M, Sanderson JK, Spieth J, Clifton SW, Latreille P, Courtney L, Porwollik S, Ali J, Dante M, Du F, et al. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature. 2001;413:852–856. doi: 10.1038/35101614. [DOI] [PubMed] [Google Scholar]
- Gorbalenya AE. Self-splicing group I and group II introns encode homologous (putative) DNA endonucleases of a new family. Protein Sci. 1994;3:1117–1120. doi: 10.1002/pro.5560030716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TIGROperons http://www.tigr.org
- Sandmeier H, Iida S, Arber W. DNA inversion regions Min of plasmid p15B and Cin of bacteriophage P1: evolution of bacteriophage tail fibre genes. J Bacteriol. 1992;174:3936–3944. doi: 10.1128/jb.174.12.3936-3944.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmeier H, Iida S, Huber P, Hiestand-Nauer R, Arber W. Gene organization in the multiple DNA inversion region min of plasmid p15B of E. coli 15T-: assemblage of a variable gene. Nucl Acids Res. 1991;19:5831–5838. doi: 10.1093/nar/19.21.5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BM, Waldor MK. Filamentous phages linked to virulence of Vibrio cholerae. Curr Opin Microbiol. 2003;6:35–42. doi: 10.1016/S1369-5274(02)00005-X. [DOI] [PubMed] [Google Scholar]
- Stromsten NJ, Benson SD, Burnett RM, Bamford DH, Bamford JK. The Bacillus thuringiensis linear double-stranded DNA phage Bam35, which is highly similar to the Bacillus cereus linear plasmid pBClin15, has a prophage state. J Bacteriol. 2003;185:6985–6989. doi: 10.1128/JB.185.23.6985-6989.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Natale DA, Garkavtsev IV, Tatusova TA, Shankavaram UT, Rao BS, Kiryutin B, Galperin MY, Fedorova ND, Koonin EV. The COG database: new developments in phylogenetic classification of proteins from complete genome. Nucl Acids Res. 2001;29:22–28. doi: 10.1093/nar/29.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst F, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. "EMBOSS: The European Molecular Biology Open Software Suite". Trends in Genet. 2000;16:276–277. doi: 10.1016/S0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucl Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Panchenko AR, Shoemaker BA, Thiessen PA, Geer LY, Bryant SH. CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucl Acids Res. 2002;30:281–283. doi: 10.1093/nar/30.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder NJ, Apweiler , Attwood TK, Bairoch A, Barrell D, Bateman A, Binns D, Biswas M, Bradley P, Bork P, et al. The InterPro Database, 2003 brings increased coverage and new features. Nucl Acids Res. 2003;31:315–318. doi: 10.1093/nar/gkg046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, Eddy SR, Griffiths-Jones S, Howe KL, Marshall M, Sonnhammer EL. The Pfam Protein Families Database. Nucl Acids Res. 2002;30:276–280. doi: 10.1093/nar/30.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci U S A. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BPROM http://www.softberry.com/berry.phtml?topic=gfindb
- Brendel V, Trifonov EN. A computer algorithm for testing potential prokaryotic terminators. Nucl Acids Res. 1984;12:4411–4427. doi: 10.1093/nar/12.10.4411. [DOI] [PMC free article] [PubMed] [Google Scholar]


