Abstract
Genomic imprinting is an epigenetic process resulting in the monoallelic parent-of-origin-specific expression of a subset of genes in the mammalian genome. The parental alleles are differentially marked by DNA methylation during gametogenesis when the genomes are in separate compartments. How methylation machinery recognizes and differentially modifies these imprinted regions in germ cells remains a key question in the field. While studies have focused on determining a sequence signature that alone could distinguish imprinted regions from the rest of the genome, recent reports do not support such a hypothesis. Rather, it is becoming clear that features such as transcription, histone modifications and higher order chromatin are employed either individually or in combination to set up parental imprints.
Introduction
Genomic imprinting affects a small number of genes in the mammalian genome and results in parent-of-origin-specific monoallelic expression [1–3]. Imprinted genes, which are typically conserved among mammals, play essential roles in the growth and development of the fetus, as well as in post-natal behavior and metabolism. Further, while many imprinted genes are ubiquitously imprinted, some exhibit tissue- or temporal-specific imprinting patterns. The best-defined class of genes that display restricted imprinting are those that are imprinted exclusively in the placenta, including Ascl2, Phlda2, Slc22a2 and Slc22a3 [4]. Notably, imprinted genes are located in approximately 1 Mb clusters throughout the genome, although singletons have been described (http://www.har.mrc.ac.uk/research/genomic_imprinting/). These clusters typically contain genes that are expressed exclusively from the maternally or paternally-inherited chromosomes. Additionally, each of these clusters is under the control of a discrete region, termed imprinting control region (ICR, also designated imprinting center or imprinting control element).
One critical attribute of imprinted genes is that they must be marked with their parental origin so that the correct allele-specific expression patterns are observed in somatic tissues. The parental-specific mark must be stable and heritable so that imprinting is maintained throughout development. They must also be erasable so that imprints can be reset from a biparental somatic pattern (germ cells are derived from the soma in mammals) to the germline-specific pattern that reflects the sex of the individual. Moreover, the most logical time for alleles to be marked is in the germline when they are in separate compartments and can be differentially modified as epigenetic reprogramming takes place (Figure 1). DNA methylation is the epigenetic modification that appears to be most integral to the marking of parental alleles, although post-translational histone modifications are clearly involved in imprinted gene expression (see below). Allele-specific DNA methylation has been described at all of the imprinted gene clusters and many other single imprinted genes. These differentially methylated regions (DMRs) are established in the germline by de novo DNA methyltransferases (DNMTs, see below) and are either maintained throughout development (primary DMRs) or arise later in development (secondary DMRs), often as a consequence of imprinted expression. Furthermore germline-specific DMRs often expand or contract during development [5*], although the reason for this is unclear. Many primary DMRs are also ICRs as gene targeting experiments have shown that deletion of the region corresponding to the DMR causes loss of imprinting of multiple genes in cis [1]. Additionally, all identified ICRs to date are marked by differential methylation. Curiously, many more maternally-methylated than paternally-methylated DMRs/ICRs have been identified. Subsequently, all DMRs referred to here are primary DMRs unless otherwise noted.
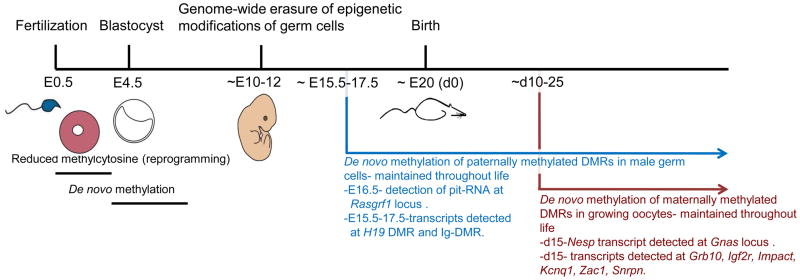
Figure 1. Timeline of epigenetic reprogramming.
The first wave of genome-wide DNA demethylation takes place shortly after fertilization, with the maternal genome passively demethylated and the methylcytosine of the paternal genome converted into hydroxymethylcytosine, which is passively eliminated. Imprinted DMRs are maintained despite this demethylation event. De novo DNA methylation occurs around implantation. In both male and female primordial germ cells another wave of DNA demethylation initiates as the cells migrate toward the genital ridge. All DMRs are also erased at this time. In male germ cells methylation imprints are acquired in prospermatogonia around E15.5–17.5. In the female germline methylation imprints are not acquired until after birth, in growing oocytes. Transcripts have been correlated with acquisition of DNA methylation in both the male and female germlines. Activities in male and female germ cells are represented in blue and red, respectively.
