Abstract
Little is known about the factors that drive the high levels of between-host variation in pathogen burden that are frequently observed in viral infections. Here, two factors thought to impact viral load variability, host genetic diversity and stochastic processes linked with viral entry into the host, were examined. This work was conducted with the aquatic vertebrate virus, Infectious hematopoietic necrosis virus (IHNV), in its natural host, rainbow trout. It was found that in controlled in vivo infections of IHNV, a suggestive trend of reduced between-fish viral load variation was observed in a clonal population of isogenic trout compared to a genetically diverse population of out-bred trout. However, this trend was not statistically significant for any of the four viral genotypes examined, and high levels of fish-to-fish variation persisted even in the isogenic trout population. A decrease in fish-to-fish viral load variation was also observed in virus injection challenges that bypassed the host entry step, compared to fish exposed to the virus through the natural water-borne immersion route of infection. This trend was significant for three of the four virus genotypes examined and suggests host entry may play a role in viral load variability. However, high levels of viral load variation also remained in the injection challenges. Together, these results indicate that although host genetic diversity and viral entry may play some role in between-fish viral load variation, they are not major factors. Other biological and non-biological parameters that may influence viral load variation are discussed.
Keywords: Variation, in vivo, viral load, entry, host diversity, isogenic
1. Introduction
Virus infections are known for high levels of viral load variability between individual infected hosts (Brault et al., 2010; Clausen et al., 2011; Desport et al., 2009; DeVincenzo, 2004; Fraser et al., 2007; Gillet et al., 2011; Gorman et al., 2006; Hirsch et al., 1995; Howey et al., 2009; Hurt et al., 2010; Mulcahy et al., 1982; Munster et al., 2009; Peñaranda et al., 2011; Piatak et al., 1993; Ten Haaft et al., 1998; Wargo and Kurath, 2011). There is a broad array of factors likely to impact the between-host viral load variation commonly observed in the field. These factors might include ecological parameters such as infection dosage, host health, host age, host size, type of exposure, and environmental conditions, (Bartholomew et al., 2003; Bonhoeffer et al., 2003; Bootland and Leong, 1999; Clausen et al., 2011; Desport et al., 2009; DeVincenzo, 2004; Garver et al., 2006; Grant et al., 2003; Hallett and Bartholomew, 2008; Howey et al., 2009; Huang et al., 2010; Inendino et al., 2005; Jorgensen et al., 2008; LaPatra, 1998; Purcell et al., 2010; Wu et al., 2008), as well as factors such as virus and host genetics (Bashirova et al., 2011; Brault et al., 2010; Fellay et al., 2007; Hurt et al., 2010; Kaslow et al., 2005; Pelak et al., 2010). However, even in laboratory challenges where ecological factors, virus genetics, and host genetic diversity are controlled, experiments demonstrate substantial between-host variability in observed virus burdens (Desport et al., 2009; Gorman et al., 2006; Hirsch et al., 1995; Howey et al., 2009; Hurt et al., 2010; Munster et al., 2009; Peñaranda et al., 2011; Ten Haaft et al., 1998; Wargo et al., 2010; Wargo and Kurath, 2011). Typically, little attention is given to the causes or impacts of viral load variation between individual hosts, and conclusions about biological phenomena are frequently based on mean trends in the data. This is surprising considering that pathogen load variation and the specific factors that regulate this variation could have important implications for disease epidemiology, evolution, and management (Alizon et al., 2009; Capparelli et al., 2009; Fraser et al., 2007; Galvani and May, 2005; Lloyd-Smith et al., 2005; Matthews et al., 2006; Mellors et al., 1996; Woolhouse et al., 1997).
Host genetic diversity and stochasticity in the viral entry process are often hypothesized as the main factors driving variation in viral burdens between hosts (Brault et al., 2010; Capparelli et al., 2009; Fellay et al., 2007; Howey et al., 2009; Kaslow et al., 2005; Matthews et al., 2006; Overturf et al., 2010; Pelak et al., 2010). However, many systems are limited in their ability to explicitly quantify the role of these factors in regulating viral load variation. For example, assessing the impacts of host genetic diversity on viral load variation requires the capability to compare genetically diverse and homogeneous host populations in vivo, while controlling for all other variables. In most systems this comparison is hindered by a lack of genetically homogeneous host populations, and where these hosts are available, they can typically only be obtained in small numbers, thus limiting the statistical power (Gorman et al., 2006). Many systems are equally challenged to assess the role of stochasticity in viral entry on viral-load variation, because they typically cannot control the infectious dosage of a pathogen through the natural route of infection, and subsequently alter this route of infection to bypass host entry. Here we utilized the host-pathogen system Infectious hematopoietic necrosis virus (IHNV) in its natural vertebrate host, rainbow trout (Oncorhynchus mykiss). In this system both host genetic diversity and the entry process can be individually controlled and altered during highly replicated in vivo experiments, thus allowing for a detailed examination of their influence on viral load variation.
IHNV is a negative-sense single-stranded RNA virus in the family Rhabdoviridae (Bootland and Leong, 1999). The virus is endemic in salmonid fishes along the Pacific Coast of North America ranging from California to Alaska (Kurath et al., 2003). Field studies indicate that IHNV viral loads can span from 102 – 107 pfu/g in single fish sampled from one infected population at the same time, with some individuals even falling outside the range of detection (Mulcahy et al., 1982). The variation in the viral burden of IHNV- infected fish in the field is often attributed to unsynchronized infections or variation in the environmental and genetic background of individual fish (Bootland and Leong, 1999; Garver et al., 2006; LaPatra, 1998; Purcell et al., 2010). However, laboratory studies of IHNV that examined fish of the same age, size, and stock, infected with the same dosage of an identical virus genotype and then held under controlled environmental conditions, consistently reveal that the quantity of virus observed at the time of peak viral load can extend over 5 orders of magnitude between individuals (Peñaranda et al., 2009; Peñaranda et al., 2011; Purcell et al., 2010; Wargo et al., 2010; Wargo and Kurath, 2011). In previous studies we demonstrated that when examining the mean viral load in these controlled IHNV experiments, factors such as host species, viral genotype virulence, and host entry can impact in vivo viral load and ultimately viral fitness (Peñaranda et al., 2009; Wargo et al., 2010; Wargo and Kurath, 2011). However, parameters that impact individual fish viral load variation when these factors are held constant have not been examined. Here, we reanalyzed some of our previously published IHNV viral load data from controlled in vivo experiments (Wargo and Kurath, 2011), as well conducted new experiments to supplement this dataset to examine how between-host viral load variation is impacted by stochastic factors associated with the process of viral entry and host genetic variability.
To examine the impact of viral entry, the viral load variation in a group of fish infected by the natural route of immersion in water containing virus was compared to the viral load variation in a group of fish infected by injecting the virus directly into the host so as to bypass the host entry step. Thus, the between-host viral load variation in immersion challenges was assumed to be influenced by stochastic processes associated with both viral entry and replication, whereas viral load variation in the injection challenges was assumed to be influenced only by processes associated with replication. The analyses comparing immersion versus injection challenges included data for two viral genotypes, HV and LV, from our previous publication (Wargo and Kurath, 2011), as well as new data generated by conducting similar experiments with two IHNV genotypes designated B and C. The examination of all four genotypes made it possible to determine how consistent the observed patterns were across a range of virus genetic backgrounds. These experiments were conducted in a standard genetically diverse trout stock (here-in labeled out-bred) and therefore estimated the effect of host-entry on viral load variation in a typical field population of trout.
As an independent strategy controlled IHNV in vivo experiments were also conducted to directly examine the impact of host genetic diversity on between-host viral load variation. As with other viral diseases, host genetics is hypothesized to be one of the main driving factors behind between-host IHNV load variation (Bashirova et al., 2011; Brault et al., 2010; Fellay et al., 2007; Kaslow et al., 2005; Overturf et al., 2010; Pelak et al., 2010) and studies suggest that there may be fish genetic markers for IHNV resistance (Barroso et al., 2008; Rodriguez et al., 2004). To determine the role of host genetics in between-fish IHNV load variation, the viral load data from the immersion challenges of the four virus genotypes conducted in out-bred trout was compared to data generated from new immersion challenges in a stock of isogenic trout (here-in labeled isogenic) produced by androgenesis, in which all trout had identical genomes (Young et al., 1996). Thus, the immersion challenges in out-bred trout provided an estimate of between-host variation in viral load when natural host genetic diversity was present, whereas the challenges in isogenic trout provided an estimate of viral load variation when host genetic diversity was absent.
In all experiments, the viral load of individual whole fish was quantified. In the immersion challenges, the amount of virus shed into surrounding water was also quantified. The study therefore examined variability in within-host viral fitness as well as transmission potential. In all cases, virus quantification occurred on day 3 post-infection when viral load typically peaks in the host (Peñaranda et al., 2009; Troyer et al., 2008). The differences in viral load variation between treatment groups were assessed by comparing coefficients of variation and conducting bootstrap tests (Amiri and Zwanzig, 2010; Amiri and Zwanzig, 2011).
