Abstract
Spore photoproduct lyase (SPL) repairs a special thymine dimer 5-thyminyl-5,6-dihydrothymine, which is commonly called spore photoproduct or SP at the bacterial early germination phase. SP is the exclusive DNA photo-damage product in bacterial endospores; its generation and swift repair by SPL are responsible for the spores’ extremely high UV resistance. The early in vivo studies suggested that SPL utilizes a direct reversal strategy to repair the SP in the absence of light. The research in the past decade further established SPL as a radical SAM enzyme, which utilizes a tri-cysteine CXXXCXXC motif to harbor a [4Fe-4S] cluster. At the 1+ oxidation state, the cluster provides an electron to the S-adenosylmethionine (SAM), which binds to the cluster in a bidentate manner as the fourth and fifth ligands, to reductively cleave the C-S bond associated with the sulfonium ion in SAM, generating a reactive 5′-deoxyadenosyl (5′-dA) radical. This 5′-dA radical abstracts the proR hydrogen atom from the C6 carbon of SP to initiate the repair process; the resulting SP radical subsequently fragments to generate a putative thymine methyl radical, which accepts a back-donated H atom to yield the repaired TpT. SAM is suggested to be regenerated at the end of each catalytic cycle; and only a catalytic amount of SAM is needed in the SPL reaction. The H atom source for the back donation step is suggested to be a cysteine residue (C141 in B. subtilis SPL), and the H-atom transfer reaction leaves a thiyl radical behind on the protein. This thiyl radical thus must participate in the SAM regeneration process; however how the thiyl radical abstracts an H atom from the 5′-dA to regenerate SAM is unknown. This paper reviews and discusses the history and the latest progress in the mechanistic elucidation of SPL. Despite some recent breakthroughs, more questions are raised in the mechanistic understanding of this intriguing DNA repair enzyme.
1. Introduction
Bacterial endospores are the longest-lived cells known on earth. They are so resistant to the harsh environment such as vacuum, heat, desiccation and radiation that they are expected to be able to survive in the outer space as hitchhikers clinging to the outside of spacecraft.1 Due to the high solar radiation flux, in particular UV radiation, the ability to protect their genomic DNA against UV radiation is suggested to be the key for spores’ survival in the outer space. Considering the facts that UV irradiation is the common sterilization means in our daily life and the number of deadly diseases associated with the spore forming bacterial strains,2 understanding how the spore genomic DNA is protected against UV light is of particular significance.
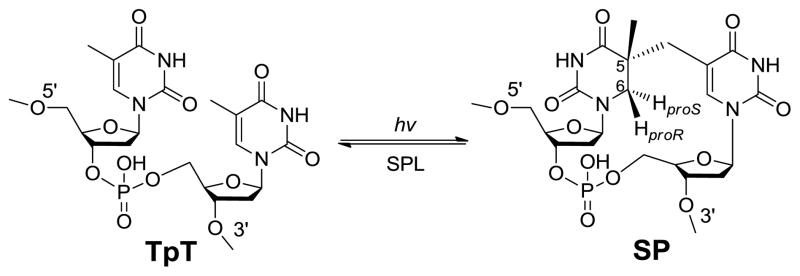
Thymine (T) is the most UV sensitive nucleobase.3–5 In duplex DNA which usually adopts a B-form conformation, the thymine photo-dimerization leads to the formation of cyclobutane pyrimidine dimer and pyrimidine (6–4) photoproduct. In contrast, the spore genomic DNA is bound by a group of DNA binding proteins named small acid-soluble proteins (SASPs) which change the DNA to an A-like conformation.6–8 As the consequence, a special thymine dimer, 5-thyminyl-5,6-dihydrothymine (commonly called spore photoproduct or SP) is produced as the exclusive UV damage product in spores (Scheme 1).9–10 Both in vitro and in vivo studies found that SP formed under UV irradiation could account for as many as 8% of the total thymine in the genomic DNA.10–11 These SP damages accumulate in the dormant spores. When spores leave the dormancy phase and start germinating, these SPs must be repaired as they have proved lethal to the germinated bacteria.12–13 The germinating spores utilize two major pathways to repair the SP dimer: the general nucleotide excision pathway14 and a spore-specific DNA repair system which involves the in situ monomerization of SP into two thymines mediated by an enzyme named spore photoproduct lyase (SPL, Scheme 1).15–18 Blocking either pathway only slightly affects the UV sensitivity of the spores; both pathways have to be interrupted before a spore species that is extremely sensitive to UV irradiation can be obtained.16,19 Between these two pathways, the faster repair rate exhibited by SPL suggests that it plays a major role in repairing the SP damage in the bacteria.16
Scheme 1.
SPL is suggested to be expressed during the sporulation process and packaged in the dormant spore.20 This is supported by the observation that the presence of either chloramphenicol or rifampin, which inhibits de novo synthesis of protein and RNA respectively during the bacterial germination, has no effect on SP repair.17 The SP photo-formation and its enzymatic repair mediated by SPL compose the unique spore SP biochemistry, which accounts for the spores’ extremely high UV resistance. The property of SP and the strong UV resistance it brings to the bacterial spores have been covered by several excellent reviews in the past several years.1,9,13,21–23 In this paper, we will focus on the mechanistic elucidation of the SP repair enzyme – SPL, which has not been specifically reviewed.
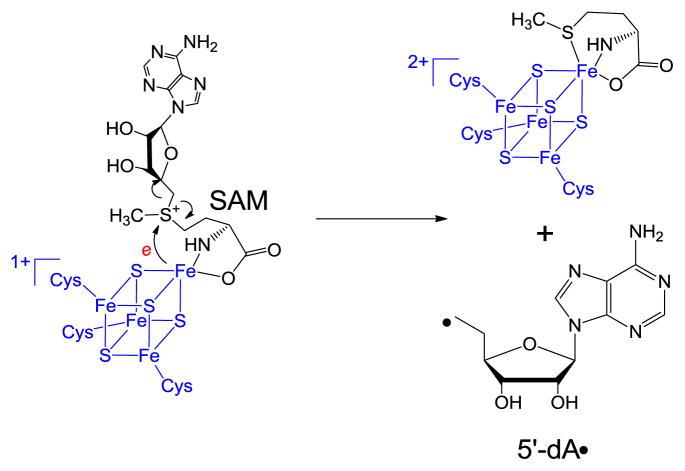
SPL is a member of the so-called radical SAM superfamily, which was defined by the characteristic CXXXCXXC motif,24 although recent evidence suggests that other three-cysteine motifs also facilitate the same radical chemistry.25–27 The three cysteine residues serve as ligands respectively for three irons in the [4Fe-4S] cluster, with the fourth iron being coordinated by the S-adenosylmethionine (SAM) in a bi-dentate manner, with its amino and carboxylate moieties serving as the fourth and fifth ligands to the cluster (Scheme 2).28 The cluster at its +1 oxidation state donates an electron to SAM to cleave its C-S bond, generating a 5′-deoxyadenosyl radical (5′-dA•) This 5′-dA radical catalyzes highly diverse biochemical reactions in animals, plants and microorganisms, including steps in metabolism, DNA/RNA modification, and the biosynthesis of vitamins, coenzymes and many antibiotics via H-atom abstraction reaction to result in controlled radical species for enzyme catalysis.24–27,29–42
Scheme 2.
2. SP formation
To probe the reaction mechanism of SPL, SP or SP analog is needed as the substrate for the enzymatic reaction. Although the SP formation in spores has been relatively thoroughly studied, to duplicate the reaction conditions in vitro and generate enough SP product via photochemistry is a tedious task. Furthermore, although SP is isolated as the exclusive DNA damage product in spores, the in vitro DNA photoreaction always produces other thymine dimers such as the cyclobutane thymine dimer as the side products,10–11,43 suggesting that the factors governing the SP formation in vivo are still not fully understood. Despite these, photochemistry is an important means to prepare SP for enzymatic studies.44–48 The unique properties of the DNA photo-reaction allows the preparation of the specifically-labeled SP substrates,44 which prove highly informative when used to probe the SPL catalysis.48 Due to the importance of the SP preparation to the SPL mechanistic studies, the progress in SP synthesis is reviewed first in the following section.
2.1 SP formation in the presence of DNA binding protein
In bacterial endospores, the genomic DNA is saturated by a group of DNA binding proteins named small acid soluble proteins (SASPs). The name of these proteins was obtained from the observation that they are readily soluble in 0.5 M acetic acid.21,48 There are three types of SASPs in B. subtilis termed α, β and γ respectively with their molecular weights ranging from 5 to 11 kDa; however only the α and β type bind and protect DNA in spores.49 All the α/β type SASPs share remarkable sequence conservation within as well as across species;49–51 their functions have proved to be interchangeable as well. Therefore a B. subtilis SASP protein named SspC was utilized in most of SASP related studies in literature.6,22,52–53 These SASPs bind to DNA and change the DNA from a normal B-form to a A-like conformation,8,54–55 which places the two thymine residues at the right positions for SP photo-dimerization.
Recently, the crystal structure of a SASP-DNA complex formed between a SspC mutant and a oligo(dG) oligo(dC)-10mer duplex DNA was obtained.6–7 The DNA was found to adopt a so-called A-B conformation: Like the A-type DNA, all the sugar puckering adopts a C3′-endo conformation; however the base planes in the crystal were found to be essentially parallel to each other and normal to the helix axis, which is a property of B-DNA. As the nucleotides used for crystallization contained no Ts for SP formation, a simulation study by substituting the 6th and 7th positions from G-C to A-T pairs was conducted. The result suggests that the methyl carbon of the 5′-T is only 3.4 Ǻ away from the C5 carbon at the 3′-T, which favors the formation of the covalent methylene bridge between the two Ts.6 In contrast, the distances between the correspondent moieties for cyclobutane dimer or (6–4) photoproduct formation ranges from 3.9 Ǻ to 4.9 Ǻ. As the thymine dimerization proves an ultrafast reaction which leaves no time for conformational change, the original DNA conformation determines the nature of the photo-dimerization product.56 Therefore, the DNA conformation in SASP-DNA complex favors the formation of SP over other thymine dimers.
The SASP-DNA complexes have been utilized to prepare the SP containing DNA strand for subsequent SPL mechanistic studies. UV irradiation of the DNA-SspC mixture yields SP in as many as 4% of total thymine in a plasmid DNA under a low salt condition; the yield of SP doubled to ~8% of total thymine when a dry film condition was applied for the photoreaction.10 Nicholson and co-workers irradiated a 34-mer duplex DNA containing a single TT step in a film reaction under 10% of relative humidity and obtained the SP as the exclusive photo-dimerization product.46 Broderick and co-workers utilized the complex formed between the SspC protein and the tritium labeled pUC18 plasmid DNA and successfully obtained the SP containing DNA via a dry-film photoreaction.45 Following the tritium migration during the SPL catalysis allowed them to propose that the SAM is regenerated after each catalytic cycle in the SPL reaction.45
2.2 SP formation without the protein partner
Although the photolysis of SspC-DNA complex produces SP in a relatively good yield, it is impossible to make SP in a large quantity due to the difficulty to produce the protein-DNA complex. In addition, 10-bp length was shown to be the smallest DNA fragment that would still binds to the SASP tightly and longer DNA strand is certainly preferred for an optimized DNA-protein interaction.7,9,53,57 Separating the SP containing DNA from the un-reacted DNA strand could thus be problematic due to the large size of the oligomer used for the photoreaction. Another route to produce SP is via the solid state DNA photolytic reaction, where single- or double-stranded DNA is photolyzed in ice or dry film.58–60 Here much shorter DNA strand can be used to facilitate product isolation as the protein partner is no longer needed to promote the SP photochemistry. How the solid state arranges the relative positions of the thymine residues and favors the SP formation is unclear at this moment.