Once imprints are set in the germline, they must be properly maintained following fertilization when the genome undergoes a period of considerable reprogramming (Figure 1). This reprogramming involves the re-setting of DNA methylation patterns, which is now known to involve conversion of methylcytosine to hydroxymethylcytosine [6*,7*], likely followed by replication dependent passive demethylation [8*]. Importantly, DNA methylation imprints must not only survive the conversion to hydroxymethylcytosine but also be maintained by the small amount of DNMT protein present in the preimplantation embryo [9,10]. Although much of this aspect of imprinting remains poorly defined, a number of factors have been described that when mutated result in the failure to maintain imprints [11].
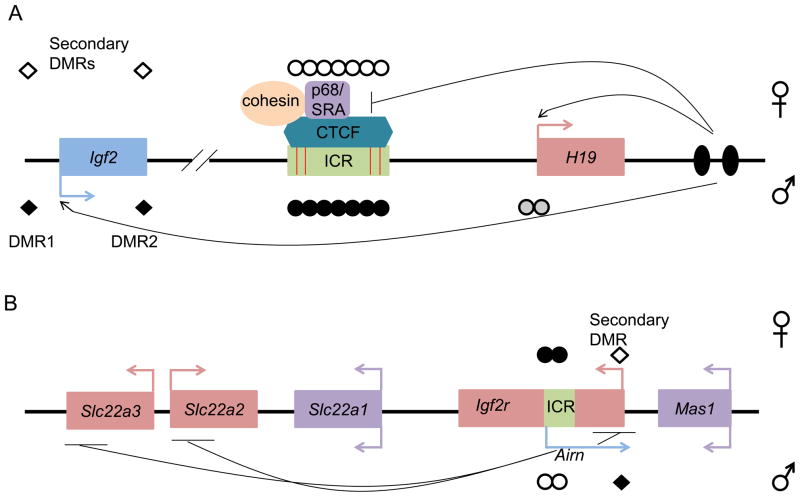
Two dominating mechanisms have been described for mediating imprinting in clusters [1–3]. Thus far, the most evolutionarily ancient [12], but seemingly least utilized mechanism is the insulator model of imprinting, which is employed by the H19/Igf2 imprinted locus. The maternally-expressed H19 gene and paternally-expressed Igf2 gene share enhancers and their reciprocal imprinting is governed by a CTCF-dependent insulator that is located between the genes. On the maternal allele in mouse, CTCF binds to 4 binding sites within the ICR, generating an insulator that prevents Igf2 from accessing the shared enhancers that are located on the H19 side of the insulator. Thus, the insulator effectively acts as an enhancer-blocker. On the paternal chromosome, methylation at the ICR not only prevents CTCF from binding, allowing Igf2 to engage the enhancers, but it is also required for methylation at the H19 promoter and silencing of H19 (Figure 2a). A more commonly employed and recently evolved mechanism of imprinting uses a long non-coding RNA (ncRNA). In this case, the ICR includes a differentially methylated promoter that regulates the expression of an ncRNA; when unmethylated the ncRNA is expressed and represses cis-linked genes. In contrast, when the ICR is methylated, the ncRNA is repressed and the cis-linked genes are expressed. The maternally-expressed Igf2r was first shown to use such a mechanism (Figure 2b). At this locus, the ncRNA Airn represses Igf2r ubiquitously and Slc22a2 and Slc22a3 in the placenta. Just how this repression is mediated is unclear but it has been proposed that Airn interacts with the Slc22a3 promoter and the H3K9 histone methyltransferase G9a in placenta, thereby epigenetically silencing transcription [13]. Alternatively, transcription through the domain has been suggested to silence genes in cis [2]. It is possible that both mechanisms are used, but in a tissue-specific manner.
Figure 2. Depiction of insulator and ncRNA mediated imprinting.
(a) Insulator-mediated imprinting at the H19/Igf2 locus. The maternal allele is represented above the line whereas the paternal allele is below the line. Depicted here are the maternally expressed H19 (pink box with pink arrow) and paternally expressed Igf2 (blue box with blue arrow) genes. On the maternal allele the ICR (green box) remains unmethylated (open circles) allowing CTCF (binding sites depicted by red bars) and its cofactors (cohesins and p68/SRA) to bind. This interaction mediates enhancer blocking allowing downstream enhancers (black ovals) to access the H19 promoter. Paternal methylation at the ICR (black circles) prevents CTCF binding and together with methylation at the H19 promoter (grey circles) allows the enhancers to access Igf2. Paternal methylation at secondary DMRs (diamonds), DMR1 and DMR2, occurs after fertilization. (b) ncRNA-mediated imprinting at the Igf2r locus. The maternal allele is represented above the line whereas the paternal allele is below the line. Depicted are the maternally expressed Slc22a3, Slc22a2, Igf2r (pink boxes with pink arrows) the paternally expressed ncRNA Airn (blue arrow) and non-imprinted Mas1 and Slc22a1 (purple boxes with purple arrows). The ICR (green box), which is hypermethylated on the maternal allele (black circles), includes the Airn promoter. The hypomethylated ICR (open circles) on the paternal allele allows Airn expression, which represses Slc222a2, Slc22a3 and Igf2r in cis. A secondary paternally methylated DMR (diamonds) is located at the Igf2r promoter. This DMR is not methylated until after transcription occurs through the region. Loci are not drawn to scale.