When examining the impact of viral entry on between-fish variation in viral load, the results showed a consistent pattern of less variability in injection compared to immersion challenges, with moderate statistical support for three of the four viral genotypes. When examining the impact of host genetic diversity on viral load variation, there was a consistent suggestive trend of less variability in isogenic compared to out-bred trout. However, this trend was not significant for any of the four genotypes, except for one genotype where the trend was significant for the shed virus population only. Despite the observed reductions in viral load variation, high levels of trout-to-trout viral load variation persisted in all treatments, including the injection challenges and isogenic trout infections. These results suggested that although viral entry and host genetic diversity may play a role in between-trout variation, they cannot account for the majority of host-to-host variation in viral load and their role in regulating this variation may be smaller than previously assumed. If these findings apply to other viral systems, this could have important implications for the factors predicted to regulate the high levels of between-host pathogen transmission variation typically observed in the field (Capparelli et al., 2009; Lloyd-Smith et al., 2005; Matthews et al., 2006; Woolhouse et al., 1997).
2. Materials and Methods
2.1. Virus
We used four genetically distinct IHNV isolates (here-in referred to as genotypes), labeled HV, LV, B, and C. These genotypes were originally isolated from farmed rainbow trout and have previously been referred to as 220:90 (HV), WRAC or 039-82 (LV), FF020-91(B), and FF030-91(C) (LaPatra et al., 1994; Troyer et al., 2008; Troyer et al., 2000). The virulence of these four genotypes, measured as virus-induced trout mortality in batch immersion challenges, has been characterized as high (HV,B and C), and low (LV) (Troyer et al., 2008; Wargo et al., 2010). The viruses were propagated on epthelioma papulosum cyprinid (EPC) cells and stored at −80°C as described elsewhere (Wargo et al., 2010). The genotypes used in these experiments underwent 4–9 passages in EPC cells after collection from the field (Fijan et al., 1983; LaPatra et al., 1994). All of the genotypes belong to the M phylogenetic clade of IHNV, which has evolved host specificity to rainbow trout (Kurath et al., 2003).
2.2. Host
Two different stocks of rainbow trout (Oncorhynchus mykiss) were used for these experiments. One stock of trout (here-in referred to as out-bred) was a genetically diverse, out-bred, stock obtained from Clear Springs Food Incorporated courtesy of Scott LaPatra, Ph.D. These out-bred trout were a standard trout farm stock and represented the fish genetic diversity found in a cultured trout farm. Although the genetic diversity of the specific lot of out-bred trout used in this study was not calculated, previous studies of other trout stocks from the same source showed an average heterozygosity score of 69.3% based on microsatellites (Silverstein et al., 2004). The out-bred trout were also obtained from the same region where the IHNV genotypes HV, LV, B, and C were originally isolated. The second stock of trout (here-in referred to as isogenic) was Arlee homozygous rainbow trout from the Washington State University Hatchery produced by androgenesis as previously described (Young et al., 1996). The isogenic trout were genetically identical to each other. All trout were pathogen free, research grade juveniles between 1–3 grams in size. The experiments described used 3 lots of out-bred trout and one lot of isogenic trout. The trout were maintained in pathogen-free water at 15°C, as previously described (Troyer et al., 2008). All animal work was approved by a University of Washington IACUC protocol.
2.3. Virus challenges
Trout were exposed to virus by one of two methods, injection or immersion challenges. For immersion challenges trout were placed in static water containing 104 pfu/ml of a single virus genotype for 12 hours as previously described (Wargo et al., 2010). For injection challenges 100 pfu of virus was injected directly into the intraperitoneal cavity as previously described (Wargo and Kurath, 2011). After viral exposure, all trout were held in flowing water to remove virus and isolated into individual tanks with 400 ml of water to prevent subsequent cross-infection. The trout were then maintained at 15°C for 3 days under static water conditions as described elsewhere (Wargo et al., 2010; Wargo and Kurath, 2011). Out-bred trout were infected by both injection and immersion challenges. Isogenic trout were only exposed by immersion challenges. For each of the three treatment regimes, 12–28 trout were exposed to single infections of the four viral genotypes (HV, LV, B, or C), and each experiment was replicated 2–3 times (table 1). This experimental design allowed us to examine the role of host genetic diversity (out-bred vs. isogenic trout) and host entry (injection vs. immersion challenges) in regulating between-trout viral load variation, independently for four viral genotypes. The data for genotypes HV and LV in out-bred fish (experiments 1–3, table 1) has been previously published using a different analysis focusing on patterns in mean viral load rather than viral load variation between individual fish (Wargo and Kurath, 2011). All other data was collected from new experiments (experiments 4–7, table 1) and has not previously been published. Unusually high levels of mortality were observed in experiment 4, likely due to low stress tolerance of isogenic trout (Table 1). In all subsequent experiments (5–7), trout were housed in larger tanks (1L compared to 0.8L) to reduce stress.
Table 1.
Number of samples from experiments. Values are given for the number of within-host and shed viral samples from individual fish in the respective experiments. Numbers enclosed in {} symbols are excluded dead trout and numbers enclosed in [] symbols are excluded virus negative samples. Bold indicates total number of samples pooled from multiple experiments used for the statistical analyses presented in the paper. Unusually high levels of mortality were observed in experiment 4, likely due to low stress tolerance of the isogenic trout (see methods).
| Genotype | Challenge | Experiment | Within-host samples | Shed samples |
|---|---|---|---|---|
| HV | Out-bred immersion Isogenic immersion Out-bred injection |
1, 2, 3 4, 5 1, 2, 3 |
22{2}, 24, 28 = 74 14{6}, 12 = 26 23, 16, 27[1] = 66 |
21{2}[1], 24, 28 = 73 14{6}, 12 = 26 N.A. |
| LV | Out-bred immersion Isogenic immersion Out-bred injection |
1, 2, 3 4, 5 1, 2, 3 |
17{5}[2], 14[6], 28 = 59 14{6}, 12 = 26 17{3}, 17[1], 25[3] = 59 |
19{5}, 17[3], 28 = 64 14{6}, 12 = 26 N.A. |
| B | Out-bred immersion Isogenic immersion Out-bred injection |
6, 7 4, 5 6, 7 |
18{1}, 27[1] = 45 17{3}, 12 = 29 21, 26[1] = 47 |
17{1}[1], 27[1] = 44 17{3}, 12 = 29 N.A. |
| C | Out-bred immersion Isogenic immersion Out-bred injection |
6, 7 4, 5 6, 7 |
28, 26[1] = 54 10{10}, 12 = 22 26{1}[1], 28 = 54 |
28, 24[3] = 52 10{10}, 12 = 22 N.A. |
2.4. Viral load quantification
To quantify within-host viral load, all of the trout were harvested 3 days post-infection, which has previously been shown to be the peak of viral replication and is typically before the onset of trout mortality (Peñaranda et al., 2009; Troyer et al., 2008; Wargo et al., 2010; Wargo and Kurath, 2011). At the time of harvest, the trout were euthanized with an overdose of tricaine methanesulfonate and stored at −80°C until further processing. We then extracted the total RNA from individual trout using guanidinium-thiocyanate as previously described (Wargo et al., 2010). A 550 ml water sample was also taken at the time of trout harvest for shed viral load quantification from all fish in the immersion challenges. Total RNA was extracted from the water samples using the QIAamp MinElute virus spin kit (Qiagen) as previously described (Wargo and Kurath, 2011).
All extracted RNA was converted to cDNA using Moloney murine leukemia virus (MMLV) reverse transcriptase, oligo(DT), and random hexamers in a 20 µl reaction as elsewhere (Wargo et al., 2010). To complete virus quantification, the cDNA was diluted 10 fold (fish samples) or 5 fold (water samples) with H2O, then subjected to quantitative PCR (qPCR) using the following genotype specific assays: qHV (see ref (Wargo and Kurath, 2011)), qLV (see ref (Wargo and Kurath, 2011)), qB (Forward primer: 5’-GCATCAGGCCTGTATGATTGAA-3’, MGB TaqMan probe: 5’ -VICCGTATACAATTCTGGATCAAA, Reverse primer: 5’- CACCACACTTCGGGAACGA-3’), and qC (Forward primer: 5’-TGCATCAGGCCTGTATGATGAC-3’, MGB TaqMan probe: 5’ -6-FAM-TCGGATACAATTCTGGAGCAG-3’, Reverse primer: 5’-CCTCACACTTCGGGAACGA-3’), targeted to the viral glycoprotein gene of the appropriate IHNV genotype. The qPCR reaction included 5 ul of the diluted cDNA along with 200 nM probe, and 900 nM of each primer in a 12 µl reaction as previously outlined (Wargo et al., 2010; Wargo and Kurath, 2011). Genotype specific glycoprotein gene transcripts (rHV-G, rLV-G, rB-G, or rC-G) were utilized in each qPCR run to generate 8-step, 10-fold dilution series standard curves, making it possible to acquire absolute viral RNA copy quantification (Wargo et al., 2010; Wargo and Kurath, 2011). Here we refer to viral RNA copies as viral load.
