1 Introduction
Diffusion tensor imaging (DTI)1 has emerged as a unique MRI-based probe of brain microstructure able to sensitively distinguish heterogeneous tissue types and detect structural abnormalities in human populations and experimental disease models. DTI maps are constructed from estimation of the diffusion tensor (DT), which describes the 3-dimensional self-diffusion of water, at every MRI voxel (Basser et al., 1994). The eigenvalues (λ1, λ2 and λ3) and eigenvectors (ε1, ε2 and ε3) of the DT can be used to characterize differences in magnitude and shape of diffusion in DTI index maps, such as fractional anisotropy (FA) and mean diffusion (MD). Although FA and MD are the primary DTI indices employed in brain research, the diffusion tensor is a rich source of structural information with the potential to provide additional metrics relevant to a range of neurobiological questions including alterations in tissue orientation properties consistent with developmental or pathological reorganizatiht on of circuitry.
In conventional analysis, the orientation of ε1 has been depicted by directionally encoded color (DEC) maps (Pajevic & Pierpaoli, 1999) enabling qualitative visual comparisons. While this type of subjective observation can provide qualitative insight into the nature of tissue abnormality, there is a general need for quantitative statistcal methods to assess the likelihood that any observed alterations differ from random variation. A range of approaches have been suggested for this purpose (Jones et al., 2002), however the existing frameworks (Wu et al., 2004; Wang et al., 2008) are based on the coherence and dispersion of ε1, and hemispheric symmetry of white matter tracts and do not make possible groupwise statistical comparison of regional orientation information.
The objective of this work was to develop a straightforward statistical approach for the quantitative analysis of directional DTI information and to interrogate robust subjective visual observations of abnormal tissue orientation on DEC maps in the hilus region of the hippocampal dentate gyrus following injury. The Fisher probability density function (Fisher, 1953), which is the spherical coordinate system analogue of the familiar Gaussian probability density function, was chosen as the basis for this framework. Historically, Fisher statistics have been widely implemented in the field of paleomagnetism (Butler & Butler, 1992; Tauxe, 2010) to characterize magnetization vectors measured from rock samples and used to determine the history of the Earth's magnetic field. Here we report for the first time the theoretical and practical application of Fisher statistics for analysis of DTI directional information and demonstrate the utility of this approach to detect directional abnormalities in the dentate gyrus following injury.
2 Theory and Calculations
2.1 Terminology and assumptions
The Fisher statistical method can be used to estimate descriptive and inferential statistics from a set of directional observations. Just as one-dimensional statistics are based on the Gaussian probability density function with true mean (μ) and variance (σ2), these directional statistics are based on the Fisher probability density function, with true direction (υ) and precision parameter (κ).
The type of directional data that can be interrogated using Fisher statistics are normally distributed sets of unit vectors where each vector has a length of 1 and originates from x=y=z=0 so that each set may be represented by a group of points on the unit sphere. For a set of N vectors, V = (v1, …, vN), a mean direction vector, vm, can be determined and each vector in the set, vi = (xi, yi, zi), can be described by it's angle, αi, relative to vm using the dot product αi,=cos−1(vi˙ vm).
Ultimately, an estimate can be made for the confidence angle, α(1-p), within which (1-p)% of all directional observations can be expected to be found and F-statistic hypothesis testing may be performed to make inferences about group differences in orientation.
2.2 The Fisher distribution
The Fisher probability density function (pdf) (Fisher, 1953) describes the probability of a point on a sphere of unit radius within an angular area dA (in steradians) centered at an angle α from υ:
| (1) |
where κ is a precision parameter that is inversely proportional to the dispersion.
Since it is more useful for this type of statistics to consider the pdf in terms of the spatial variables α and φ, the azimuthal angle, and because dA=sin(α)dαdϕ, the pdf is more often written as:
| (2) |
which gives the probability of finding a direction vector in the circular strip on the unit sphere between the angles α and α+dα from the true direction.
2.3 Descriptive statistics
The Fisher probability distribution is the basis for the estimation of descriptive and inferential statistics presented in this section.
