Abstract
Marburg virus (MARV) and Ebola virus (EBOV) constitute the family Filoviridae of enveloped viruses (filoviruses) that cause severe hemorrhagic fever. Infection by MARV is required for fusion between the host cell and viral membranes, a process that is mediated by the two subunits of the envelope glycoprotein GP1 (surface subunit) and GP2 (transmembrane subunit). Upon viral attachment and uptake, it is believed that the MARV viral fusion machinery is triggered by host factors and environmental conditions found in the endosome. Next, conformational rearrangements in the GP2 ectodomain result in the formation of a highly stable six-helix bundle; this refolding event provides the energetic driving force for membrane fusion. Both GP1 and GP2 from EBOV have been extensively studied, but there is little information available for the MARV glycoproteins. Here we have expressed two variants of the MARV GP2 ectodomain in Escherichia coli and analyzed their biophysical properties. Circular dichroism indicates that the MARV GP2 ectodomain adopts an α-helical conformation, and one variant sediments as a trimer by equilibrium analytical ultracentrifugation. Denaturation studies indicate the α-helical structure is highly stable at pH 5.3 (unfolding energy, ΔGunf H2O, of 33.4 ± 2.5 kcal/mol and melting temperature, Tm, of 75.3 ± 2.1 °C for one variant). Furthermore, we found the α-helical stability to be strongly dependent on pH with higher stability under lower pH conditions (Tm values ranging from ~92 °C at pH 4.0 to ~38 °C at pH 8.0). Mutational analysis suggests two glutamic acid residues (E579 and E580) are partially responsible for this pH-dependent behavior. Based on these results, we hypothesize that pH-dependent folding stability of the MARV GP2 ectodomain provides a mechanism to control conformational preferences such that the six-helix bundle ‘post-fusion’ state is preferred under conditions of appropriately matured endosomes.
Marburg virus (MARV) and the related Ebola virus (EBOV) belong to the family Filoviridae of enveloped viruses that cause a rapidly progressing hemorrhagic fever with human case fatalities of 50-90% (1, 2). Similar to other enveloped viruses, infection by MARV requires coordinated conformational changes in the envelope glycoproteins that ultimately result in fusion between the viral and cellular membranes (3 – 6). The envelope glycoprotein spike of MARV and EBOV consists of three copies each of a surface subunit (GP1) and a transmembrane subunit (GP2) that anchors the spike to the viral membrane (3, 5, 7 – 11). Structural and biochemical work EBOV GP1/GP2 has demonstrated that, prior to membrane fusion, the viral particle must first be internalized into cellular endosomes or lysosomes where host cysteine proteases cathepsins L and B (CatL and CatB) remove all but a small (~17 kDa) portion of GP1 (7, 8, 12 – 14). Host factors are then proposed to trigger the fusion reaction, possibly by interaction with the remaining fragment of GP1. Recent reports have demonstrated that the endosomal cholesterol transporter Niemann Pick C1 (NPC-1) is critical for EBOV entry, and other unidentified factors may also be required (15, 16). The GP2 subunit contains two heptad repeat regions in the ectodomain (N- and C-terminal, NHR and CHR, respectively) that fold into a stable six-helix bundle. Therefore, the postulated EBOV fusion mechanism is similar to that of HIV-1 gp41and other members of the structurally defined ‘class I’ family of envelope glycoproteins (3 – 6, 9 – 11). In this model, the N-terminal portion of GP2, which contains a fusion loop, embeds in the host cell membrane following receptor binding to cleaved GP1. This conformational change in GP2 gives rise to a transient intermediate known as the ‘prehairpin’ or ‘extended’ intermediate in which the NHR and CHR are exposed to the extraviral environment. Next, folding of the GP2 six-helix bundle provides the driving force for membrane fusion by pulling the virus and host cell membranes into proximity. The events leading to MARV membrane fusion are presumably similar, since sequences of GP1 and GP2 are highly conserved among these two viruses (17, 18). However, few biochemical studies on the MARV glycoproteins have been reported to date.
The envelope glycoproteins of many viruses that enter the cell via the endosome contain structural elements that drastically affect stability of the pre-fusion conformation (the spike) or the post-fusion conformation in a pH-dependent manner (4 – 6, 19 – 26). In these cases, protonation of one or more side chains drives conformational preferences toward active fusogenic states at low pH (21 – 24). This feature provides a general mechanism for controlling membrane fusion such that it is triggered only when the virus is present in an appropriately matured endosome. For example, low pH-induced conformational changes in the envelope glycoprotein of influenza A virus (HA) results in the exposure of the fusion loop and ultimately its insertion into the host membrane (19, 20). However, the precise residues that are responsible for this transformation vary among strains (27). In the alphavirus Semliki Forest Virus (SFV), the prefusion spike consists of two glycoproteins, E1 and E2, which form a fusion-inactive heterodimer (22 – 24). Protonation of conserved histidine residues in both subunits destabilize the E1-E2 homodimer and promote formation of an E1 trimer, the active fusogenic form of the envelope glycoprotein. In EBOV entry, acidic pH is required to activate CatL and CatB, which have optimal catalytic activity at pH ~5 and whose proteolysis of GP1 is an essential step for infection (12, 13). We recently demonstrated that the folding stability of designed proteins modeled after the six-helix bundle of EBOV GP2 is dependent on pH, with higher stability at lower pH, suggesting that pH alters conformational preferences of the NHR and CHR regions (28). It has been recently reported that peptides corresponding to the fusion loop of EBOV GP2 undergo pH-dependent structural changes that result in increase fusogenic activity at low pH (29). Furthermore, the proteolytically primed form of the GP1/GP2 spike assembly can be activated to bind lipids by treatment with acidic pH or mild reductants (30). Together, these findings suggest a model for EBOV membrane fusion in which the acidification of endosomal compartments triggers the fusogenic machinery of the viral envelope spike.
Here we describe the expression, purification, and initial biophysical characterization of the GP2 ectodomain from MARV. We found that the MARV GP2 ectodomain forms an α-helical trimer, suggesting a membrane fusion mechanism similar to that of EBOV. Furthermore, we found the stability of the MARV α-helical bundle to be pH-sensitive, with much higher stability under acidic conditions. We performed site-directed mutagenesis to identify residues that are responsible for this behavior. Variants containing mutations to several acidic residues in the NHR core trimer and on the external CHR α-helices were characterized and found to have altered pH-dependent stability. These results suggest that membrane fusion in MARV may also be controlled by pH-dependent stability of the post-fusion conformation.
