Abstract
Holocarboxylase synthetase (HCS) catalyzes the binding of biotin to lysine (K) residues in histones H3 and H4. Histone biotinylation marks are enriched in repressed loci, including retrotransposons. Preliminary studies suggested that K16 in histone H4 is a target for biotinylation by HCS. Here we tested the hypotheses that H4K16bio is a real histone mark in human chromatin, and that H4K16bio is overrepresented in repressed gene loci and repeat regions. Polyclonal rabbit anti-human H4K16bio was generated and affinity purified. An extensive series of testing with synthetic and natural targets confirmed that this new antibody is specific for H4K16bio. Using anti-H4K16bio and chromatin immunoprecipitation assays, we demonstrated that H4K16bio is overrepresented in repeat regions [pericentromeric alpha satellite repeats and long terminal repeats (LTR)] compared with euchromatin promoters. H4K16bio was also enriched in the repressed interleukin-2 gene promoter in human lymphoid cells; transcriptional activation of the interleukin-2 gene by mitogens and phorbol esters coincided with a depletion of the H4K16bio mark at the gene promoter. The enrichment of H4K16bio depended on biotin supply; the enrichment at LTR22 and promoter 1 of the sodium-dependent multivitamin transporter (SMVT) was greater in biotin-supplemented cells compared with biotin-normal and biotin-deficient cells. The enrichment of H4K16bio at LTR15 and SMVT promoter 1 was significantly lower in fibroblasts from an HCS-deficient patient compared with an HCS wild-type control. We conclude that H4K16bio is a real phenomenon and that this mark, like other biotinylation marks, is overrepresented in repressed loci where it marks HCS docking sites.
Keywords: biotin, histone, holocarboxylase synthetase, human
1. Introduction
Histones mediate the folding of DNA into chromatin [1]. Amino acid residues in the N-terminal tails and, to a lesser extent, other domains of histones are targets for covalent modifications such as acetylation and methylation [2]. These posttranscriptional modifications play important roles in gene regulation, chromatin remodeling, mitosis, and DNA repair. For example, lysine (K)-9 acetylated histone H3 (H3K9ac) and K4-trimethylated histone H3 (H3K4me3) are overrepresented in transcriptionally active euchromatin, whereas hypoacetylated histones and K9-dimethylated histone H3 (H3K9me2) are overrepresented in heterochromatin and repressed genes.
Wolf and co-workers provided evidence that histones are also modified by covalent attachment of the vitamin biotin, based on in vitro studies with purified histones and the histone biotinyl ligase, biotinidase [3]. Subsequently, biotinylated histones were detected in chromatin from human and other metazoans by using radiotracers, streptavidin, anti-biotin, biotinylation site-specific antibodies, and mass spectrometry [4–10]. The majority of biotinylation takes place at K4, K9, K18, and perhaps K23 in histone H3 [11, 12], and K8 and K12 in histone H4 [6], but a weak biotinylation signal can also be detected in histone H2A [4, 13]. These previous studies also suggest that holocarboxylase synthetase (HCS) is more important than biotinidase for catalyzing biotinylation of histones in vivo [7, 12, 14]. Phenotypes of HCS knockdown include a short life span and low heat survival in Drosophila melanogaster [7] and aberrant gene regulation in humans [15, 16].
Biotinylation of histones is a fairly rare event, i.e., less than 0.1% of histones in bulk extracts are biotinylated in samples of human origin [10, 17]. However, the comparably low level of global histone biotinylation is not representative of the level of biotinylation in confined regions of chromatin. For example, about one of three histone H4 molecules in telomeric repeats is biotinylated at K12 [18]. Biotinylation of histones has important functions in gene repression and genome stability. Previous studies suggest that K9- and K18-biotinylated histone H3 (H3K9bio, H3K18bio) and K8- and K12-biotinylated histone H4 (H4K8bio, H4K12bio) are enriched in repeat regions and participate in gene repression [15, 16, 18–20]. The abundance of histone biotinylation marks depends on both biotin supply and HCS activity [15, 16, 20].
Preliminary studies provide evidence that histone H4 is also biotinylated at K16 (H4K16bio) [13]. Here, we tested the hypotheses that H4K16 is a true biotinylation mark in human chromatin and that H4K16bio, like other biotinylation signatures, is overrepresented in repeat regions and participates in the repression of transcriptionally competent chromatin.
2. Materials and Methods
2.1. Cell culture
Jurkat human lymphoma cells (ATCC, Manassas, VA) were cultured in commercial RPMI-1640 (Thermo Scientific, Waltham, MA) using standard cell culture techniques [21]. Regular commercial RPMI-1640 contains 820 nmol/l biotin, which is >3,000 times the biotin concentration in human plasma [22]. In some experiments, cells were cultured in biotin-deficient (0.025 nmol/l biotin), biotin-physiological (0.25 nmol/l biotin), and biotin- pharmacological (10 nmol/l biotin) media for 12 d prior to analysis [21]. These concentrations represent levels observed in plasma from biotin-deficient individuals, biotin-normal individuals, and individuals taking over-the-counter biotin supplements [22, 23]. Biotin-defined media were prepared using customized RPMI-1640 and biotin-depleted fetal bovine serum [21]. Efficacy of treatment was confirmed by probing biotinylated carboxylases with IRDye-streptavidin [20]. Where indicated, expression of interleukin-2 (IL-2) was stimulated by treating Jurkat cells with phorbol-12-myristate-13-acetate (PMA) and phytohemaglutinin (PHA) [21] for 3 h.