Here, we will focus on current developments in the field of imprinting. Although the mechanisms underlying genomic imprinting are slowly being elucidated, some of the most significant developments have centered on characterizing the earliest steps in the imprinting process, the recognition and marking of imprinted regions in germ cells.
Methylation machinery and recognition of DMRs in germ cells
Experiments by Jaenisch and colleagues, which deleted the maintenance methyltransferase Dnmt1, first supported a central role for DNA methylation for imprinted gene expression [14]. Studies of conditional knockouts of the de novo methyltransferases Dnmt3a and Dnmt3b in germ cells provided evidence that DNMT3a is required for de novo methylation of all DMRs (both maternal and paternal) except for the paternally-methylated Rasgrf1 ICR, which uses both DNMT3a and DNMT3b [15,16]. A defect in the establishment of methylation of germline DMRs was also observed in mice deficient for DNMT3-like (DNMT3L) [17,18]. DNMT3L, which lacks methyltransferase activity, has the ability to interact with both DNMT3a and DNMT3b. Oocytes deficient for DNMT3L lacked methylation at maternal DMRs. Additionally, Dnmt3L null females exhibited a maternal lethal phenotype, with embryos dying around E9.5. These embryos were hypomethylated at all maternally-methylated DMRs but global methylation did not appear reduced [17]. DNMT3L also plays a critical role in establishment of paternal methylation imprints. Though reports vary on the extent that paternal DMRs were affected in DNMT3L deficient male germ cells, ICRs at H19, Rasgrf1 and Gtl2 (Ig-DMR) require DNMT3L for full methylation [15,16,18,19].
Though the DNA methylation machinery is now well-established, it remains to be determined how this machinery is recruited to specific CpGs. Recently, a link has been found between histone methylation and DNA methylation. DNMT3L has binding affinity for nucleosomes containing unmethylated H3K4, which is abolished with the addition of methyl groups to this residue [20]. These results suggest that patterns of histone methylation could dictate patterns of DNA methylation, which could then be stably inherited. Furthermore, oocytes that lack lysine demethylase 1B (KDM1B), which functions as an H3K4 demethylase, were shown to be hypomethylated at a number of imprinted loci [21*], suggesting that an unmethylated H3K4 is necessary for DNA methylation. Consistently, male germ cells assayed at E15.5 (the onset of de novo methylation) revealed high levels of H3K4me3 at the maternally-methylated Snrpn ICR and KvDMR but absence of H3K4me3 at paternally-methylated H19 ICR and Ig-DMR, suggesting that H3K4me3 prevents maternally-methylated DMRs from acquiring DNA methylation in the male germline [22*]. Interestingly, the association of DNA methylation with an unmethylated H3K4 and the protection from DNA methylation with methylation of H3K4 is not limited to imprinted loci, but has been observed genome-wide [23,24].
Even with the observation that DNA methylation requires a favorable histone environment, it is still unclear what (if any) sequence signature distinguishes DMRs from other CpG rich regions in the genome. One such signature has been proposed to be the spacing of CpGs at DMRs. Molecular modeling of the DNMT3a/DNMT3L complex indicated an optimal periodicity of CpGs for methylation at about 8–10 base pairs [25]. Interestingly, Jia and colleagues [25] reported this periodicity in 12 maternal DMRs. However, recent studies were unable to detect this trend at DMRs in both oocytes and sperm [5*] or were unable to detect a difference in CpG spacing between methylated and unmethylated CpG islands (all CpG rich regions in the genome, of which DMRs are only a small subset) in oocytes and sperm [26*]. Overall, the role of CpG spacing as a signature for germline DMRs remains unclear.
Another proposed sequence feature that could distinguish DMRs from the rest of the genome is the composition of repetitive elements at imprinted loci [27]. Sequence analysis reveals a depletion of SINE elements [28,29] but an enrichment of tandem repeats [30] at imprinted regions. Thus, it has been hypothesized that the presence of certain repetitive elements or the absence of others could be the key distinguishing trait recognized by methylation machinery [27]. Recently, it has been confirmed that DMRs have a higher overall concentration of tandem repeats than CpG islands [5*,30]. However, this comparative analysis could be skewed by the Rasgrf1 ICR, which is exceptionally repeat rich. Moreover, there are still a substantial number of CpG islands that contain more tandem repeats than many DMRs [5*]. Genome-wide analysis of CpG islands found no correlation between frequency of tandem repeats and methylation status [26*]. Thus, although DMRs are located in regions within or adjacent to tandem repeats, it does not seem that this is their key distinguishing feature, or necessarily a feature that would attract methylation machinery. Rather than there being a single sequence trait of all DMRs, each DMR may contain a locus-specific signature that differentiates it from the rest of the genome.