Without the binding protein to control the DNA conformation, the yield of SP is even lower in the solid state photoreaction and significant amount of side reactions also occur. For instance, the cis-syn cyclobutane thymine dimer becomes the major product in solid state DNA photolysis and the yield of SP is only around 1% in dinucleotide TpT photolysis.44 Despite the low yield, the SP can be generated in a better controlled environment and the product readily isolated by HPLC. Under this approach, the SP containing single-stranded GGSPGG 6-mer was prepared by Carell and coworkers from the GGTTGG photolysis and used for SPL activity studies.47 By exposing the dry films of TpT to UVC radiations, Fontecave and coworkers prepared the dinucleotide SP TpT and proved it to be a SPL substrate.61 The photolysis of thymidine produces the dinucleoside SP in solid state photoreaction as well,44,59–60,62 and the enzyme assay proves that without the phosphodiester bond in between to maintain the “right” conformation, such a SP species is an extremely poor SPL substrate.61,63–64
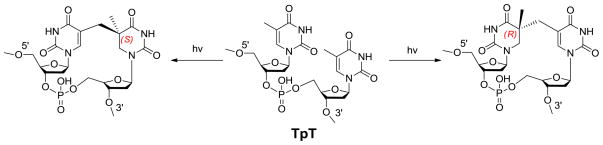
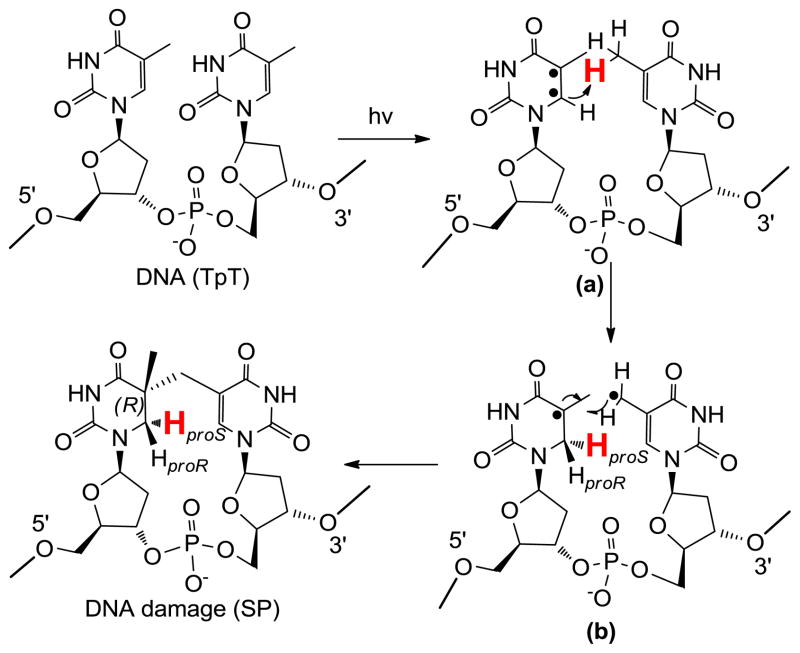
The dinucleotide SP TpT possesses the size of a small organic molecule, which determines that every carbon-associated hydrogen atom on the dinucleotide SP can be closely monitored by NMR spectroscopy. Due to the right-handed double-helical structure of the genomic DNA, the C5 chiral center in SP should adopt an R configuration if the methyl group from the 3′-T forms the methylene bridge to the C5 carbon at the 5′-T (Scheme 3). Similarly, a chiral center with an S configuration should be formed if the reaction involves the methyl group of the 5′-T and the C5 carbon of the 3′-T.21,62 Using the solid state photolysis of the dinucleotide TpT to generate SP and analyzing the SP structure via 1D and 2D NNR spectroscopy, Mantel and coworkers concluded that the former reaction is what occurs in nature; the formed SP reveals an R configuration.62 Furthermore, the SP obtained from the dinucleotide TpT photolysis and the SP generated via the photolysis of plasmid DNA followed by enzyme digestion exhibited identical physical properties,61 suggesting that the SP species used in the NMR studies is the truly biologically relevant species.
Scheme 3.
Additionally, the dinucleotide TpT photochemistry provides a powerful tool to study the reaction mechanism of SP photo-formation. The number of H atoms in TpT and SP TpT remains unchanged, suggesting that SP may form via an intramolecular H-atom transfer reaction with one of the H atoms in the methyl moiety at the 3′-T of TpT migrating to the C6 carbon of the 5′-T in the formed SP. Due to the ultrafast nature of the thymine photo-dimerization reaction,56 the reaction intermediate(s) cannot live long enough to allow any conformational change. Therefore, the SP product must be stereo-specific.
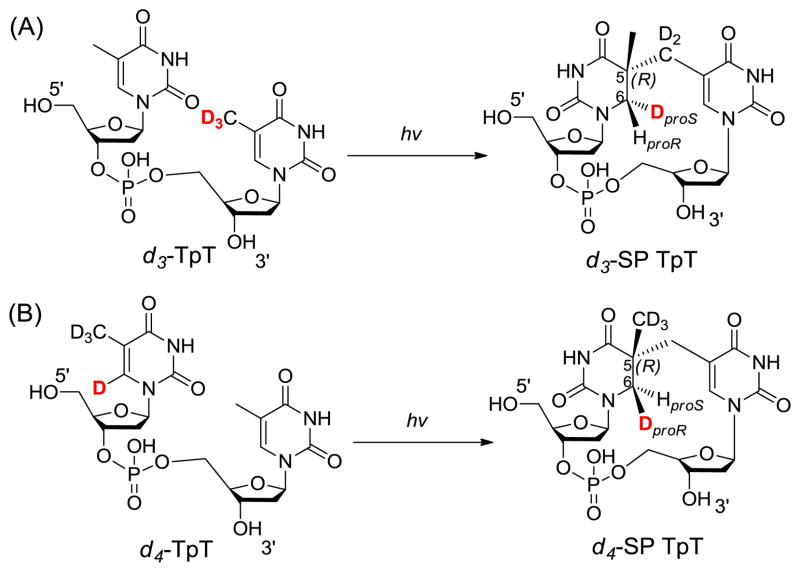
To test the hypothesis, two dinucleotide TpT substrates with either the methyl group at the 3′-T (d3-TpT, Scheme 4) or the hydrogen atom attached to the C6 carbon of the 5′-T labeled by deuterium (d4-TpT, Scheme 4) were prepared. Photolysis of the d3-TpT resulted in an exclusive deuterium transfer to the H6proS position as revealed by NMR spectroscopy. The migrated atom became a protium when the d4-TpT which possesses a -CH3 moiety on the 3′-T was employed (Scheme 4).44 The H-transfer reaction was found to be 3.5 times faster than the D-transfer reaction, suggesting that the hydrogen atom abstraction is involved in the rate-determining step.
Scheme 4.
Taken together, these results support a reaction mechanism shown in Scheme 5. UV light first excites the C=C double bond of the 5′-T into a pair of radicals (intermediate a). The C6 radical abstracts a hydrogen atom from the methyl group of the 3′-T, leading to the formation of a 5-α-thyminyl and a 5,6-dihydrothymin-5-yl radical (intermediate b). This step is potentially rate-limiting as indicated by the observed primary isotope effect. The resulting two radicals subsequently recombine to yield the final product SP.44 It is worth to point out that Varghese suggested that the SP forms through a consecutive mechanism via the recombination of a 5-α-thyminyl and a 5,6-dihydrothymin-5-yl radical about 40 year ago;59,65 however, he failed to explain how these two species are generated. The mechanism proposed above thus filled the critical gap in his mechanistic proposal. The H/D transfer pattern in SP photochemistry can also be explained using a concerted mechanism as proposed by Cadet that the cleavage of the methyl C-H and C=C bonds and the formation of the new C-C and C-H bonds occur at the same time.4 To distinguish these two mechanisms is extremely difficult due to the ultrafast nature of the thymine photo-dimerization reaction.
Scheme 5.
The selectively labeled SPs shown in Scheme 4 has proved to be highly useful substrates in probing the SPL catalysis.48 Furthermore, as explained below, the mechanism revealed in Scheme 5 provides a novel means to prepare SP analogs as either SPL substrates or inhibitors.
2.3 SP preparation via organic synthesis
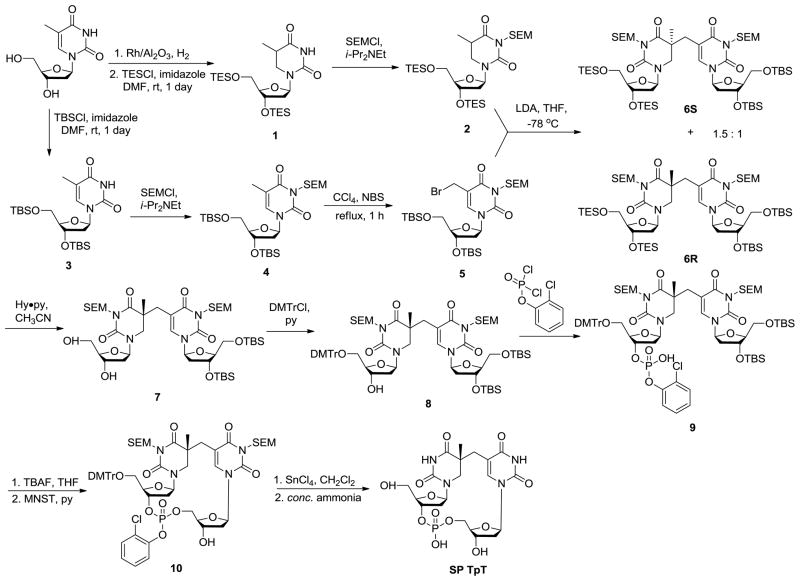
The low yield of SP from DNA photoreaction determines that it is very difficult to be produced in a relatively large quantity. Fortunately, organic synthesis provides another route for SP preparation. The synthesis of SP TpT was first achieved by Kim and Begley in 1995 through ~ 15 reaction steps.66 As shown in Scheme 6, the synthesis was conducted by the formation of the methylene bridge between the two thymine bases followed by the insertion of the phosphodiester linker between the two deoxyriboses. The overall yield of SP is still lower than 10%; however, such a synthesis can be carried out via a series of one-flask reactions, thus paving the way for SP preparation in milligram scale.
Scheme 6.