Human WG2215 HCS-deficient skin fibroblast (Montreal Children’s Hospital Cell Repository, Montreal, Canada) were cultured in Dulbecco’s modified Eagle’s medium containing 10 nmol/l biotin to investigate effects of HCS deficiency on histone biotinylation [20]. Human HCS wild-type IMR-90 fibroblasts (ATCC, Manassas, VA) were used as controls. HCS activity in IMR-90 and WG2215 fibroblasts was assessed by using biotinylated carboxylases as markers in streptavidin blots [20].
2.2. Antibodies
Rabbit anti-human H4K16bio was generated as described [6]. Briefly, an H4-based, K16-biotinylated synthetic peptide (GGAK(bio)RHRKVLRD, Anaspec; San Jose,CA) was conjugated to keyhole limpet hemocyanin (KLH; Pierce, IL) and used as antigen for injection into New Zealand rabbits. Specificity tests revealed that the anti-serum cross-reacted with K16-acetylated histone H4, using the synthetic peptide GGAK(ac)RHRKVLRD (H4K16ac) as a target (data not shown). To eliminate cross-reactivity with H4K16ac, the antiserum was affinity purified by using immobilized peptide H4K16ac and HiTrap NHS-Activated Sepharose High Performance Column (cat.# 17-0717-01, GE Healthcare; Piscataway, NJ) following the manufacturer’s protocol. Identity and purity of all synthetic peptides were confirmed by mass spectrometry and HPLC, respectively, by the manufacturer.
Biotinylation site specificity of anti-H4K16bio was confirmed by using synthetic histone-based peptides, histone bulk extracts, biotin-depleted histones, and competition studies as described before [6]. Equal loading and transfer was confirmed by staining membranes with Ponceau S [20].
Polyclonal rabbit anti-human antibodies to H4K16bio, H4K12bio and H3K9me2 were generated in a commercial facility (Cocalico Biologicals, Reamstown PA) as described [6, 11, 20], while antibodies to the C-terminus in histone H3 (ab1791) and H3K9ac (ab10812) were purchased from Abcam (Cambridge, MA). H3K9me2 and H4K12bio were used as gene repression and heterochromatin marks, whereas H3K9ac was used as a euchromatin mark. Rabbit polyclonal anti-human pyruvate carboxylase (PC) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA) and PC in transblots was probed as described before [20].
2.3. Micro chromatin immunoprecipitation (μChIP) assay
The enrichment of H4K16bio in distinct loci in human chromatin was assessed by micro chromatin immunoprecipitation (μChIP) assay as described [20, 24] with the following modifications. Chromatin form Jurkat cells was precipitated using 14μl of anti-H4K12bio and anti-H3K9me2, and 28 μl of anti-H4K16bio sera.
2.4. Quantitative real-time PCR (qRT-PCR)
The abundance of DNA in immunoprecipitated chromatin was quantified by qRT-PCR using Perfecta SYBR Green FastMix ROX (VWR; West Chester, PA) and PCR primers for long terminal repeats (LTR) 15 and 22, pericentromeric alpha satellite repeats in chromosome 4, and promoters in genes coding for IL-2, sodium-dependent multivitamin transporter (SMVT), aldehyde dehydrogenase (ADH) 5, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) [20]. Amplicons were quantified using the cycle threshold values method [25]. GAPDH was used to normalize qRT-PCR data for amplification efficiency. The relative enrichment of histone marks in precipitated chromatin was calculated as percent of input DNA [26, 27]. Enrichment data were normalized for nucleosomal occupancy by precipitating samples with an antibody to the (non-modified) C-terminus in histone H3. Non-specific rabbit IgG (Santa Cruz, Santa Cruz, CA) was used as a control for background noise and precipitated negligible amounts of chromatin (<5% of compared with anti-H4K16bio), consistent with previous observations [20].
The abundance of mRNA coding for IL-2, LTR22, and SMVT was quantified using Absolute QPCR SYBR Green fluorescein mix (ABgene; Rochester, NY) as described[6, 20].
2.5. Statistical analysis
Bartlett’s test was used to test for homogeneity of variances [28]. Data were log transformed if variances were heterogeneous. Significance of differences among more than two groups was tested by one-way ANOVA and Fisher’s Protected Least Significant Difference procedure for post hoc testing. Student’s paired t-test was used for pairwise comparisons of data from Jurkat cells before and after stimulation with PMA and PHA. μChIP data from WG2215 and IMR-90 fibroblasts were not normally distributed and were analyzed by using the Mann-Whitney U test. StatView 5.0.1 (SAS Institute, Cary NC, USA) was used to perform all calculations. Differences were considered statistically significant if P < 0.05. Data are expressed as mean ± S.D.