2.3.1 Mean direction and resultant vector
The mean direction vector, vm, describing the average of a set of N unit vectors can be calculated as follows:
where
| (3) |
and R is the length of the resultant vector sum of all observations, which is given by:
| (4) |
2.3.2 The precision parameter
The precision parameter, κ, is a population parameter that describes the concentration of the pdf around the true direction. As κ approaches 0, the distribution becomes uniform over the unit sphere and as κ approaches∞, the distribution becomes singular at the true direction. The sample estimate of κ is k (Fisher, 1953; McFadden, 1980) given by:
| (5) |
where N is the number of unit vectors in the sample and R is the length of the resultant vector calculated in (4).
2.3.3 Confidence angles
For a confidence level of (1-p)% (typically 95% or p=0.05), a circle can be defined by all points at an angle of α(1-p) from the calculated mean direction, vm. α(1-p) is given by:
| (6) |
where N is the number of unit vectors in the sample, R is the resultant vector calculated in (4) and p is defined according to confidence level.
2.5 Hypothesis testing
In order to test the null hypothesis - that sample observations from 2 or more groups are taken from the same population - the mean direction vectors and confidence angles may be compared across groups or inferential statistics may be calculated.
An intuitive comparison of group directional information is to use the circle of confidence defined by α95. The clearest inferences can be made when: 1) α95 circles for the groups do not overlap, then the null hypothesis is unlikely or 2) the mean direction vector from one group lies inside α95 of the other, then the null hypothesis is likely. A less clear case occurs when α95 circles for the groups do overlap, but the mean from each group lies outside the α95-circle of the other.
A more quantitative way to perform hypothesis testing is by calculation of the F statistic. The following equation was derived by Watson (Watson, 1956), to compare two groups with N1 and N2 observed unit vectors respectively and resultant vectors of length R1 and R2 respectively:
| (7) |
where N=N1+N2 and R is the length of the resultant vector for the pooled direction vector observations from both groups. The larger the value of F, the more different the two group mean directions and, a p-value may be obtained using the appropriate degrees of freedom (2 and 2(N-2) respectively). Equation 7 also extends to hypothesis testing of more than 2 groups.
2.5 Special considerations for DTI data
Although Fisher statistics are generalizable to the analysis of many types of directional data sets, it is important to consider certain inherent attributes of DTI data for accurate directional analysis. Here, it is suggested that the potential pitfalls of DTI data be avoided or accounted for prior to statistical analysis so that the input for Fisher statistics are groups of direction vectors where each vector accurately represents the tissue orientation within a given region of interest (ROI) for one brain sample and is collected and calculated consistently across all samples and groups.
2.5.1 Sources of experimental error
Sources of variance that may contribute to erroneous directional measurement include: positioning of the sample within the scanner reference frame, gross brain structure abnormalities (e.g. by uneven fixation) and ROI mask placement variations. To the extent possible, these should be reduced by experimental practices or corrected for by post-processing techniques, for example DTI-appropriate registration to standard space (Jones et al., 2002).
2.5.2 DEC ambiguity and lateralization
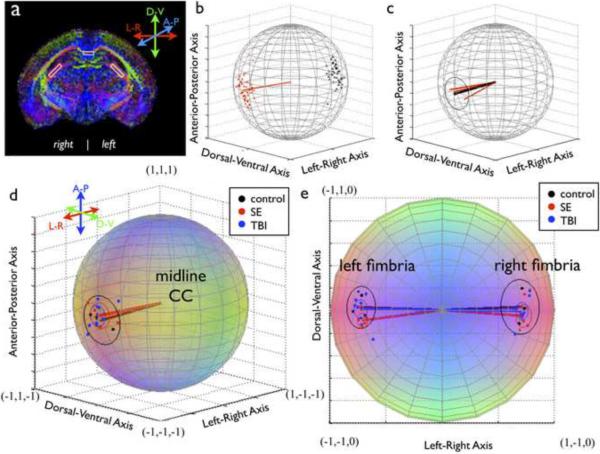
Most DEC maps report directional coloration based on an absolute value algorithm, which creates maps that are qualitatively intuitive and do not suffer from discontinuity artifacts (Pajevic & Pierpaoli, 1999), however this approach suffers from ambiguity in that each color represents 4 unique directions. For example, if <R,G,B> = < |x|, |y|, |z|>, then the directions: x,y,z = (0.6667, 0.3333, 0.6667), (−0.6667, 0.3333, 0.6667), (0.6667, −0.3333, 0.6667) and (−0.6667, −0.3333, 0.6667) are all different from one another, but coded for by the same shade of “pink”. This is particularly evident for bilateral structures, such as the fimbria (see figures 1a and e), which are encoded for by the same color in both brain hemispheres when in fact the ε1 vectors for symmetric anatomical locations are oriented differently, albeit symmetrically. Consequently, care should be taken to consider ROIs from different hemispheres separately or carefully transform directional data from anatomically symmetric regions to a single hemisphere.