This synthetic procedure has inspired researchers to prepare other SP analogs via organic synthesis. Carell and coworkers skipped the phosphate insertion steps and produced the dinucleoside SP with no phosphodiester linkage after de-protecting the compounds 6S and 6R respectively shown in Scheme 6.64 Between the two isomers obtained, the SPL only repairs the R isomer,63 suggesting that the enzyme has a strict requirement about the steric configuration of its substrate. Such a dinucleoside SP dimer is repaired at a rate which is about one sixtieth of that when SP TpT is used as the enzyme substrate, suggesting that the phosphodiester linker likely constrains the conformational freedom of the substrate, making it easier to fit into the enzyme binding pocket.61
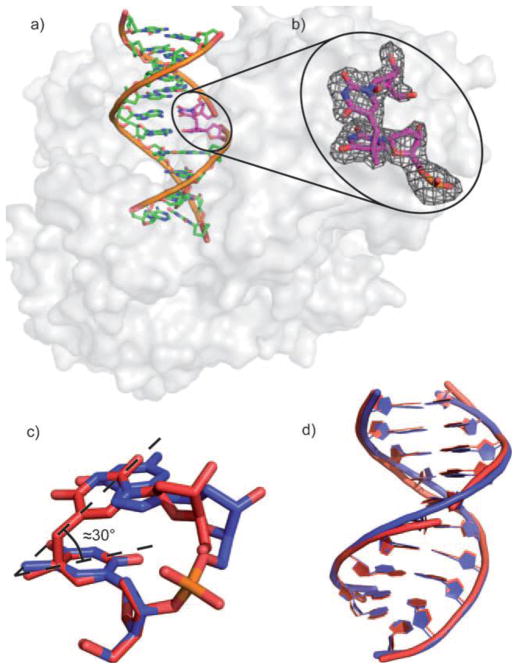
The phosphoramidite derivative of the dinucleoside SP was also synthesized, which enabled the SP incorporation into a 12-mer DNA strand.67 Here, the methylene bridge between the two thymine bases of SP but not the phosphodiester backbone connects the DNA strand together. Such a SP containing duplex was co-crystallized with the DNA polymerase from Geobacillus stearothermophilus by Carell and coworkers.67 As shown in Figure 1, the SP with an R chiral center fits perfectly into the framework of the duplex DNA, with the two SP thymine residues forming standard H-bonds with the two adenines on the complementary strand. Due to the lack of the phosphodiester bond in SP, the conformational distortion created by the methylene bridge between the two thymine bases of SP could be released. It is thus unknown if the structure observed truly reflects the SP conformation in the spore genomic DNA at this point. Nevertheless, this study is the first attempt to incorporate SP into the DNA oligomer via standard synthesis and represents a significant step forward in SP biochemical studies. Such a SP containing single-stranded DNA is also a SPL substrate.67 However, the repair reaction appeared to be very slow although no exact reaction rate was reported.
Figure 1.
a) Crystal structure of the dinucleoside SP-containing duplex in complex with B. st. DNA polymerase. b) Zoom-in-view of the dinucleoside SP which contains an R chiral center at the C5 carbon. c) Overlay of the SP lesion (red) and of an undamaged TpT segment (blue). d) Overlay of the undamaged DNA and the SP- containing DNA bound to the polymerase. (The figure is copied from reference 66 with permission from John Wiley & Sons)
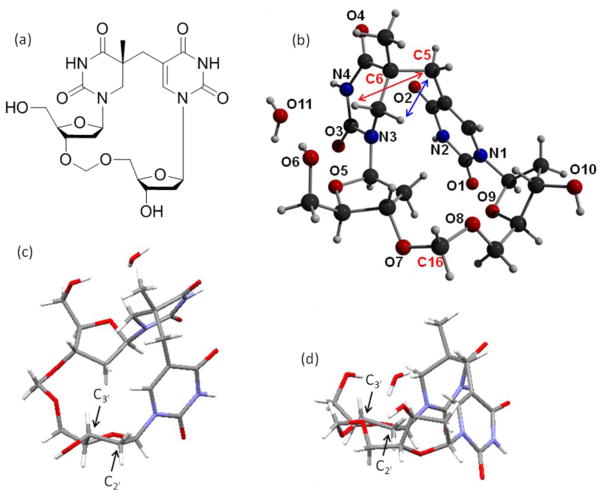
At the same time, Li and coworkers adopted a different approach to crystallize SP.68 They focused on the dinucleotide SP TpT and treated it as a small organic molecule. Moreover, from the experience learned in the crystallization studies of cyclobutane thymine dimers, the authors rationalized that the negatively charged phosphate moiety in natural DNA may hinder the crystallization process, as reflected by the fact that all the dimers crystallized to date contain either no or a neutral linker between the two thymine residues.69–72 A formacetal linker (-CH2-) was thus chosen to replace the phosphate for the SP crystallographic studies. The SP TCH2T was synthesized via a similar procedure shown in Scheme 6: the methylene bridge between the two thymine bases was installed first before the -CH2- linker was inserted between the two deoxyriboses.68 However, the synthesis is not a simple repetition of Begley’s work due to the changes in the protection and the de-protection strategies to make those compatible with the presence of the formacetal linker. As expected, the -CH2- linker greatly improves the crystallization property of SP; a SP crystal was readily obtained after solvent evaporation and the structure solved with a high resolution due to its small molecule nature (Figure 2).
Figure 2.
a) The ChemDraw figure of the dinucleotide SP analog containing the formacetal linker. (b) X-ray structure of the 5R-CH2SP with the nitrogen and oxygen atoms labeled. For clarity purpose, only the bridging methylene carbon between the two thymine bases (C5), the newly formed chiral center at the 5′-T (C6) and the formacetal carbon (C16) are labeled among the 21 carbon atoms in the structure. The structure clearly shows that the chiral center (C6) adopts an R configuration. The distance between the H6proS and the C5 carbon (shown in blue arrow) is found to be 2.63 Ǻ and that between the H6proR and the C5 carbon (shown in red arrow) is found to be 3.36 Ǻ. c) The C3′-endo conformation exhibited by the deoxyribose connected to the 3′-thymine in SP. d) The C3′-endo conformation exhibited by the deoxyribose connected to the 5′-thymine in SP.
The 2D-ROESY spectroscopic and DFT computational studies prove that the formacetal linker containing SP possesses a very similar structure to the SP TpT. Therefore, the structure shown in Figure 2 should be biologically relevant. The 5′-T base is no longer planar due to the loss of aromaticity after the addition of the methylene bridge. The distance between the methylene carbon (C5) and the H6proS (shown in blue arrow) was found to be 2.63 Ǻ and that between the C5 and the H6proR (shown in red arrow) to be 3.36 Ǻ. This observation agrees with the mechanistic proposal that the H6proS originates from the methyl group of the 3′-T during the photoreaction;44 relatively minimal conformational change is generated in SP formation. Both deoxyriboses were found to adopt a C3′-endo conformation, a feature of the A-form DNA. As the spore genomic DNA is suggested to adopt an A-like conformation due to the binding of the small acid soluble proteins,6,8,10 the observation here further suggests that after the SP formation, the methylene bridge in SP forces the two deoxyriboses to maintain an A-like conformation even though the DNA binding proteins are absent.
Such a formacetal containing SP is also a SPL substrate. Incubating the SP TCH2T with SPL reveals the Vmax to be half of that when phosphate containing SP TpT is used.68 Comparing with the 60-fold rate decrease exhibited by the dinucleoside SP,61 this observation suggests that the major function of the linker during the enzyme catalysis is to constrain the substrate conformation so that it can fit into the SPL binding pocket. As the same time, the phosphate may be recognized by the enzyme via the electrostatic interaction, as reflected by the 50% repair rate reduction from the SP TpT reaction.
3. SP repair via SPL
As aforementioned, the SP damage is repaired at the bacterial early germination phase mainly by the enzyme spore photoproduct lyase (SPL). SPL catalyzes the SP repair reaction via a radical mediated direct reverse mechanism and belongs to the radical SAM superfamily.
3.1 substrate recognition
Electrostatic interaction is indicated as a major substrate recognition element in nucleic acid modification enzymes: the enzyme binding pocket contains a network of positively charged protein residues such as lysine and arginine, which interact with the negatively charged phosphodiester backbone of the nucleic acid to achieve the optical substrate recognition. In photolyase which repairs the cyclobutane pyrimidine dimer (T<>T) via light-harvesting flavin cofactors, the six-bp region around the T<>T was found to be protected by the enzyme from damaging by the hydroxyl radicals.73–74 Analyzing the enzyme-substrate interaction reveals that besides the cyclobutane structure as the major recognition element, the enzyme also interacts with the phosphate that is immediately 5′ to the T<>T and the three phosphates that are 3′ to the T<>T.73–75 The photolyase takes the dinucleotide T<>T as a substrate; however the repair rate is much slower due to the weaker enzyme-substrate interaction caused by the loss of these electrostatic interactions.76 The photolyase does not contact the phosphate between the two Ts in T<>T and the phosphate can be ethylated without changing the substrate’s binding affinity to the enzyme.75 This phosphodiester linkage is thus considered to mainly play a structural role to constrain the T<>T conformation. The photolyase binds and repairs the T<>T poorly at a basal level if the phosphodiester linkage is missing.76
DNA photolyase and SPL both repair the DNA damage through in situ monomerization reactions to reverse the dimer into two thymine residues via radical mediated reactions. These two enzymes share a lot of common features in their catalyses. Nicholson and coworkers compared the amino acid sequences between the SPL and the DNA photolyase from a number of microorganisms and identified a region of sequence homology near the carboxyl-termini of these proteins, suggesting that these enzymes could have descended from a common ancestral protein.15 DNase I digestion experiment found that SPL protects at least 9 nucleotides in the SP containing DNA strand with 5 nucleotides 3′ to and 2 nucleotides 5′ to the SP damage,46 suggesting that the phosphates included in this region may be involved in the binding interaction with SPL.
As shown in Table 1, the SPL recognizes and repairs a range of SP containing DNA substrates, ranging from dinucleotide and dinucleoside SP to SP containing single- and double-stranded DNA. The fastest reaction rate observed to date employed a single-stranded SP containing GGSPGG 6-mer as the substrate,47 while the double-stranded SP-containing plasmid DNA also supports a fast repair reaction.45 Under the assumption that the enzymes used in these studies were equally active, such an observation suggests that the SPL may dissociate the local duplex DNA structure into two single strands before the repair process is conducted. The observation that the SP containing oligomer is repaired by SPL at a much faster rate than dinucleotide SP TpT strongly suggests that similar to the DNA photolyase, SPL recognizes the phosphate moieties upstream and/or downstream the SP damage as well. The fact that dinucleotide SP is repaired by SPL also suggests that SPL is likely to utilize the base flipping strategy, that the damaged dinucleotide is flipped out from the DNA helix into an extrahelical position, to recognize and repair the SP damage. Base flipping mechanism has been shown to occur in many DNA modification enzymes.80 For instance, the T<>T repair by photolyase as well as the DNA alkylation repair by iron-containing enzymes like AlkB are all conducted under this mechanism.81–83 The phosphodiester linker in the dinucleotide SP is more likely to function as the conformational holder as that in T<>T, as reflected by the slightly decreased enzyme reaction rate using SP with a formacetal linker as the substrate68 and the much reduced rate when the linkage is absent.61,63–64
Table 1.