Active transcription as a signal for de novo methylation
Many reports have recently uncovered a link between transcription and methylation, which at first glance is counter to expectations given the general observation that DNA methylation is associated with gene repression. For example, as described above, KDM1B is critical for establishment of methylation of many maternal DMRs [21*]. Interestingly, the human orthologue of KDM1B, LSD2, is associated with gene bodies of actively transcribed genes [31]. Additionally, the PWWP domain of DNMT3a binds H3K36me3, a mark of transcriptional elongation, which increases the methyltransferase activity of DNMT3a in vitro [32]. These observations suggest that de novo methylation is targeted to sites of active transcription. Analysis of CpG islands in day 10 oocytes, the time at which de novo methylation is first apparent, has provided further evidence for the requirement of transcription for methylation. Intragenic methylated CpG islands are more likely to be within active transcription units than unmethylated intragenic CpG islands. Moreover, methylated CpG islands within annotated promoters more frequently overlap transcripts than unmethylated CpG islands within promoters [26*]. One possible explanation for the link between active transcription and methylation is that transcription may help maintain open chromatin, which in turn allows methylation machinery to access DNA.
Multiple imprinted loci have been shown to require transcription for establishment of DNA methylation; the first of these being the Gnas locus [33*]. This locus encodes numerous transcripts; the protein coding transcripts Gnas, Gnasx1 and Nesp and the non-coding transcripts Nespas and 1A. This region contains two maternally-methylated DMRs, one that encompasses the Gnasx1 and Nespas promoters, which acts as the ICR, and another that covers the 1A promoter. Chotalia and colleagues made a targeted mutation in the mouse that truncated the Nesp transcript, the furthest upstream transcript, and observed hypomethylation (in varying degrees) at all DMRs at the Gnas locus in newborn pups [33*]. Furthermore, analysis of mutant oocytes revealed a loss of methylation at the DMRs, suggesting a defect in methylation establishment. Interestingly, transcripts were also detected in growing oocytes at the maternally-methylated DMRs of the Grb10, Igf2r, Impact, Kcnq1, Zac1 and Snrpn imprinted loci [33*,34] (Figure 1).
The imprinted Rasgrf1 locus, which harbors a paternally-methylated ICR, also requires transcription for methylation establishment [35*]. In spermatogonia of mice mutant for various proteins in the piRNA pathway, including MILI, MIWI2 and MITOPLD, the Rasgrf1 ICR, but not other paternally-methylated DMRs, exhibited reduced methylation. Further analysis revealed that a non-coding RNA (named pit-RNA) transcribed from the Rasgrf1 ICR was targeted by piRNAs, causing cleavage of this RNA. The authors propose a model in which targeting of piRNAs to pit-RNA is an important step in sequence specific methylation at the Rasgrf1 locus [35*].
Though paternal-specific methylation of the H19 ICR and Ig-DMR does not require the piRNA pathway [35*], transcription has been detected at both of these ICRs [22*]. In contrast to the absence of transcription at these ICRs in somatic cells or E13.5 male germ cells (prior to de novo methylation), high levels of RNA can be detected starting at E15.5 (the onset of de novo methylation), through E17.5 in male germ cells (Figure 1). Transcription was detected throughout both ICRs and in the region between H19 and its ICR, as well as downstream of the H19 promoter, and between Gtl2 and its ICR [22*]. Transgenic experiments that incorporate H19 sequences at an ectopic locus are consistent with the idea that sequences outside of the ICR are required for DNA methylation establishment in the germline. For example, when the H19 ICR is targeted to an ectopic locus, it fails to establish methylation in sperm [36,37]. In contrast, when the H19 ICR plus upstream sequences (ranging from −4.4kb to +2.9kb from the H19 transcriptional start site) is inserted, a subset of spermatozoa were fully methylated at this exogenous site [37]. Further, a BAC transgene, including sequences from −7kb to +140kb to the H19 transcriptional start site, properly established methylation at the H19 ICR [37]. Consistently, numerous cis-acting ICR mutations that disrupt imprinted gene expression or maintenance of methylation during embryogenesis, still properly establish methylation in sperm [38–40]. Thus, it is possible that these additional sequences contain transcripts necessary for de novo methylation, though this hypothesis is untested. Together, these studies provide evidence for the requirement of transcription at both maternally- and paternally-methylated ICRs.
