Abstract
Cancer is a disease caused by the accumulation of genetic alterations in association with successive waves of clonal expansion. Mapping of the human genome sequence, in conjunction with technical advances in the ability to sequence entire genomes, have provided new insight into the mutational spectra and genetic events associated with clonal evolution of cancer. Moving forward, a clearer understanding of those alterations that undergo positive and negative selection throughout carcinogenesis and leading to metastatic dissemination would provide a boon not only to our understanding of cancer evolution, but to the development of potential targets for therapeutic intervention as well.
Keywords: next generation sequencing, metastasis, clonal evolution, high throughput
1. INTRODUCTION
Cancer remains a major public health concern both in the United States and worldwide. Although the estimated number of cancer-related deaths is declining in the United States, cancer is still the leading cause of death among those younger than 85 years of age [1]. It is now widely accepted that cancer is a set of diseases that results from the accumulation of genetic alterations [2]. Particularly in the last decade, as the decreasing cost and increasing feasibility of sequencing methodologies have made the technique more applicable to high-throughput analyses, we have witnessed an explosion in the amount of genetic information available about many forms of cancer. The whole genomes of breast and colorectal cancers, pancreatic adenocarcinoma, glioblastoma multiforme, acute myeloid leukemia, mesothelioma, ovarian clear cell carcinoma, medulloblastoma, pancreatic neuroendocrine tumors, head and neck squamous cell carcinomas, and multiple myeloma have now been reported [3–12]. The many alterations identified include mutations in oncogenes and tumor suppressor genes, gene amplifications and deletions, and chromosomal rearrangements; the effects of these genetic alterations combine to provide a proliferative and survival advantage to cells. Additional molecular changes may also occur that allow the resulting neoplasm to invade into surrounding tissue and, eventually, metastasize to other organs.
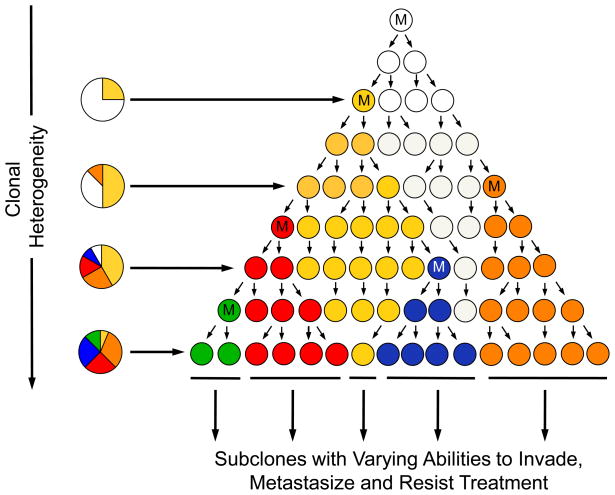
Despite the wealth of sequencing data available for many tumor types, the genetic evolution of these cancers remains, largely, a mystery. Tumors begin to evolve separately from their surrounding normal tissue when a mutation confers a proliferative advantage on a single cell. The progeny of this cell undergo additional mutations, some of which are acted upon by positive Darwinian selection to produce another clonal expansion [13]. This continuous process persists throughout the lifetime of the tumor, eventually yielding clones that have the abilities to invade surrounding tissue, seed and survive in distant locations, and resist treatment (Figure 1). The generation of and selection for these clones during cancer evolution, in addition to the heterogeneity that develops as a result, are major challenges faced when attempting to combat these diseases in the clinic.
Figure 1. Schematic of intratumoral clonal evolution.
Neoplasia begins when a genetic alteration (denoted by M) occurs in a normal cell that confers a selective growth advantage. Over time, additional alterations accumulate in the progeny of this cell, leading to successive waves of clonal expansion and the eventual formation of subclones with differing abilities to metastasize and evade treatment. Subclonal evolution is believed to follow Darwinian selection in that some subclones will become extinguished by others with greater abilities to survive in the tenuous microenvironment of the neoplasm.
However, the evolution of cancer is not as straightforward as a stepwise series of mutations. As a result of genetic instability that develops within the tumor cells, cancers are often a heterogeneous mix of genomes. The goal of many cancer geneticists is to understand this genetic complexity at the molecular level. While the sequencing and analysis of many cancer genomes provides an excellent first step in this process, little can be gleaned about the evolution of various cancers from such data. The advent of next-generation sequencing has provided cancer researchers with a powerful new tool in their arsenal to study the evolution of cancer. This review will discuss the history of research into cancer evolution, as well as the recent strides made possible by more advanced sequencing technology.