SP substrates used to probe the SPL reaction
3.2 Previously hypothesized SPL reaction mechanism
SPL catalyzes the SP repair reaction via a direct reversal strategy with no light needed. Such a conclusion was first reached by Donnellan and Stafford in 1968,12 three years after SP was discovered in UV irradiated endospores.84 After labeling the thymine residues by tritium in spore genomic DNA and generating the SP via photolysis, the authors found that no tritium leaked into the media after these SPs were repaired in the germinated spores.12 In contrast, if an excision repair mechanism is involved, one would expect the labeled SP damages to be released into the media. Later, Rupert et. al. proved that the radioactivity disappearing from the SP peak appeared to be stoichiometrically recovered in the thymine peak,18 further supporting this direct-reversal repair hypothesis.
The first in vitro studies of SPL was conducted by Nicholson and coworkers in 1998.77 They engineered the SPL enzyme by attaching a 6-His tag at either the N-terminus or the C-terminus of the protein to facilitate the protein purification from the dormant B. subtilis spores using nickel-nitrilotriacetic acid (Ni-NTA) agarose affinity chromatography. They also over-expressed the SPL from E. Coli and purified the protein by Ni-NTA chromatography via an engineered 10-His tag. The protein purified via either route possessed a reddish-brown color and exhibited a UV absorption band ~ 400 nm, which is the typical UV absorption for an [4Fe-4S] cluster. Additionally, they found that the enzyme shares sequence similarity in its [4Fe-4S] binding motif with the activating enzymes of class III ribonucleotide reductase and pyruvate-formate lyase respectively; the SPL activity is dependent upon the reducing conditions and the presence of the S-adenosylmethionine as a cofactor.
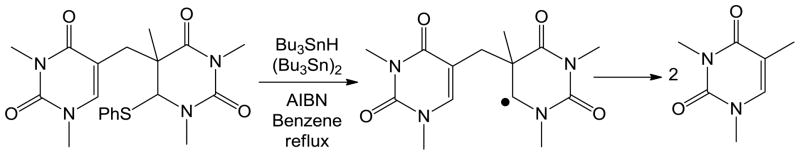
The first mechanistic insight for this SP repair reaction was obtained via a modeling studies conducted by Mehl and Begley in 1999.85 As shown in Scheme 7, via chemical synthesis, a SP analog with a weak C-S bond attached to the C6 carbon was prepared. Under the radical initiating conditions, such an analog readily fragmented into two 1, 3-dimethylthymine molecules. This observation suggests that the SP repair process is likely to be initiated via a hydrogen atom abstraction reaction occurring at the C6 position of SP. The reaction is mediated by the 5′-dA radical generated via a potential SAM cleavage reaction, which is followed by the β scission of the methylene linkage between the two thymines. An H-atom back transfer from the 5′-dA to the resulting thyminyl radical would complete the repair process.
Scheme 7.
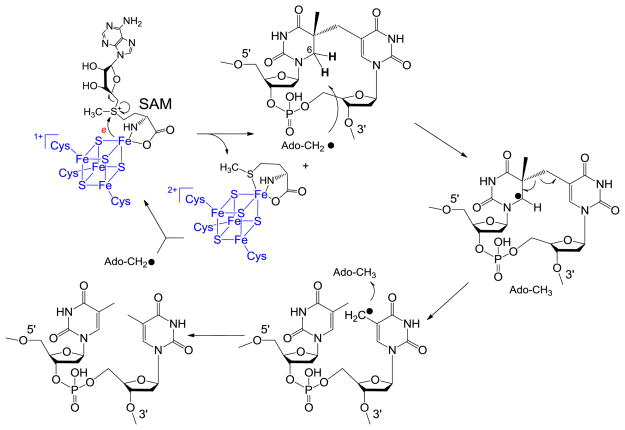
In 2001, Sofia and coworkers defined the radical SAM superfamily using the tri-cysteine CXXXCXXC motif.24 As shown in Scheme 2, SAM redox chemistry leads to the C-S bond cleavage of the sulfonium ion, generating the 5′-dA radical. The H atom abstraction by the 5′-dA radical from SP should yield 5′-dA, which was later observed by Rebeil and Nicholson via an in intro enzymatic assay.78 The remaining question at that point was how the catalytic cycle was constructed, and this question was answered by a tritium labeling experiment conducted by Cheek and Broderick.45 After selectively labeling the thymine residues in a plasmid DNA with tritium and generating the SP dimer via photochemistry, they recovered the tritium-labeled SAM once the SP was repaired via the SPL reaction. Moreover, only a catalytic amount of SAM was needed for the catalysis. These observations were similar to what were observed in another radical SAM enzyme – the lysine 2,3-aminomutase, where a SAM regeneration after each catalytic cycle was proposed.86–89
To accommodate these observations in SPL reaction, a reaction mechanism was proposed as shown in Scheme 8.45 This mechanism contains a minimal number of intermediates yet is able to interpret all the experimental data obtained at that time. It was therefore considered as a reasonable working hypothesis for this SPL enzyme.
Scheme 8.
3.3 Modified SPL reaction mechanism
Such a mechanism was first challenged by a theoretical calculation.90–91 As shown in Scheme 8, the 5′-dA is involved in both hydrogen atom transfer steps: the 5′-dA radical abstracts a H atom at the 5′-thymine of SP, and it has to move down for one nucleotide, roughly 3.4 Ǻ under the assumption that the SP containing DNA adopts a regular B-form conformation in the repair process, to donate the H atom back to the methyl radical at the 3′-thymine, yielding the repaired TpT. Such a movement requires a dramatic protein conformational change and is unfavored energetically. To solve this problem, the calculation inserted an extra H atom exchange step between the two thymine methyl groups to return the radical to the 5′-thymine before the hydrogen back-donation step between the 5′-dA and the thymine radical occurs. As described below, such an assumption proves to be unnecessary with the involvement of the protein residue(s) in SPL catalysis.
The first experimental evidence which questions the hypothesized SPL mechanism was obtained by an in vivo mutagenic study conducted by Nicholson and coworkers.92 The B. subtilis SPL contains four conserved cysteine residues, C91, C95, C98, and C141. The first three cysteines belong to the radical SAM CXXXCXXC motif. Mutating any of these cysteines would drastically destabilize the [4Fe-4S] cluster and thus deactivate the enzyme. Surprisingly, Nicholson found that the C141A mutant has an equal effect in deactivating the SPL enzyme. Spores carrying any of these C→A mutations in their SPL enzyme are very sensitive to UV irradiation.92 These observations suggest that the C141 residue must be involved in the enzyme catalysis; however, its role is not reflected by the hypothesized mechanism.
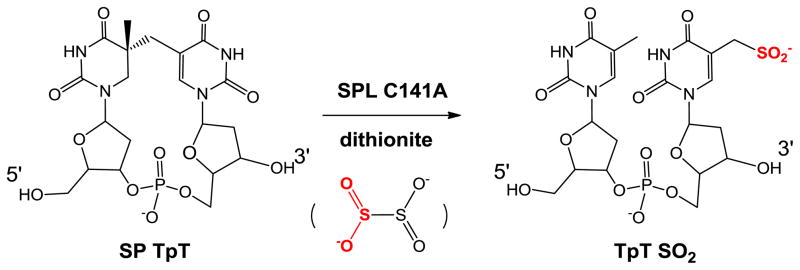
Fontecave and co-workers later re-examined this B. subtilis SPL C141A mutant in an in vitro enzymatic study using dinucleotide SP TpT as the substrate.93 After using excess of sodium dithionite to reduce the [4Fe-4S] cluster from the 2+ to 1+ oxidation state to initiate the radical SAM reaction, they identified a TpTSO2 species with the SO2 moiety attached to the methyl group of the 3′-T as the major product from the SP repair reaction (Scheme 9). The -SO2 moiety was found to originate from the homolytic cleavage of the S-S bond in dithionite; the resulting •SO2 was subsequently added to the methyl radical of the 3′-thymine to yield the observed TpTSO2. Such an observation demonstrates that the C141A mutation severely disturbs the H-atom back donation step in the SP repair process.
Scheme 9.
Both observations strongly suggest the involvement of protein residue(s) in the enzyme catalysis, which has been further proved by a recent SPL mechanistic study conducted by Li and coworkers using diastereoselectively labeled dinucleotide SP TpT substrates.48 As shown in Scheme 1, the SP C6 carbon possesses two H atoms and is pro-chiral. Given that enzymatic reactions are usually highly stereoselective, if the SP repair reaction is initiated by H-abstraction at C6, which H-atom is taken?
To answer this question requires the preparation of SP substrates with the two 6-H atoms being selectively labeled. Such substrates cannot be obtained via organic synthesis shown in Scheme 6, as the synthesis includes a hydrogenation step catalyzed by Rh/Al2O3. If D2 is used in the hydrogenation step to introduce the deuterium label into SP, the D2 addition to the C=C bond would occur from either side of the thymine plane, resulting in a mixture of two configurational isomers with deuterium occupying either the H6proR or the H6proS position. These isomers cannot be separated via chromatographic means, and thus cannot provide any useful mechanistic information when used to probe the SPL reaction.
In contrast, photochemistry proves a reliable means to prepare the SP substrate with an exclusive stereoselectivity.44 As shown in Scheme 4, starting from the selectively labeled dinucleotide TpT, dinucleotide SP TpT with either the H6proR or the H6proS position labeled by deuterium can be readily produced after the solid state photolysis. Using these substrates to probe the enzyme reaction would reveal the H atom abstracted by the 5′-dA radical to initiate the SP repair reaction.
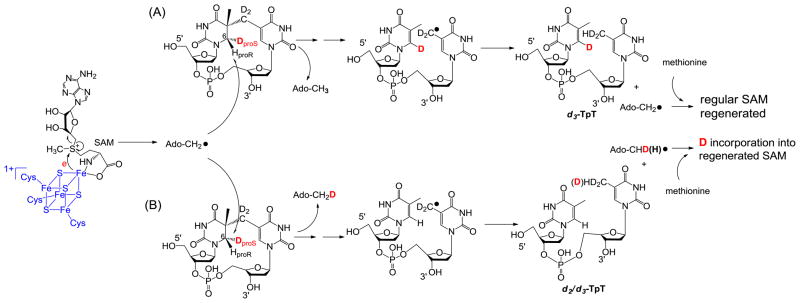
Figure 3 summarizes the expected reactions when d3-SP TpT is used as the enzyme substrate under the reaction mechanism proposed in Scheme 8. (1) H-atom abstraction by the 5′-dA radical generates the 5′-dA as the product. Analyzing the 5′-dA gives the most direct evidence on which H atom is taken. (2) If a protium at the proR position is taken during the initiation step, all the deuteriums from the SP substrate will be retained in the TpT product. In contrast, if the deuterium at the proS position is taken, a d2/d3 mixture in the TpT is expected due to the fact that the two protiums and one deuterium in the methyl group of the 5′-dA all have chances to be abstracted by the TpT radical. (3) Consequently, a d0/d1 mixture is expected for the regenerated SAM if the deuterium at the proS position is abstracted by the 5′-dA radical.