Locus-specific features at the H19 ICR are necessary for proper epigenetic landscape and genomic organization during spermatogenesis
Accumulating evidence argues against a single signature for all DMRs, but rather, there are likely to be locus-specific features. As described above, the H19/Igf2 locus utilizes a CTCF-dependent insulator to repress Igf2 maternally and methylation of the ICR to repress H19 paternally [41–44] (Figure 2a). Interestingly, asymmetric acquisition of DNA methylation in the germline has been observed at the H19 ICR, with the paternal allele being methylated first [22*,45,46*], indicating that the two alleles retain somatic cell memory. Recent studies in male germ cells suggest that CTCF binding may coordinate this somatic memory mark, as functional CTCF binding sites are required for allele-specific histone modifications and inter-chromosomal interactions. Lee and colleagues observed a slight enrichment of H3K4me2 at the maternal H19 ICR and reciprocally, a slight enrichment of H3K9me3 at the paternal H19 ICR in E13.5–14.5 prospermatogonia [46*]. However, when the maternal H19 ICR was mutated in such a way that CTCF could not bind, the biased enrichment was no longer detected. Furthermore, delayed de novo methylation at the H19 ICR of the maternal allele was no longer detectable when the maternal allele was mutated at the CTCF sites [46*], suggesting that maternal H3K4me2 (which requires CTCF binding) protects this allele from DNA methylation and needs to be demethylated before de novo DNA methylation can take place. These data indicate that intact CTCF sites coordinate allele specific histone modifications that facilitate marking of the parental origin of each allele.
Inter-chromosomal interactions also appear to be dependent on CTCF binding sites at the H19 ICR in male germ cells. Physical interactions of the H19/Igf2 region with other imprinted regions such as Copg2, Htr2a and Dlk1 have been observed in spermatogonia. However, these interactions were not detectable when a mutated H19 ICR that does not bind CTCF was maternally inherited [47]. Recent reports have indicated that CTCF interacts with cofactors such as cohesins [48–52], the DEAD-box RNA-binding protein p68 (p68), and the steroid receptor RNA activator (SRA) [53] at the H19 ICR. Though analysis of cofactor interaction with CTCF has not been performed in germ cells, it is possible that these various cofactors facilitate higher order chromatin at the H19 locus. The loss of CTCF binding suggests that these factors may no longer mediate inter-chromosomal interactions or allele specific histone modifications. Nevertheless, it still remains to be determined if and how inter-chromosomal interactions are involved in germline reprogramming.
Conclusions
Significant questions remain in the study of how imprinted loci are marked with their parental origin, escape post-fertilization reprogramming and are appropriately regulated in the soma. Accumulating evidence suggests that locus-specific sequences, transcription, histone modifications and higher order chromatin structure are employed either individually or in combination for these processes. Here we focused on the process of marking the parental origin of imprinted genes. Interestingly, there is a correlation between transcription at DMRs and methylation establishment. Whereas the Gnas and Rasgrf1 loci clearly require transcription for DNA methylation establishment, cause and effect is less clear at other imprinted loci. The presence of RNA is not sufficient to prove that DNA methylation machinery requires active transcription. For example, Henckel and colleagues detected transcriptional activity at a number of maternally-methylated DMRs in male germ cells [22*], which is contrary to the idea that transcripts are necessary for methylation (as these regions remain unmethylated in male germ cells). Experiments that truncate or delete individual transcripts will be required to determine the role of these transcripts in methylation establishment.
In the future it will be important to interrogate chromatin structure and dynamics during methylation establishment in germ cells. Currently, such experiments are not feasible due to the large number of cells required and the difficulty in obtaining sufficient numbers of germ cells. Nevertheless, as technology advances and becomes more sensitive, elucidation of the mechanism and dynamics of reprogramming in the germline will become possible.