3. Results
3.1. Specificity of anti-H4K16bio
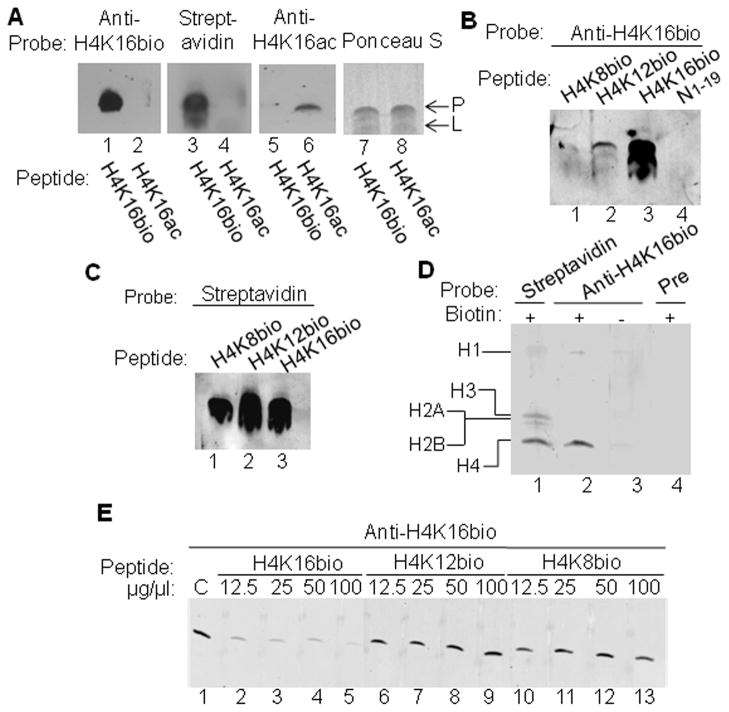
Affinity-purified anti-H4K16bio is specific for its designated target, based on the following lines of observations. In a first series of experiments we compared the affinity of anti-H4K16bio for the synthetic peptides H4K16bio and H4K16ac (Fig. 1A). Purified anti-H4K16bio produced a strong signal with peptide H4K16bio (lane 1), but not signal with peptide H4K16ac (lane 2). The presence of biotinylation and acetylation marks was confirmed by probing transblots with IRDye-streptavidin (lanes 3 and 4) and anti-H4K16ac (lanes 5 and 6). Equal loading and transfer was confirmed by staining transblots with Ponceau S (lanes 7and 8). Note the non-specific signal in the Ponceau S-stained membrane, caused by interference of the dye in the sample loading buffer (denoted “L”).
Fig. 1.
Validation of anti-H4K16bio. (A) K16-biotinylated and –acetylated peptides were probed with anti-H4K16bio, IRDye-streptavidin, anti-H4K16ac, and Ponceau S. (B) Synthetic H4-based peptides biotinylated at K8 (N6-15bioK8), K12 (N6-15bioK12), and K16 (N13-24bioK16), and non-biotinylated peptide N1-19 were probed with anti-H4K16bio. (C) Peptides H4K8bio, H4K12bio, and H4K16bio from panel B were probed with IRDye-streptavidin. (D) Bulk extracts of Jurkat cell histones were probed with IRDye-streptavidin, anti-H4K16bio and pre-immune serum; “-“ denotes biotin-depleted histone samples. (E) Bulk extracts of Jurkat cell histones were probed with anti-H4K16bio in the presence of increasing amounts of synthetic peptides H4K16bio, H4K12bio, and H4K8bio. The control sample (“C”) was assayed in the absence of peptide competitors. Some gels were electronically re-arranged to facilitate comparisons. Abbreviations: L, loading buffer; P, peptide.
In a second series of experiments, we assessed the biotinylation site specificity of anti-H4K16bio by comparing its binding to synthetic peptides biotinylated at K8 (N6-15bioK8), K12 (N6-15bioK12), or K16 (N13-24bioK16), where subscripts denoted amino acid residues in histone H4. The non-biotinylated peptide N1-19 was used as negative control. Anti-H4K16bio produced a strong signal with peptide H4K16bio, whereas the signals with peptides H4K8bio and H4K12bio were hardly detectable (Fig. 1B, lanes 1-3). No signal was detectable with non-biotinylated peptide (lane 4). Biotinylation of synthetic peptides was confirmed by probing transblots with IRDye-streptavidin (Fig. 1C, lanes 1–3). Loading of the non-biotinylated peptide N1-19 was adjusted gravimetrically.
In a third series of experiments, we identified H4K16bio in histone bulk extracts from Jurkat cells. First, we probed histones with streptavidin to demonstrate that the majority of biotinylation marks resides in histones H3 and H4, and that biotinylation of histones H1, H2A, and H2B is quanitatively minor (Fig. 1D, lane 1), consistent with our previous results [4]. Equal loading and integrity of proteins was confirmed by staining with Commassie blue (not shown). When histones were probed with anti-H4K16bio the antibody produced a signal only with histone H4, but not with other classes of histones (lane 2). This signal was specifically caused by the biotinylated fraction of histone H4, because biotin-depleted histones did not produce a signal (lane 3). When transblots were probed with pre-immune serum, no signal was detectable (lane 4).
In a fourth and final series of antibody testing, bulk extracts of Jurkat cell histones were probed with anti-H4K16bio in the presence of increasing amounts of synthetic peptides. As expected, peptide H4K16bio outcompeted histone H4 for binding by anti-H4K16bio compared with peptide-free control (Fig. 1E, lanes 1-5). In contrast, peptides H4K12bio (lanes 6-9) and H4K8bio (lanes 10-13) did not compete for binding.
3.2. Relative enrichment of H4K16bio at selected genomic loci
H4K16bio is overrepresented in pericentromeric alpha satellite repeats (heterochromatin) in chromosome 4 compared the promoters of the two euchromatin genes GAPDH and ADH5 in Jurkat cells (Fig. 2A). The enrichment of H4K16bio in the promoter of GAPDH gene was similar to that of ADH5. The known repression marks H4K12bio and H3K9me2 [19, 20] were used as positive controls and showed a pattern similar to that of H4K16bio (Fig. 2B and C). In contrast, the known activation mark H3K9ac (negative control) was greatly enriched in euchromatin promoters compared with pericentromeric alpha satellite repeats (Fig. 2D).
Fig. 2.