MATERIALS AND METHODS
Cloning, Expression, Purification, and Refolding of MARV GP2 Ectodomain
A synthetic DNA fragment encoding ‘MarVGP2-C’ (see Results) with an N-terminal hexahistidine tag was obtained from a commercial supplier (Genewiz, South Plainfield, NJ). This gene was cloned into pET28a (Novagen, Madison, WI) with NdeI and XhoI restriction sites to produce the expression plasmid pJH4. Site-directed mutagenesis was performed introduce a C557S mutation to yield the expression plasmid pJH5 (encoding ‘MarVGP2-S’). Expression, purification, and refolding of MarVGP2-C and MarVGP2-S were similar. E. coli BL21 (Invitrogen, Carlsbad, CA) cells harboring pJH4 or pJH5 were grown in LB broth at 37 °C to OD600 ~0.6, and protein expression induced by addition of 1 mM isopropyl β-D-1 thiogalactopyranoside (IPTG). The culture was incubated at 37 °C an additional 14-16 hrs. Cells were harvested by centrifugation, lysed by stirring in 6 M guanidine HCl (GdnHCl) for 3 hrs at room temperature. The cell debris was pelleted by ultracentrifugation and the supernatant was applied directly to Ni-NTA resin (Qiagen, Valencia, CA). The resin was washed with 10 column volumes of phosphate-buffered saline (PBS) containing 6 M GdnHCl and 50 mM imidazole, then the MarVGP2-C or MarVGP2-S protein eluted with PBS containing 6 M GdnHCl and 250 mM imidazole. The eluted protein was concentrated, and then further purified by reverse-phase HPLC on a Vydac (Hesperia, CA) C18 column (10 ΔM, 250 × 21.2 mm) with water/acetonitrile/trifluoroacetic acid mobile phases. Fractions containing pure protein were pooled, lyophilized, and redissolved in 6 M GdnHCl. The proteins were refolded by step-wise dialysis first into 100 mM glycine HCl pH 3.5, then into 10 mM sodium acetate pH 5.3. Protein concentrations were kept below 0.5 mg/mL during the refolding process. The precipitated material was removed by centrifugation and the refolded protein used immediately for analysis or flash frozen and stored at − 80 °C.
Circular Dichroism (CD) Spectroscopy
Measurements were performed on a Jasco J-815 spectrometer with a 1 cm quartz cuvette. Protein concentrations ranged from 1 – 4 μM as determined by absorbance at 280 nm. Circular dichroism wavelength scans were obtained with an 1 nm step size and a 2 sec averaging time. The signal was converted to mean molar ellipticity (θ) using the equation θ = 100 ε / (n l [P]tot), where ε is the raw CD signal in degrees, n is the number of amide chromophores, l is the pathlength, and [P]tot is the protein concentration (31).
Chemical and thermal denaturation was monitored at 222 nm (θ222nm). For chemical denaturation, protein samples were diluted into the appropriate analysis buffer containing varying amounts of GdnHCl, the solution allowed to equilibrate for 1 min, and then θ222nm determined. The data were plotted as a function of [GdnHCl], corrected for baseline in folded and unfolded states, then converted to fraction unfolded (Funf) using the equation [θ222nm – θU] / [[θF – θU], where θF and θU are the CD signals for the folded and unfolded states, respectively. The ΔGunf at each [GdnHCl] concentration was calculated using a monomer-trimer model (3 MU ↔ TF, where MU is the unfolded monomer and TF is the folded trimer): ΔGunf = RT ln {3 F 3 unf [P] 2 tot / (1 – Funf) } where R is the gas constant and T is the absolute temperature (32). The unfolding energy in water (ΔGunf, H2O) was estimated by linear extrapolation to [GdnHCl] = 0 using points in the transition region (33). Thermal denaturation data were obtained with a 3 – 5 °C step size and 2 min equilibration at each temperature. The θ222nm was plotted as a function of temperature, corrected for baseline, and converted to Funf. The data were fit to a standard four-parameter logistic equation and the melting temperature, Tm, obtained from the inflection point of the curve.
Equilibrium Analytical Ultracentrifugation (AU)
Analysis was performed on a Beckman XL-1 analytical ultracentrifuge with a Ti60 rotor. Samples of MarVGP2-S were loaded into 1.2 cm cells at a protein concentration of 20 μM, and storage buffer that did not contain protein was used in the reference cell. Equilibrium sedimentation experiments were carried out at rotor speeds of 12 and 19 kRPM. At each rotor speed, samples were allowed to equilibrate for 24 hrs and the radial spectra collected at 230 nm. Comparison of overlaid spectra indicated that there was no mass depletion. The data from both rotor speeds were fit globally to a single ideal species model using the program Heteroanalysis (Biotechnology/Bioservices Center, University of Connecticut, Storrs, CT). Non-linear regression was performed in accordance with the expression cr = co exp [M (1 – νρ) ω2 (r2 – r 2o) / 2RT] + base, where cr is the concentration (in absorbance units) at radial position r, co is the concentration at an arbitrary reference position ro near the meniscus, ν is the partial specific volume, ρ is the solvent density, ω is the rotor speed, R is the gas constant, T is the absolute temperature, and base is a baseline absorbance correction to account for non-sedimenting species (34). Molecular weight estimates were obtained from the parameter M. Fits were judged to be adequate if there was no systematic deviation of residuals. A partial specific volume of 0.7337 mL/g was calculated for MarVGP2-S based on amino acid composition, and a solvent density of 1.00182 g/mol was estimated using the program Sednterp (University of New Hampshire).
Structural Modeling and Site-Directed Mutagenesis
A structural homology model of the MARV GP2 ectodomain was generated using the SWISS-MODEL server (http://swissmodel.expasy.org/) with the alignment shown in Figure 1A and PDB ID 2EBO as a template (11). This structural model was used to identify positions for analysis by site-directed mutagenesis. Mutant MarVGP2-C or MarVGP2-S clones were prepared using the following protocol. Briefly, pJH4 or pJH5 DNA served as the template for oligonucleotide-based plasmid replication using the primers containing the desired substitutions. Following DNA synthesis, the template DNA was destroyed by digestion with DpnI for 3 hrs at 37 °C, and E. coli XL1-Blue (Stratagene, LaJolla, CA) transformed with the resulting mixture. Clones were screened by sequence analysis and those that contained the desired mutations were used for protein production. Preparation of mutant proteins was similar to the wild-type MarVGP2-C and MarVGP2-S (described above) and generally with comparable yields.
Figure 1.
(A) Sequence alignment of GP2 from Zaire ebolavirus (EBOV), ASLV, and MARV (residues that differ from EBOV shown colored gray). The amino acid numbering for EBOV GP2 and MARV GP2 differ by one; the EBOV numbering is shown above the sequence alignment and the MARV numbering shown below. The regions corresponding to the NHR, CHR, and helix-turn-helix regions of EBOV GP2 are indicated with cylinders whose colors match the structural elements depicted in panel B. Established disulfide bond connectivities are shown, and glycosylation sites are indicated with an ‘o’. The T565 (EBOV numbering) residue that gives rise to the unusual 3-4-4-3 hydrophobic stutter in the NHR is indicated with an ‘x’, other residues in the NHR that form core positions are denoted a and d. The C556 (EBOV numbering) residue in NHR is indicated with an asterix (below). (B) Crystal structures of the EBOV GP2 ectodomain reported by Wiley and coworkers (ref. 10, PDB ID 1EBO), and Kim and coworkers (ref. 11, PDB ID 2EBO). In the Wiley structure, the trimeric GCN4 tag is shown in cyan.