Acknowledgments
We apologize to those whose work could not be mentioned due to space limitations. We thank members of the laboratory for critical reading of the manuscript. Work in the Bartolomei lab is supported by the NIH (GM051279, HD042026 and HD068157).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
- 1.Bartolomei MS. Genomic imprinting: employing and avoiding epigenetic processes. Gene Dev. 2009;23:2124–2133. doi: 10.1101/gad.1841409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barlow DP. Genomic Imprinting: A Mammalian Epigenetic Discovery Model. Annu Rev Genet. 2011;45:379–404. doi: 10.1146/annurev-genet-110410-132459. [DOI] [PubMed] [Google Scholar]
- 3.Ferguson-Smith AC. Genomic imprinting: the emergence of an epigenetic paradigm. Nat Rev Genet. 2011;12:565–575. doi: 10.1038/nrg3032. [DOI] [PubMed] [Google Scholar]
- 4.Frost JM, Moore GE. The importance of imprinting in the human placenta. PLoS Genet. 2010;6:e1001015. doi: 10.1371/journal.pgen.1001015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5*.Tomizawa Si, Kobayashi H, Watanabe T, Andrews S, Hata K, Kelsey G, Sasaki H. Dynamic stage-specific changes in imprinted differentially methylated regions during early mammalian development and prevalence of non-CpG methylation in oocytes. Development. 2011;138:811–820. doi: 10.1242/dev.061416. 15 germline DMRs were examined in mouse sperm, oocytes and embryos and it was determined that maternal gametic DMRs appeared as unmethylated islands in male germ cells, the extent of gametic DMRs differs significantly in germ cells compared to embryos and substantial non-CpG DNA methylation was evident in oocytes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6*.Iqbal K, Jin SG, Pfeifer GP, Szabo PE. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc Natl Acad Sci U S A. 2011;108:3642–3647. doi: 10.1073/pnas.1014033108. This work demonstrated that the paternal genome in zygotes is not actively demethylated, but rather, methycytosine is converted to hydroxymethylcytosine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7*.Wossidlo M, Nakamura T, Lepikhov K, Marques CJ, Zakhartchenko V, Boiani M, Arand J, Nakano T, Reik W, Walter J. 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat Commun. 2011;2 doi: 10.1038/ncomms1240. This group also showed that the paternal genome in zygotes is not actively demethylated. Instead methycytosine is converted to hydroxymethylcytosine. [DOI] [PubMed] [Google Scholar]
- 8*.Inoue A, Zhang Y. Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science. 2011;334:194. doi: 10.1126/science.1212483. This work demonstrated that the loss of hydroxymethylacytosine of the paternal genome during early embryogenesis is replication dependent. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cirio MC, Martel J, Mann M, Toppings M, Bartolomei M, Trasler J, Chaillet JR. DNA methyltransferase 1o functions during preimplantation development to preclude a profound level of epigenetic variation. Dev Biol. 2008;324:139–150. doi: 10.1016/j.ydbio.2008.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cirio MC, Ratnam S, Ding F, Reinhart B, Navara C, Chaillet JR. Preimplantation expression of the somatic form of Dnmt1 suggests a role in the inheritance of genomic imprints. BMC Dev Biol. 2008:8. doi: 10.1186/1471-213X-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weaver JR, Susiarjo M, Bartolomei MS. Imprinting and epigenetic changes in the early embryo. Mamm Genome. 2009;20:532–543. doi: 10.1007/s00335-009-9225-2. [DOI] [PubMed] [Google Scholar]
- 12.Smits G, Mungall AJ, Griffiths-Jones S, Smith P, Beury D, Matthews L, Rogers J, Pask AJ, Shaw G, VandeBerg JL, McCarrey JR, et al. Conservation of the H19 noncoding RNA and H19-IGF2 imprinting mechanism in therians. Nat Genet. 2008;40:971–976. doi: 10.1038/ng.168. [DOI] [PubMed] [Google Scholar]
- 13.Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R, Fraser P. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322:1717–1720. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- 14.Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366:362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- 15.Kaneda M, Okano M, Hata K, Sado T, Tsujimoto N, Li E, Sasaki H. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature. 2004;429:900–903. doi: 10.1038/nature02633. [DOI] [PubMed] [Google Scholar]
- 16.Kato Y, Kaneda M, Hata K, Kumaki K, Hisano M, Kohara Y, Okano M, Li E, Nozaki M, Sasaki H. Role of the Dnmt3 family in de novo methylation of imprinted and repetitive sequences during male germ cell development in the mouse. Hum Mol Genet. 2007;16:2272–2280. doi: 10.1093/hmg/ddm179. [DOI] [PubMed] [Google Scholar]
- 17.Bourc’his D, Xu GL, Lin CS, Bollman B, Bestor TH. Dnmt3L and the establishment of maternal genomic imprints. Science. 2001;294:2536–2539. doi: 10.1126/science.1065848. [DOI] [PubMed] [Google Scholar]