2. EARLY STUDIES OF CANCER EVOLUTION
The concept of clonal evolution in cancer was originally proposed by Peter Nowell, who theorized that acquired genetic instability increases as a neoplasm progresses, resulting in heterogeneity [14]. Consequently, numerous genetically distinct subclones develop within the neoplasm, resulting in intratumoral diversity. As sublines evolve from the original primary clone, a Darwinian selection process occurs to allow the cancer to become more malignant [13]; subclones with a mutation resulting in a selective advantage (i.e. enhanced tumor growth, and the abilities to invade and attract blood vessels) persist within the tumor, whereas those subclones without such beneficial properties may eventually become overgrown. This “survival of the fittest” within cancers, and cancer cell heterogeneity in general, have been studied with increasing sensitivity as technology has developed.
2.1 Cell-Based Studies
Evidence that distinct cell populations within a tumor progress along separate evolutionary paths originally came from studies examining differences in metastatic capability of sublines generated from a single tumor. One of these early clues stemmed from the observation that B16 melanoma cell clones have differential survival capabilities when injected into the bloodstream, with a vast minority of cells (0.2%) surviving [15]. Follow-up studies were performed using sublines generated from the parental B16 cell line. After intravenous injection of these clones, it was found that, compared to the parental cell line, the clones were inconsistent in their metastatic behavior, both in terms of the number and location of metastases formed, suggesting that the individual clones arose along separate evolutionary paths [16]. Comparable studies showed analogous differences in metastatic capability among clones of UV-2237 fibrosarcoma cells [17] and that clones that yielded a higher metastatic burden tended to have a faster rate of genetic mutation than clones with low metastatic burden[18]. Furthermore, the heterogeneity of metastatic potential became more pronounced after a metastatic lesion had formed, again suggesting a temporal relationship between metastasis and increasing genomic instability [19]. Once the concept of heterogeneity of metastatic potential within a primary tumor was established, the question was posed whether separate metastases within a patient are derived from the same cell. Karyotypic analysis of K-1735 melanoma cell lines injected into mice indicated that individual metastatic lesions can be derived from different cells within the primary tumor [20]. This result provided an important piece to the puzzle of cancer’s evolution – heterogeneity within the primary tumor leads to variable abilities to metastasize.
2.2 Cytogenetic Analysis
Another method that has been commonly used for the study of cancer evolution is comparative genomic hybridization (CGH). This technique is able to detect small-scale copy number changes [21]. Evolution of tumor samples can be predicted from CGH data based on the frequency of these chromosomal gains or losses: should a particular region be altered in most samples of a tumor, it is inferred that that genetic event occurred early in the evolution of the tumor and was subsequently passed onto its progeny.
CGH has been applied in describing the evolution of metastases from the primary site of cancer. In breast cancer, colorectal cancer, pancreatic cancer, and squamous cell carcinoma of the lung, gross chromosomal gains and losses occur more frequently in distant metastases than in the primary cancers [22–26]. Furthermore, direct comparisons of breast cancer CGH data by hierarchical cluster analysis indicated that lymph node metastases tend to be more similar to the primary cancer than are distant metastases, albeit in a small patient cohort [26]. These results can be attributed to different selection pressures that are faced during separate metastatic events, resulting in divergent evolution from the parental clone. Additional evidence for this phenomenon was seen by Nishizaki et al., who noted certain large-scale losses to be associated preferentially with lymph node metastases in breast cancer [24]. Furthermore, particular gains and losses seen in metastases from squamous cell carcinomas of the lung were consistently observed in patients, with a similar circumstance occurring in pancreatic cancer, suggesting that certain alterations are selected for during the metastatic evolution of these cancers [22,25].
CGH has also been utilized to address whether different metastatic lesions arise as a result of similar evolutionary events. It appears that, in some tumor types, different metastases evolve separately. For instance, in colorectal cancer patients, lung metastases harbored more chromosomal changes than liver metastases, suggesting a greater evolutionary timeline for lung lesions to develop [27]. In addition, CGH was used to show that in ovarian cancer, bilateral ovarian tumors arise as metastases during the latter stages of these cancers’ development [28]. However, CGH has yielded murky results in determining the evolutionary lineages of metastases. In breast cancer, a combination of CGH and fluorescent in situ hybridization (FISH) demonstrated that metastases surgically removed at different times from the same patient arise from one precursor clone in the primary tumor [29]. By contrast, analysis of multiple metastases per melanoma patient indicated that some metastases share a common progenitor clone, whereas others have evidence of separate evolutionary paths [30]. Perhaps further advanced technology that allows more detailed genetic analysis will yield insight of greater resolution into the clonality of multiple metastases, although the possibility exists that the patterns of clonality observed in various cancer types will be dependent on individual disease biology.