RESULTS
Predicted Six-Helix Bundle of the Marburg Virus GP2 Ectodomain
To define the MARV GP2 six-helix bundle, we compared its ectodomain sequence to that of EBOV (Zaire strain) GP2 and the structurally related avian sarcoma/leukosis virus (ASLV) Env ectodomains (Figure 1A) (9 – 11, 35, 36). The crystal structures for two variants of the EBOV GP2 ectodomain in the post-fusion state were described previously by Wiley and coworkers (PDB ID 1EBO) (10), and Kim and coworkers (PDB ID 2EBO) (Figure 1B) (11). The EBOV GP2 ectodomain adopts a six-helix bundle with a long, central trimeric coiled-coil core consisting of the NHR segment and three shorter CHR α-helices arranged in an antiparallel configuration about the periphery of the NHR core trimer. An intervening loop region contains a short helix-turn-helix segment that is stabilized by an intramolecular disulfide bond between C601 and C608. The Wiley EBOV GP2 structure contained a trimeric GCN4 segment N-terminal to the NHR to promote solubility and expression (9, 10). Based on the alignment with EBOV GP2 and analysis of these structures, we predicted that a fragment consisting of residues 553-633 of MARV GP2 would adopt a stable six-helix bundle with residues 553-596 forming the NHR core trimer, residues 597-615 forming the loop, and residues 616-633 forming CHR. (Note the amino acid numbering for EBOV GP2 and MARV GP2 differs by one; the numbering for both proteins is shown in Figure 1A). This fragment encompasses the majority of the ectodomain but lacks the fusion loop and the membrane-proximal external region.
Previous work with EBOV GP2 and ASLV Env established disulfide bond connectivity of the three cysteines located at positions 601, 608, and 609 (positions 602, 609, and 610 in MARV GP2) (9 – 11, 35, 36). The residues C601 and C608 in EBOV GP2 form the intramolecular linkage that stabilizes the helix-turn-helix region of the loop between the NHR and CHR, and C609 participates in an intermolecular bridge that tethers GP2 to the surface subunit (GP1) (8). However, both C608 and C609 can both form disulfide bonds with C601 in the isolated EBOV GP2 ectodomain (9 – 11). Therefore, we incorporated a C610S mutation (this position is predicted to participate in the disulfide bond with GP1) in our MARV GP2 design to prevent heterogeneous disulfide bond formation. The residue C556 is predicted to lie in the α-helical NHR region in the EBOV GP2 post-fusion conformation (10), but this residue participates in a disulfide bridge with C511 to stabilize an antiparallel β-sheet in the pre-fusion conformation (8). The segment between C511 and C556 (EBOV GP2 numbering) is well-conserved among filoviruses and is thought to correspond to the fusion loop (8). The residue C556 was replaced by aspartic acid in the Wiley EBOV GP2 construct and by alanine in the Kim EBOV GP2 construct (10, 11). We prepared two constructs of the MARV GP2 ectodomain: one in which the analogous wild-type C557 side chain was preserved (MarVGP2-C), and a second containing a C557S mutation (MarVGP2-S).
A ‘stutter’ in the periodicity of the NHR heptad repeat pattern was observed in the EBOV GP2 post-fusion conformation (9 – 11). The residue T565 points toward the core of the NHR trimer but occupies a position that, along with the surrounding core NHR residues, results in an unusual 3-4-4-3 hydrophobic repeat (the canonical heptad repeat contains 3-4-3-4 periodicity). This stutter causes a slight distortion of the EBOV NHR α-helix; the alignment in Figure 1A indicates that such a stutter also exists in MARV GP2 (T566) and is included in both MarVGP2-C and MarVGP2-S constructs.
Purification and Characterization of MarVGP2-C and MarVGP2-S
MarVGP2-C and MarVGP2-S proteins were isolated from E. coli and refolded by step-wise dialysis first into 100 mM glycine HCl pH 3.5, then into 10 mM sodium acetate buffer (pH 5.3). A portion of protein in each case precipitated during the refolding process; this aggregation could be controlled to some extent by maintaining low protein concentrations (< 0.5 mg/mL). Despite this aggregation, we recovered a reasonable yield of soluble, refolded material through this procedure (~0.1 mg/L of culture). The final refolding buffer did not contain reducing agent in order to allow disulfide bond formation between C602 and C609 (which stabilizes the helix-turn-helix segment). SDS-PAGE analysis under reducing and non-reducing conditions indicated a significant proportion of MarVGP2-C existed as disulfide-bonded oligomers, likely via crosslinks between the unpaired C557 residue on individual chains (see Supporting Information). In contrast, such crosslinked species were not abundantly observed in MarVGP2-S.
Circular dichroism indicates that both MarVGP2-C and MarVGP2-S are α-helical with double minima at 208 nm and 222 nm in 10 mM sodium acetate, pH 5.3 (Figure 2A). Analytical ultracentrifugation studies under similar conditions at 20 ΔM indicate that MarVGP-S sediments as a single ideal species with a molecular weight of 34,250 ± 280 Da (Figure 2B), consistent with a stable trimer (expected trimer molecular weight 33,260 Da). We therefore conclude that MarVGP2-S forms a six-helix bundle, similar to the corresponding regions of EBOV GP2 post-fusion conformation (9 – 11). To estimate the folding stability of MarVGP2-S, we performed chemical denaturation with GdnHCl (Figure 2C). This analysis yielded a ΔGunf, H2O of 33.4 ± 2.5 kcal/mol when fit to a monomer-trimer model at pH 5.3 (32). To determine whether disulfide bonding between the C602 and C609 contributed to this stability, we performed the GdnHCl denaturation in the presence of the reducing agent tris(hydroxypropyl)phosphine (THP, 2 mM) and found that the protein was markedly destabilized under reducing conditions. The denaturant midpoint, Cm, was 4.7 ± 0.2 M GdnHCl without THP, and 1.4 ± 0.1 with THP. This result indicates that the MARV GP2 C602-609 disulfide bond was oxidized during the refolding protocol and is consistent with observations that the EBOV GP2 ectodomain is stabilized by disulfide bridges between the corresponding C601 and C608 residues (11). In EBOV GP2, the established disulfide bond connectivity of C601-C608 was determined by stability studies with a C609A mutant (which would form an obligate C601-C608 disulfide) and a C608A mutant (obligate C601-C609 disulfide) (11). The C601-C608 disulfide variant displayed enhanced stability relative to the C601-C609 disulfide variant. We did not examine a C602-C610 disulfide variant of MarVGP2-S; however, the highly stable nature of the MarVGP2-S α-helical structure suggests this disulfide bonding pattern is also relevant in MARV GP2.
Figure 2.
Biophysical characterization of the MARV GP2 ectodomain. (A) CD spectra of MarVGP2-S in 10 mM sodium acetate, pH 5.3 and 10 mM sodium phosphate, pH 7.0 (B) Representative analytical ultracentrifugation at pH 5.3. High-resolution data were obtained at 12 and 19 krpm rotor speeds and fit to a single ideal species model to yield a molecular weight estimate consistent with a trimer (see text). (c) Guanidine HCl denaturation of MarVGP2-S in 10 mM NaOAc pH 5.3 in the absence and presence of the reducing agent THP, and in 20 mM NaH2PO4, pH 7.0. The denaturant midpoint (Cm) values are 4.7 ± 0.2 M GdnHCl at pH 5.3, 1.4 ± 0.1 M GdnHCl at pH 5.3 with THP, and 2.4 ± 0.1 M GdnHCl at pH 7.0. Linear extrapolation on the pH 5.3 data yielded ΔGunf,H2O = 33.4 ± 2.5 kcal/mol (inset).
pH-Dependent Stability
We examined the CD spectra of MarVGP2-S and MarVGP2-C under various buffering conditions and found that the ratio of peaks at 208 nm and 222 nm varied with pH for both proteins (wavelength scans at pH 5.3 and pH 7.0 are shown in Figure 2A). Strong 222 nm signals were observed at pH 5.3 and pH 7.0, indicating significant α-helical character under both of these conditions. However, the relative intensities of the 208 nm and 222 nm peaks varied with pH, suggesting differences in quaternary packing of α-helical segments (37).