Figure 1.
Determination of DTI directional information. (a) Representative ROIs for the CC and fimbria are shown on a DEC map. The approach for determination of single sample (b) and group (c) directional information for the CC is shown, where red points are included eigenvector measurements, black points are excluded (antipodal) measurements, red lines are single sample direction vectors and the black line is the group mean direction vector. Group analysis is shown for the CC (d) and the fimbria (e) where each colored point represents one sample direction vector and the colored circles correspond to α95 for each group. All plots are on the unit sphere and color coding is consistent with DEC maps (red-left/right; green-dorsal/ventral; blue-anterior/posterior)
2.5.3 Antipodal symmetry
Perhaps the most substantial issue in the analysis of directional DTI data arises from the arbitrary sign of the DTI eigenvectors (i.e. ε1=−ε1) which implies that the collection of eigenvectors in an ROI may be clustered about two points, known as antipodes, that are exactly opposite one another on the unit sphere (illustrated in figure 1b). Uncorrected, this could lead to severe misestimation of the direction vector. To avert this, it is possible to map all vectors to a single antipode before determining the direction vector. Similarly, direction vectors for all observations within and across groups should be taken from antipodes in the same hemisphere, although choice of the hemisphere will depend on the orientation of the structure in question.
3. Methods
3.1 DTI
3.1.1 Brain samples
Ex-vivo brain samples from two rat models of brain injury, status epilepticus (SE) and traumatic brain injury (TBI), were compared to control samples. DTI images from these models were selected specifically for the development of the Fisher statistics method based on qualitative identification of abnormalities on a DEC map. All rats were from the Sprague Dawley strain and all animals were housed and treated according to national guidelines and institutional oversight and approval.
SE was induced by kainic acid administered intraperitoneally (8–10 mg/kg) to juvenile (P=35, n=4) and adult (P70, n=2) male rats according to conventional procedures (Ben-Ari, 1985). For inclusion, rats must have demonstrated sustained class V seizure behavior and the groups were combined based on similarity of the brain images and identified abnormalities on the DEC map. TBI was induced in adult rats (n=6 male and 4 female) by controlled cortical impact (CCI) according to conventional procedures (Dixon et al., 1991) using a stereotaxic impactor with 3mm cylindrical bit placed over the temporo-parietal cortex. CCI was performed with a velocity of 4 m/s and penetration depth between 1–3 mm. Controls (n=3) were normal adult male rats.
At greater than 6 months following SE or CCI each rat was transcardially perfused with 4% paraformaldehyde (PFA) and the brain was removed from the skull and stored in PFA for a minimum of 2 weeks. Prior to imaging, brains were rinsed in 0.9% saline for 48 hours. During imaging, brain samples were placed in a custom built sample holder and immersed in Fluorinert (FC-3283; 3M, St. Paul, MN).
3.1.2 Acquisition
For each 7 hour and 2 minute ex-vivo DTI acquisition, 3 brains were simultaneously imaged using a 4.7 T Agilent MRI system and 3.5cm diameter quadrature volume RF coil. A series of multi-slice, diffusion-weighted, spin echo images were acquired with three non-weighted (b~0) and 30 diffusion weighted (b~1200 s/mm2, δ=4.5 ms, Δ=11.2 ms and G=23.84 G/cm), using non-colinear weighting directions (selected from the GE medical systems tensor.dat direction set by Bryan Mock). Other imaging parameters were TE/TR=24.17/2000ms, FOV = 30×30 mm2, matrix = 192×192 reconstructed to 256×256, slice thickness = 0.5mm, number of slices = 35 and 2 signal averages.