Contrary to CGH, loss of heterozygosity (LOH) studies analyze loss of specific alleles in the genome. Investigations of colorectal, bladder, and prostate cancer indicated that allelic losses increase in frequency during progression from primary cancer to metastasis, although no such differences were detected between primary breast cancer and lymph node metastases [31–34]. Furthermore, in some cases a putative order of allelic changes has been linked to disease progression, while others have associated specific LOH events with invasive cancer and metastasis [34–36].
2.3 Sanger (“First Generation”) Sequencing
While cytogenetic studies such as karyotyping, CGH, and LOH were important in identifying the regions of the cancer genome that undergo important changes necessary for cancer evolution, the genes involved in the process remained largely unknown. First-generation (Sanger) sequencing technology allowed researchers to analyze the clonality and evolution of several different cancer types at single base-pair resolution.
Based on sequencing analysis followed by three-dimensional reconstruction of the tumor, most colon cancers examined could be divided into one major clone, with several minor clones throughout [37]. Sequencing has also shown that the presence of mutations in KRAS and TP53 varies throughout primary colorectal cancers; however, heterogeneity of these particular mutations tends to fade over time after the clones containing the altered gene products undergo positive selection [38]. A similar instance of reduction in heterogeneity over time was observed in B cell chronic lymphocytic leukemia (CLL) by the sequencing of VHDJH rearrangements [39]. These results suggest that heterogeneity precedes positive selection of particular clones in the evolution of cancer, and disappears as positive selection ensues.
Sanger sequencing has also provided important information about the progression from primary cancer to metastasis. A broad view of the mutational landscape of colon cancer showed that the total number of point mutations across the genome increased as the lesions evolved from adenoma to colon cancer and, ultimately, to metastasis [40]. Similar results were obtained in pancreatic cancer, albeit in greater detail. For example, our laboratory sequenced distinct regions of primary pancreatic tumors as well as multiple metastatic lesions obtained from patients at rapid autopsy [41]. We showed that genetically discrete subclones exist within the primary pancreatic cancer, and that each metastatic lesion arises from a different subclone at different evolutionary timepoints. Furthermore, based on a calculated proliferative rate, the known rates of mutation, and the number of mutations identified by whole-exome sequencing of multiple subclones within the primary tumor and distant metastases, a timeline for pancreatic cancer evolution was proposed: the major clone (parental clone) develops over the course of 11.7 years, metastatic subclones arise within the primary carcinoma after an additional 6.8 years, and lethal distant metastases arise within 2.7 years after. A similar study of colorectal cancers yielded a comparable evolutionary timeline: it takes 17 years for an advanced carcinoma to arise from a large adenoma, but only 1.8 years for the carcinoma to metastasize [40]. These results emphasize that the positive selection events necessary for cancer initiation and evolution occur over the course of several decades. Consequently, there is a large window of opportunity for cancer detection and treatment.
3. SECOND-GENERATION SEQUENCING IN CANCER
3.1 Advantages of Second-Generation Sequencing in Cancer Applications
Despite the major advances made in deciphering how cancers develop by methods such as CGH, LOH, and Sanger sequencing, there are several advantages to next-generation sequencing technologies that make it preferable to the aforementioned techniques for the study of cancer evolution. Next-generation sequencing circumvents the problem of reduced DNA quality that results from areas of necrosis within the tumor [42]. Furthermore, specific assays have been developed such that it is easier to detect alterations in DNA other than point mutations; to date, next-generation sequencing has been applied to the study of chromosomal rearrangements, copy number changes, and RNA sequencing [43–45]. Finally, it is extremely rare for a cancer sample – be it from a biopsy or surgical resection – to be purely cancerous. Normal genomes dilute the cancer genomes such that rare genetic variants can be difficult to detect with Sanger sequencing. Next-generation sequencing technologies allow detection of genomes at much lower frequencies than Sanger sequencing [42,46]. Given the large amount of data generated by next-generation sequencing studies, the challenge for researchers has been to determine the evolutionary significance of the genetic aberrations identified in these analyses.