Figure 3.
Using d3-SP TpT as the enzyme substrate to illustrate the expected reaction products according to the current SPL mechanism shown in Scheme 3. (A): The 5′-dA radical abstracts the protium at the H6proR position to initiate the reaction. All three deuterium atoms in compound 2 should be retained in the repair product TpT; thus d3-TpT as well as regular 5′-dA and SAM are expected after the reaction. (B): The 5′-dA radical abstracts the deuterium at the H6proS position to initiate the reaction. Correspondingly, a deuterium is incorporated into the resulting 5′-dA and d2/d3-TpT and d1/d0-SAM mixtures are expected by the end of the reaction due to the H or D abstraction from the methyl group of 5′-dA by the TpT radical in the H atom back donation step.
ESI-MS analysis of the 5′-dA generated via the d3-SP TpT repair reaction revealed no sign of deuterium incorporation. In contrast, the vast majority of the 5′-dA generated via the d4-SP TpT repair reaction, which contains a deuterium at the H6proR position, exhibits a +1 signal relative to the mass of the naturally occurring 5′-dA in the MS spectrum, suggesting that a deuterium is abstracted in the radical initiation step. Furthermore, the d4-SP TpT repair reaction is 2.8 ± 0.3 times slower than the d3-SP TpT reaction, corresponding to an apparent kinetic isotope effect of 2.8 for this SPL reaction. All these observations suggest that it is the H6proR atom being abstracted by the 5′-dA radical and the SPL reaction is truly stereoselective.
Such an observation also suggests that the SP repair process is not a simple reversal of its formation, as indicated by the equation in Scheme 1. During the SP photo-formation, an H atom migrates from the methyl group of the 3′-thymine to the H6proS position on the 5′-thymine of SP, making the C6 carbon pro-chiral.44 In contrast, during the repair process, the SPL enzyme abstracts the H6proR atom to initiate the repair process. This conclusion also provides an intriguing possibility to prepare the mechanism based SPL inhibitors by introducing a substitution group to the C6 carbon at the 5′-thymine of a TT sequence. During the SP photo-formation process, such a substitution will be pushed to the R position of the formed SP analog while the migrating H atom occupies the S position. The resulting SP analog thus no longer supports the stereoselective H-atom abstraction step, resulting in the inhibition of the SPL enzyme. Experimental work is needed in the future to test this intriguing hypothesis.
If the H atom abstraction step is as expected, the result from the H-atom back donation step is quite surprising.48 Since no deuterium is involved in the catalytic cycle in d3-SP TpT reaction, all three deuterium atoms were found to be retained in the repaired TpT. However, the TpT product isolated from the d4-SP TpT repair reaction was found to contain only three deuteriums as well. This observation suggests that the deuterium abstracted by the 5′-dA radical is not returned to the repaired TpT; a protium from an unknown resource is instead incorporated into the product.
Since the enzyme reaction is carried out in the aqueous solution, the most abundant protium source is the aqueous buffer. A reaction was thus conducted in D2O and indeed a deuterium was found to be incorporated into the repaired TpT.48 Since the SP repair process is conducted via a radical mechanism, it is impossible for the radical intermediate to abstract a hydrogen atom directly from the water molecule as that would leave a highly detrimental hydroxyl radical behind. The only explanation is that the H atom incorporation is conducted by a proton exchange process via the acid/base chemistry. Such a chemistry cannot happen on the thymine base, but can occur through protein residues such as cysteine and tyrosine. These residues possess a SH and an OH moiety respectively that can mediate acid/base exchange reaction as well as the radical reaction. Considering the facts that both in vivo and in vitro studies proved the cysteine residue 141 to be involved in the enzyme catalysis,92–93 it is almost certain that C141 participates in the H atom back donation step and is the most likely H atom donor to the methyl radical in thymine. Cysteine mediated H atom exchange with the aqueous buffer was observed in a number of radical enzymes such as the class II ribonucleotide reductase,94–96 the pyruvate formate lyase,94,97–98 and benzylsuccinate synthase.99 The observation of the H/D exchange with the aqueous buffer is a clear indication that the cysteine residue is involved in the enzyme catalysis.
The cysteine 141 instead of the 5′-dA to serve as the direct H atom donor is more reasonable considering the difference in bond strength between the S-H and the C-H bonds. A calculation predicts the C-H bond dissociation energy (BDE) of 5′-dA to be 12.2 kcal/mol higher than that for the methyl moiety of thymine (98.2 vs 86.0 kcal/mol); in contrast, the BDE of the S-H bond in cysteine is suggested to be around 82 kcal/mol;90–91 which is ~3–4 kcal/mol weaker than the allylic C-H bond. It is therefore plausible for SPL to use the cysteine instead of the C-H bond of 5′-dA as the direct H-atom donor, which greatly lowers the energy barrier for the H-atom back-transfer step.
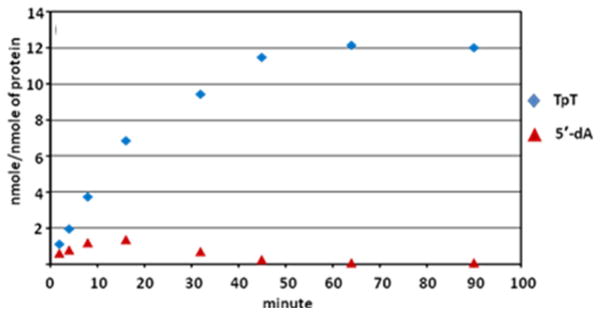
Although the 5′-dA has been observed during the SPL catalysis, such a species does not accumulate in the reaction system.48 As shown in Figure 4, prolonged reaction yields no 5′-dA in the reaction solution. Furthermore, the quantity of 5′-dA isolated seems to be directly correlated to the reaction rate, suggesting the 5′-dA observed is a reaction intermediate, and not the abortive product as commonly observed in other radical SAM enzymes.78,93,100–101 The facts that only a catalytic amount of SAM is needed in SPL reaction and the prolonged reaction leads to the re-consumption of all the 5′-dA intermediate supports the assumption that SAM is regenerated after each catalytic cycle. However, no deuterium incorporation into SAM was observed in d4-SP TpT reaction. This is contradictory to what was observed in the previous tritium labeling experiments,45 but instead suggests that the SAM regeneration process seems to be more complicated than originally thought.
Figure 4.
5′-dA and TpT formation observed in the acid-quenched SPL reaction at various time points using dinucleotide SP TpT as the enzyme substrate.
The fact that C141 serves as the potential H atom donor to the thymine methyl radical indicates that a thiyl radical is subsequently generated on this protein residue. This thiyl radical thus must be involved in the SAM regeneration process. The simplest model to accommodate this rationale is to insert this cysteine residue between the thymine allyl radical and the 5′-dA in the reaction pathway. Although the thiyl radical is more stable than the allylic radical in thymine, making the direct H-atom transfer from 5′-dA to the thiyl radical even more unfavorable, coupling the H atom abstraction by the thiyl radical with the SAM regeneration process which is heavily favored thermodynamically may provide a rationale for the SPL reaction.102–103 A similar strategy was used to explain the adenosylcobalamin regeneration in the class II ribonucleotide reductase (RNR),104 where the energy cost in the H atom abstraction by the catalytic thiyl radical is compensated by the favored addition reaction between the resulting 5′-dA• and Cob(II)alamin species.
Although it is possible that SAM is regenerated via this single cysteine residue, a more complex reaction pathway which involves more protein residue(s) cannot be excluded. Actually, the SPL enzyme seems to possess some “error-proof” function: the externally added thiol complexes cannot severely disturb the enzyme catalysis, suggesting that the protein harbored thiyl radical is well protected against competition from these small thiol compounds. Such a protection is difficult to achieve by a single cysteine residue, but becomes possible via a cysteine-containing electron transfer pathway. If it is true, the SPL enzyme may then share some similarity with the Class I ribonucleotide reductase.105
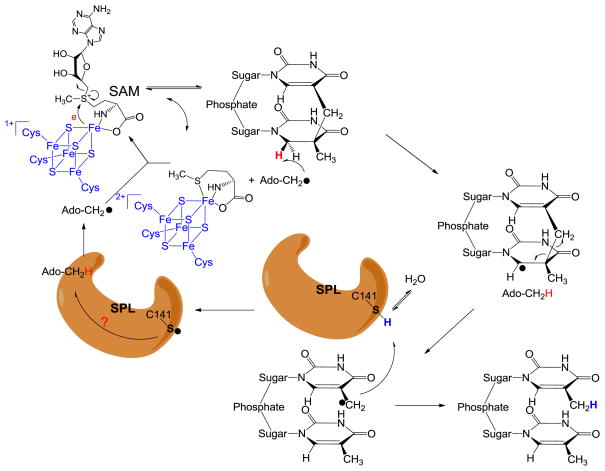
To the consistent with these experimental observations, a modified SPL mechanism was recently proposed as shown in Figure 5. After the reductive SAM cleavage reaction with an electron provided by the [4Fe-4S]1+ cluster, the resulting 5′-dA radical abstracts the H6proR atom to yield the SP radical on the C6 carbon. Subsequent fragmentation of SP leads to the formation of a delocalized methyl radical on the 3′-T. Instead of abstracting an H atom back from the 5′-dA, the TpT radical draws an H atom from an unidentified protein residue, with the C141 the most likely candidate, to yield the repaired TpT. The resulting thiyl radical on this cysteine could abstract an H atom back from 5′-dA as what occurs in class II ribonucleotide reductase; or it may oxidize its neighboring residue(s) to form another protein radical species before it reacts with 5′-dA, which reminds us the electron transfer network exhibited by the class I ribonucleotide reductase. Recombination of the 5′-dA radical and the methionine regenerates SAM at the end of each catalytic cycle. The involvement of the protein residue(s) in catalysis perfectly solves the space problem raised by the theoretical calculation: The H atom pathway is relayed via a protein network, which makes the H atom exchange step between the two methyl groups unnecessary.90–91
Figure 5.
Newly proposed reaction mechanism for SPL catalyzed SP dimer repair. The 5′-dA radical generated from SAM reductive cleavage reaction takes the 6-HproR atom to yield a C6 radical on SP, the SP methylene bridge subsequently undergoes a hemolytic cleavage to give a thymine methyl radical. (Note: This allyl radical is likely to delocalize to the thymine aromatic ring; the current drawing as a methyl radical is a simplified model to facilitate discussion) This radical abstracts an H atom back from an unknown protein residue, presumably C141, to generate a thiyl radical, releasing the repaired TpT. This thiyl radical either takes an H-atom back from the 5′-dA directly, or it reacts with other protein residues before the 5′-dA is re-oxidized to the radical form again. The resulting 5′-dA radical recombines with methionine to regenerate SAM and finish one catalytic cycle.