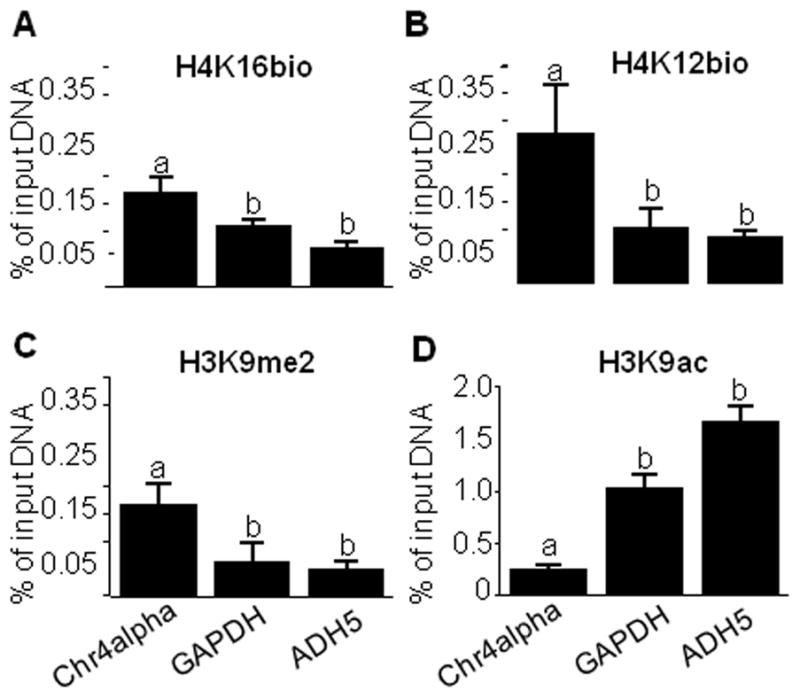
H4K16bio is overrepresented in pericentromeric alpha satellite repeats (heterochromatin) compared with promoters in euchromatin. Chromatin was immunoprecipitated using antibodies to H4K16bio, H4K12bio, H3K9me2, and H3K9ac, and qRT-PCR was used to quantify the relative enrichment of alpha satellite repeats in chromosome 4 (Chr4alpha), and promoters of the GAPDH and ADH5 genes. Bars without a common letter are significantly different (n = 4, P< 0.05).
H4K16bio was overrepresented in the transcriptionally repressed promoter in the IL-2 gene prior to stimulation with PMA and PHA compared with 3 h after activation with PMA and PHA (Fig. 3A). qRT-PCR was used to confirm efficacy of de-repression of the IL-2 gene by treatment with PMA and PHA (arbitrary units): 1.0 ± 0.2 before stimulation vs. 28 ± 2.7 after stimulation (n=4; P< 0.05). The controls showed the expected pattern. While the repression marks H4K12bio and H3K9me2 were removed from the IL-2 gene promoter in response to stimulation with PMA and PHA (Fig. 3B and C), the relative enrichment of the activation mark H3K9ac was greater after than before gene activation (Fig. 3D).
Fig. 3.
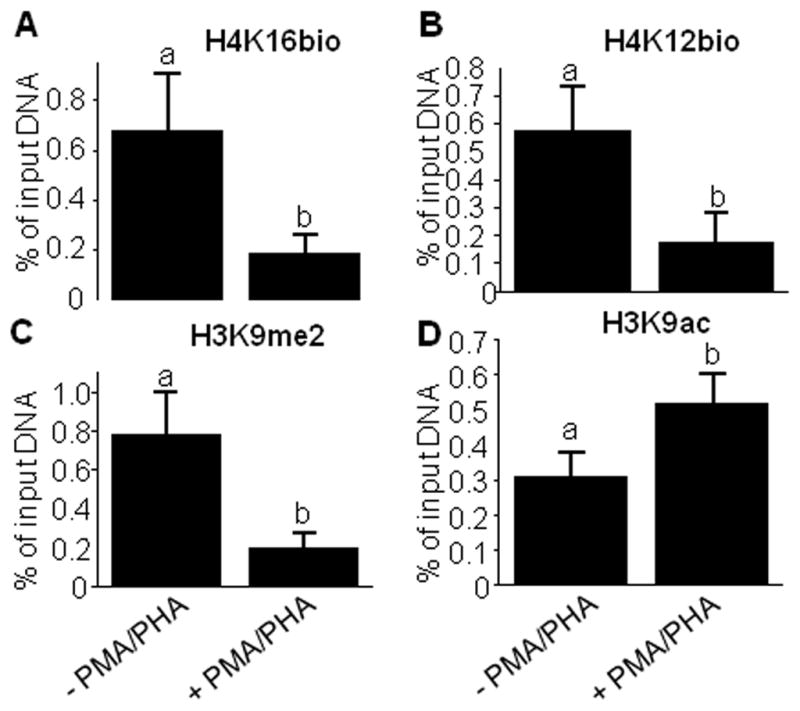
Differential enrichment of histone marks in the IL-2 gene promoter in Jurkat cells. were stimulated with PMA/PHA for 3h. Chromatin was immunoprecipitated with antibodies to H4K16bio, H4K12bio, H3K9me2, and H3K9ac before and after stimulation with phorbol-12-myristate-13-acetate (PMA) and phytohemagglutinin (PHA). qRT-PCR was used to quantify the relative enrichment of IL-2 promoter sequences. Bars without a common letter are significantly different (n = 5, P < 0.05).