2.5. Statistics
To compare mean viral load differences between groups we employed general linear model (GLM) analyses with the dependent variable in the models set to viral load and the factors in the models set to treatment (out-bred immersion, isogenic immersion, or out-bred injection), and genotype (HV, LV, B, or C). In cases where multiple comparisons were made, Tukey’s tests were employed to determine where significant differences between groups was supported (P<0.05). Since water samples were not taken from injection challenges, this treatment was not included when analyzing mean shed viral load. Viral load was log transformed in the GLM analyses to meet the model assumptions of normality and variance homogeneity.
The viral load analysis revealed significant mean viral load differences between the treatment groups. Since the standard deviation is a function of the mean, it was not appropriate to compare the variability of groups with a different mean using the standard deviation. The variability between groups was therefore compared using the coefficient of variation (CV), defined here as the standard deviation/mean, making it a unitless parameter. Because the data was not normally distributed, the coefficient of variation of treatment groups was compared using a distribution-free bootstrap method proposed by Amiri and Zwanzig (Amiri and Zwanzig, 2010; Amiri and Zwanzig, 2011). Briefly, the p-value was calculated with the formula#:{Tb(Y1b*,Y2b*) > T(Y1,Y2)}/(B+1). The terms in this formula are defined as follow:T(Y1,Y2) equals the test statistic CV(Y1)–CV(Y2), where Yi equals the actual data representing the treatment group of interest and CV equals the coefficient of variation. Tb(Y1b*,Y2b*) equals the test statistic CV(Y1b*,)–CV(Y2b*), where Yib* equals the data for the treatment group of interest generated by pooling Y1 and Y2 and then sampling the pool with replacement to the original sample size of Yi. B equals the number of bootstrap replicates for which Tb(Y1b*,Y2b*) is generated, herein, B was set to 10000. This analysis resulted in 24 pairwise tests requiring the use of a Bonferroni correction in which comparisons were considered significant where the p-value < (0.05/24 =0.0021).
For all analyses, the data within treatment groups from the different experiments was pooled to obtain sufficient statistical power and minimize the type I error rate due to multiple comparisons. To verify the validity of pooling the data, the analysis was also run without pooling the data and the same patterns were observed. Only the results from the analyses on pooled data are presented. All dead trout (37/616 trout) were excluded from the analyses presented since the exact time of death could not be determined and viral replication kinetics as well as stability are believed to be different in dead compared to live trout (Wargo and Kurath, 2011). However, the analyses were also run including the dead trout and the same results were obtained (data not shown). Since this research was inherently concerned with quantifying differences in between-trout variation in viral load, and variability can only be examined when viral load values are available, we excluded all samples from the analyses presented in which virus could not be detected (26/944 processed samples). We also reran the analyses, setting negative samples to the minimum detection threshold of the qPCR assays (1000 viral copies), and this did not change the results. The final analyses included data from 7 separate experiments involving 897 samples from 566 fish (table 1).
3. Results
3.1. Individual trout viral load
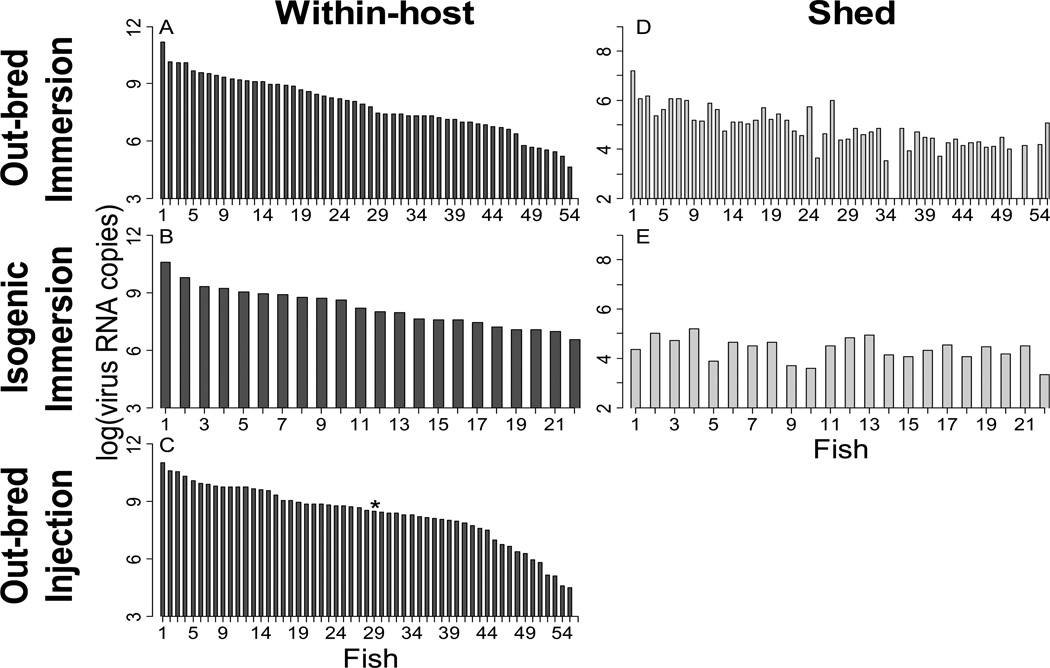
The viral load of individual trout across all experiments ranged from 1.3 × 104 to 2.1 × 1011 viral RNA copies/g trout in within-host samples and 5.0 × 102 to 1.5 × 108 viral RNA copies/ml H20 in shed virus samples (Table 2, Figs. 1, S1, S2, S3). For within-host samples, the range of viral load differed between the three challenge types such that out-bred immersion trout showed the largest range of viral load values, isogenic immersion samples showed the lowest range, and out-bred injection trout showed an intermediate range (Table 2). A similar ranking was found for shed virus, such that the water samples from the out-bred immersion challenges showed a greater viral load range than samples from isogenic immersion challenges (Table 2). Water samples were not taken from injection trout so the range in viral load of shed virus in injection challenges was not calculated. There was no clear pattern when comparing the viral load range of genotypes HV, LV, B, or C across the different challenge types. For example, genotype HV had the highest range of within-host viral load values in out-bred immersion challenges but the lowest in isogenic immersion challenges (Table 2). Thus, the ordering of the four genotypes was inconsistent in the different challenge types.
Table 2.
The range of quantified within-host and shed viral load in each of the treatments. Virus negative samples are excluded from this range and the range is across all of the experiments within that treatment.
| Genotype | Challenge | Within-host (virus copies/g fish) |
Shed (virus copies/ml H20) |
|---|---|---|---|
| HV | Out-bred immersion Isogenic immersion Out-bred injection |
2.8 × 104 – 2.1 × 1011 1.4 × 106 – 3.7 × 109 2.0 × 106 – 1.3 × 1011 |
8.7 × 103 – 3.4 × 107 2.1 × 103 – 1.2 × 105 N.A. |
| LV | Out-bred immersion Isogenic immersion Out-bred injection |
1.3 × 104 – 4.0 × 1010 6.2 × 104 – 9.3 × 108 1.3 × 105 – 1.2 × 1011 |
5.0 × 102 – 1.5 × 108 8.1 × 102 – 4.4 × 106 N.A. |
| B | Out-bred immersion Isogenic immersion Out-bred injection |
1.4 × 105 – 2.2 × 1010 1.2 × 105 – 8.6 × 109 5.4 × 104 – 6.2 × 109 |
2.9 × 103 – 5.3 × 105 7.1 × 102 – 7.0 × 104 N.A. |
| C | Out-bred immersion Isogenic immersion Out-bred injection |
4.5 × 104 – 1.5 × 1011 3.7 × 106 – 3.7 × 1010 3.1 × 104 – 1.0 × 1011 |
3.6 × 103 – 1.6 × 107 2.2 × 103 – 1.6 × 105 N.A. |
Figure 1.
Viral load of individual trout exposed to IHNV genotype C. Panels A, B, and C show the within-host log transformed viral loads [log(virus RNA copies/g trout)] of individual trout from out-bred immersion, isogenic immersion, and out-bred injection trout, respectively. Panels D and E show the log transformed shed viral loads [log(virus RNA copies/ml of H2O)] from the out-bred immersion and isogenic immersion trout respectively. Water samples were not taken from out-bred injection trout so shed virus load was not calculated for these trout. Note that panels A–C have a different Y-axis scale than panels D–E. Trout are arranged from highest to lowest within-host viral load. Since the shed samples come from the same trout as the within-host trout, they are arranged in the order of the within-host trout. Trout with a symbol (*) above the bar died during the experiment and were excluded from all further figures and analyses. Dead isogenic immersion challenged fish were not processed so no virus quantity values are shown for these fish. Trout with no detectable virus (missing bar) were also excluded from all further figures and analyses. Minimum Y-axis value represents the minimum detection limit of the genotype specific qPCR assays. Figures showing the individual trout viral loads of genotypes HV, LV, and B are presented in the supplemental materials.