- 18.Bourc’his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–99. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- 19.Webster KE. Meiotic and epigenetic defects in Dnmt3L-knockout mouse spermatogenesis. Proc Natl Acad Sci U S A. 2005;102:4068–4073. doi: 10.1073/pnas.0500702102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ooi SKT, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin S-P, Allis CD, Cheng X, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21*.Ciccone DN, Su H, Hevi S, Gay F, Lei H, Bajko J, Xu G, Li E, Chen T. KDM1B is a histone H3K4 demethylase required to establish maternal genomic imprints. Nature. 2009;461:415–418. doi: 10.1038/nature08315. This study demonstrated the requirement for KDM1B, a histone H3K4 demethylase, for de novo methylation at a number of imprinted DMRs in oocytes. [DOI] [PubMed] [Google Scholar]
- 22*.Henckel A, Chebli K, Kota SK, Arnaud P, Feil R. Transcription and histone methylation changes correlate with imprint acquisirion in male germ cells. EMBO J. doi: 10.1038/emboj.2011.425. In press. This work investigated histone methylation and DNA methylation patterns in primordial germ cells. Additionally, transcripts were detected at paternal DMRs at the time of methylation establishment. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 24.Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, Gnirke A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jia D, Jurkowska RZ, Zhang X, Jeltsch A, Cheng X. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature. 2007;449:248–251. doi: 10.1038/nature06146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26*.Smallwood SA, Tomizawa S-i, Krueger F, Ruf N, Carli N, Segonds-Pichon A, Sato S, Hata K, Andrews SR, Kelsey G. Dynamic CpG island methylation landscape in oocytes and preimplantation embryos. Nat Genet. 2011;43:811–814. doi: 10.1038/ng.864. This study investigated over 1000 methylated CpG islands in oocytes to determine features that dictate methylation status and found correlation between transcription and methylation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walter J, Hutter B, Khare T, Paulsen M. Repetitive elements in imprinted genes. Cytogenet Genome Res. 2006;113:109–115. doi: 10.1159/000090821. [DOI] [PubMed] [Google Scholar]
- 28.Ke X, Thomas NS, Robinson DO, Collins A. The distinguishing sequence characteristics of mouse imprinted genes. Mamm Genome. 2002;13:639–645. doi: 10.1007/s00335-002-3038-x. [DOI] [PubMed] [Google Scholar]
- 29.Greally JM. Short interspersed transposable elements (SINEs) are excluded from imprinted regions in the human genome. Proc Natl Acad Sci U S A. 2002;99:327–332. doi: 10.1073/pnas.012539199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hutter B, Helms V, Paulsen M. Tandem repeats in the CpG islands of imprinted genes. Genomics. 2006;88:323–332. doi: 10.1016/j.ygeno.2006.03.019. [DOI] [PubMed] [Google Scholar]
- 31.Fang R, Barbera AJ, Xu Y, Rutenberg M, Leonor T, Bi Q, Lan F, Mei P, Yuan G-C, Lian C, Peng J, et al. Human LSD2/KDM1b/AOF1 Regulates Gene Transcription by Modulating Intragenic H3K4me2 Methylation. Mol Cell. 2010;39:222–233. doi: 10.1016/j.molcel.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dhayalan A, Rajavelu A, Rathert P, Tamas R, Jurkowska RZ, Ragozin S, Jeltsch A. The Dnmt3a PWWP Domain Reads Histone 3 Lysine 36 Trimethylation and Guides DNA Methylation. J Biol Chem. 2010;285:26114–26120. doi: 10.1074/jbc.M109.089433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33*.Chotalia M, Smallwood SA, Ruf N, Dawson C, Lucifero D, Frontera M, James K, Dean W, Kelsey G. Transcription is required for establishment of germline methylation marks at imprinted genes. Genes Dev. 2009;23:105–117. doi: 10.1101/gad.495809. First report showing the requirement of transcription for de novo methylation at an imprinted DMR. This work demonstrated the requirement of the Nesp transcript for establishment of methylation at DMRs in the Gnas locus. Additionally, transcripts were detected at numerous other imprinted DMRs in oocytes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mapendano CK, Kishino T, Miyazaki K, Kondo S, Yoshiura K, Hishikawa Y, Koji T, Niikawa N, Ohta T. Expression of the Snurf-Snrpn IC transcript in the oocyte and its putative role in the imprinting establishment of the mouse 7C imprinting domain. J Hum Genet. 2006;51:236–243. doi: 10.1007/s10038-005-0351-8. [DOI] [PubMed] [Google Scholar]