3.3. Biotin dependence
Previous studies suggest that the abundance of H3K9bio, H3K18bio, H4K8bio, and H4K12bio at LTR15, LTR22, and SMVT promoter 1 depends on the concentration of biotin in culture media and dietary biotin intake in various model organisms including healthy adults [15, 16, 20]. Here, we observed a similar scenario for H4K16bio. The H4K16bio mark was 13-fold and 4-fold more abundant in the LTR22 locus in cells cultured in medium containing 10 nmol/l compared with cells cultured in 0.025 nmol/l, and 0.25 nmol/l biotin, respectively (Fig. 4A). Similar patterns were observed for the repression marks H4K12bio (Fig. 4B), and H3K9me2 (Fig.4C), while the enrichment of H3K9ac was not affected by biotin (Fig. 4D). When the abundance of H4K16bio was low at LTRs, the transcriptional activity increased compared with cells where H4K16bio was high. The abundance of LTR transcripts originating in the U5 region [15] was 2.9 ± 0.6 in cells cultured in medium containing 0.025 nmol/l, 0.9 ± 0.06 in cells cultured in medium containing 0.25 nmol/l, and 0.2 ± 0.02 in cells cultured in medium containing 10 nmol/l (arbitrary units, n=4, P < 0.05 among all treatment groups),
Fig. 4.
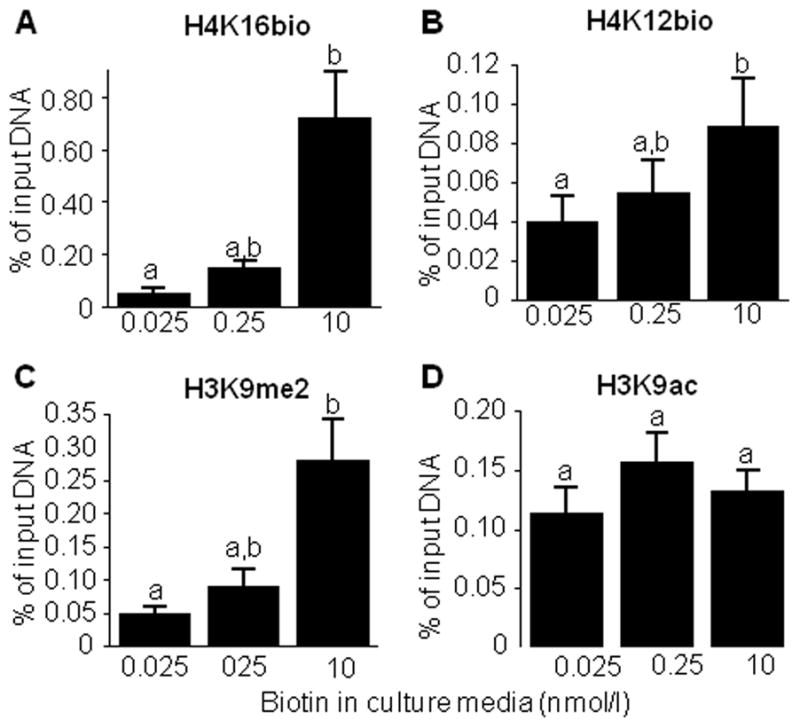
The abundance of H4K16bio at the LTR22 locus depends on the concentration of biotin in culture media. Cells were cultured in biotin-defined media for 12 d, and chromatin was precipitated with antibodies to H4K16bio, H4K12bio, H3K9me2, and H3K9ac; qRT-PCR was used to quantify the relative enrichment of LTR22 sequences. Bars without a common letter are significantly different (n = 4–5, P < 0.05).
The enrichment of H4K16bio, H4K12bio, and H3K9me2 at the SMVT promoter 1locus was comparable to that at LTR22, e.g., the H4K16bio mark was 1.7-fold and 1.4-fold more abundant in cells cultured in medium containing 10 nmol/l compared with cells cultured in 0.025 nmol/l, and 0.25 nmol/l biotin, respectively. However, the differences among the different biotin concentrations did not reach statistical significance for the SMVT promoter (P = 0.21).
The efficacy of biotin treatment was confirmed by probing biotinylated holocarboxylases with IRDye-streptavidin (Fig. 5, upper gel). Clearly, the abundance of holocarboxylases depended on the concentration of biotin in culture media. Equal loading of lanes was confirmed by using anti-PC as a probe for total PC (holo & apo; Fig. 5, lower gel). The levels of biotinylated acetyl-CoA carboxylases 1 and 2 are below the limit of detection on Jurkat cells, consistent with previous observations [21].
Fig. 5.
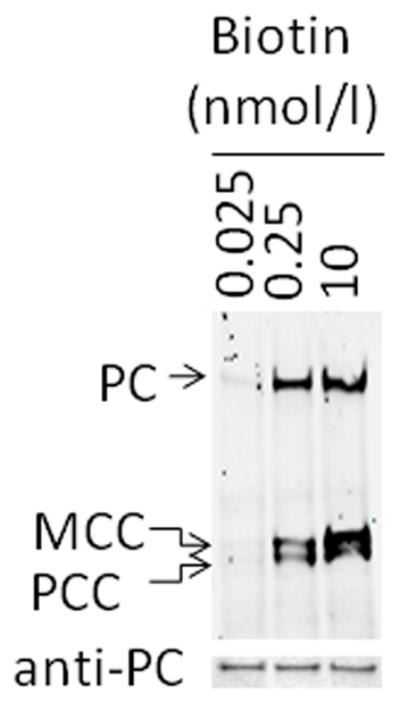
Biotinylation of carboxylases depends on the concentration of biotin in culture media. Jurkat cells were culture in biotin-defined media for 12 d, and caroxylase-bound biotin was probed with IRDye-streptavidin in pyruvate carboxylase (PC), 3-methylcrotonyl-CoA carboxylase (MCC), and propionyl-CoA carboxylase (PCC) (upper gel). Equal loading of lanes was confirmed by using anti-PC as a probe (lower gel).