To further explore pH-dependent structural effects, we performed thermal denaturation of MarVGP2-S and MarVGP2-C under buffering conditions ranging from pH 4.0 to pH 8.0 using the 222 nm CD signal to monitor folding (Figure 3 and Table 1). We found that the melting temperature (Tm), which provides an estimate of overall folding stability, was sensitive to pH in this range. In 10 mM sodium acetate, MarVGP2-S could not be completely unfolded at pH 4.0 and 4.7 (Tms estimated to be ~92 and ~88 °C, respectively, based on partial unfolding curves), consistent with a highly stable α-helical bundle. However, at pH 5.3 full denaturation was observed with a Tm of 75.3 ± 2.1 °C, indicating that the unfolding stability is much lower at this pH than at pH 4.0 and 4.7. At pH 6.1 (10 mM NaOAc buffer) and pH 7.0 (20 mM NaH2PO4 buffer), the Tm values were significantly lower (58.8 ± 1.1 °C and 51.8 ± 0.4 °C, respectively). Similar trends were observed for MarVGP2-C, with Tm values ranging from 89.0 ± 0.2 °C at pH 4.0 to ~38 °C at pH 8.0 (Figure 3B and Table 1). Figure 3C shows Tm as a function of pH for both MarVGP2-S and MarVGP2-C. Overall, the ectodomain structural stability is significantly higher at lower pHs, with a ~50 °C difference in Tm over the range of pH 4.0 to pH 8.0. The pH-dependent stability of MarVGP2-S was confirmed by GdnHCl denaturation (Figure 2C); the Cm was significantly higher at pH 5.3 (Cm = 4.7 ± 0.2 M GdnHCl) than at pH 7.0 (Cm = 2.4 ± 0.1 M GdnHCl).
Figure 3.
pH-Dependent thermal stability of the MARV GP2 α-helical bundle as monitored by [θ]222. Thermal denaturation curves of MarVGP2-S (A) and MarVGP2-C (B) under various buffering conditions. (C) Plot of Tm vs. pH.
Table 1.
Stability of MarVGP2-S and MarVGP2-C under Various Buffering Conditions.
| Buffering condition | Tm (°C)A |
|---|---|
| MarVGP2-S | |
| 10 mM NaOAc, pH 4.0 | ~92B |
| 10 mM NaOAc, pH 4.7 | ~88B |
| 10 mM NaOAc, pH 5.3 | 75.3 ± 2.1 |
| 10 mM NaOAc, pH 5.7 | 65.1 ± 1.0 |
| 10 mM NaOAc, pH 6.1 | 58.8 ± 1.1 |
| 20 mM NaHPO4, pH 7.0 | 51.8 ± 0.4 |
| MarVGP2-C | |
| 10 mM NaOAc, pH 4.0 | 89.0 ± 0.2 |
| 10 mM NaOAc, pH 4.8 | 86.9 ± 0.3 |
| 10 mM NaOAc, pH 5.3 | 72.4 ± 1.4 |
| 10 mM NaOAc, pH 6.1 | 47.0 ± 1.5 |
| 20 mM NaHPO4, pH 7.0 | 51.6 ± 1.3 |
| 20 mM NaHPO4, pH 8.0 | ~38C |
| 10 mM NaOAc, pH 5.3 + 500 mM NaCl | 78.2 ± 3.0 |
| 20 mM NaHPO4, pH 7.0 + 500 mM NaCl | 59.3 ± 1.0 |
Errors listed here represent 95% confidence interval from data fitting.
Complete unfolding was not observed at 100 °C, an estimate for Tm is provided based on fitting the partial unfolding curve.
A broad thermal transition prevented accurate determination of Tm.
In the GP2 ectodomains structures from EBOV, a conserved central asparagine (N586) in the NHR points toward the center of the core and binds monovalent anions (10, 11). This anion-binding pocket was also observed in the ectodomain of murine moloney leukemia virus (MoMLV), which is structurally similar to EBOV GP2 despite being from a phylogenetically unrelated virus (38). The sequence alignment suggests a similar pocket exists in MARV GP2 (N587). In EBOV GP2, the presence and nature of the monovalent ion were reported to have moderate effects on thermal stability (the reported Tms were 84 °C in the presence of 5 mM NaCl or NaBr, 79 °C in 5 mM NaF, and 77 °C without salt) (11). Although the data in Figure 3 and Table 1 were obtained in buffer that did not contain salt, and protein samples were exhaustively dialyzed during the refolding process, we cannot rule out the possibility that trace monovalent anions affected the Tm determinations under various buffering conditions. However, the observed Tm values in various buffering conditions spanned a range of ~50 °C, a much larger range than previously observed with salt effects in EBOV GP2 (range of ~8 oC) (11). At both pH 5.3 and pH 7.0, we observed an increase in Tm for MarVGP-C upon addition of 500 mM NaCl to the solution (ΔTm of 5.8 °C at pH 5.3 and 7.7 °C at pH 7.0, Figure 3C and Table 1). However, the stabilization was relatively minor in comparison to the change in stability among the various pH conditions. Furthermore, the Tm at pH 5.3 in the presence of 500 mM NaCl was 18.9 °C higher than that at pH 7.0 with an equivalent amount of salt (78.2 ± 3.0 °C vs. 59.3 ± 1.0 °C). Therefore, we conclude that the salt effects on stability are relatively minor in comparison to the effects of pH.
These results suggest that stability of the MARV GP2 six-helix bundle is sensitive to pH, possibly by protonation of side chain groups. The stabilization of the structure in high salt concentrations may also indicate that charge screening increases stability of the six-helix bundle, further implicating the interaction between ionic groups controls the pH-dependent stability. The pH range of the transition from highly stable (pH 4) to moderately stable (pH 7) occurs in a range that is consistent with pH-dependent stability mediated by acidic residues (Glu and Asp) or histidine residues in the influenza, SFV, and VSV glycoproteins (19 – 26). Similar phenomena were observed in designed protein mimics of the EBOV GP2 α-helical bundle; in this case, the pH-dependent stability was in part due to acidic residues that, when deprotonated, are predicted to disfavor the α-helical bundle formation (28). Mutational studies on the disulfide-containing loop of ASLV Env suggest that this region may play a role in pH-dependent conformational changes for that system (36). Furthermore, the recently reported structure of GP2 from an arenavirus suggest that many intrachain salt bridges stabilize the post-fusion conformation (39).