3.2. Mean viral load
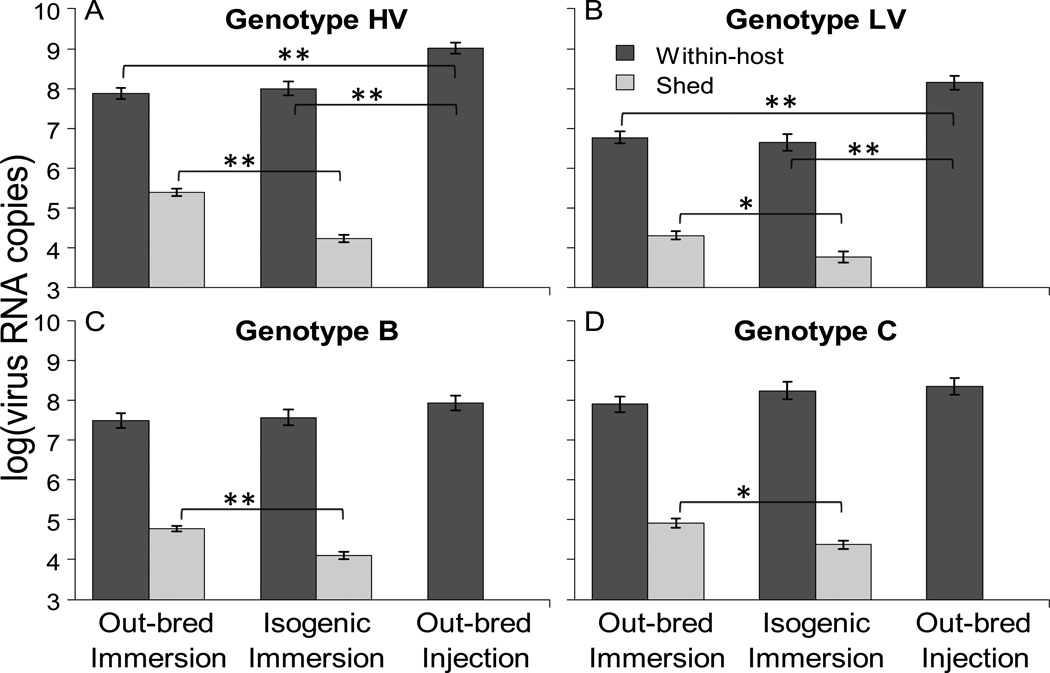
The four viral genotypes (HV, LV, B, and C) were independently analyzed to examine if there were differences in the amount of virus produced in out-bred immersion, isogenic immersion, and out-bred injection treatments. When examining within-host viral load, more virus was produced in out-bred injection challenges compared to out-bred immersion and isogenic immersion challenges for all genotypes, with the trend being significant for genotype HV and LV (Fig. 2; HV: F1,141 = 23.6, P<0.001; LV: F1,163 = 19.0, P<0.001; B: F1,118 = 1.6, P=0.21; C: F1,128 = 1.6, P=0.22). However, there was no significant difference in within-host viral load between out-bred immersion and isogenic immersion treatments for any of the four genotypes. In regards to the quantity of virus shed into water samples, significantly more virus was detected in out-bred immersion treatments compared to isogenic immersion treatments, for all four genotypes (Fig. 2; HV: F1,97 = 53.1, P<0.001; LV: F1,88 = 8.4, P=0.005; B: F1,71= 29.9, P<0.001; C: F1,72 = 1.6, P=0.003) .
Figure 2.
Mean viral load in the three treatments for four different viral genotypes. Bars represent the mean virus RNA copies/g of trout from within-host (black bars) or mean virus RNA copies/ml H2O from shed (gray bars) virus populations, ±1 standard error of the mean(S.E.M). Values for genotype HV (panel A), LV (panel B), B (panel C), and C (panel D) pooled across 2–3 experiments (table 1) within the out-bred immersion, isogenic immersion, and out-bred injection challenges, are shown. All panels have the same Y and X axes. Bars exclude dead trout as well as samples where no virus was detected (see methods). No water samples were taken from out-bred injection trout so the mean virus shed from this treatment could not be calculated. Significant differences by GLM analysis are indicated with brackets and symbol (*: P≤0.005; **: P<0.001).
3.3. Coefficient of variation
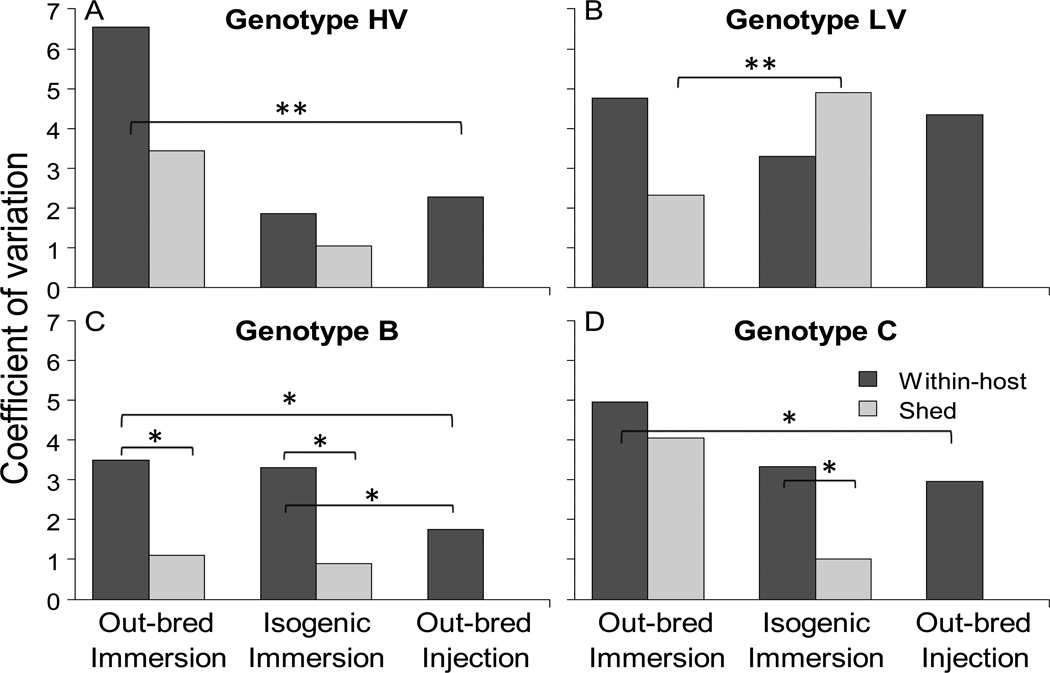
The coefficient of variation in viral load from out-bred immersion, isogenic immersion, and out-bred injection treatments, was compared independently for each of the four genotypes (HV, LV, B, and C). This analysis provided an estimate of the impact of host genetic diversity (out-bred immersion versus isogenic immersion), virus entry (out-bred immersion versus out-bred injection), and the relative importance of host genetic diversity and entry (isogenic immersion versus out-bred injection), on the between-host variation in viral load. When examining within-host viral load, a trend of less variability was observed in out-bred injection challenges compared to out-bred immersion challenges for all four genotypes, with this trend being significant for genotypes B and C at the α=0.05 cut-off and genotype HV at the Bonferroni corrected α=0.0021 cut-off (Table 3, fig. 3). It was also found that there was a suggestive trend of less within-host viral load variability in isogenic immersion challenges compared to out-bred immersion challenges, however this trend was not statistically significant for any of the four genotypes (Table 3, fig. 3). A less consistent pattern was observed when comparing the within-host viral load variability of isogenic immersion to out-bred injection challenges, in that more variability was found in out-bred injection challenges for genotypes HV and LV and less variability for genotypes B and C. Of these observed patterns, genotype B was the only genotype showing a significant difference between the isogenic immersion and out-bred injection challenges (Table 3, fig. 3).
Table 3.