3.2 Analysis of Primary Cancer Heterogeneity and Evolution using Next-Generation Sequencing
As mentioned, a major advantage of second-generation sequencing technology is the ability to successfully sequence genetic material from lower-frequency samples[46]. Consequently, uncommon subclones may be examined in various cancer types. Multiple clones were identified in one-quarter of CLL patients using 454 sequencing of the IGH locus [47]. Similarly, in chronic myelomonocytic leukemia (CMML) and myelodysplastic syndrome (MDS) patients, deep sequencing of the TET2 gene revealed the presence of scarce clones in several patients, in which a dominant clone could also be identified [48]. It should be noted that many of these patients’ TET2 mutations went unnoticed by Sanger sequencing. Heterogeneity in ploidy of breast cancers was also observed by single-cell sequencing [49]. That more advanced sequencing technology is able to identify these rare populations of cancer cells is truly remarkable, especially when the frequency of the clones is considered – Campbell and colleagues were able to identify clones that comprised as low as 0.02% of the cells within the cancer[47].
In addition to detection of exceedingly rare subclones within cancers, researchers have begun to use next-generation sequencing to propose mechanisms by which mutational selection and evolution occur within cancers. Sequencing of a non-small cell lung cancer revealed two interesting patterns [50]. First, expressed genes had fewer mutations than silenced genes and the mutations in expressed genes were less likely to be found in the transcribed copy. Second, regions of the genome directly upstream of transcription start sites also had reduced mutation rates. These results imply that, despite the widespread genomic instability present within cancers, negative selection may exist to protect against mutations in genes important for basic cell functions. A similar phenomenon was observed when the IGH gene was sequenced in CLL patients [47]. Of the alterations detected in the regions of IGH important for heavy chain structure, an unexpectedly high number of them were silent mutations, indicating that changes to this particular locus are also selected against. While this concept (that genes important for cellular function are not often mutated in cancers) was previously assumed to be true, not until the age of next-generation sequencing was the idea confirmed.
When considering the phylogeny of a cancer’s evolution, it is common to assume that clones undergo divergent evolution. The results of some next-generation sequencing studies indicate that convergent, or at least redundant, evolution may be occurring. In particular, deep sequencing of a non-small cell lung cancer revealed multiple genetic events occurring in the same pathways [50]. Strikingly, the EGFR MAP-kinase pathway was affected by mutation or amplification of many different pathway members. While the existence of these mutations in different subclones of the tumor was not described in the study, it is interesting to speculate that the tumor may have evolved several different mechanisms toward “pathway addiction” rather than via addiction to one oncogene in the pathway. This concept is particularly daunting when attempting to target these signaling networks in treating cancers.
Next-generation sequencing has also allowed researchers to determine the overarching trend for the mechanism of cancer evolution. Paired-end sequencing of chromosomal rearrangements in multiple cancer types identified massive genomic rearrangements occurring over relatively small regions of the genome [51]. Remarkably, these rearrangements did not greatly alter copy numbers of the affected genes. The authors propose that this phenomenon be called “chromothripsis,” or “chromosome shattering into pieces.” Particularly due to the relative consistency of copy numbers after the complex rearrangements of these genomic regions, the authors suggest that the rearrangement resulted from a single event of chromosome destruction that was repaired by non-homologous end joining, and that the resulting mosaic chromosome provides the tumor cell with an evolutionary advantage, since, under normal circumstances, such a drastic genetic hit would cause the apoptosis machinery to engage.
Such an event would give credence to a punctuated equilibrium model of cancer evolution. The theory of punctuated equilibrium was originally proposed by Stephen Jay Gould and Niles Eldredge as a theory of the mechanisms through which species evolve [52]. In contrast to a more gradual model of evolution, punctuated equilibrium is characterized by long intervals of “stasis,” or stability, followed by rare events that cause evolutionary change. Certainly, one instance of chromosome destruction could be considered a rare event in the evolution of many cancer types. However, more evidence is needed to further study whether cancer evolution, in general, falls under the punctuated equilibrium model or if a more gradual model of evolution is more accurate.