3.1.3 Post-processing
DTI maps were created offline using a combination of FSL software and custom Matlab code (version 7.8, R2009a). The diffusion tensor was determined at each voxel using a non-linear least squares fitting algorithm (Koay et al., 2006). FA and DEC maps were generated as well as images containing the Cartesian vector coordinates for ε1.
3.1.4 Experimental error considerations
To address the sources of experimental error described in section 2.5.1, no brains were included with compromised perfusion surgeries and design of the sample holder and positioning of the samples in the scanner reference frame were intended to minimize variations in sample orientation. No registration of the images was performed to make post-processing corrections for sample misalignment. ROI generation was carefully planned and performed to ensure consistent placement.
3.2 Direction analysis
3.2.1 ROI eigenvector analysis
Multi-slice ROI masks for the left and right hilus of the dorsal hippocampal dentate gyrus, midline corpus callosum (CC) and left and right fimbria were manually created for each sample in the native space of the acquired DTI images based on anatomical landmarks as defined by standard coordinates of a rat brain atlas (Watson & Paxinos, 1986). The ROI masks were then used to extract ε1 for every voxel in the ROI.
Based on the suggestions made in 2.5.3, all ε1 measurements were mapped to a single antipode and a single unit direction vector for each ROI for each sample was determined as follows. First, a reference “pole” was defined automatically by pooling all ε1 measurements and their −ε1 pairs across all voxels and all subjects for a given ROI. Then, cluster analysis was performed to determine a pair of necessarily opposite vectors and the vector in the positive z hemisphere was selected for use as the “pole”. Next, each (ε1, −ε1) pair in the ROI was evaluated and only the vector nearest to the “pole” was included. Finally, the unit direction vector describing average direction in the ROI for each sample was defined as the vector average of the mapped eigenvectors within the ROI and the collection of sample unit direction vectors across experimental groups were subjected to Fisher statistical analysis.
3.2.2 Fisher statistics implementation
All statistical analysis and generation of spherical plots was performed using custom Matlab code. For each group, the resultant vector length and mean direction vector were calculated from the collection of observed direction vectors according to equations (3) and (4). For visualization and hypothesis testing, the mean direction vector and circle defined by α95, calculated using equation (6), were plotted for each group and the direction vectors for each sample in all three groups were plotted. The F-statistic was calculated for group comparisons according to equation (7) and a p-value was determined from the F distribution table.
4. Results
Sets of directional DTI data were measured and analyzed for the CC, fimbria and hilus of rats following SE, TBI and normal controls. The technique and results are illustrated in figures 1 and 2.
Figure 2.
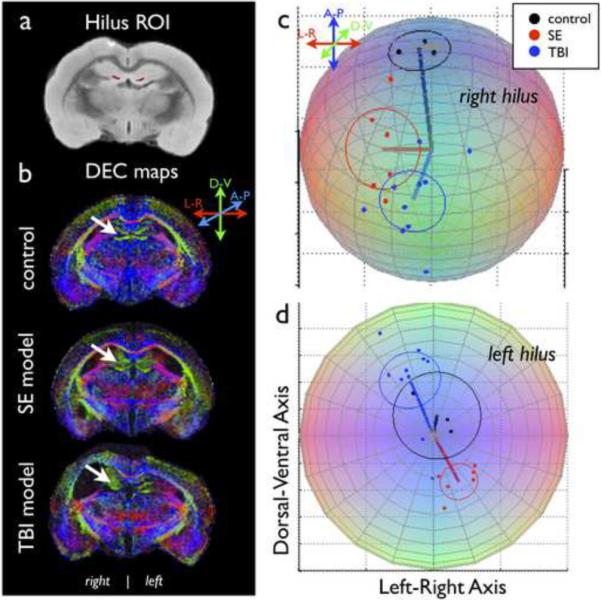
Orientation differences in the hilus of the hippocampal dentate gyrus. ROI analysis of the hilus (red mask, a) was used to confirm subjective observations (white arrows, b) of altered tissue orientation following SE or TBI. Spherical plots of the mean direction vectors (lines) and α95 (circles) for each experimental group and colored points representing the observed direction vectors for all samples in each of the groups are shown on the unit sphere for the right (c) and left (d) hilus.