Statistical results from coefficient of variation (CV) bootstrap test (Amiri and Zwanzig, 2010; Amiri and Zwanzig, 2011). The analysis tests whether the null hypothesis CV1=CV2 can be rejected. See figure 3 for graphical interpretation. Values shown are p-values.
| Within-host | Shed | Within-host Vs. Shed | |||||
|---|---|---|---|---|---|---|---|
| Genotype |
Out-bred Immersion Vs. Isogenic Immersion |
Out-bred Immersion Vs. Out-bred Injection |
Isogenic Immersion Vs. Out-bred Injection Immersion Immersion |
Out-bred Immersion Vs. Isogenic Immersion |
Out-bred Immersion |
Isogenic Immersion |
|
| HV | 0.13 | 0** | 0.41 | 0.15 | 0.17 | 0.13 | |
| LV | 0.33 | 0.38 | 0.42 | 0.0005** | 0.07 | 0.11 | |
| B | 0.52 | 0.025* | 0.005* | 0.32 | 0.049* | 0.02* | |
| C | 0.33 | 0.036** | 0.17 | 0.09 | 0.31 | 0.02** | |
A single * denotes significance at the α=0.05 cut-off, double ** denotes significance at the multiple test Bonferroni corrected α=0.0021 cut-off (0.05/24 tests). All dead trout and samples negative for virus were excluded from the analyses, but the same quantitative results were obtained when these trout were included (data not shown). The results presented are from the analyses on data pooled across experiments within treatments. The same patterns were observed when the analyses were run on unpooled data (data not shown).
Figure 3.
Coefficient of variation. Bars show the untransformed coefficient of variation (defined as the standard deviation/mean) in within-host (black bars) or shed (gray bars) viral load. Values for genotype HV (panel A), LV (panel B), B (panel C), and C (panel D) are shown for out-bred immersion, isogenic immersion, and out-bred injection challenges. All panels have the same Y and X axes. The Y axis has no units because the coefficient of variation is a unitless parameter. Bars exclude dead trout as well as samples where no virus was detected (see methods). No water samples were taken from out-bred injection trout so the coefficient of variation for this treatment was not calculated. Significant differences by bootstrap method are indicated with brackets and symbol (*: P<0.05; **: P<0.0021, cut-off Bonferroni correction).
The pattern of variability in the amount of virus shed into water samples was largely consistent with that observed for the within-host viral load data. Genotypes HV, B, and C all showed a trend of less viral load variability in virus shed from isogenic immersion challenges compared to out-bred immersion challenges, although this trend was not significant for any of the three genotypes. Genotype LV however was not consistent with this pattern in that significantly more variability was found in the viral load of shed samples from isogenic immersion compared to out-bred immersion challenges. When directly comparing the variability in the quantity of shed virus to within-host viral load within each treatment, a consistent pattern of less variation in the quantity of shed virus was observed, with the pattern being significant for genotype B in out-bred immersion and isogenic immersion challenges, and genotype C in isogenic immersion challenges, at the α=0.05 cut-off (Table 3, fig. 3). The only genotype not consistent with this pattern was genotype LV in isogenic immersion challenges where more viral load variability was observed in shed compared to within-host samples, however, this trend was not statistically significant. Since no samples were taken from out-bred injection challenges, no comparisons to these challenges could be made for shed virus.
4. Discussion
It is typically assumed that host genetic diversity is one of the primary driving forces behind between-host variation in pathogen burdens, despite limited empirical support (Fellay et al., 2007; Kaslow et al., 2005; Pelak et al., 2010). Here it was observed that although there was a suggestive trend of reduced viral load variation in a genetically clonal stock of isogenic trout compared to a genetically diverse out-bred stock, this trend was not statistically significant for any of four viral genotypes examined. In fact, on average 65% of the between-host viral load variation persisted even in the genetically homogeneous isogenic trout. These results led us to conclude that for the virus IHNV, although host genetic diversity may play a role in between-host viral load variation, it is a minor component of the factors that regulate this variability. This finding is in agreement with previous studies of IHNV, which demonstrated high levels of between-fish variation in the innate interferon immune response in controlled IHNV infections in isogenic trout (Purcell et al., 2009b). These combined results therefore contradict current dogma about the driving force of between-host variability in pathogen burden (Brault et al., 2010; Fellay et al., 2007; Kaslow et al., 2005; Overturf et al., 2010; Pelak et al., 2010).
In an effort to examine another potential source of between-host viral load variation, groups of trout were infected by direct injection of the virus so as to bypass the host entry process. We hypothesized that although all trout are exposed to the same challenge dosage in immersion challenges, there may be heterogeneity in the amount of virus that enters each trout and initiates the infection, or the timing when entry occurs, due to stochastic processes associated with crossing the fish skin and mucosal barrier. This stochasticity in entry could ultimately result in variability in peak viral load (Peñaranda et al., 2009; Wargo and Kurath, 2011). The results from the current study indicated that indeed injecting virus directly into the host reduced between-host viral load variation for all four viral genotypes examined, with a significant reduction in 3 of the genotypes. Furthermore, given that viral load was higher in injection challenges compared to the other challenge types, our results suggest that injection challenges did indeed bypass some aspects of the viral replication barrier associated with host entry. Therefore, we concluded that stochastic factors associated with the process of virus entry into the host do contribute to variation in subsequent viral loads. However, since on average 59% of between-host viral load variation remained in the injection challenges compared to immersion challenges, we also concluded that entry is not the major factor to regulate viral load variability.
The strategy for this study was to independently assess two distinct factors that are generally hypothesized to be major influences responsible for between-host viral load variation during the natural infections. To do this, we varied only a single factor in each experiment, and our results indicated clearly that neither host genetic heterogeneity, nor stochastic events during host entry were responsible for the majority of the variation observed. We did not examine the combined impact of using clonal fish with challenges by injection because interpretation of results would be confounded by having two variables relative to the natural infection system of interest. Comparisons of the viral load variability in isogenic immersion challenges and out-bred injection challenges revealed an inconsistent pattern, in that the variability was higher in injection challenges for two virus genotypes but lower for the other two virus genotypes. The inconsistency of this pattern, combined with the finding that between-fish viral load variation was significantly reduced in injection challenges but not isogenic fish, suggests that stochastic events in viral entry play an equal if not greater role in between-fish viral load variation than host genetic diversity. Regardless of which factor is more important, the conclusions drawn from the study remain, that host genetic diversity and stochastic processes associated with entry are not the primary factors regulating between-fish viral load variation.
A common limitation to in vivo studies is lack of statistical power due to small sample sizes. In the present study sample sizes, ranging from 22–74 trout, were considerably larger than those typically used for experimental in vivo vertebrate virus studies (Desport et al., 2009; Howey et al., 2009; Hurt et al., 2010; Munster et al., 2009; Peñaranda et al., 2011; Ten Haaft et al., 1998; Wargo and Kurath, 2011), greatly enhancing the statistical power. However, sample size limitations could explain the finding that for genotype HV there was a significant difference in the coefficient of variation between out-bred immersion and out-bred injection trout, but not out-bred immersion and isogenic immersion trout, despite a greater reduction in variability (fig. 3). In this analysis both out-bred immersion and injection trout had larger group sizes (74 and 66 trout respectively) than the isogenic trout treatment group (26 trout) due to limitations in the number of of clonal fish available. Although the statistical tests used were robust against comparing groups with different samples sizes, like any test, reduced sample sizes can impact their statistical power (Amiri and Zwanzig, 2010; Amiri and Zwanzig, 2011). Regardless of the statistical outcome, the finding holds true that there were substantial levels of between-host variation in the isogenic trout treatment groups (coefficient of variation = 200 – 350%).
The obvious question that remains is what is the source of between-host viral load variation for IHNV in rainbow trout? One possibility is stochastic parameters inherent in the IHNV infection process other than entry into the host. For example, stochastic factors at the cellular entry level such as cell type infected, cell division cycle, or the timing of the cellular immune response may introduce variability. Additionally, stochastic processes or bottlenecks during the spread of the virus throughout host tissues and organs could contribute to variation in viral loads. The present study was not designed to test these hypotheses. However, if cellular entry or viral spread contributed to between-fish variation, this implies that there was some type of mechanism to drive the variation which does not involve host genetics. One such mechanism could be epigenetic effects.
The role of epigenetics in disease has received increasing attention (Barros and Offenbacher, 2009; Holliday, 2006; Rando and Verstrepen, 2007). In the present study, it is possible that although the isogenic trout were genetically identical, epigenetic factors during development could have lead to differences in disease susceptibility or immune function between individuals. Similar phenomena have been hypothesized to explain phenotypic heterogeneity observed in clonal populations of zebrafish (Massicotte et al., 2011). Little work has been done to examine how epigenetic effects might impact host resistance to infectious diseases in the context of viral replication. There is increasing evidence that epigenetic effects play an important role in the heterogeneity of clinical outcome to infection with oncogenic infectious agents, and it seems reasonable to assume similar processes could occur with non-cancer causing viruses (Fernandez and Esteller, 2010). Epigenetic effects could be particularly relevant to viruses like IHNV, if disease outcome (death or survival) is determined early in infection by a race between the host innate immune response and viral replication (Peñaranda et al., 2009; Purcell et al., 2009a; Purcell et al., In press). It is conceivable that epigenetic variation between hosts impacts this race, by generating variability in timing of the innate immune responses between fish. Previous IHNV studies indicate that for inducible innate immune genes that have been examined, the host immune response appears to track viral load rather than vice versa, with a strong association between the fish immune response and IHNV load (Peñaranda et al., 2009; Purcell et al., 2009a; Purcell et al., 2010). Furthermore, the viral load outcome in the host is determined very early in infection and therefore, if immune variation is the driver behind between-host viral load variation, the constitutive rather than inducible immune response is the most probable source of this variation (Peñaranda et al., 2009; Purcell et al., 2009a; Purcell et al., 2010).