Structural Homology Model and Mutagenesis
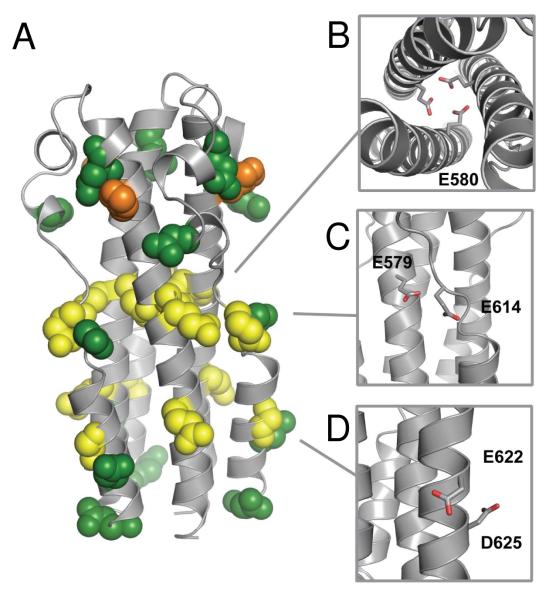
We generated a structural homology model of the MARV GP2 ectodomain based on the structure and alignment of the EBOV ectodomain in an attempt to determine which residues might be responsible for the pH-dependent stability (Figure 4). The MARV GP2 ectodomain contains six total glutamic acid residues, five aspartic acid residues, and a single histidine residue (these ionizable side chains are shown in spacefill in Figure 4A). We identified three putative side chain-side chain interactions that could potentially contribute to pH-dependent behavior (Figures 4B-4D). The residue E580 in MARV corresponds to a heptad repeat core residue L579 in HR1 of EBOV; the MARV GP2 structural homology model predicts the side chain of this glutamic acid residue is oriented toward the core of the trimer (Figure 4B) (10, 11). We hypothesized that this residue may be responsible for the pH-dependent stability since, if deprotonated, juxtaposition of E580 anionic side chains from opposing monomers to form the central NHR coiled-coil would be unfavorable. Similar interactions between acidic residues are observed at the low pH-dependent trimer interface of VSV G (26). Variants of the homodimeric coiled-coil GCN4 containing an aspartic acid residue at a core a position exhibit similar pH-dependent effects (40). At the surface of the model, the side chain of E579 on the NHR (E578 in EBOV) is predicted to be near E614 in the loop region (H613 in EBOV) (Figure 4C). Contacts between acidic side chains are also observed in the low pH coiled-coils of VSV G and influenza HA (20, 26) Furthermore, E622 (D621 or N621 in EBOV, depending on strain) and D625 (D624 in EBOV) on the short CHR segment have the capacity to form i→i+3 intrahelical interactions (Figure 4D). We previously reported that similar side chain-side chain anionic residues accounted for the pH-dependent stability of the EBOV GP2 six-helix bundle (28). While surface ionic interactions in coiled-coils generally have modest effects, it has been shown in several systems that additive effects of multiple interactions can play a large role in coiled-coil interactions (41 – 45).
Figure 4.
(A) Structural homology model of MARV GP2 ectodomain based on the crystal structure of EBOV GP2 (ref. 11, PDB ID 2EBO). Glutamic acid (yellow), aspartic acid (green), and histidine (orange) residues are shown in spacefill. (B-C) Predicted interactions that could destabilize the six-helix bundle, shown in stick and colored by atom for clarity: (B) E580 interactions in the NHR core trimer; (C) E579-E614 interaction between opposing monomers; (D) E622-D625 i→i+3 intrahelical interaction.
We generated point mutants of MarVGP2-S and MarVGP2-C at positions 579 and 580, and examined their thermal stability to explore the role of these positions in the pH-dependent stability. The results from these studies are summarized in Table 2. A variant of MarVGP2-C that contained a E579Q mutation at the position predicted to face inward in the core NHR trimer (MarVGP2-CpEQ) was highly unstable at pH 7.0 (Tm ~23 °C); and a variant containing a E578Q mutation (MarVGP2-CpQE) did not exhibit strong α-helical signal under these conditions (this position is predicted to interact with E614). These results indicate that neither the E579Q nor the E580Q mutations alone were sufficient to increase the stability at pH 7.0. However, a double mutant containing Glu→Gln mutations at both positions (MarVGP2-CpQQ) had an unusual thermal denaturation profile with at least two transition phases at pH 7.0 (Figure 5). Fitting to a multiparameter logistic equation provided two transition temperatures (a minor transition, T1, and a major transition, T2) of 27.6 ± 0.5 °C and 74.0 ± 0.2 °C respectively. The structural basis of this biphasic thermal unfolding is not clear; in the cases of OmpC and tropomyosin, similar behavior has been interpreted to indicate non-cooperative unfolding with each transition corresponding to unfolding of a discrete structural element (46, 47). The first transition in MarVGP2-CpQQ corresponds to approximately 30% of the CD signal, which is equal to the contribution of the CHR segments to total α-helical content of the EBOV GP2 post-fusion structure (10, 11). It is therefore possible that the minor transition corresponds to unfolding of the peripheral CHR segments, and the major transition corresponds to unfolding of the NHR core trimer. At pH 4.0, the MarVGP2-CpQQ clone underwent a single cooperative transition with Tm of 84.1 ± 0.9 °C. Similar behavior was observed with a double mutant (E579Q/E580Q) of MarVGP2-S (MarVGP2-SpQQ), though the transition temperatures were somewhat different. These results are consistent with a model in which the E579Q and E580Q mutations serve to stabilize the core NHR trimer at pH 7.0, but have little or no stabilizing effect on the CHR segments.
Table 2.
Summary of Results from Analysis of MarVGP2-S and MarVGP2-C Mutants
| Variant | Mutation(s) | Tm(pH ~4.7) (°C)A | Tm(pH 7.0) (°C)A | ΔTm (°C) |
|---|---|---|---|---|
| MarVGP2-C (WT) | N/A | 86.9 ± 0.3 | 51.6 ± 1.3 | 35.3 |
| MarVGP2-CpQE | E579Q | ND | (not stable) | -- |
| MarVGP2-CpEQ | E580Q | ND | ~ 23 °C | -- |
| MarVGP2-CpQQ | E579Q/E580Q | 84.1 ± 0.9 | T1, 27.6 ± 0.5 T2, 74.0 ± 0.2 |
-- |
|
| ||||
| MarVGP2-S (WT) | N/A | ~88 | 51.8 ± 0.4 | ~36 |
| MarVGP2-SpQQ | E579Q/E580Q | 81.5 ± 3.5 | T1, 32.2 ± 3.8 T2, 69.9 ± 0.6 |
-- |
| MarVGP2-SpKE | E579K | 76.9 ± 3.2 | ~ 39B | ~38 |
| MarVGP2-SpEK | E580K | 54.8 ± 1.1 | ~ 20B | ~35 |
| MarVGP2-SppQN | E622Q/D625N | 83.5 ± 0.4 | 41.6 ± 2.1 | 41.9 |
Errors listed here represent 95% confidence interval from data fitting.
N/A, not applicable; ND, not determined.
A broad thermal transition prevented accurate determination of Tm.
Figure 5.
Denaturation profiles of MarVGP2-CpQQ and MarVGP2-SpQQ at pH 4.7 and pH 7.0.
To further examine the role of residues E579 and E580, we prepared two additional mutants of MarVGP-S: one with an E579K mutation and another with an E580K mutation (MarVGP2-SpEK and MarVGP2-SpKE, respectively). We expected that incorporation of a lysine residue at position 580, which should maintain a positive charge across a broad pH range, should be more destabilizing to global folding stability than an E579K mutation. Indeed, MarVGP2-SpEK was less stable than both MarVGP2-SpKE (ΔTm = 21.5 °C) and MarVGP2-S (ΔTm = 31.0 °C) at pH 4.7. However, both mutants were also less stable than MarVGP2-S at pH 7.0. Together, these results suggest that pH-dependent stability of MARV GP2 six-helix bundle is mediated to some degree by E579 and E580.