3.3 Studying Evolution of Metastatic Capability by Second-Generation Sequencing
A major question that remains in cancer research is whether there exist mutations that promote the positive selection of metastatic clones. While the mechanism for normal cells to divide is typically active and becomes dysregulated in cancer, a separate set of processes must be deregulated on for a cell to gain the ability to successfully metastasize. Next-generation sequencing has been utilized to determine differences in genetic profiles between primary cancers and their matched metastases. Using paired-end sequencing, a lobular breast cancer and its matched metastasis were compared [53]. Thirty-two total coding mutations were identified in the metastatic lesion, of which nineteen were not detected in the primary cancer. Of the remaining mutations identified in the metastasis, five were present in most cells in the primary tumor, and six were detected in 1–13% of cells. These results highlight two concepts: the intrinsic heterogeneity that develops within cancers as they evolve due to genetic instability as well as the power of next-generation sequencing to detect extremely rare clones of cells within a population. Additional analysis of matched primary and metastatic pancreatic adenocarcinomas by massively parallel paired-end sequencing revealed similar patterns [54]. Each patient had certain genomic rearrangements that were present in the primary cancer and all of its corresponding metastases, whereas some rearrangements were present in only a subset of lesions or only in one metastatic tumor. Furthermore, lung metastases were determined to have evolved further than metastases to abdominal organs, due to the number of rearrangements identified in each lesion. These results confirm that individual metastases evolve separately within a cancer, a result reached by Yachida et al. in a similar study using Sanger sequencing [41,54].
Next-generation sequencing has yielded some interesting, though inconclusive, results in the push to identify metastasis-promoting mutations. Sequencing of a matched primary breast cancer, xenograft, and brain metastasis revealed that while most mutations that were identified were present in all three lesions, two mutations (in SNED1 and FLNC) existed only in the metastasis [55]. Additionally, a putative lung-specific alteration in the PARK2 gene was identified in a pancreatic cancer patient [54]. Two distinct alterations were identified in this gene, one of which was found exclusively in the lung lesions, while the other was only observed in peritoneal, liver, and omental metastases. As aforementioned, CGH studies revealed that certain areas of the genome are consistently gained or lost as squamous cell lung carcinomas and pancreatic adenocarcinomas metastasize [22,25]. The next-generation sequencing projects described herein provide evidence that mutations in specific genes occur in association with metastasis. These undertakings confirm the CGH findings, although at much greater resolution. This comparison illustrates the true advantages of next-generation sequencing compared to older techniques in the pursuit of metastasis-driving genetic alterations. Although these results are exciting, as they suggest that certain genes may be associated with organ-specific metastases, it is important to emphasize that these results were obtained from only one patient, and a larger cohort must be sequenced in order to verify putative metastasis-promoting genes as well as mechanistic studies of the mutated genes.
Next-generation sequencing has also been applied to determining the clonal evolution of metastasis, that is, whether multiple metastases arise from the same region of the primary cancer. In this case, studies have yielded mixed results. While next-generation sequencing of a femur metastasis, rib metastasis, and adrenal metastasis from a prostate cancer patient revealed that the three lesions were clonal in origin, each metastatic lesion had genetic abnormalities that were distinct from the other two [56]. Along the same lines, two independent studies of breast cancer suggested that the primary tumors and one metastasis per patient have common origins [49,55]. While these analyses have yielded very detailed pictures of the genetic landscapes of the patients’ cancers, it is impossible to use this information to make generalized statements about how metastases develop. However, larger-scale applications of next-generation sequencing have suggested that independent metastases arise from separate clonal entities in the primary cancer [54]. By examining the patterns of rearrangements identified by massively parallel paired-end sequencing in primary pancreatic cancer and multiple metastases per patient, Campbell and colleagues determined that the metastases examined must have arisen from at least two discrete regions of the primary tumor, again in line with the results of Yachida et al. [41,54]. Moreover, rearrangement patterns were observed that support the idea that metastases can seed other metastases [54].
Consequently, despite the advances in the amount of sequencing data that can be obtained per patient using next-generation methods, larger patient cohorts will be necessary in order to determine the dynamics of cancer metastasis, both in terms of causal mutations and clonal evolution of the tumors.
4. Conclusions and Future Implications
It is still relatively early in next-generation sequencing era; yet, there has been a wealth of information made available to date as a result of such new technologies, especially in the field of cancer genetics. Understanding the events that undergo positive and negative selection throughout carcinogenesis would provide a boon to our understanding of cancer evolution. It is clear that next-generation sequencing has the potential to unlock these mysteries, and further studies are likely to shed additional light onto this area.