In addition to epigenetics factors, viral genetic diversity could have also played a role in the observed between-fish variability in viral load (Bashirova et al., 2011; Brault et al., 2010; Hurt et al., 2010). The virus stocks used in this study were all natural quasispecies (Domingo 1985) known to contain one dominant IHNV consensus genotype. Therefore it is plausible that there was stochastic variation in which genetic type from the quasispecies swarm became dominant in a given host. However, previous studies characterizing the mutant spectrum of IHNV populations showed relatively low genetic diversity within in vivo infections from the field, as well as in vitro viral stocks used to initiate infections (Emmenegger et al., 2003). Additionally, given that isogenic trout still maintained high levels of viral load variation, if the quasispecies phenomenon is a driving force behind this variation, it suggests stochasticity rather than host genetics is more important for determining the dominant viral variant in a given host.
A non-biological source of variation in this study worth mentioning could be that introduced as a result of the precision of the sample processing protocols. Both the reverse transcription and qPCR methodologies are well-documented as having greater than 95% precision, and are therefore unlikely sources of between-sample variation in results (Peñaranda et al., 2011; Purcell et al., 2006; Wargo et al., 2010; Wargo and Kurath, 2011). More probable sources of error during sample processing are the fish and water RNA extraction protocols. To estimate the precision of the sample processing protocols, the total RNA in individual extracted samples was quantified. The extraction protocols are designed such that all samples start with either the same concentration of host (within-host virus samples) or carrier RNA (shed virus samples). Given that the majority of RNA is host or carrier rather than virus RNA, the final concentration of RNA after extraction offers a measure of consistency between samples. Within-host sample RNA concentrations were found to have a coefficient of variation of 0.81 and shed virus samples were found to have a coefficient of variation of 0.16. This level of variation was substantially less than that observed in qPCR quantified viral load (within-host: 1.75–6.5, shed: 0.9–4.9), and therefore the effect of variation in the extraction protocol is unlikely to have impacted the viral load variation to a significant degree. Furthermore, any impact of sample processing on viral load variability, should have affected all samples by the same amount, regardless of treatment.
An interesting finding was that less variability was observed in the shed virus load compared to the within-host viral load from the same fish within various treatment groups in 7 out of 8 cases, even when controlling for the difference in mean viral load. Although the higher precision of the water extraction compared to trout extraction protocol may have had some impact on this, it is unlikely that this was the only cause of the reduced variability in shed virus. Another possible explanation is that viral shedding involves some sort of bottleneck, restricting the amount of virus exiting the host to a tighter range, thus reducing variability. Little is known about the process of IHNV shedding from fish. The virus is primarily believed to be transmitted through the gills and urine, offering potential bottleneck sites. Our finding that the mean shed viral load was consistently less in isogenic compared to out-bred fish, whereas mean within-host viral load was not different between the two fish stocks, provides further support for the possible existence of a shedding bottleneck and suggests that host genetics may play a role in regulating such a bottleneck.
Any of the possible sources of between-host viral load variation discussed above could be relevant to the high variability in between-host viral load observed in other systems (Capparelli et al., 2009; Lloyd-Smith et al., 2005; Matthews et al., 2006; Woolhouse et al., 1997). However, this list of possible mechanisms is by no means exhaustive and the importance of these and other factors will depend heavily on the biological system investigated. Our goal here was to assess the importance of host genetic diversity, as well as stochastic processes associated with viral entry, on between-host viral load variability when challenge dosage and environmental conditions were controlled. Even when host genetic variation was eliminated and the viral entry step was bypassed, substantial levels of between-fish viral load variation were observed. Therefore, we conclude that host genetic diversity and viral entry alone cannot explain between-host viral load variation.
Our finding that between-host pathogen-load variation is observed even in cases where host genetic diversity is absent, is congruent with studies of other pathogens in highly inbreed host populations such as mice and agricultural animal species (Brault et al., 2010; Desport et al., 2009; Gorman et al., 2006; Howey et al., 2009; Hurt et al., 2010; Munster et al., 2009; Peñaranda et al., 2011; Wargo and Kurath, 2011). Our results also shed light on correlative and quantitive genetics studies aimed at determining host genetic markers of disease resistance that often find that host genetic diversity explains less than 50% of the observed viral-load heterogeneity (Barroso et al., 2008; Fellay et al., 2007; Rodriguez et al., 2004). If this pathogen-load variation translates to variation in overall transmission between hosts, this could explain the ‘super-spreader’ phenomenon observed in viruses such as SARS, where a few host individuals are responsible for the majority of transmission events (Capparelli et al., 2009; Lloyd-Smith et al., 2005; Matthews et al., 2006; Woolhouse et al., 1997). The result observed here of large amounts of shedding heterogeneity between-hosts supports the hypothesis that heterogeneity in transmission of that shed virus to new hosts would also be observed. If this is true, our shedding results indicate that, for IHNV, host genetic diversity is not the primary driver of transmission heterogeneity observed across individuals in a population. To our knowledge the relationship between host-to-host transmission heterogeneity and host genetic diversity has not been experimentally examined in IHNV or other viral systems. Ultimately, heterogeneity in transmission between individuals can drastically affect the accuracy of estimates of important epidemiological parameters such as the mean number of infections caused by an infected individual at a population level, or Ro (Capparelli et al., 2009; Fraser et al., 2007; Galvani and May, 2005; Lloyd-Smith et al., 2005; Matthews et al., 2006; Woolhouse et al., 1997). As such, the results from this study support the work of other groups suggesting that the processes regulating natural between-host variation in pathogen burdens are more complex than previously assumed, and epidemiological models of pathogen transmission may benefit from incorporating pathogen-load variation, even in host populations with low genetic diversity (Capparelli et al., 2009; Fraser et al., 2007; Lloyd-Smith et al., 2005; Matthews et al., 2006; Woolhouse et al., 1997).
Acknowledgements
We thank B. Kerr, M. Purcell, R. Life, L. Villarreal, A. Read, S. Amiri, and J. Winton for valuable discussion and T. Thompson, B. Batts, and P. Wheeler for technical assistance. We are grateful to S.E. LaPatra of Clearsprings Foods, Inc., for providing research grade rainbow trout. This work was supported by the USGS Western Fisheries Research Center, and funding for A. Wargo was provided by the National Institute of Health R.L. Kirschestein NRSA University of Washington institutional service award 5 T32 AI007509 and National Science Foundation Ecology of Infectious Diseases grant 0812603. The project was also supported in part by a USDA Special Research Grant titled “Aquaculture Washington and Idaho” to Gary Thorgaard. The funding agencies had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Mention of trade names does not imply U.S. Government endorsement.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Andrew R. Wargo, Email: awargo@uw.edu.
Alison M. Kell, Email: alisonmkell@yahoo.com.
Robert J. Scott, Email: rjscott1@uw.edu.
Gary H. Thorgaard, Email: thorglab@wsu.edu.
Gael Kurath, Email: gkurath@usgs.gov.