3.4. HCS deficiency
Consistent with a previous report [20], the enrichment of H4K16bio and the repression marks H4K12bio and H3K9me2 decreased at the LTR22 locus in HCS-deficient fibroblasts (WG2215) compared with HCS-normal fibroblasts (IMR-90) (Fig. 6. A-C). The same pattern was obtained if H4K16bio was studied at SMVT promoter 1 and LTR15 loci (data not shown). HCS activity in both types of fibroblasts was confirmed by probing transblots of biotinylated carboxylases with IRDye-streptavidin (Fig. 6D, upper gel). Biotinylated carboxylases were easily detectable in IMR-90 fibroblasts, whereas no signal was detectable in WG2215 fibroblasts. Anti-PC was used to confirm that the decrease of biotinylation signal in WG2215 cells was due to the inability of mutant HCS to biotinylate carboxylases rather than low apocarboxylases expression or differences in the loading or integrity of the proteins (Fig. 6D, lower gel).
Fig. 6.
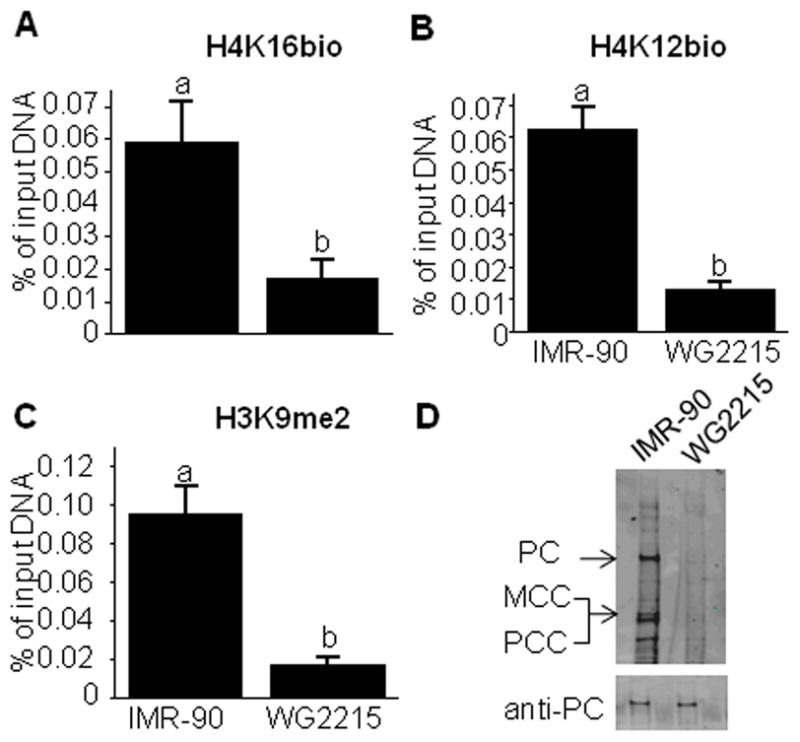
The enrichment of H4K16bio at LTR22 depends on HCS in primary fibroblasts. (A–C) HCS-normal fibroblasts (IMR-90) and HCS-deficient fibroblasts (WG2215) were cultured in medium containing 10 nmol/l biotin. Chromatin was precipitated with antibodies to H4K16bio, H4K12bio, and H3K9me2, and qRT-PCR was used to quantify the relative enrichment of LTR22. Bars without a common letter are significantly different (n = 3–4, P < 0.05). (D) Biotin in PC, MCC, and PCC was probed with IRDye-streptavidin (upper gel); equal loading was confirmed by using anti-PC as a probe (lower gel).
4. Discussion
Here, we demonstrate that K16 in human histone is a target for covalent biotinylation, and we provide first insights into possible biological functions of H4K16bio in pericentromeric heterochromatin and gene repression. A novel antibody was generated and its specificity for H4K16bio was demonstrated in an extensive series of testing. Importantly, we report that the abundance of the H4K16bio mark depends on both biotin supply and HCS activity.
H4K16bio seems to be a mark for gene repression, based on the following observations. First, the enrichment of H4K16bio in the IL-2 gene promoter greatly decreases in response to gene activation with phorbol ester and mitogen. Second, the enrichment of H4K16bio at the retroelement LTR22 depends on the concentration of biotin in culture media. Depletion of H4K16bio co-incides with an increased abundance of LTR transcript. This observation is particularly important given that previous in-depth studies of retroelement repression suggest an increase in chromosomal abnormalities and, therefore, cancer risk in biotin-depleted cells [15]. Third, the enrichment of H4K16bio in the SMVT gene promoter increases in response to biotin supplementation. In previous studies we provided evidence that increased biotinylation of histones in the SMVT promoter is an important regulatory mechanism to repress the expression of the SMVT gene in biotin-supplemented cells [16]. It appears that the roles of histone biotinylation in gene repression are not specific for distinct biotinylation sites, as all biotinylation sites studies to date are overrepresented in transcriptionally repressed chromatin [19, 20].