- 35*.Watanabe T, Tomizawa Si, Mitsuya K, Totoki Y, Yamamoto Y, Kuramochi-Miyagawa S, Iida N, Hoki Y, Murphy PJ, Toyoda A, Gotoh K, et al. Role for piRNAs and Noncoding RNA in de Novo DNA Methylation of the Imprinted Mouse Rasgrf1 Locus. Science. 2011;332:848–852. doi: 10.1126/science.1203919. This study reports that piRNAs are required for establishment of methylation at the Rasgrf1 ICR. The authors propose that a non-coding RNA at the Rasgrf1 locus is targeted by these piRNAs and that cleavage of this RNA is necessary for de novo methylation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park KY, Sellars EA, Grinberg A, Huang SP, Pfeifer K. The H19 differentially methylated region marks the parental origin of a heterologous locus without gametic DNA methylation. Mol Cell Biol. 2004;24:3588–3595. doi: 10.1128/MCB.24.9.3588-3595.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gebert C, Kunkel D, Grinberg A, Pfeifer K. H19 Imprinting Control Region Methylation Requires an Imprinted Environment Only in the Male Germ Line. Mol Cell Biol. 2009;30:1108–1115. doi: 10.1128/MCB.00575-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thorvaldsen JL, Duran KL, Bartolomei MS. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev. 1998;12:3693–3702. doi: 10.1101/gad.12.23.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Engel N, West AG, Felsenfeld G, Bartolomei MS. Antagonism between DNA hypermethylation and enhancer-blocking activity at the H19 DMD is uncovered by CpG mutations. Nat Genet. 2004;36:883–888. doi: 10.1038/ng1399. [DOI] [PubMed] [Google Scholar]
- 40.Ideraabdullah FY, Abramowitz LK, Thorvaldsen JL, Krapp C, Wen SC, Engel N, Bartolomei MS. Novel cis-regulatory function in ICR-mediated imprinted repression of H19. Dev Biol. 2011;355:349–357. doi: 10.1016/j.ydbio.2011.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Engel N, Thorvaldsen JL, Bartolomei MS. CTCF binding sites promote transcription initiation and prevent DNA methylation on the maternal allele at the imprinted H19/Igf2 locus. Hum Mol Genet. 2006;15:2945–2954. doi: 10.1093/hmg/ddl237. [DOI] [PubMed] [Google Scholar]
- 42.Schoenherr CJ, Levorse JM, Tilghman SM. CTCF maintains differential methylation at the Igf2/H19 locus. Nat Genet. 2002;33:66–69. doi: 10.1038/ng1057. [DOI] [PubMed] [Google Scholar]
- 43.Pant V, Mariano P, Kanduri C, Mattsson A, Lobanenkov V, Heuchel R, Ohlsson R. The nucleotides responsible for the direct physical contact between the chromatin insulator protein CTCF and the H19 imprinting control region manifest parent of origin-specific long-distance insulation and methylation-free domains. Genes Dev. 2003;17:586–590. doi: 10.1101/gad.254903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fedoriw AM, Stein P, Svoboda P, Schultz RM, Bartolomei MS. Transgenic RNAi reveals essential function for CTCF in H19 gene imprinting. Science. 2004;303:238–240. doi: 10.1126/science.1090934. [DOI] [PubMed] [Google Scholar]
- 45.Davis TL, Yang GJ, McCarrey JR, Bartolomei MS. The H19 methylation imprint is erased and re-established differentially on the parental alleles during male germ cell development. Hum Mol Genet. 2000;9:2885–2894. doi: 10.1093/hmg/9.19.2885. [DOI] [PubMed] [Google Scholar]
- 46*.Lee DH, Singh P, Tsai SY, Oates N, Spalla A, Spalla C, Brown L, Rivas G, Larson G, Rauch TA, Pfeifer GP, et al. CTCF-dependent chromatin bias constitutes transient epigenetic memory of the mother at the H19-Igf2 imprinting control region in prospermatogonia. PLoS Genet. 2010;6:e1001224. doi: 10.1371/journal.pgen.1001224. Enrichment of H3K4me2 at the maternal H19 ICR and H3K9me3 at the paternal H19 ICR in prospermatogonia was reported. The timing correlates with asymmetric acquisition of DNA methylation on the two parental alleles (the paternal allele is methylated first), all of which required intact CTCF sites on the maternal allele. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sandhu KS, Shi C, Sjolinder M, Zhao Z, Gondor A, Liu L, Tiwari VK, Guibert S, Emilsson L, Imreh MP, Ohlsson R. Nonallelic transvection of multiple imprinted loci is organized by the H19 imprinting control region during germline development. Genes Dev. 2009;23:2598–2603. doi: 10.1101/gad.552109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nativio R, Wendt KS, Ito Y, Huddleston JE, Uribe-Lewis S, Woodfine K, Krueger C, Reik W, Peters JM, Murrell A. Cohesin is required for higher-order chromatin conformation at the imprinted IGF2-H19 locus. PLoS Genet. 2009;5:e1000739. doi: 10.1371/journal.pgen.1000739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, Yahata K, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 50.Xiao T, Wallace J, Felsenfeld G. Specific sites in the C terminus of CTCF interact with the SA2 subunit of the cohesin complex and are required for cohesin-dependent insulation activity. Mol Cell Biol. 2011;31:2174–2183. doi: 10.1128/MCB.05093-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin S, Ferguson-Smith AC, Schultz RM, Bartolomei MS. Nonallelic Transcriptional Roles of CTCF and Cohesins at Imprinted Loci. Mol Cell Biol. 2011;31:3094–3104. doi: 10.1128/MCB.01449-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stedman W, Kang H, Lin S, Kissil JL, Bartolomei MS, Lieberman PM. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J. 2008;27:654–666. doi: 10.1038/emboj.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yao H, Brick K, Evrard Y, Xiao T, Camerini-Otero RD, Felsenfeld G. Mediation of CTCF transcriptional insulation by DEAD-box RNA-binding protein p68 and steroid receptor RNA activator SRA. Genes Dev. 2010;24:2543–2555. doi: 10.1101/gad.1967810. [DOI] [PMC free article] [PubMed] [Google Scholar]