References
- Alizon S, Hurford A, Mideo N, Van Baalen M. Virulence evolution and the trade-off hypothesis: history, current state of affairs and the future. J. Evol. Biol. 2009;22(2):245–259. doi: 10.1111/j.1420-9101.2008.01658.x. [DOI] [PubMed] [Google Scholar]
- Amiri S, Zwanzig S. An Improvement of the nonparametric bootstrap test for the comparison of the coefficient of variations. Commun. Stat. B-Simul. 2010;39(9):1726–1734. [Google Scholar]
- Amiri S, Zwanzig S. Assessing the coefficient of variations of chemical data using bootstrap method. J. Chemometr. 2011;25:295–300. [Google Scholar]
- Barros SP, Offenbacher S. Epigenetics: Connecting environment and genotype to phenotype and disease. J. Dent. Res. 2009;88(5):400–408. doi: 10.1177/0022034509335868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barroso R, Wheeler P, LaPatra S, Drew R, Thorgaard G. QTL for IHNV resistance and growth identified in a rainbow (Oncorhynchus mykiss) x Yellowstone cutthroat (Oncorhynchus clarki bouvieri) trout cross. Aquaculture. 2008;277(3–4):156–163. [Google Scholar]
- Bartholomew JL, Lorz HV, Sollid SA, Stevens DG. Susceptibility of juvenile and yearling bull trout to Myxobolus cerebralis and effects of sustained parasite challenges. J. Aquat. Anim. Health. 2003;15(3):248–255. [Google Scholar]
- Bashirova AA, Thomas R, Carrington M. HLA/KIR restraint of HIV: Surviving the fittest. Annu. Rev. Immunol. 2011;Vol. 29:295–317. doi: 10.1146/annurev-immunol-031210-101332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonhoeffer S, Funk GA, Gunthard HF, Fischer M, Muller V. Glancing behind virus load variation in HIV-1 infection. Trends Microbiol. 2003;11(11):499–504. doi: 10.1016/j.tim.2003.09.002. [DOI] [PubMed] [Google Scholar]
- Bootland LM, Leong JC. Infectious hematopoietic necrosis virus. In: Woo PTK, Bruno DW, editors. Fish diseases and disorders. Vol. 3. CAB International; 1999. pp. 57–112. [Google Scholar]
- Brault SA, Bird BH, Balasuriya UBR, MacLachlan NJ. Genetic heterogeneity and variation in viral load during equid herpesvirus-2 infection of foals. Vet. Microbiol. 2010;147(3–4):253–261. doi: 10.1016/j.vetmic.2010.06.031. [DOI] [PubMed] [Google Scholar]
- Capparelli R, Parlato M, Iannaccone M, Roperto S, Marabelli R, Roperto F, Iannelli D. Heterogeneous shedding of Brucella abortus in milk and its effect on the control of animal brucellosis. J.Appl. Microbiol. 2009;106(6):2041–2047. doi: 10.1111/j.1365-2672.2009.04177.x. [DOI] [PubMed] [Google Scholar]
- Clausen LN, Weis N, Astvad K, Schonning K, Fenger M, Krarup H, Bukh J, Benfield T. Interleukin-28B polymorphisms are associated with hepatitis C virus clearance and viral load in a HIV-1-infected cohort. J. Viral Hepatitis. 2011;18(4):e66–e74. doi: 10.1111/j.1365-2893.2010.01392.x. [DOI] [PubMed] [Google Scholar]
- Desport M, Ditcham WGF, Lewis JR, McNab TJ, Stewart ME, Hartaningsih N, Wilcox GE. Analysis of Jembrana disease virus replication dynamics in vivo reveals strain variation and atypical responses to infection. Virology. 2009;386(2):310–316. doi: 10.1016/j.virol.2009.01.014. [DOI] [PubMed] [Google Scholar]
- DeVincenzo JP. Natural infection of infants with respiratory syncytial virus subgroups A and B: A study of frequency, disease severity, and viral load. Pediatric Research. 2004;56(6):914–917. doi: 10.1203/01.PDR.0000145255.86117.6A. [DOI] [PubMed] [Google Scholar]
- Emmenegger EJ, Troyer RM, Kurath G. Characterization of the mutant spectra of a fish RNA virus within individual hosts during natural infections. Virus Res. 2003;96:15–25. doi: 10.1016/s0168-1702(03)00169-2. [DOI] [PubMed] [Google Scholar]
- Fellay J, Shianna KV, Ge DL, Colombo S, Ledergerber B, Weale M, Zhang KL, Gumbs C, Castagna A, Cossarizza A, Cozzi-Lepri A, De Luca A, Easterbrook P, Francioli P, Mallal S, Martinez-Picado J, Miro JM, Obel N, Smith JP, Wyniger J, Descombes P, Antonarakis SE, Letvin NL, McMichael AJ, Haynes BF, Telenti A, Goldstein DB. A whole-genome association study of major determinants for host control of HIV-1. Science. 2007;317(5840):944–947. doi: 10.1126/science.1143767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez AF, Esteller M. Viral epigenomes in human tumorigenesis. Oncogene. 2010;29(10):1405–1420. doi: 10.1038/onc.2009.517. [DOI] [PubMed] [Google Scholar]
- Fijan N, Sulimanovic E, Bearzotti M, Muzinic D, Zwillenberg LO, Chilmonczyk S, Vautherot JF, de Kinkelin P. Some properties of the epithelioma papulosum cyprini (EPC) cell line from carp Cyprinus carpi. Ann. Inst. Pasteur Vir. 1983;134(2):207–220. [Google Scholar]
- Fraser C, Hollingsworth TD, Chapman R, de Wolf F, Hanage WP. Variation in HIV-1 set-point viral load: Epidemiological analysis and an evolutionary hypothesis. PNAS. 2007;104(44):17441–17446. doi: 10.1073/pnas.0708559104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvani AP, May RM. Epidemiology - Dimensions of superspreading. Nature. 2005;438(7066):293–295. doi: 10.1038/438293a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garver K, Batts W, Kurath G. Virulence comparisons of infectious hematopoietic necrosis virus U and M genogroups in sockeye salmon and rainbow trout. J. Aquat. Anim. Health. 2006;18(4):232–243. doi: 10.1577/H05-038.1. [DOI] [PubMed] [Google Scholar]
- Gillet NA, Malani N, Melamed A, Gormley N, Carter R, Bentley D, Berry C, Bushman FD, Taylor GP, Bangham CRM. The host genomic environment of the provirus determines the abundance of HTLV-1-infected T-cell clones. Blood. 2011;117(11):3113–3122. doi: 10.1182/blood-2010-10-312926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman S, Harvey N, Moro D, Lloyd M, Voigt V, LM S, Lawson M, Shellam G. Mixed infection with multiple strains of murine cytomegalovirus occurs following simultaneous or sequential infection of immunocompetent mice. J. Gen. Virol. 2006;87:1123–1132. doi: 10.1099/vir.0.81583-0. [DOI] [PubMed] [Google Scholar]
- Grant EC, Philipp DP, Inendino KR, Goldberg TL. Effects of temperature on the susceptibility of largemouth bass to largemouth bass virus. J. Aquat. Anim. Health. 2003;15(3):215–220. [Google Scholar]
- Hallett SL, Bartholomew JL. Effects of water flow on the infection dynamics of Myxobolus cerebralis. Parasitology. 2008;135(3):371–384. doi: 10.1017/S0031182007003976. [DOI] [PubMed] [Google Scholar]
- Hirsch VM, Dapolito G, Johnson PR, Elkins WR, London WT, Montali RJ, Goldstein S, Brown C. Induction of AIDS by simian immunodeficiency virus from an African-Green Monkey-species specific variation in pathognecity correlates with the extent of in-vivo replication. J. Virol. 1995;69(2):955–967. doi: 10.1128/jvi.69.2.955-967.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday R. Epigenetics: A historical overview. Epigenetics. 2006;1(2):76–80. doi: 10.4161/epi.1.2.2762. [DOI] [PubMed] [Google Scholar]
- Howey R, Quan M, Savill NJ, Matthews L, Alexandersen S, Woolhouse M. Effect of the initial dose of foot-and-mouth disease virus on the early viral dynamics within pigs. J. Roy. Soc. Interface. 2009;6(39):835–847. doi: 10.1098/rsif.2008.0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HH, Shih WL, Li YH, Wu CF, Chen PJ, Lin CL, Liu CJ, Liaw YF, Lin SM, Lee SD, Yu MW. Hepatitis B viraemia: its heritability and association with common genetic variation in the interferon gamma signalling pathway. Gut. 2010;60(1):99–107. doi: 10.1136/gut.2010.207670. [DOI] [PubMed] [Google Scholar]
- Hurt AC, Nor'e SS, McCaw JM, Fryer HR, Mosse J, McLean AR, Barr IG. Assessing the viral fitness of oseltamivir-resistant influenza viruses in ferrets, using a competitive-mixtures model. J.Virol. 2010;84(18):9427–9438. doi: 10.1128/JVI.00373-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inendino KR, Grant EC, Philipp DP, Goldberg TL. Effects of factors related to water quality and population density on the sensitivity of juvenile largemouth bass to mortality induced by viral infection. J. Aquat. Anim. Health. 2005;17(4):304–314. [Google Scholar]