A recent report suggests that some of the commercial antibodies to biotinylated histones cross-react with acetylated histones [29]. In response to that report, we re-examined our in-house antibodies to H3K9bio, H3K18bio, H4K8bio, and H4K12bio and did not detect any cross-reactivity with acetylation marks [10, 14]. We propose that these conflicting results are caused by the targets used for specificity testing. Healy et al. used recombinant histones that were enzymatically acetylated or biotinylated [29]; the extent acetylation and biotinylation and the actual modification sites (e.g., K8 vs. K12) remained unknown. In contrast, in our studies we used synthetic peptides as targets, where both the extent and location of the modification were unambiguous [10, 14]. However, in contrast with the other in-house antibodies, the batch of polyclonal anti-H4K16bio tested here showed some cross-reactivity with H4K16ac. It seems prudent to individually validate each batch of polyclonal antibodies to biotinylated histones.
Our studies of histone biotinylation in HCS mutant fibroblasts lend further strength to the notion that histone biotinylation is a real phenomenon. The abundance of biotinylation marks was substantially lower in HCS mutant WG2215 fibroblasts compared with IMR-90 fibroblasts. Similar observations were made in previous studies [11, 15, 16], along with the observation that recombinant human HCS and its microbial ortholog BirA biotinylate histones in vitro [12, 14]. There is still some level of uncertainty as to whether effects of HCS on gene regulation are mediated by biotinylation of histones or biotinylation of proteins such as carboxylases. However, the phenotypes of HCS knockdown are vastly different than phenotypes of carboxylase knockdown [7, 30], suggesting that aberrant biotinylation of histones in HCS-deficient cells impairs gene regulation.
Biotinylation of histones is a rare event, occurring in <0.1% of histones [10, 17]. One report suggested that histone biotinylation is an in vitro artifact [29], but that report has been rebutted [10]. Importantly, evidence is emerging that HCS is an integral part of a multiprotein gene repressor complex in human chromatin, and that the rare biotinylation marks seen in the human epigenome are mere marks for HCS docking sites [10]. This theory would explain why all known histone biotinylation are enriched in repressed loci, including the H4K16bio mark reported here. Further mechanistic studies in whole organisms await the arrival of a conditional HCS knockout mouse, which is an ongoing effort in our laboratory.
Abbreviations
- ADH
aldehyde dehydrogenase
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- H3K4me3
K4-trimethylated histone H3
- H3K9ac
K9-acetylated histone H3
- H3K9bio
K9-biotinylated histone H3
- H3K9me2
K9-dimethylated histone H3
- H3K18bio
K18-biotinylated histone H3
- H4K8bio
K8-biotinylated histone H4
- H4K12bio
K12-biotinylated histone H4
- H4K16bio
K16-biotinylated histone H4
- HCS
holocarboxylase synthetase
- IL-2
interleukin-2
- K
lysine
- KLH
keyhole limpet hemocyanin
- LTR
long terminal repeats
- MCC
3-methylcrotonyl-CoA carboxylase
- PC
pyruvate carboxylase
- PCC
propionyl-CoA carboxylase
- qRT-PCR
quantitative real-time polymerase chain reaction
- SMVT
sodium-dependent multivitamin transporter
Footnotes
Grants: A contribution of the University of Nebraska Agricultural Research Division, supported in part by funds provided through the Hatch Act. Additional support was provided by NIH grants DK063945, DK077816 and ES015206, USDA grant 2006-35200-17138, and by NSF grant EPS- 0701892.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wolffe A. Chromatin. 3. Academic Press; San Diego, CA: 1998. [Google Scholar]
- 2.Kouzarides T, Berger SL. Chromatin modifications and their mechanism of action. In: Allis CD, Jenuwein T, Reinberg D, editors. Epigenetics. Cold Spring Harbor Press; Cold Spring Harbor, NY: 2007. pp. 191–209. [Google Scholar]
- 3.Hymes J, Fleischhauer K, Wolf B. Biotinylation of histones by human serum biotinidase: assessment of biotinyl-transferase activity in sera from normal individuals and children with biotinidase deficiency. Biochem Mol Med. 1995;56:76–83. doi: 10.1006/bmme.1995.1059. [DOI] [PubMed] [Google Scholar]
- 4.Stanley JS, Griffin JB, Zempleni J. Biotinylation of histones in human cells: effects of cell proliferation. Eur J Biochem. 2001;268:5424–29. doi: 10.1046/j.0014-2956.2001.02481.x. [DOI] [PubMed] [Google Scholar]
- 5.Peters DM, Griffin JB, Stanley JS, Beck MM, Zempleni J. Exposure to UV light causes increased biotinylation of histones in Jurkat cells. Am J Physiol Cell Physiol. 2002;283:C878–C84. doi: 10.1152/ajpcell.00107.2002. [DOI] [PubMed] [Google Scholar]
- 6.Camporeale G, Shubert EE, Sarath G, Cerny R, Zempleni J. K8 and K12 are biotinylated in human histone H4. Eur J Biochem. 2004;271:2257–63. doi: 10.1111/j.1432-1033.2004.04167.x. [DOI] [PubMed] [Google Scholar]
- 7.Camporeale G, Giordano E, Rendina R, Zempleni J, Eissenberg JC. Drosophila holocarboxylase synthetase is a chromosomal protein required for normal histone biotinylation, gene transcription patterns, lifespan and heat tolerance. J Nutr. 2006;136:2735–42. doi: 10.1093/jn/136.11.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chew YC, Raza AS, Sarath G, Zempleni J. Biotinylation of K8 and K12 co-occurs with acetylation and mono-methylation in human histone H4. FASEB J. 2006;20:A610. [abstract] [Google Scholar]