There was no significant difference in CC mean direction between controls and SE model animals (F=0.198, p=0.823) or TBI model animals (F=0.228, p=0.798). There were also no directional differences found in the left or right fimbria between controls and SE (left: F=3.325, p=0.066; right: F=0.782, p=0.476) or TBI (left: F=0.041, p=0.960; right: F=0.196, p=0.823) groups.
Significant directional differences were found in the hilus between the control group and the SE model group (left: F=16.520, p<0.001; right: F=15.189, p<0.001) as well as the TBI model group on the side of injury (right: F=24.8560, p<0.001), but not the contralateral side (left: F=2.230, p=0.131).
5. Discussion
The Fisher statistical method was applied to obtain descriptive statistics of DTI directional information in three groups of ex-vivo rat brain samples and enabled quantitative inferences about directional differences in 3 different brain structures. No directional abnormalities were found following injury in the CC or fimbria, but a significant difference was found for the hilus that confirmed subjective visual observations of ε1 directional shift from anterior-posterior to dorsal-ventral. Although the structural substrates of this directional alteration remain to be shown, the hilus is known to be highly susceptible to cell loss and aberrant reorganization following injury and during epileptogenesis (Dudek et al., 2002). Directional analysis using Fisher statistics appears to be sensitive to tissue change following both TBI and SE and may complement and extend the findings of other post-SE DTI investigations, which have shown increased FA in the dentate gyrus of the hippocampus (Laitinen et al., 2010) and correlations of DTI measures with histological measures of mossy fiber sprouting (Kuo et al., 2008; Laitinen et al., 2010). This approach also quantitatively distinguished the expected spatial patterns of directional abnormalities between the two models by showing significant bilateral differences from control values in the SE model and differences in the TBI model that were restricted to the side of injury.
Fisher statistics are potentially useful for the comparison of direction vectors between experimental groups, however they may not be suitable for all types of directional DTI data. The requirement that directional data are normally distributed about the mean direction vector and independently observed precludes analysis within a single sample (e.g. eigenvectors in an ROI). For data that do not conform to the requirements of Fisher statistics, other approaches exist, such as the Kent distribution for elliptically distributed directions, the Bingham distribution for bimodal data or non-parametric methods (a useful review of these methods exists in Tauxe, 2010).
6. Conclusion
A framework for the statistical description and inference of directional DTI data has been presented based on the Fisher statistical method and was demonstrated to be effective for distinguishing ROIs with altered directional properties following injury. Quantitative investigation of directional properties of brain tissue are potentially useful in both animal and human studies to identify regions of tumor infiltration, white matter tract disruption, developmental structural organization or any other process that affects the directional microstructure of brain tissue.
Acknowledgements
The authors thank Sue Osting and Craig Levenick for their technical assistance with animal surgeries and sample preparation. This work was funded by the following sources: NINDS R01 25020 and Department of Defense DR080424. EH is funded in part by an award from the Lily's Fund for Epilepsy Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Abbreviations: Diffusion tensor imaging, DTI; diffusion tensor, DT; fractional anisotropy, FA; mean diffusion, MD; directionally encoded color maps, DEC maps; probability density function, pdf; region of interest, ROI; status epilepticus, SE; traumatic brain injury, TBI; controlled cortical impact, CCI; corpus callosum, CC.