A variant of MarVGP2-S containing the two mutations E622Q and D625N on the CHR segment (MarVGP2-SppQN) was also examined (Table 2). This variant had similar pH-dependent stability to the MarVGP2-S, but was less stable at neutral pH. These results suggest that alteration of these two acidic side chains to their neutral analog has little effect o the overall pH-dependent stability. Similar surface-exposed side chain-side chain residues were postulated to be important for pH-dependent stability of α-helical bundle proteins designed from the EBOV GP2 NHR and CHR segments (28). Therefore, we conclude such interactions play less of a role in the pH-dependent stability MarVGP2-S.
DISCUSSION
Characterization of the MARV GP2 Six-Helix Bundle
Here we describe the first characterization of the MARV GP2 ectodomain; we find this segment adopts a stable trimeric α-helical bundle, consistent with the known structures of the phylogenetically related EBOV GP2 and structurally related MoMLV TM ectodomains (9 – 11, 38). Chemical denaturation indicates that the MARV GP2 six-helix bundle has a folding stability of 33.5 ± 2.5 kcal/mol, which is comparable to the stability of other six-helix bundles involved in viral membrane fusion (48, 49). In the model for membrane fusion involving ‘class I’ envelope glycoproteins (i.e., those with α-helical ectodomains), folding of a six-helix bundle by the NHR and CHR segments provides the energetic driving force to overcome the barriers associated with fusing the host and viral membranes (4 – 6). Here we show that the relatively short α-helical bundle region of MARV GP2 provides a significant amount of folding energy for this task. We further found that the disulfide bond between C602 and C609 is required for the overall folding stability and therefore likely critical for function.
pH-Dependent Stability: Implications for Membrane Fusion
The folding stability of the MARV GP2 ectodomain six-helix bundle, as estimated by Tm, was greatly dependent on pH, with higher stability at lower pH. We previously reported similar pH-dependent stability of α-helical bundle proteins designed from the EBOV GP2 NHR and CHR segments (28). The sensitivity of the six-helix bundle folding to environmental pH may play a role in the process of membrane fusion. It is thought that the post-fusion α-helical bundle is the lowest energy (ground) state conformation. In the pre-fusion state, the ectodomain is prevented from adopting the α-helical bundle by interactions with a surface subunit (e.g., gp120 in HIV-1 or HA in influenza A virus) (3 – 6, 21, 50). A triggering event releases the constraints on the ectodomain when the viral particle is in a suitable environment for membrane fusion to result in productive viral infection (this model is known as the ‘spring-loaded clamp’ model). The precise molecular events that lead to triggering in HIV-1 gp41 and influenza HA have been established; however the details of the EBOV fusion pathway are still under investigation. Furthermore, little is known about MARV entry – it is likely the molecular events are similar to EBOV given the similarity of their envelope glycoprotein sequences, but biological studies indicate there may be some differences in the entry mechanisms (17, 18). For EBOV, it has recently been shown that NPC1 is critical for entry, and this protein may play a role in deployment of the GP2 fusion machinery (15, 16).
We and others have suggested that an additional trigger may be endosomal pH, which serves to increase the stability of the post-fusion six-helix bundle (28, 36). Furthermore, CatL and CatB have optimal activity at pH ~ 5 and recent studies also suggest that the fusion loop may itself have some fusogenic properties that are enhanced in lower pH environments (29, 30). Together, these results suggest that multiple factors involving endosomal resident proteins (NPC1, CatL, and CatB) and endosomal conditions (low pH) work in concert to release constraints on the GP1/GP2 pre-fusion structure, promote formation of the GP2 post-fusion structure, and activate the fusion loop. However, unlike the pH-sensitive transformations observed in influenza HA and SFV E1/E2, in which protonation of specific residues serves to destabilize the pre-fusion conformation (19 – 24), it appears specific protonation events increase the stability of the post-fusion conformation. The results shown here provide the first evidence that a similar mechanism for pH-dependent stabilization of the post-fusion conformation exists in MARV. Furthermore, mutational analysis suggests the pH-dependent stability is controlled to some degree by the acidic residues E579 and E580; the juxtaposition of these anionic residues near other negatively charged residues at neutral pH is predicted to destabilize the post-fusion conformation based on the homology model. An accurate description of the side chain-side chain interactions that stabilize and destabilize the post-fusion conformation awaits a high-resolution crystal structure of the MARV GP2 ectodomain.
Models for Thermodynamic and Kinetic Control of pH-Dependent Conformational Changes
The dramatic structural rearrangement in the central stalk of influenza HA provides a model to distinguish mechanisms of pH-dependent conformational changes (19 – 21, 50, 51). A 36-residue segment, which contains a heptad repeat pattern and is predicted to form a trimeric coiled-coil, is essentially unstructured and monomeric at neutral pH but adopts a highly stable α-helical bundle at low pH (21). Baker and Agard proposed two models to describe how environmental pH could have such a dramatic consequence on the overall fold (Figure 6) (51). In the ‘thermodynamic control’ model, the pH affects the relative stabilities of the lowest energy conformation – at pH 7, the pre-fusion structure represents the ground state. However, at lower pH, the ‘fusion active’ conformation becomes preferred. A defining feature of this model is that the transition from pre-fusion to fusion active conformations is reversible, as would be the effects of pH on the equilibrium. In the ‘kinetic control’ model, the fusion active conformation is the lowest energy structure but a high kinetic barrier maintains the pre-fusion conformation. Exposure to low pH decreases the kinetic barrier, facilitating transition to the lowest energy (fusion active) conformation. The conformational rearrangements in the kinetic model would be irreversible. Early experiments with HA indicate that exposure to low pH does in fact trigger the pH-dependent conformation on the virus; however, this process is irreversible which has led to the conclusion that HA obeys the kinetic control model (3 – 6, 19 – 21). Furthermore, the fusion active conformation is adopted spontaneously by synthetic peptides corresponding to the core 36-residue segment at low pH (21), and the fusogenic subunit of HA (HA2) can be refolded into the post-fusion structure at neutral pH suggesting that it is the lowest energy conformation (52). These observations suggest the post-fusion is the lowest energy conformer across all pH conditions.
Figure 6.
Thermodynamic control and kinetic control models for pH-dependent conformational rearrangements.
Other viral membrane fusion proteins, however, appear to exhibit features that are reminiscient of the thermodynamic control model. For example, the envelope glycoprotein G from vesicular stomatitis virus undergoes a pH-dependent transition that appears to be reversible (25, 26). Structural studies suggest that protonation of His residues that are clustered together in the pre-fusion conformation, and of Asp residues that are clustered together in the post-fusion conformation shift the equilibrium toward the post-fusion conformation (25, 26). The core structural feature of the VSV G low pH structure is a six-helix bundle (similar to the class I membrane fusion proteins) and contains a number of acidic residues in proximity. In EBOV, exposure to low pH and mild reductants can trigger the fusogenic properties of the ectodomain on liposomes (30). However, it is unclear if these in vitro triggers fully recapitulate the in vivo fusion cascade, and it is currently unknown whether such treatments are reversible. We have shown that α-helical bundle proteins corresponding to the ectodomains of EBOV and MARV GP2 have pH-dependent stability (28), but these studies have been performed on the isolated proteins alone and it has not been demonstrated that this behavior is relevant to fusion on a viral surface. Therefore, further experiments on the viral surface are required to establish a model for control of the filovirus GP2 conformational changes associated with membrane fusion.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Michael Brenowitz for assistance with the analytical ultracentrifugation experiments.
