Abstract
Background
The mouse Fv1 (friend virus) susceptibility gene inhibits the development of the murine leukaemia virus (MLV) by interacting with its capsid (CA) protein. As no structures are available for these proteins we have constructed molecular models based on distant sequence similarity to other retroviral capsid proteins.
Results
Molecular models were constructed for the amino terminal domains of the probable capsid-like structure for the mouse Fv1 gene product and the capsid protein of the MLV. The models were based on sequence alignments with a variety of other retrovirus capsid proteins. As the sequence similarity of these proteins with MLV and especially Fv1 is very distant, a threading method was employed that incorporates predicted secondary structure and multiple sequence information. The resulting models were compared with equivalent models constructed using the sequences of the capsid proteins of known structure.
Conclusions
These comparisons suggested that the MLV model should be accurate in the core but with significant uncertainty in the loop regions. The Fv1 model may have some additional errors in the core packing of its helices but the resulting model gave some support to the hypothesis that it adopts a capsid-like structure.
Keywords: virus capsid protein models threading, Fv1, MLV
Background
The Fv1 gene is one of a series of mouse genes that control the susceptibility of mice to murine leukaemia virus (MLV) [1-3]. The gene acts in the cell to restrict virus replication [4] through a mechanism that is still uncertain. Genetic studies suggest that the target for the Fv1 gene product is the capsid protein (CA) of MLV [5,6] and it is thought to interact with CA after entry of the virus into the cell but before integration and formation of the provirus.
When cloned and sequenced [7], the Fv1 gene was found to have sequence similarity to endogenous retroviruses of the HERV-L and MuERV-L families [7,8]. Based on its position within the Gag gene of these endogenous elements, it appears that Fv1 encodes a capsid-like protein. This structural assignment of the Fv1 gene is consistent with its function as it can be postulated that the gene product might act as a dominant negative mutation and interfere with the MLV capsid function [9]. Sequence alignments have been made between Fv1 and other retroviral capsid proteins [8] but besides one region of clear similarity, called the Major Homology Region (MHR), there is otherwise little that is conserved across the full family of retroviral (and related lenti-virus) CA sequences.
There are now several known structures for retroviral capsid proteins in the Protein Databank (PDB). While some of these are only fragmentary, a selection can be extracted that gives a reasonable phylogenetic spread across the retroviruses, with examples from three out of six genera of orthoretroviruses. In all the known structures, the CA protein has an all-α type structure consisting of two domains: a larger N-terminal and smaller C-terminal domain with a short extended linker-region between them. As this linker enters the C-terminal domain it incorporates the MHR. There is considerable variation in the orientation of the domains and in the conformation of the loop-regions between α-helices, particularly in the N-terminal domain.
In this work, we have exploited these multiple structures to construct consensus molecular models using threading methods both for the Fv1 gene product (FV1) and its target protein, the MVL CA. As threading takes known and predicted structure into account, it should provide better alignments for the regions that lie outside the MHR. However, as these methods are still far from perfect, we have constructed a model based on each known structure and the degree to which these agree has been used to assess the confidence of different parts of the model. As the threading method we have used has some 'free' parameters (such as the gap-penalty) we have introduced a novel modelling strategy in which the parameters are varied to give maximum agreement among the resulting models.
Results and Discussion
The sequence alignments compiled on the proteins of known structure using the Ψ-BLAST/QUEST search strategy (Methods Sectn.) were used in the modelling of both the MLV CA and FV1 sequences. These varied from 4 to 7 sequences (Table 1). Not unexpectedly, the alignments are to some extent similar, in particular each contains the sequence of the HIV-1 CA structure [1e6jP]. (QUEST is biased to retain sequences of known structure). Greatest overlap exists between the sequence sets of the two human viruses, with the HTLV-I sequences being a subset of the HIV-1 sequences (Table 0(d) and Table 0(c)). While it would be possible to extend and realign these sub-families based on structure comparisons, they were left unaltered so as to be equivalent to the MLV and FV1 alignments described below. This allows control modelling tests to be directly comparable to those performed for sequences of unknown structure.