Along the same lines, while many mutations have been identified as contributing to cancer progression as a result of the high-throughput nature of next-generation sequencing, it is important to identify which mutations are specifically selected for during clonal evolution. As a result of genetic instability within the cancer, “passenger” mutations accumulate alongside the “driver” mutations. While mutations that fall under the latter category have important functions throughout cancer evolution, those in the former undergo neither positive nor negative selection, but persist throughout the lifespan of the tumor, regardless. Thus, sorting of mutations into either of these classes will be necessary for learning how cancers evolve and, particularly, how metastases form.
Highlights.
Cancer progression is typified by subclonal evolution.
Subclonal evolution can be discerned using a variety of methods.
Next generation sequencing technologies have revolutionized study of cancer genomes.
Next generation methods provide the technical resolution needed to decipher clonal evolution.
Acknowledgments
Financial Support: Supported by National Institutes of Health grants CA140599 and P50 CA62924.
Footnotes
The authors have no financial conflicts of interest related to this work.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Siegel R, Ward E, Brawley O, Jemal A. Cancer Statistics, 2011. Cancer. 2011;61:212–36. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- 2.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–67. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 3.Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–7. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bueno R, De Rienzo A, Dong L, Gordon GJ, Hercus CF, Richards WG, et al. Second generation sequencing of the mesothelioma tumor genome. PloS One. 2010;5:e10612. doi: 10.1371/journal.pone.0010612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471:467–72. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiao Y, Shi C, Edil BH, de Wilde RF, Klimstra DS, Maitra A, et al. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science. 2011;331:1199–203. doi: 10.1126/science.1200609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones S, Wang T-L, Shih I-M, Mao T-L, Nakayama K, Roden R, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010;330:228–31. doi: 10.1126/science.1196333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones S, Zhang X, Parsons DW, Lin JC-H, Leary RJ, Angenendt P, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–6. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–66. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parsons DW, Jones S, Zhang X, Lin JC-H, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–12. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parsons DW, Li M, Zhang X, Jones S, Leary RJ, Lin JC-H, et al. The genetic landscape of the childhood cancer medulloblastoma. Science. 2011;331:435–9. doi: 10.1126/science.1198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–13. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 13.Cahill DP, Kinzler KW, Vogelstein B, Lengauer C. Genetic instability and darwinian selection in tumours. Trends Cell Biol. 1999;9:M57–60. [PubMed] [Google Scholar]
- 14.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–8. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 15.Fidler IJ. Metastasis: quantitative analysis of distribution and fate of tumor emboli labeled with 125-I-5-Iodo-2′-deoxyuridine. J Natl Cancer Inst. 1971;45:773–82. [PubMed] [Google Scholar]
- 16.Fidler IJ, Kripke ML. Metastasis results from preexisting variant cells within a malignant tumor. Science. 1977;197:893–5. doi: 10.1126/science.887927. [DOI] [PubMed] [Google Scholar]
- 17.Kripke ML, Gruys E, Fidler IJ. Metastatic heterogeneity of cells from an ultraviolet light-induced murine fibrosarcoma of recent origin. Cancer Res. 1978;38:2962–7. [PubMed] [Google Scholar]
- 18.Cifone MA, Fidler IJ. Increasing metastatic potential is associated with increasing genetic instability of clones isolated from murine neoplasms. Proc Natl Acad Sci U S A. 1981;78:6949–52. doi: 10.1073/pnas.78.11.6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poste G, Tzeng J, Doll J, Greig R, Rieman D, Zeidman I. Evolution of tumor cell heterogeneity during progresive growth of individual lung metastases. Proc Natl Acad Sci U S A. 1982;79:6574–8. doi: 10.1073/pnas.79.21.6574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Talmadge JE, Wolman SR, Fidler IJ. Evidence for the clonal origin of spontaneous metastases. Science. 1982;217:361–3. doi: 10.1126/science.6953592. [DOI] [PubMed] [Google Scholar]