ABBREVIATIONS
- MARV
Marburg virus
- EBOV
Ebola virus
- CatL
Cathepsin L
- CatB
Cathepsin B
- NPC-1
Neimann-Pick C1
- NHR
N-heptad repeat
- CHR
C-heptad repeat
- SFV
Semliki Forest Virus
- IPTG
isopropyl-β-D-thiogalactopyranoside
- GdnHCl
guanidine HCl
- CD
circular dichroism
- AU
analytical ultracentrifugation
- ASLV
avian sarcoma leukosis virus
- THP
tris(hydroxypropyl)phosphine
- MoMLV
murine moloney leukemia virus
Footnotes
SUPPORTING INFORMATION AVAILABLE SDS-PAGE analysis of purified MarVGP2-S and MarVGP2-C. This material is available free of charge via the Internet at http://pubs.acs.org.
This work was funded by the Albert Einstein College of Medicine, and the National Institutes of Health (R01-AI088027 to K. C. and R01-AI090249 to J. R. L.). J. S. H. was supported in part by NIH Molecular Biophysics Training Grant T32-GM008572, and J. F. K. by NIH Medical Scientist Training Grant T32-GM007288.
Note: The authors declare no competing financial interest.
REFERENCES
- 1.Feldmann H, Geisbert TW. Ebola haemorrhagic fever. The Lancet. 2011;377:849–862. doi: 10.1016/S0140-6736(10)60667-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuhn JH, Becker S, Ebihara H, Geisbert TW, Johnson KM, Kawaoka Y, Lipkin WI, Negredo AI, Netesov SV, Nichol ST, Palacios G, Peters CJ, Tenorio A, Volchkov VE, Jahrling PB. Proposal for a revised taxonomy of the family Filoviridae: classification, names of taxa and viruses, and virus abbreviations. Arch. Virol. 2010;155:2083–2103. doi: 10.1007/s00705-010-0814-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee JE, Saphire E,O. Ebolavirus glycoprotein structure and mechanism of entry. Future Virol. 2009;4:621–635. doi: 10.2217/fvl.09.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harrison SC. Viral membrane fusion. Nat. Struct. Mol. Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.White JM, Delos SE, Brecher M, Schornberg K. Structures and mechanisms of viral membrane fusion proteins: Multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 2008;43:189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eckert DM, Kim PS. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 2001;70:777–810. doi: 10.1146/annurev.biochem.70.1.777. [DOI] [PubMed] [Google Scholar]
- 7.Lee JE, Fusco ML, Hessell AJ, Oswald WB, Burton DR, Saphire EO. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature. 2008;454:177–182. doi: 10.1038/nature07082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dias JM, Kuehne AI, Abelson DM, Bale S, Wong AC, Halfmann P, Muhammad MA, Fusco ML, Zak SE, Kang E, Kawaoka Y, Chandran K, Dye JM, Saphire EO. A shared structural solution for neutralizing ebolaviruses. Nat. Struct. Mol. Biol. 2011;18:1424–1427. doi: 10.1038/nsmb.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weissenhorn W, Calder LJ, Wharton SA, Skehel JJ, Wiley DC. The central structural feature of the membrane fusion protein subunit from the Ebola virus glycoprotein is a long triple-stranded coiled coil. Proc. Natl. Acad. Sci. USA. 1998;95:6032–6036. doi: 10.1073/pnas.95.11.6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weissenhorn W, Carfí A, Lee KH, Skehel JJ, Wiley DC. Crystal structure of the Ebola virus membrane fusion subunit, GP2, from the envelope glycoprotein ectodomain. Mol. Cell. 1998;2:605–616. doi: 10.1016/s1097-2765(00)80159-8. [DOI] [PubMed] [Google Scholar]
- 11.Malashkevich VN, Schneider BJ, McNally ML, Milhollen MA, Pang JX, Kim PS. Core structure of the envelope glycoprotein GP2 from Ebola virus at 1.9-A resolution. Proc. Natl. Acad. Sci. USA. 1999;96:2662–2667. doi: 10.1073/pnas.96.6.2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chandran K, Sullivan NJ, Felbor U, Whelan SP, Cunningham JM. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science. 2005;308:1643–1645. doi: 10.1126/science.1110656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schornberg K, Matsuyama S, Kabsch K, Delos S, Bouton A, White J. Role of endosomal cathepsins in entry mediated by the Ebola virus glycoprotein. J. Virol. 2006;80:4174–4178. doi: 10.1128/JVI.80.8.4174-4178.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dube D, Brecher MB, Delos SE, Rose SC, Park EW, Schornberg KL, Kuhn JH, White JM. The primed ebolavirus glycoprotein (19-kilodalton GP1,2): sequence and residues critical for host cell binding. J. Virol. 2009;83:2883–2891. doi: 10.1128/JVI.01956-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carette JE, Raaben M, Wong AC, Herbert AS, Obernosterer G, Mulherkar N, Kuehne AI, Kranzusch PJ, Griffin AM, Ruthel G, Dal Cin P, Dye JM, Whelan SP, Chandran K, Brummelkamp TR. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature. 2011;477:340–343. doi: 10.1038/nature10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Côté M, Misasi J, Ren T, Bruchez A, Lee K, Filone CM, Hensley L, Li Q, Ory D, Chandran K, Cunningham J. Small molecule inhibitors reveal Niemann-Pick C1 is essential for Ebola virus infection. Nature. 2011;477:344–348. doi: 10.1038/nature10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Battacharyya S, Hope TJ, Young JA. Differential requirements for clathrin endocytic pathway components in cellular entry by Ebola and Marburg glycoprotein pseudovirions. Virology. 2011;419:1–9. doi: 10.1016/j.virol.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsuno K, Kishida N, Usami K, Igarashi M, Yoshida R, Nakayama E, Shimojima M, Feldmann H, Irimura T, Kawaoka Y, Takada A. Different potential of C-type lectin-mediated entry between Marburg virus strains. J. Virol. 2010;84:5140–5147. doi: 10.1128/JVI.02021-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bullough PA, Hughson FM, Skehel JJ, Wiley DC. Structure of the influenza haemaglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 20.Wilson IA, Skehel JJ, Wiley DC. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3Å resolution. Nature. 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 21.Carr CM, Kim PS. A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell. 1993;73:823–832. doi: 10.1016/0092-8674(93)90260-w. [DOI] [PubMed] [Google Scholar]
- 22.Sánchez-San Martín C, Liu CY, Kielian M. Dealing with low pH: entry and exit of alphaviruses and flaviviruses. Trends Microbiol. 2009;17:514–521. doi: 10.1016/j.tim.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fritz R, Stiasny K, Heinz FX. Identification of specific histidines as pH sensors in flavivirus membrane fusion. J. Cell Biol. 2008;183:353–361. doi: 10.1083/jcb.200806081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qin ZL, Zheng Y, Kielian M. Role of conserved histidine residues in the low pH dependence of the Semliki Forest virus fusion protein. J. Virol. 2009;83:4670–4677. doi: 10.1128/JVI.02646-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roche S, Rey FA, Gaudin Y, Bressanelli S. Structure of the prefusion form of the vesicular stomatitis virus glycoprotein G. Science. 2007;315:843–848. doi: 10.1126/science.1135710. [DOI] [PubMed] [Google Scholar]