- Jorgensen SM, Afanasyev S, Krasnov A. Gene expression analyses in Atlantic salmon challenged with infectious salmon anemia virus reveal differences between individuals with early, intermediate and late mortality. BMC Genomics. 2008;9:16. doi: 10.1186/1471-2164-9-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaslow RA, Dorak T, Tang J. Influence of host genetic variation on susceptibility to HIV type 1 infection. J. Infect. Dis. 2005;191:S68–S77. doi: 10.1086/425269. [DOI] [PubMed] [Google Scholar]
- Kurath G, Garver KA, Troyer RM, Emmenegger EJ, Einer-Jensen K, Anderson ED. Phylogeography of infectious hematopoietic necrosis virus in North America. J. Gen. Virol. 2003;84(4):803–814. doi: 10.1099/vir.0.18771-0. [DOI] [PubMed] [Google Scholar]
- LaPatra SE. Factors affecting pathogenicity of infectious hematopoietic necrosis virus (IHNV) for salmonid fish. J. Aquat. Anim. Health. 1998;10(2):121–131. [Google Scholar]
- LaPatra SE, Lauda KA, Jones GR. Antigenic variants of infectious hematopoietic necrosis virus and implications for vaccine development. Dis. Aquat.Organ. 1994;20(2):119–126. [Google Scholar]
- Lloyd-Smith JO, Schreiber SJ, Kopp PE, Getz WM. Superspreading and the effect of individual variation on disease emergence. Nature. 2005;438(7066):355–359. doi: 10.1038/nature04153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massicotte R, Whitelaw E, Angers B. DNA methylation A source of random variation in natural populations. Epigenetics. 2011;6(4):421–427. doi: 10.4161/epi.6.4.14532. [DOI] [PubMed] [Google Scholar]
- Matthews L, Low JC, Gally DL, Pearce MC, Mellor DJ, Heesterbeek JAP, Chase-Topping M, Naylor SW, Shaw DJ, Reid SWJ, Gunn GJ, Woolhouse MEJ. Heterogeneous shedding of Escherichia coli O157 in cattle and its implications for control. PNAS. 2006;103(3):547–552. doi: 10.1073/pnas.0503776103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellors JW, Rinaldo CR, Gupta P, White RM, Todd JA, Kingsley LA. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science. 1996;272(5265):1167–1170. doi: 10.1126/science.272.5265.1167. [DOI] [PubMed] [Google Scholar]
- Mulcahy D, Burke J, Pascho R, Jenes CK. Pathogenesis of infectious hematopoietic necrosis virus in adult sokeye salmon (Oncorhynchus nerka) Can. J. Fish. Aquat. Sci. 1982;39(8):1144–1149. [Google Scholar]
- Munster VJ, de Wit E, van den Brand JMA, Herfst S, Schrauwen EJA, Bestebroer TM, van de Vijver D, Boucher CA, Koopmans M, Rimmelzwaan GF, Kuiken T, Osterhaus A, Fouchier RAM. Pathogenesis and transmission of swine-origin 2009 A(H1N1) influenza virus in ferrets. Science. 2009;325(5939):481–483. doi: 10.1126/science.1177127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overturf K, LaPatra S, Towner R, Campbell N, Narum S. Relationships between growth and disease resistance in rainbow trout, Oncorhynchus mykiss (Walbaum) J. Fish Dis. 2010;33(4):321–329. doi: 10.1111/j.1365-2761.2009.01124.x. [DOI] [PubMed] [Google Scholar]
- Pelak K, Goldstein DB, Walley NM, Fellay J, Ge D, Shianna KV, Gumbs C, Gao X, Maia JM, Cronin KD, Hussain SK, Carrington M, Michael NL, Weintrob AC. Host determinants of HIV-1 control in African Americans. J. Infect. Dis. 2010;201(8):1141–1149. doi: 10.1086/651382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peñaranda MMD, Purcell MK, Kurath G. Differential virulence mechanisms of infectious hematopoietic necrosis virus (IHNV) in rainbow trout (Oncorhynchus mykiss) include host entry and virus replication kinetics. J. Gen. Virol. 2009;90(9):2172–2182. doi: 10.1099/vir.0.012286-0. [DOI] [PubMed] [Google Scholar]
- Peñaranda MMD, Wargo AR, Kurath G. In vivo fitness correlates with host-specific virulence of Infectious hematopoietic necrosis virus (IHNV) in sockeye salmon and rainbow trout. Virology. 2011;417(2):312–319. doi: 10.1016/j.virol.2011.06.014. [DOI] [PubMed] [Google Scholar]
- Piatak M, Saag MS, Yang LC, Clark SJ, Kappes JC, Luk KC, Hahn BH, Shaw GM, Lifson JD. High-levels of HIV-1 plasma during all stages of infection determined by competitive PCR. Science. 1993;259(5102):1749–1754. doi: 10.1126/science.8096089. [DOI] [PubMed] [Google Scholar]
- Purcell M, Hart S, Kurath G, Winton J. Strand-specific, real-time RT-PCR assays for quantificaation of genomic and positive-sense RNAs of the fish rhabdovirus, infectious hematopoietic necrosis virus. J. Virol. Methods. 2006;132(1–2):18–24. doi: 10.1016/j.jviromet.2005.08.017. [DOI] [PubMed] [Google Scholar]
- Purcell MK, Garver KA, Conway C, Elliott DG, Kurath G. Infectious haematopoietic necrosis virus genogroup-specific virulence mechanisms in sockeye salmon, Oncorhynchus nerka (Walbaum), from Redfish Lake, Idaho. J. Fish Dis. 2009a;32(7):619–631. doi: 10.1111/j.1365-2761.2009.01045.x. [DOI] [PubMed] [Google Scholar]
- Purcell MK, Laing K, Winton JR. Immunity to fish Rhabdoviruses. Viruses. doi: 10.3390/v4010140. (In press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell MK, Laing K, Woodson J, Thorgaard G, Hansen J. Characterization of the interferon genes in homozygous rainbow trout reveals two novel genes, alternate splicing and differential regulation of duplicated genes. Fish Shellfish Immun. 2009b;26(2):293–304. doi: 10.1016/j.fsi.2008.11.012. [DOI] [PubMed] [Google Scholar]
- Purcell MK, Lapatra SE, Woodson JC, Kurath G, Winton JR. Early viral replication and induced or constitutive immunity in rainbow trout families with differential resistance to Infectious hematopoietic necrosis virus (IHNV) Fish Shellfish Immun. 2010;28(1):98–105. doi: 10.1016/j.fsi.2009.10.005. [DOI] [PubMed] [Google Scholar]
- Rando OJ, Verstrepen KJ. Timescales of genetic and epigenetic inheritance. Cell. 2007;128(4):655–668. doi: 10.1016/j.cell.2007.01.023. [DOI] [PubMed] [Google Scholar]
- Rodriguez M, LaPatra S, Williams S, Famula T, May B. Genetic markers associated with resistance to infectious hematopoietic necrosis in rainbow trout and steelhead trout (Oncorhynchus mykiss) backcrosses. Aquaculture. 2004;241(1–4):93–115. [Google Scholar]
- Silverstein JT, Rexroad CE, King TL. Genetic variation measured by microsatellites among three strains of domesticated rainbow trout (Oncorhynchus mykiss, Walbaum) Aquaculture Research. 2004;35(1):40–48. [Google Scholar]
- Ten Haaft P, Verstrepen B, Uberla K, Rosenwirth B, Heeney J. A pathogenic threshold of virus load defined in simian immunodeficiency virus- or simian-human immunodeficiency virus-infected macaques. J. Virol. 1998;72(12):10281–10285. doi: 10.1128/jvi.72.12.10281-10285.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troyer RM, Garver KA, Ranson JC, Wargo AR, Kurath G. In vivo virus growth competition assays demonstrate equal fitness of fish rhabdovirus strains that co-circulate in aquaculture. Virus Res. 2008;137(2):179–188. doi: 10.1016/j.virusres.2008.07.018. [DOI] [PubMed] [Google Scholar]
- Troyer RM, LaPatra SE, Kurath G. Genetic analyses reveal unusually high diversity of infectious haematopoietic necrosis virus in rainbow trout aquaculture. J. Gen. Virol. 2000;81(12):2823–2832. doi: 10.1099/0022-1317-81-12-2823. [DOI] [PubMed] [Google Scholar]
- Wargo AR, Garver KA, Kurath G. Virulence correlates with fitness in vivo for two M group genotypes of Infectious hematopoietic necrosis virus (IHNV) Virology. 2010;404(1):51–58. doi: 10.1016/j.virol.2010.04.023. [DOI] [PubMed] [Google Scholar]
- Wargo AR, Kurath G. In vivo fitness associated with high virulence in a vertebrate virus is a complex trait regulated by host entry, replication, and shedding. J. Virol. 2011;85(8):3959–3967. doi: 10.1128/JVI.01891-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolhouse MEJ, Dye C, Etard JF, Smith T, Charlwood JD, Garnett GP, Hagan P, Hii JLK, Ndhlovu PD, Quinnell RJ, Watts CH, Chandiwana SK, Anderson RM. Heterogeneities in the transmission of infectious agents: Implications for the design of control programs. PNAS. 1997;94(1):338–342. doi: 10.1073/pnas.94.1.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CF, Yu MW, Lin CL, Liu CJ, Shih WL, Tsai KS, Chen CJ. Long-term tracking of hepatitis B viral load and the relationship with risk for hepatocellular carcinoma in men. Carcinogenesis. 2008;29(1):106–112. doi: 10.1093/carcin/bgm252. [DOI] [PubMed] [Google Scholar]
- Young WP, Wheeler PA, Fields RD, Thorgaard GH. DNA fingerprinting confirms isogenicity of androgenetically derived rainbow trout lines. J. Hered. 1996;87(1):77–81. doi: 10.1093/oxfordjournals.jhered.a022960. [DOI] [PubMed] [Google Scholar]