- 21.Kallioniemi A, Kallioniemi O, Sudar D, Rutovitz D, Gray JW, Waldman F, et al. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science. 1992;258:818–21. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- 22.Armengol G, Capella G, Farre L, Peinado MA, Miro R, Caballin MR. Genetic evolution in the metastatic progression of human pancreatic cancer studied by CGH. Lab Invest. 2001;81:1703–7. doi: 10.1038/labinvest.3780383. [DOI] [PubMed] [Google Scholar]
- 23.Jiang J-K, Chen Y-J, Lin C-H, Yu I-T, Lin J-K. Genetic changes and clonality relationship between primary colorectal cancers and their pulmonary metastases--an analysis by comparative genomic hybridization. Genes Chromosomes Cancer. 2005;43:25–36. doi: 10.1002/gcc.20167. [DOI] [PubMed] [Google Scholar]
- 24.Nishizaki T, DeVries S, Chew K, Goodson WH, Ljung BM, Thor A, et al. Genetic alterations in primary breast cancers and their metastases: direct comparison using modified comparative genomic hybridization. Genes Chromosomes Cancer. 1997;19:267–72. doi: 10.1002/(sici)1098-2264(199708)19:4<267::aid-gcc9>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 25.Petersen S, Aninat-Meyer M, Schluns K, Gellert K, Dietel M, Petersen I. Chromosomal alterations in the clonal evolution to the metastatic stage of squamous cell carcinomas of the lung. Br J Cancer. 2000;82:65–73. doi: 10.1054/bjoc.1999.0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmidt-Kittler O, Ragg T, Daskalakis A, Granzow M, Ahr A, Blankenstein TJF, et al. From latent disseminated cells to overt metastasis: genetic analysis of systemic breast cancer progression. Proc Natl Acad Sci U S A. 2003;100:7737–42. doi: 10.1073/pnas.1331931100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knosel T, Schluns K, Dietel M, Petersen I. Chromosomal alterations in lung metastases of colorectal carcinomas: associations with tissue specific tumor dissemination. Clin Exp Met. 2005;22:533–8. doi: 10.1007/s10585-005-5239-7. [DOI] [PubMed] [Google Scholar]
- 28.Micci F, Haugom L, Ahlquist T, Abeler VM, Trope CG, Lothe RA, et al. Tumor spreading to the contralateral ovary in bilateral ovarian carcinoma is a late event in clonal evolution. J Oncol. 2010;2010:646340. doi: 10.1155/2010/646340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuukasjarvi T, Karhu R, Tanner M, Nupponen N, Pennanen S, Kallioniemi A, et al. Genetic heterogeneity and clonal evolution underlying development of asynchronous metastasis in human breast cancer. Cancer Res. 1997;29:1597–604. [PubMed] [Google Scholar]
- 30.Harbst K, Staaf J, Masback A, Olsson H, Ingvar C, Vallon-Christersson J, et al. Multiple metastases from cutaneous malignant melanoma patients may display heterogeneous genomic and epigenomic patterns. Melanoma Research. 2010;20:381–91. [PubMed] [Google Scholar]
- 31.Blaker H, Graf M, Rieker RJ, Otto HF. Comparison of losses of heterozygosity and replication errors in primary colorectal carcinomas and corresponding liver metastases. J Pathol. 1999;188:258–62. doi: 10.1002/(SICI)1096-9896(199907)188:3<258::AID-PATH350>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 32.Bonsing BA, Corver WE, Fleuren GJ, Cleton-Jansen A, Devilee P, Cornelisse CJ. Allelotype analysis of flow-sorted breast cancer cells demonstrates genetically related diploid and aneuploid subpopulations in primary tumors and lymph node metastases. Genes Chromosomes Cancer. 2000;28:173–83. doi: 10.1002/(sici)1098-2264(200006)28:2<173::aid-gcc6>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 33.Jones TD, Carr MD, Eble JN, Wang M, Lopez-Beltran A, Cheng L. Clonal origin of lymph node metastases in bladder carcinoma. Cancer. 2005;104:1901–10. doi: 10.1002/cncr.21466. [DOI] [PubMed] [Google Scholar]
- 34.Saric T, Braknac Z, Troyer DA, Padalecki SS, Sarodsy M, Williams K, et al. Genetic pattern of prostate cancer progression. Int J Cancer. 1999;81:219–24. doi: 10.1002/(sici)1097-0215(19990412)81:2<219::aid-ijc9>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 35.Barrett MT, Sanchez CA, Prevo LJ, Wong DJ, Galipeau PC, Paulson TG, et al. Evolution of neoplastic cell lineages in Barrett oesophagus. Nat Genet. 1999;22:106–9. doi: 10.1038/8816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Radford DM, Phillips NJ, Fair KL, Ritter JH, Holt M, Donis-Keller H. Allelic loss and the progression of breast cancer. Cancer Res. 1995;55:5180–3. [PubMed] [Google Scholar]