- 26.Roche S, Bressanelli S, Rey FA, Gaudin Y. Crystal structure of the low-pH form of the vesicular stomatitis virus glycoprotein G. Science. 2006;313:187–191. doi: 10.1126/science.1127683. [DOI] [PubMed] [Google Scholar]
- 27.Daniels RS, Downie JC, Hay AJ, Knossow M, Skehel JJ, Wang ML, Wiley DC. Fusion mutants of the influenza virus hemagglutinin glycoprotein. Cell. 1985;40:431–439. doi: 10.1016/0092-8674(85)90157-6. [DOI] [PubMed] [Google Scholar]
- 28.Harrison JS, Higgins CD, Chandran K, Lai JR. Designed protein mimics of the Ebola virus glycoprotein GP2 α-helical bundle: Stability and pH effects. Protein Sci. 2011;20:1587–1596. doi: 10.1002/pro.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gregory SM, Harada E, Liang B, Delos SE, White JM, Tamm LK. Structure and function of the complete internal fusion loop from Ebolavirus glycoprotein 2. Proc. Natl. Acad. Sci. USA. 2011;108:11211–11216. doi: 10.1073/pnas.1104760108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brecher M, Schornberg KL, Delos SE, Fusco ML, Saphire EO, White JM. Cathepsin cleavage potentiates the Ebola virus glycoprotein to undergo a subsequent fusion-relevant conformational change. J. Virol. 2012;86:364–372. doi: 10.1128/JVI.05708-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cantor CR, Schimmel PR. Biophysical Chemistry, Part II: Techniques for the study of biological structure and function. W. H. Freeman and Company; New York, NY: 1998. pp. 426–428. [Google Scholar]
- 32.Boice JA, Dieckmann GR, Degrado WF, Fairman R. Thermodynamic analysis of a designed three-stranded coiled coil. Biochemistry. 1996;35:14480–14485. doi: 10.1021/bi961831d. [DOI] [PubMed] [Google Scholar]
- 33.Pace CN. Determination and analysis of urea and guanidine hydrochloride denaturation curves. Methods Enzymol. 1986;131:266–280. doi: 10.1016/0076-6879(86)31045-0. [DOI] [PubMed] [Google Scholar]
- 34.Schuster TM, Laue TM, editors. Modern analytical ultracentrifugation. Birkhauser; Boston, MA: 1994. pp. 3–15. [Google Scholar]
- 35.Gallaher WR. Similar structural models of the transmembrane proteins of Ebola and Avian Sarcoma viruses. Cell. 1996;85:477–478. doi: 10.1016/s0092-8674(00)81248-9. [DOI] [PubMed] [Google Scholar]
- 36.Delos SE, La B, Gilmartin A, White JM. Studies of the "chain reversal regions" of the avian sarcoma/leukosis virus (ASLV) and ebolavirus fusion proteins: analogous residues are important, and a His residue unique to EnvA affects the pH dependence of ASLV entry. J. Virol. 2010;84:5687–5694. doi: 10.1128/JVI.02583-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lau SY, Taneja AK, Hodges RS. Synthesis of a model protein of defined secondary and quaternary structure: Effect of chain length on the stabilization and formation of two-stranded α-helical coiled-coils. J. Biol. Chem. 1984;259:13253–13261. [PubMed] [Google Scholar]
- 38.Fass D, Harrison SC, Kim PS. Retrovirus envelope domain at 1.7 angstrom resolution. Nat. Struct. Biol. 1996;3:465–469. doi: 10.1038/nsb0596-465. [DOI] [PubMed] [Google Scholar]
- 39.Igonet S, Vaney MC, Vonhrein C, Bricogne G, Stura EA, Hengartner H, Eschli B, Rey FA. X-ray structure of the arenavirus glycoprotein GP2 in its postfusion hairpin conformation. Proc. Natl. Acad. Sci. USA. 2011;108:19967–19972. doi: 10.1073/pnas.1108910108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lau WL, Degrado WF, Roder H. The effects of pKa tuning on the thermodynamics and kinetics of folding: design of a solvent-shielded carboxylate pair at the a-position of a coiled-coil. Biophys. J. 2010;99:2299–2308. doi: 10.1016/j.bpj.2010.07.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kohn WD, Cyril MK, Hodges RS. Protein destabilization by electrostatic repulsions in the two-stranded α-helical coiled-coil/leucine zipper. Protein Sci. 1995;4:237–250. doi: 10.1002/pro.5560040210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scholtz JM, Qian H, Robbins VH, Baldwin RL. The energetics of ion-pair and hydrogen-bonding interactions in a helical peptide. Biochemistry. 1993;32:9668–9676. doi: 10.1021/bi00088a019. [DOI] [PubMed] [Google Scholar]
- 43.O’Shea EK, Lumb KJ, Kim PS. Peptide ‘Velcro’: design of a heterodimeric coiled-coil. Curr, Biol. 1993;3:658–667. doi: 10.1016/0960-9822(93)90063-t. [DOI] [PubMed] [Google Scholar]
- 44.O’Shea EK, Rutkowski R, Kim PS. Mechanism of specificity in the Fos-Jun oncoprotein heterodimer. Cell. 1992;68:699–708. doi: 10.1016/0092-8674(92)90145-3. [DOI] [PubMed] [Google Scholar]
- 45.Lumb KJ, Kim PS. Measurement of interhelical electrostatic interactions in the GCN4 leucine zipper. Science. 1995;268:436–439. doi: 10.1126/science.7716550. [DOI] [PubMed] [Google Scholar]
- 46.Keegan N, Ridley H, Lakey JH. Discovery of biphasic thermal unfolding of OmpC with implications for surface loop stability. Biochemistry. 2010;49:9715–9721. doi: 10.1021/bi100877y. [DOI] [PubMed] [Google Scholar]
- 47.Heller MJ, Nili M, Homsher E, Tobacman LS. Cardiomyopathic tropomyosin mutations that increase thin filament Ca2+ sensitivity and tropomyosin N-domain flexibility. J. Biol. Chem. 2003;278:41742–41748. doi: 10.1074/jbc.M303408200. [DOI] [PubMed] [Google Scholar]
- 48.Jelesarov I, Lu M. Thermodynamics of trimer-of-hairpins formation by the SIV gp41 envelope glycoprotein. J. Mol. Biol. 2001;307:637–656. doi: 10.1006/jmbi.2001.4469. [DOI] [PubMed] [Google Scholar]
- 49.Marti DN, Bjelic S, Lu M, Bosshard HR, Jelesarov I. Fast folding of the HIV-1 and SIV gp41 six-helix bundles. J. Mol. Biol. 2004;336:1–8. doi: 10.1016/j.jmb.2003.11.058. [DOI] [PubMed] [Google Scholar]
- 50.Carr CM, Chaudry C, Kim PS. Influenza hemagglutinin is spring-loaded by a metastable native conformation. Proc. Natl. Acad. Sci. USA. 1997;94:14306–14313. doi: 10.1073/pnas.94.26.14306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baker D, Agard DA. Influenza hemagglutinin: Kinetic control of protein function. Structure. 1994;2:907–910. doi: 10.1016/s0969-2126(94)00091-3. [DOI] [PubMed] [Google Scholar]
- 52.Swalley SE, Baker BM, Calder LJ, Harrison SC, Skehel JJ, Wiley DC. Full-length influenza hemagglutinin HA2 refulds into the trimeric low-pH-induced conformation. Biochemistry. 2004;43:5902–5911. doi: 10.1021/bi049807k. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.