4. Summary and outlook
Despite the facts that SP was discovered nearly a half a century ago and the correct prediction by the early in vivo studies that the SP is repaired via a direct reversal strategy with no light needed, little mechanistic insight was gained about this repair process during the first four decades of SP biochemical studies. The major obstacles hindering the SPL study were the stability of the protein due to oxidation of the [4Fe-4S] cluster in the air and the difficulty to prepare enough SP substrate.
Tremendous progress has been made over the last decade in solving these two issues. The discovery of the radical SAM enzyme superfamily and the radical mechanism involved in the enzyme catalysis instructs the researchers to adopt a strictly anaerobic environment for the SPL reaction. Under this condition, the enzyme remains active for at least several hours.48,63,67 Besides using the traditional photochemical approach to generate the SP, synthetic chemistry have been developed to prepare the dinucleotide and dinucleoside SP as well as incorporate SP into the oligomer strand.44,64,66–68,106 Although the syntheses are lengthy and the overall yields are low, SP preparation in milligram scale has become a realistic expectation. The progress in the SP synthesis resulted in the first structural characterizations of SP.67–68 The progress in the mechanistic elucidation of SP photo-formation also has a significant impact to SPL mechanistic studies. The photochemistry mechanism enables the selective labeling of the SP substrate, which turned out to be highly informative when used to explore the SPL catalysis. Such a mechanism also suggests a promising strategy in preparing SP analogs to potentially inhibit the SPL catalysis; which requires to be tested in the future.
The mechanistic studies of SPL have established several facts to date: (1) SPL is a radical SAM enzyme; it utilizes the [4Fe-4S] cluster and the SAM as a cofactor to generate the catalytic 5′-dA radical.24,78 (2) The 5′-dA radical abstracts the H6proR atom from the SP substrate to initiate the repair process.48 (3) the 5′-dA is NOT the direct H atom donor to the thymine based radical to yield the repaired TpT, some protein residue(s) with the C141 the most likely candidate serves this role.48 (4) Only a catalytic amount of SAM is needed for the SPL catalysis, suggesting a possible SAM regeneration after each catalysis cycle.45,48
Currently, the most intriguing question left to be answered in SPL catalysis is: How is SAM regenerated at the end of each catalytic cycle? The involvement of C141 as the direct H atom donor leaves a thiyl radical on the protein and this radical must interact with the 5′-dA directly or indirectly to abstract an H atom and regenerate the 5′-dA radical before it recombines with methionine to produce the SAM. Is it a one step reaction like what occurs in class II ribonucleotide reductase, or a two or even multiple step reaction similar to what occurs in class I ribonucleotide reductase? More work is clearly demanded in the future to understand this intriguing DNA repair enzyme.
Highlights.
Spore photoproduct lyase (SPL) repairs the special thymine dimer 5-thyminyl-5,6-dihydrothymine, also called spore photoproduct or SP.
SPL utilizes the radical SAM chemistry to catalyze the SP repair reaction.
Latest evidence suggests that at least one protein radical is involved in catalysis.
Future work should focus on the characterization of the catalytic pathway in protein.
Acknowledgments
The author thanks the National Institutes of Environmental Health Sciences (R00ES017177) and the IUPUI startup fund for financial support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nicholson W, Schuerger A, Setlow P. The solar UV environment and bacterial spore UV resistance: considerations for Earth-to-Mars transport by natural processes and human spaceflight. Mutat Res Fundam Mol Mech Mutagen. 2005;571:249–264. doi: 10.1016/j.mrfmmm.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 2.Prescott LM, Harley JP, Klein DA. Microbiology. 6. McGraw-Hill Higher Education; Dubuque, IA: 2005. [Google Scholar]
- 3.Lamola AA. Triplet photosensitization and the photobiology of thymine dimers in DNA. Pure Appl Chem. 1970;24:599–610. doi: 10.1351/pac197024030599. [DOI] [PubMed] [Google Scholar]
- 4.Cadet J, Vingy P. Photochemistry and the nucleic acids. Vol. 1 Wiley; New York: 1990. [Google Scholar]
- 5.Durbeej B, Eriksson LA. On the Formation of Cyclobutane Pyrimidine Dimers in UV-irradiated DNA: Why are Thymines More Reactive? Photochem Photobiol. 2003;78:159–167. doi: 10.1562/0031-8655(2003)078<0159:otfocp>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 6.Lee KS, Bumbaca D, Kosman J, Setlow P, Jedrzejas MJ. Structure of a protein-DNA complex essential for DNA protection in spores of Bacillus species. Proc Nat Acad Sci. 2008;105:2806–2811. doi: 10.1073/pnas.0708244105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bumbaca D, Kosman J, Setlow P, Henderson RK, Jedrzejas MJ. Crystallization and preliminary X-ray analysis of the complex between a Bacillus subtilis [alpha]/[beta]-type small acid-soluble spore protein and DNA. Acta Crystallogr Sect F. 2007;63:503–506. doi: 10.1107/S1744309107022750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mohr SC, Sokolov NV, He CM, Setlow P. Binding of small acid-soluble spore proteins from Bacillus subtilis changes the conformation of DNA from B to A. Proc Nat Acad Sci. 1991;88:77–81. doi: 10.1073/pnas.88.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Setlow P. Resistance of spores of Bacillus species to ultraviolet light. Environ Mol Mutagen. 2001;38:97–104. doi: 10.1002/em.1058. [DOI] [PubMed] [Google Scholar]
- 10.Nicholson WL, Setlow B, Setlow P. Ultraviolet irradiation of DNA complexed with alpha/beta-type small, acid-soluble proteins from spores of Bacillus or Clostridium species makes spore photoproduct but not thymine dimers. Proc Nat Acad Sci. 1991;88:8288–8292. doi: 10.1073/pnas.88.19.8288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Setlow B, Setlow P. Dipicolinic acid greatly enhances production of spore photoproduct in bactrial-spores upon UV irradiation. Appl Environ Microb. 1993;59:640–643. doi: 10.1128/aem.59.2.640-643.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donnellan JE, Stafford RS. The Ultraviolet Photochemistry and Photobiology of Vegetative Cells and Spores of Bacillus megaterium. Biophys J. 1968;8:17–28. doi: 10.1016/S0006-3495(68)86471-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Setlow P. Mechanisms for the Prevention of Damage to DNA in Spores of Bacillus Species. Annu Rev Microbiol. 1995;49:29–54. doi: 10.1146/annurev.mi.49.100195.000333. [DOI] [PubMed] [Google Scholar]
- 14.Munakata N. Mapping of the genes controlling excision repair of Pyrimidine photoproducts in Bacillus subtilis. Molecular and General Genetics MGG. 1977;156:49–54. [Google Scholar]
- 15.Fajardo-Cavazos P, Salazar C, Nicholson WL. Molecular cloning and characterization of the Bacillus subtilis spore photoproduct lyase (spl) gene, which is involved in repair of UV radiation-induced DNA damage during spore germination. J Bacteriol. 1993;175:1735–1744. doi: 10.1128/jb.175.6.1735-1744.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Munakata N, Rupert CS. Genetically Controlled Removal of “Spore Photoproduct”. from Deoxyribonucleic Acid of Ultraviolet-Irradiated Bacillus subtilis Spores. J Bacteriol. 1972;111:192–198. doi: 10.1128/jb.111.1.192-198.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Munakata N, Rupert CS. Dark repair of DNA containing “spore photoproduct” in Bacillus subtilis. Molecular and General Genetics MGG. 1974;130:239–250. doi: 10.1007/BF00268802. [DOI] [PubMed] [Google Scholar]
- 18.Wang TCV, Rupert CS. Evicence for the monomerization of spore photoproduct to two thymines by the light-dependent “spore repair”. process in bacillus subtilis. Photochem Photobiol. 1977;25:123–127. doi: 10.1111/j.1751-1097.1977.tb07432.x. [DOI] [PubMed] [Google Scholar]
- 19.Munakata N. Genetic analysis of a mutant of Bacillus subtilis producing ultraviolet-sensitive spores. Molecular and General Genetics MGG. 1969;104:258–263. doi: 10.1007/BF02539290. [DOI] [PubMed] [Google Scholar]
- 20.Pedraza-Reyes M, Gutierrez-Corona F, Nicholson WL. Temporal regulation and forespore-specific expression of the spore photoproduct lyase gene by sigma-G RNA polymerase during Bacillus subtilis sporulation. J Bacteriol. 1994;176:3983–3991. doi: 10.1128/jb.176.13.3983-3991.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Desnous CL, Guillaume D, Clivio P. Spore Photoproduct: A Key to Bacterial Eternal Life. Chem Rev. 2010;110:1213–1232. doi: 10.1021/cr0781972. [DOI] [PubMed] [Google Scholar]
- 22.Setlow P. Small, acid-soluble spore proteins of Bacillus species: structure, synthesis, genetics, function, and degradation. Annu Rev Microbiol. 1988;42:319–338. doi: 10.1146/annurev.mi.42.100188.001535. [DOI] [PubMed] [Google Scholar]
- 23.Nicholson WL, Munakata N, Horneck G, Melosh HJ, Setlow P. Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments. Microbiol Mol Biol Rev. 2000;64:548–572. doi: 10.1128/mmbr.64.3.548-572.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, Miller NE. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 2001;29:1097–1106. doi: 10.1093/nar/29.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marsh ENG, Patterson DP, Li L. Adenosyl Radical: Reagent and Catalyst in Enzyme Reactions. ChemBioChem. 2010;11:604–621. doi: 10.1002/cbic.200900777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chatterjee A, Li Y, Zhang Y, Grove TL, Lee M, Krebs C, Booker SJ, Begley TP, Ealick SE. Reconstitution of ThiC in thiamine pyrimidine biosynthesis expands the radical SAM superfamily. Nat Chem Biol. 2008;4:758–765. doi: 10.1038/nchembio.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paraskevopoulou C, Fairhurst SA, Lowe DJ, Brick P, Onesti S. The elongator subunit Elp3 contains a Fe4S4 cluster and binds S-adenosylmethionine. Mol Microbiol. 2006;59:795–806. doi: 10.1111/j.1365-2958.2005.04989.x. [DOI] [PubMed] [Google Scholar]