- 9.Takechi R, Taniguchi A, Ebara S, Fukui T, Watanabe T. Biotin deficiency affects the proliferation of human embryonic palatal mesenchymal cells in culture. J Nutr. 2008;138:680–84. doi: 10.1093/jn/138.4.680. [DOI] [PubMed] [Google Scholar]
- 10.Kuroishi T, Rios-Avila L, Pestinger V, Wijeratne SSK, Zempleni J. Biotinylation is a natural, albeit rare, modification of human histones. Mol Genet Metab. 2011 doi: 10.1016/j.ymgme.2011.08.030. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kobza K, Camporeale G, Rueckert B, Kueh A, Griffin JB, Sarath G, et al. K4, K9, and K18 in human histone H3 are targets for biotinylation by biotinidase. FEBS J. 2005;272:4249–59. doi: 10.1111/j.1742-4658.2005.04839.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kobza K, Sarath G, Zempleni J. Prokaryotic BirA ligase biotinylates K4, K9, K18 and K23 in histone H3. BMB Reports. 2008;41:310–15. doi: 10.5483/bmbrep.2008.41.4.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chew YC, Camporeale G, Kothapalli N, Sarath G, Zempleni J. Lysine residues in N- and C-terminal regions of human histone H2A are targets for biotinylation by biotinidase. J Nutr Biochem. 2006;17:225–33. doi: 10.1016/j.jnutbio.2005.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bao B, Pestinger V, IHY, Borgstahl GEO, Kolar C, Zempleni J. Holocarboxylase synthetase is a chromatin protein and interacts directly with histone H3 to mediate biotinylation of K9 and K18. J Nutr Biochem. 2011;22:470–75. doi: 10.1016/j.jnutbio.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chew YC, West JT, Kratzer SJ, Ilvarsonn AM, Eissenberg JC, Dave BJ, et al. Biotinylation of histones represses transposable elements in human and mouse cells and cell lines, and in Drosophila melanogaster. J Nutr. 2008;138:2316–22. doi: 10.3945/jn.108.098673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gralla M, Camporeale G, Zempleni J. Holocarboxylase synthetase regulates expression of biotin transporters by chromatin remodeling events at the SMVT locus. J Nutr Biochem. 2008;19:400–08. doi: 10.1016/j.jnutbio.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bailey LM, Ivanov RA, Wallace JC, Polyak SW. Artifactual detection of biotin on histones by streptavidin. Anal Biochem. 2008;373:71–77. doi: 10.1016/j.ab.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 18.Wijeratne SS, Camporeale G, Zempleni J. K12-biotinylated histone H4 is enriched in telomeric repeats from human lung IMR-90 fibroblasts. J Nutr Biochem. 2010;21:310–16. doi: 10.1016/j.jnutbio.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Camporeale G, Oommen AM, Griffin JB, Sarath G, Zempleni J. K12-biotinylated histone H4 marks heterochromatin in human lymphoblastoma cells. J Nutr Biochem. 2007;18:760–68. doi: 10.1016/j.jnutbio.2006.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pestinger V, Wijeratne SSK, Rodriguez-Melendez R, Zempleni J. Novel histone biotinylation marks are enriched in repeat regions and participate in repression of transcriptionally competent genes. J Nutr Biochem. 2011;22:328–33. doi: 10.1016/j.jnutbio.2010.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manthey KC, Griffin JB, Zempleni J. Biotin supply affects expression of biotin transporters, biotinylation of carboxylases, and metabolism of interleukin-2 in Jurkat cells. J Nutr. 2002;132:887–92. doi: 10.1093/jn/132.5.887. [DOI] [PubMed] [Google Scholar]
- 22.Mock DM, Lankford GL, Mock NI. Biotin accounts for only half of the total avidin-binding substances in human serum. J Nutr. 1995;125:941–46. doi: 10.1093/jn/125.4.941. [DOI] [PubMed] [Google Scholar]
- 23.Zempleni J, Helm RM, Mock DM. In vivo biotin supplementation at a pharmacologic dose decreases proliferation rates of human peripheral blood mononuclear cells and cytokine release. J Nutr. 2001;131:1479–84. doi: 10.1093/jn/131.5.1479. [DOI] [PubMed] [Google Scholar]
- 24.Dahl JA, Collas P. A rapid micro chromatin immunoprecipitation assay (microChIP) Nature protocols. 2008;3:1032–45. doi: 10.1038/nprot.2008.68. [DOI] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–08. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Blanchard R, Gu X, Burnette A, Huang J, Zeng X. Genome-Wide, Promoter-Tilling, Real-Time PCR Assays for Analyzing Chromatin Immunoprecipitation Samples. 2007. [Google Scholar]
- 27.Haring M, Offermann S, Danker T, Horst I, Peterhansel C, Stam M. Chromatin immunoprecipitation: optimization, quantitative analysis and data normalization. Plant methods. 2007;3:11. doi: 10.1186/1746-4811-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Statview Institute. StatView Reference. 3. SAS Publishing; Cary, NC: 1999. [Google Scholar]
- 29.Healy S, Perez-Cadahia B, Jia D, McDonald MK, Davie JR, Gravel RA. Biotin is not a natural histone modification. Biochim Biophys Acta. 2009;1789:719–33. doi: 10.1016/j.bbagrm.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 30.Camporeale G, Zempleni J, Eissenberg JC. Susceptibility to heat stress and aberrant gene expression patterns in holocarboxylase synthetase-deficient Drosophila melanogaster are caused by decreased biotinylation of histones, not of carboxylases. J Nutr. 2007;137:885–89. doi: 10.1093/jn/137.4.885. [DOI] [PMC free article] [PubMed] [Google Scholar]