References
- Basser PJ, Mattiello J, LeBihan D. MR diffusion tensor spectroscopy and imaging. Biophysical journal. 1994;66(1):259–267. doi: 10.1016/S0006-3495(94)80775-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Ari Y. Limbic seizure and brain damage produced by kainic acid: mechanisms and relevance to human temporal lobe epilepsy. Neuroscience. 1985 Feb;14(2):375–403. doi: 10.1016/0306-4522(85)90299-4. [DOI] [PubMed] [Google Scholar]
- Butler RF, Butler RF. Paleomagnetism: magnetic domains to geologic terranes. Blackwell Scientific Publications; Boston: 1992. [Google Scholar]
- Dixon CE, Clifton GL, Lighthall JW, Yaghmai AA, Hayes RL. A controlled cortical impact model of traumatic brain injury in the rat. J Neurosci Methods. 1991 Oct;39(3):253–62. doi: 10.1016/0165-0270(91)90104-8. [DOI] [PubMed] [Google Scholar]
- Dudek FE, Hellier JL, Williams PA, Ferraro DJ, Staley KJ. The course of cellular alterations associated with the development of spontaneous seizures after status epilepticus. Progress in Brain Research. 2002;135:53–65. doi: 10.1016/S0079-6123(02)35007-6. [DOI] [PubMed] [Google Scholar]
- Fisher RA. Dispersion on a sphere. Proceedings of the Royal Society of London. Series A. Mathematical and Physical Sciences. 1953;217(1130):295. [Google Scholar]
- Jones DK, Griffin LD, Alexander DC, Catani M, Horsfield MA, Howard R, Williams SCR. Spatial normalization and averaging of diffusion tensor MRI data sets. NeuroImage. 2002;17(2):592–617. [PubMed] [Google Scholar]
- Koay CG, Chang LC, Carew JD, Pierpaoli C, Basser PJ. A unifying theoretical and algorithmic framework for least squares methods of estimation in diffusion tensor imaging. J Magn Reson. 2006 Sep;182(1):115–125. doi: 10.1016/j.jmr.2006.06.020. [DOI] [PubMed] [Google Scholar]
- Kuo LW, Lee CY, Chen JH, Wedeen VJ, Chen CC, Liou HH, Tseng WYI. Mossy fiber sprouting in pilocarpine-induced status epilepticus rat hippocampus: A correlative study of diffusion spectrum imaging and histology. Neuroimage. 2008;41(3):789–800. doi: 10.1016/j.neuroimage.2008.03.013. [DOI] [PubMed] [Google Scholar]
- Laitinen T, Sierra A, Pitkanen A, Grohn O. Diffusion tensor MRI of axonal plasticity in the rat hippocampus. Neuroimage. 2010;51(2):521–530. doi: 10.1016/j.neuroimage.2010.02.077. [DOI] [PubMed] [Google Scholar]
- McFadden P. The best estimate of Fisher's precision parameter k. Geophysical Journal of the Royal Astronomical Society. 1980;60(3):397–407. [Google Scholar]
- Pajevic S, Pierpaoli C. Color schemes to represent the orientation of anisotropic tissues from diffusion tensor data: application to white matter fiber tract mapping in the human brain. Magnetic Resonance in Medicine. 1999;42(3):526–540. [PubMed] [Google Scholar]
- Sutula TP, Dudek FE. Unmasking recurrent excitation generated by mossy fiber sprouting in the epileptic dentate gyrus: an emergent property of a complex system. Prog Brain Res. 2007;163:541–563. doi: 10.1016/S0079-6123(07)63029-5. [DOI] [PubMed] [Google Scholar]
- Tauxe L. Essentials of paleomagnetism. University of California Press; 2010. [Google Scholar]
- Wang JJ, Lin YC, Wai YY, Liu HL, Lin CP, Huang YZ. Visualization of the coherence of the principal diffusion orientation: An eigenvector-based approach. Magnetic Resonance in Medicine. 2008;59(4):764–770. doi: 10.1002/mrm.21458. [DOI] [PubMed] [Google Scholar]
- Watson C, Paxinos G. The rat brain in stereotaxic coordinates. Academic Press; 1986. [DOI] [PubMed] [Google Scholar]
- Watson G. Analysis of dispersion on a sphere. Geophysical Journal International. 1956;7:153–159. [Google Scholar]
- Wu YC, Field AS, Chung MK, Badie B, Alexander AL. Quantitative analysis of diffusion tensor orientation: theoretical framework. Magnetic Resonance in Medicine. 2004;52(5):1146–1155. doi: 10.1002/mrm.20254. [DOI] [PubMed] [Google Scholar]