- 28.Walsby CJ, Ortillo D, Yang J, Nnyepi MR, Broderick WE, Hoffman BM, Broderick JB. Spectroscopic approaches to elucidating novel iron-sulfur chemistry in the “radical-Sam”. protein superfamily. Inorg Chem. 2005;44:727–741. doi: 10.1021/ic0484811. [DOI] [PubMed] [Google Scholar]
- 29.Vey JL, Drennan CL. Structural Insights into Radical Generation by the Radical SAM Superfamily. Chem Rev. 2011;111:2487–2506. doi: 10.1021/cr9002616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruszczycky MW, Choi SH, Liu HW. Stoichiometry of the Redox Neutral Deamination and Oxidative Dehydrogenation Reactions Catalyzed by the Radical SAM Enzyme DesII. J Am Chem Soc. 2010;132:2359–2369. doi: 10.1021/ja909451a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Szu PH, Ruszczycky MW, Choi SH, Yan F, Liu HW. Characterization and Mechanistic Studies of DesII: A Radical S-Adenosyl-l-methionine Enzyme Involved in the Biosynthesis of TDP-d-Desosamine. J Am Chem Soc. 2009;131:14030–14042. doi: 10.1021/ja903354k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mulder DW, Boyd ES, Sarma R, Lange RK, Endrizzi JA, Broderick JB, Peters JW. Stepwise [lsqb]FeFe[rsqb]-hydrogenase H-cluster assembly revealed in the structure of HydA[Dgr]EFG. Nature. 2010;465:248–251. doi: 10.1038/nature08993. [DOI] [PubMed] [Google Scholar]
- 33.Okada Y, Yamagata K, Hong K, Wakayama T, Zhang Y. A role for the elongator complex in zygotic paternal genome demethylation. Nature. 2010;463:554–558. doi: 10.1038/nature08732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Driesener R, Challand M, Mcglynn S, Shepard E, Boyd E, Broderick J, Peters J, Roach P. [FeFe]-Hydrogenase Cyanide Ligands Derived From S-Adenosylmethionine-Dependent Cleavage of Tyrosine. Angew Chem Int Ed. 2010;49:1687–1690. doi: 10.1002/anie.200907047. [DOI] [PubMed] [Google Scholar]
- 35.Grove TL, Ahlum JH, Sharma P, Krebs C, Booker SJ. A Consensus Mechanism for Radical SAM-Dependent Dehydrogenation? BtrN Contains Two [4Fe-4S] Clusters. Biochem. 2010;49:3783–3785. doi: 10.1021/bi9022126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yan F, Lamarre JM, Röhrich R, Wiesner J, Jomaa H, Mankin AS, Fujimori DG. RlmN and Cfr are Radical SAM Enzymes Involved in Methylation of Ribosomal RNA. J Am Chem Soc. 2010;132:3953–3964. doi: 10.1021/ja910850y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wecksler SR, Stoll S, Tran H, Magnusson OT, Wu SP, King D, Britt RD, Klinman JP. Pyrroloquinoline Quinone Biogenesis: Demonstration That PqqE from Klebsiella pneumoniae Is a Radical S-Adenosyl-l-methionine Enzyme. Biochem. 2009;48:10151–10161. doi: 10.1021/bi900918b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grove TL, Lee K-H, St Clair J, Krebs C, Booker SJ. In Vitro Characterization of AtsB, a Radical SAM Formylglycine-Generating Enzyme That Contains Three [4Fe-4S] Clusters. Biochem. 2008;47:7523–7538. doi: 10.1021/bi8004297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frey PA, Magnusson OT. S-Adenosylmethionine: a wolf in sheep’s clothing, or a rich man’s adenosylcobalamin? Chem Rev. 2003;103:2129–2148. doi: 10.1021/cr020422m. [DOI] [PubMed] [Google Scholar]
- 40.Frey PA, Hegeman AD, Ruzicka FJ. The radical SAM superfamily. Crit Rev Biochem Mol Biol. 2008;43:63–88. doi: 10.1080/10409230701829169. [DOI] [PubMed] [Google Scholar]
- 41.Farrar CE, Siu KKW, Howell PL, Jarrett JT. Biotin Synthase Exhibits Burst Kinetics and Multiple Turnovers in the Absence of Inhibition by Products and Product-Related Biomolecules. Biochem. 2011;49:9985–9996. doi: 10.1021/bi101023c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grove TL, Benner JS, Radle MI, Ahlum JH, Landgraf BJ, Krebs C, Booker SJ. A Radically Different Mechanism for S-Adenosylmethionine Dependent Methyltransferases. Science. 2011;332:604–607. doi: 10.1126/science.1200877. [DOI] [PubMed] [Google Scholar]
- 43.Fairhead H, Setlow P. Binding of DNA to alpha/beta-type small, acid-soluble proteins from spores of Bacillus or Clostridium species prevents formation of cytosine dimers, cytosine-thymine dimers, and bipyrimidine photoadducts after UV irradiation. J Bacteriol. 1992;174:2874–2880. doi: 10.1128/jb.174.9.2874-2880.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin G, Li L. Elucidation of Spore-Photoproduct Formation by Isotope Labeling. Angew Chem Int Ed. 2010;49:9926–9929. doi: 10.1002/anie.201005228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheek J, Broderick J. Direct H atom abstraction from spore photoproduct C-6 initiates DNA repair in the reaction catalyzed by spore photoproduct lyase: Evidence for a reversibly generated adenosyl radical intermediate. J Am Chem Soc. 2002;124:2860–2861. doi: 10.1021/ja017784g. [DOI] [PubMed] [Google Scholar]
- 46.Slieman TA, Rebeil R, Nicholson WL. Spore photoproduct (SP) lyase from Bacillus subtilis specifically binds to and cleaves SP (5-thyminyl-5,6-dihydrothymine) but not cyclobutane pyrimidine dimers in UV-irradiated DNA. J Bacteriol. 2000;182:6412–6417. doi: 10.1128/jb.182.22.6412-6417.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pieck J, Hennecke U, Pierik A, Friedel M, Carell T. Characterization of a new thermophilic spore photoproduct lyase from geobacillus stearothermophilus (SplG) with defined lesion containing DNA substrates. J Biol Chem. 2006;281:36317–36326. doi: 10.1074/jbc.M607053200. [DOI] [PubMed] [Google Scholar]
- 48.Yang L, Lin G, Liu D, Dria KJ, Telser J, Li L. Probing the Reaction Mechanism of Spore Photoproduct Lyase (SPL) via Diastereoselectively Labeled Dinucleotide SP TpT Substrates. J Am Chem Soc. 2011;133:10434–10447. doi: 10.1021/ja110196d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Setlow P. Small, acid-soluble spore proteins of Bacillus species: structure, synthesis, genetics, function, and degradation. Annu Rev Microbiol. 1988;42:319–338. doi: 10.1146/annurev.mi.42.100188.001535. [DOI] [PubMed] [Google Scholar]
- 50.Mason JM, Setlow P. Different small, acid-soluble proteins of the alpha/beta type have interchangeable roles in the heat and UV radiation resistance of Bacillus subtilis spores. J Bacteriol. 1987;169:3633–3637. doi: 10.1128/jb.169.8.3633-3637.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tovar-Rojo F, Setlow P. Effects of mutant small, acid-soluble spore proteins from Bacillus subtilis on DNA in vivo and in vitro. J Bacteriol. 1991;173:4827–4835. doi: 10.1128/jb.173.15.4827-4835.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Frenkiel-Krispin D, Sack R, Englander J, Shimoni E, Eisenstein M, Bullitt E, Horowitz-Scherer R, Hayes CS, Setlow P, Minsky A, Wolf SG. Structure of the DNA-SspC complex: implications for DNA packaging, protection, and repair in bacterial spores. J Bacteriol. 2004;186:3525–3530. doi: 10.1128/JB.186.11.3525-3530.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hayes DS, Peng ZY, Setlow P. Equilibrium and kinetic binding interactions between DNA and a group of novel, nonspecific DNA-binding proteins from spores of Bacillus and Clostridium species. J Biol Chem. 2000;275:35040–35050. doi: 10.1074/jbc.M005669200. [DOI] [PubMed] [Google Scholar]
- 54.Nicholson WL, Setlow B, Setlow P. Binding of DNA in vitro by a small, acid-soluble spore protein from Bacillus subtilis and the effect of this binding on DNA topology. J Bacteriol. 1990;172:6900–6906. doi: 10.1128/jb.172.12.6900-6906.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Griffith J, Makhov A, Santiagolara L, Setlow P. Electron-Microscopic Studies of the Interaction between a Bacillus-Subtilis Alpha/Beta-Type Small, Acid-Soluble Spore Protein with DNA - Protein-Binding Is Cooperative, Stiffens the DNA, and Induces Negative Supercoiling. Proc Nat Acad Sci. 1994;91:8224–8228. doi: 10.1073/pnas.91.17.8224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schreier WJ, Schrader TE, Koller FO, Gilch P, Crespo-Hernandez CE, Swaminathan VN, Carell T, Zinth W, Kohler B. Thymine Dimerization in DNA Is an Ultrafast Photoreaction. Science. 2007;315:625–629. doi: 10.1126/science.1135428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hayes CS, Alarcon-Hernandez E, Setlow P. N-terminal Amino Acid Residues Mediate Protein-Protein Interactions between DNA-bound alpha/beta-Type Small, Acid-soluble Spore Proteins from Bacillus Species. J Biol Chem. 2001;276:2267–2275. doi: 10.1074/jbc.M007858200. [DOI] [PubMed] [Google Scholar]
- 58.Varghese AJ. 5-Thyminyl-5,6-dihydrothymine from DNA irradiated with ultraviolet light. Biochem Biophys Res Comm. 1970;38:484–490. doi: 10.1016/0006-291x(70)90739-4. [DOI] [PubMed] [Google Scholar]
- 59.Varghese AJ. Photochemistry of thymidine in ice. Biochem. 1970;9:4781–4787. doi: 10.1021/bi00826a023. [DOI] [PubMed] [Google Scholar]
- 60.Varghese AJ. Photochemistry of thymidine as a thin solid film. Photochem Photobiol. 1971;13:357–364. [Google Scholar]
- 61.Chandor A, Berteau O, Douki T, Gasparutto D, Sanakis Y, Ollagnier-De-Choudens S, Atta M, Fontecave M. Dinucleotide spore photoproduct, a minimal substrate of the DNA repair spore photoproduct lyase enzyme from Bacillus subtilis. J Chem Biol. 2006;281:26922–26931. doi: 10.1074/jbc.M602297200. [DOI] [PubMed] [Google Scholar]
- 62.Mantel C, Chandor A, Gasparutto D, Douki T, Atta M, Fontecave M, Bayle PA, Mouesca JM, Bardet M. Combined NMR and DFT studies for the absolute configuration elucidation of the spore photoproduct, a UV-induced DNA lesion. J Am Chem Soc. 2008;130:16978–16984. doi: 10.1021/ja805032r. [DOI] [PubMed] [Google Scholar]