- 37.Baisse B, Bouzourene H, Saraga EP, Bosman FT, Benhattar J. Intratumor genetic heterogeneity in advanced human colorectal adenocarcinomas. Int J Cancer. 2001;93:346–52. doi: 10.1002/ijc.1343. [DOI] [PubMed] [Google Scholar]
- 38.Losi L, Baisse B, Bouzourene H, Benhattar J. Evolution of intratumoral genetic heterogeneity during colorectal cancer progression. Carcinogenesis. 2005;26:916–22. doi: 10.1093/carcin/bgi044. [DOI] [PubMed] [Google Scholar]
- 39.Bagnara D, Callea V, Stelitano C, Morabito F, Fabris S, Neri A, et al. IgV gene intraclonal diversication and clonal evolution in B-cell chronic lymphocytic leukaemia. Br J Haematol. 2006;133:50–8. doi: 10.1111/j.1365-2141.2005.05974.x. [DOI] [PubMed] [Google Scholar]
- 40.Jones S, Chen W-D, Parmigiani G, Diehl F, Beerenwinkel N, Antal T, et al. Comparative lesion sequencing provides insights into tumor evolution. Proc Natl Acad Sci U S A. 2008;105:4283–8. doi: 10.1073/pnas.0712345105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yachida S, Jones S, Bozic I, Antal T, Leary R, Fu B, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–7. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meyerson M, Gabriel S, Getz G. Advances in understanding cancer genomes through second-generation sequencing. Nat Rev Genet. 2010;11:685–96. doi: 10.1038/nrg2841. [DOI] [PubMed] [Google Scholar]
- 43.Campbell PJ, Stephens PJ, Pleasance ED, O’Meara S, Li H, Santarius T, et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat Genet. 2008;40:722–9. doi: 10.1038/ng.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chiang DY, Getz G, Jaffe DB, Kelly MJTO, Zhao X, Carter SL, et al. High-resolution mapping of copy-number alterations with massively parallel sequencing. Nat Methods. 2009;6:99–103. doi: 10.1038/nmeth.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mortazavi A, Williams BA, Mccue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:1–8. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 46.Thomas RK, Nickerson E, Simons JF, Janne PA, Tengs T, Yuza Y, et al. Sensitive mutation detection in heterogeneous cancer specimens by massively parallel picoliter reactor sequencing. Nat Med. 2006;12:852–5. doi: 10.1038/nm1437. [DOI] [PubMed] [Google Scholar]
- 47.Campbell PJ, Pleasance ED, Stephens PJ, Dicks E, Rance R, Goodhead I, et al. Subclonal phylogenetic structures in cancer revealed by ultra-deep sequencing. Proc Natl Acad Sci U S A. 2008;105:13081–6. doi: 10.1073/pnas.0801523105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith AE, Mohamedali AM, Kulasekararaj A, Lim Z, Gaken J, Lea NC, et al. Next-generation sequencing of the TET2 gene in 355 MDS and CMML patients reveals low-abundance mutant clones with early origins, but indicates no definite prognostic value. Blood. 2010;116:3923–32. doi: 10.1182/blood-2010-03-274704. [DOI] [PubMed] [Google Scholar]
- 49.Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–4. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee W, Jiang Z, Liu J, Haverty PM, Guan Y, Stinson J, et al. The mutation spectrum revealed by paired genome sequences from a lung cancer patient. Nature. 2010;465:473–7. doi: 10.1038/nature09004. [DOI] [PubMed] [Google Scholar]
- 51.Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gould SJ, Eldredge N. Punctuated equilibria: the tempo and mode of evolution reconsidered. Paleobiology. 1977;3:115–51. [Google Scholar]
- 53.Shah SP, Morin RD, Khattra J, Prentice L, Pugh T, Burleigh A, et al. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature. 2009;461:809–13. doi: 10.1038/nature08489. [DOI] [PubMed] [Google Scholar]
- 54.Campbell PJ, Yachida S, Mudie LJ, Stephens PJ, Pleasance ED, Stebbings LA, et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467:1109–13. doi: 10.1038/nature09460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ding L, Ellis MJ, Li S, Larson DE, Chen K, Wallis JW, et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464:999–1005. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Robbins CM, Tembe WA, Baker A, Sinari S, Moses TY, Beckstrom-Sternberg S, et al. Copy number and targeted mutational analysis reveals novel somatic events in metastatic prostate tumors. Genome Res. 2011;21:47–55. doi: 10.1101/gr.107961.110. [DOI] [PMC free article] [PubMed] [Google Scholar]