- 63.Chandra T, Silver SC, Zilinskas E, Shepard EM, Broderick WE, Broderick JB. Spore Photoproduct Lyase Catalyzes Specific Repair of the 5R but Not the 5S Spore Photoproduct. J Am Chem Soc. 2009;131:2420–2421. doi: 10.1021/ja807375c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Friedel M, Berteau O, Pieck J, Atta M, Ollagnier-De-Choudens S, Fontecave M, Carell T. The spore photoproduct lyase repairs the 5S- and not the 5R-configured spore photoproduct DNA lesion. Chem Comm. 2006:445–447. doi: 10.1039/b514103f. [DOI] [PubMed] [Google Scholar]
- 65.Wang SY. Photochemistry and photobiology of nucleic acids. Academic Press; New York: 1976. [Google Scholar]
- 66.Kim SJ, Lester C, Begley TP. Synthesis of the Dinucleotide Spore Photoproduct. J Org Chem. 1995;60:6256–6257. [Google Scholar]
- 67.Heil K, Kneuttinger AC, Schneider S, Lischke U, Carell T. Crystal Structures and Repair Studies Reveal the Identity and the Base-Pairing Properties of the UV-Induced Spore Photoproduct DNA Lesion. Chem Eur J. 2011;17:9651–9657. doi: 10.1002/chem.201100177. [DOI] [PubMed] [Google Scholar]
- 68.Lin G, Chen CH, Pink M, Pu J, Li L. Chemical Synthesis, Crystal Structure and Enzymatic Evaluation of a Dinucleotide Spore Photoproduct Analogue Containing a Formacetal Linker. Chem Eur J. 2011;17:9658–9668. doi: 10.1002/chem.201101821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hruska FE, Voituriez L, Grand A, Cadet J. Molecular structure of the cis-syn photodimer of d(TpT) (cyanoethyl ester) Biopolymers. 1986;25:1399–1417. doi: 10.1002/bip.360240512. [DOI] [PubMed] [Google Scholar]
- 70.Butenandt J, Eker APM, Carell T. Synthesis, crystal structure, and enzymatic evaluation of a DNA-photolesion isostere. Chem Eur J. 1998;4:642–654. [Google Scholar]
- 71.Camerman N, Camerman A. Crystal and Molecular Structure of Photodimer-a of 1,3-Dimethylthymine (Isomer in Irradiated Deoxyribonucleic Acid) J Am Chem Soc. 1970;92:2523. doi: 10.1021/ja00711a050. [DOI] [PubMed] [Google Scholar]
- 72.Leonard NJ, Golankiewicz K, Mccredie RS, Johnson SM, Paul IC. Synthetic spectroscopic models related to coenzymes and base pairs. III. A 1,1′-trimethylene-linked thymine photodimer of cis-syn structure. J Am Chem Soc. 1969;91:5855–5862. [Google Scholar]
- 73.Baer M, Sancar GB. Photolyases from Saccharomyces-Cerevisiae and Escherichia-Coli Recognize Common Binding Determinants in DNA Containing Pyrimidine Dimers. Mol Cell Biol. 1989;9:4777–4788. doi: 10.1128/mcb.9.11.4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kiener A, Husain I, Sancar A, Walsh C. Purification and Properties of Methanobacterium-Thermoautotrophicum DNA Photolyase. J Biol Chem. 1989;264:13880–13887. [PubMed] [Google Scholar]
- 75.Sancar A. Structure and Function of DNA Photolyase and Cryptochrome Blue-Light Photoreceptors. Chem Rev. 2003;103:2203–2238. doi: 10.1021/cr0204348. [DOI] [PubMed] [Google Scholar]
- 76.Kim ST, Sancar A. Effect of Base, Pentose, and Phosphodiester Backbone Structures on Binding and Repair of Pyrimidine Dimers by Escherichia-Coli DNA Photolyase. Biochem. 1991;30:8623–8630. doi: 10.1021/bi00099a019. [DOI] [PubMed] [Google Scholar]
- 77.Rebeil R, Sun Y, Chooback L, Pedraza-Reyes M, Kinsland C, Begley TP, Nicholson WL. Spore photoproduct lyase from Bacillus subtilis spores is a novel iron-sulfur DNA repair enzyme which shares features with proteins such as class III anaerobic ribonucleotide reductases and pyruvate-formate lyases. J Bacteriol. 1998;180:4879–4885. doi: 10.1128/jb.180.18.4879-4885.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rebeil R, Nicholson WL. The subunit structure and catalytic mechanism of the Bacillus subtilis DNA repair enzyme spore photoproduct lyase. Proc Nat Acad Sci. 2001;98:9038–9043. doi: 10.1073/pnas.161278998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Silver S, Chandra T, Zilinskas E, Ghose S, Broderick W, Broderick J. Complete stereospecific repair of a synthetic dinucleotide spore photoproduct by spore photoproduct lyase. J Biol Inorg Chem. 2010;15:943–955. doi: 10.1007/s00775-010-0656-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Roberts RJ, Cheng X. Base flipping. Annu Rev Biochem. 1998;67:181–198. doi: 10.1146/annurev.biochem.67.1.181. [DOI] [PubMed] [Google Scholar]
- 81.Vande Berg BJ, Sancar GB. Evidence for Dinucleotide Flipping by DNA Photolyase. J Biol Chem. 1998;273:20276–20284. doi: 10.1074/jbc.273.32.20276. [DOI] [PubMed] [Google Scholar]
- 82.Christine KS, Macfarlane AW, Yang K, Stanley RJ. Cyclobutylpyrimidine Dimer Base Flipping by DNA Photolyase. J Biol Chem. 2002;277:38339–38344. doi: 10.1074/jbc.M206531200. [DOI] [PubMed] [Google Scholar]
- 83.Yang C-G, Garcia K, He C. Damage Detection and Base Flipping in Direct DNA Alkylation Repair. ChemBioChem. 2009;10:417–423. doi: 10.1002/cbic.200800580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Donnellan JE, Jr, Setlow RB. Thymine Photoproducts but not Thymine Dimers Found in Ultraviolet-Irradiated Bacterial Spores. Science. 1965;149:308–310. doi: 10.1126/science.149.3681.308. [DOI] [PubMed] [Google Scholar]
- 85.Mehl RA, Begley TP. Mechanistic studies on the repair of a novel DNA photolesion: The spore photoproduct. Org Lett. 1999;1:1065–1066. doi: 10.1021/ol9908676. [DOI] [PubMed] [Google Scholar]
- 86.Frey P, Magnusson O. S-Adenosylmethionine: A wolf in sheep’s clothing, or a rich man’s adenosylcobalamin? Chem Rev. 2003;103:2129–2148. doi: 10.1021/cr020422m. [DOI] [PubMed] [Google Scholar]
- 87.Chen D, Walsby C, Hoffman BM, Frey PA. Coordination and mechanism of reversible cleavage of S-adenosylmethionine by the [4Fe-4S] center in lysine 2,3-aminomutase. J Am Chem Soc. 2003;125:11788–11789. doi: 10.1021/ja036120z. [DOI] [PubMed] [Google Scholar]
- 88.Moss M, Frey PA. The role of S-adenosylmethionine in the lysine 2,3-aminomutase reaction. J Biol Chem. 1987;262:14859–14862. [PubMed] [Google Scholar]
- 89.Baraniak J, Moss ML, Frey PA. Lysine 2,3-aminomutase. Support for a mechanism of hydrogen transfer involving S-adenosylmethionine. J Biol Chem. 1989;264:1357–1360. [PubMed] [Google Scholar]
- 90.Guo J, Luo Y, Himo F. DNA repair by spore photoproduct lyase: A density functional theory study. J Phys Chem B. 2003;107:11188–11192. [Google Scholar]
- 91.Himo F. C-C bond formation and cleavage in radical enzymes, a theoretical perspective. BBA-Bioenergenices. 2005;1707:24–33. doi: 10.1016/j.bbabio.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 92.Fajardo-Cavazos P, Rebeil R, Nicholson W. Essential cysteine residues in Bacillus subtilis spore photoproduct lyase identified by alanine scanning mutagenesis. Curr Microbiol. 2005;51:331–335. doi: 10.1007/s00284-005-0052-8. [DOI] [PubMed] [Google Scholar]
- 93.Chandor-Proust A, Berteau O, Douki T, Gasparutto D, Ollagnier-De-Choudens S, Fontecave M, Atta M. DNA repair and free radicals, new insights into the mechanism of spore photoproduct lyase revealed by single amino Acid substitution. J Biol Chem. 2008;283:36361–36368. doi: 10.1074/jbc.M806503200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Stubbe J, Van Der Donk WA. Protein Radicals in Enzyme Catalysis. Chem Rev. 1998;98:705–762. doi: 10.1021/cr9400875. [DOI] [PubMed] [Google Scholar]
- 95.Beck WS, Abeles RH, Robinson WG. Transfer of hydrogen from cobamide coenzyme to water during enzymatic ribonucleotide reduction. Biochem Biophys Res Comm. 1966;25:421–425. doi: 10.1016/0006-291x(66)90222-1. [DOI] [PubMed] [Google Scholar]
- 96.Hogenkamp HPC, Ghambeer RK, Brownson C, Blakley RL, Vitols E. Cobamides and Ribonucleotide Reduction. J Biol Chem. 1968;243:799–808. [PubMed] [Google Scholar]
- 97.Knappe J, Sawers G. A Radical-Chemical Route to Acetyl-Coa - the Anaerobically Induced Pyruvate Formate-Lyase System of Escherichia-Coli. Fems Microbiol Rev. 1990;75:383–398. doi: 10.1111/j.1574-6968.1990.tb04108.x. [DOI] [PubMed] [Google Scholar]
- 98.Knappe J, Volker Wagner AF. In: Methods in Enzymology. Judith PK, editor. Vol. 258. Academic Press; 1995. pp. 343–362. [DOI] [PubMed] [Google Scholar]
- 99.Marsh ENG, Li L. Mechanism of benzylsuccinate synthase probed by substrate and isotope exchange. J Am Chem Soc. 2006;128:16056–16057. doi: 10.1021/ja067329q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Duschene KS, Broderick JB. The antiviral protein viperin is a radical SAM enzyme. FEBS Lett. 2010;584:1263–1267. doi: 10.1016/j.febslet.2010.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Padovani D, Thomas F, Trautwein AX, Mulliez E, Fontecave M. Activation of class III ribonucleotide reductase from E-coli. The electron transfer from the iron-sulfur center to S-adenosylmethionine. Biochem. 2001;40:6713–6719. doi: 10.1021/bi002936q. [DOI] [PubMed] [Google Scholar]
- 102.Wang SC, Frey PA. S-adenosylmethionine as an oxidant: the radical SAM superfamily. Trends Biochem Sci. 2007;32:101–110. doi: 10.1016/j.tibs.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 103.Wang SC, Frey PA. Binding energy in the one-electron reductive cleavage of S-adenosylmethionine in lysine 2,3-aminomutase, a radical SAM enzyme. Biochem. 2007;46:12889–12895. doi: 10.1021/bi701745h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lawrence CC, Stubbe J. The function of adenosylcobalamin in the mechanism of ribonucleoside triphosphate reductase from Lactobacillus leichmannii. Curr Opin Chem Biol. 1998;2:650–655. doi: 10.1016/s1367-5931(98)80097-5. [DOI] [PubMed] [Google Scholar]
- 105.Stubbe J, Nocera DG, Yee CS, Chang MCY. Radical initiation in the class I ribonucleotide reductase: Long-range proton-coupled electron transfer? Chem Rev. 2003;103:2167–2201. doi: 10.1021/cr020421u. [DOI] [PubMed] [Google Scholar]
- 106.Friedel M, Pieck J, Klages J, Dauth C, Kessler H, Carell T. Synthesis and stereochemical assignment of DNA spore photoproduct analogues. Chem Eur J. 2006;12:6081–6094. doi: 10.1002/chem.200600169. [DOI] [PubMed] [Google Scholar]