Table 1.
Template sequences selected for alignment. The Ψ-BLAST/QUEST search strategy (Methods Sectn.) when started with the probe sequence of the PDB structure indicated by "SEED" selected the sequences indicated in each subtable: (a) EIAV [1eia], (b) RSV [1d1dA], (c) HIV-1 [1e6jP] and (d) HTLV-I [1qrjA]. The sequences are identified by their gene identification (gi) number (first column) and their local source databank identifier. The sequence fragment (automatically extracted by QUEST) is given as a range of residue numbers.
| (a) EIAV | (b) RSV | ||||
| SEED | 1eia | 12084543 | pdb-1E6J | 0–210 | |
| 6358699 | gb-AAF07324 | 131–342 | 8072301 | gb-AAF71968 | 0–155 |
| 6815746 | gb-AAF28696 | 0–173 | 6649692 | gb-AAF21520 | 5–224 |
| 12084543 | pdb-1E6J | 3–210 | 120850 | sp-P18041 | 97–362 |
| 27803398 | gb-AAO21890 | 120–281 | 294961 | gb-AAA74706 | 116–381 |
| 5106563 | gb-AAD39752 | 81–346 | |||
| SEED | 1d1dA | ||||
| (c) HIV-1 | (d) HTLV-I | ||||
| 6358699 | gb-AAF07324 | 129–340 | SEED | 1qrjA | |
| 22037894 | gb-AAM90230 | 148–359 | 12084543 | pdb-1E6J | 0–210 |
| SEED | 1e6jP | 22037894 | gb-AAM90230 | 144–370 | |
| 532325 | gb-AAA99545 | 50–224 | 9886907 | gb-AAG01643 | 0–222 |
| 9886907 | gb-AAG01643 | 0–211 | |||
MLV Modelling
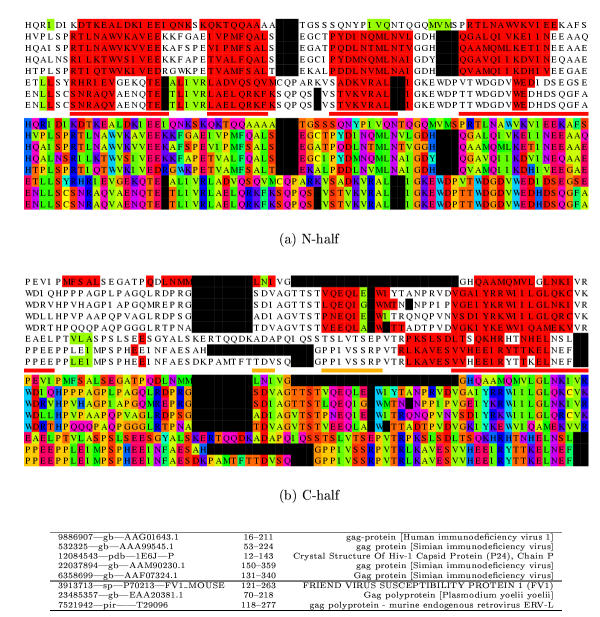
The databank search using the MLV sequence as a probe provided a useful collection of six sequences (Table 1(a)) which, with the MLV probe sequence itself, were passed through the PsiPred secondary structure prediction protocol (Methods Sectn.). When the predicted secondary structures were viewed on the aligned sequences, several distinct α-helices were apparent, consistent with the protein having an all-α type structure similar to the other capsid proteins of known structure (Figure 1).
Figure 1.
MLV sequence alignment with the sequence of 1qrjA and related sequences. The alignment is displayed twice in different colouring schemes. In the top two panels, (a) the amino half is coloured firstly by predicted secondary structure (red = α, green = β) and then below using a different colour for each amino acid type (Taylor, 1997; black = gap). The known helices (N1–N5) marked as red lines (minor helices are orange) between the two blocks. In the lower two panels (b) the carboxy half of the alignment is shown in a similar way. The sequence identifiers are given below the alignment in the same order as they are aligned. The mid-line divides the 1qrjA sub-family from the MLV sub-family.
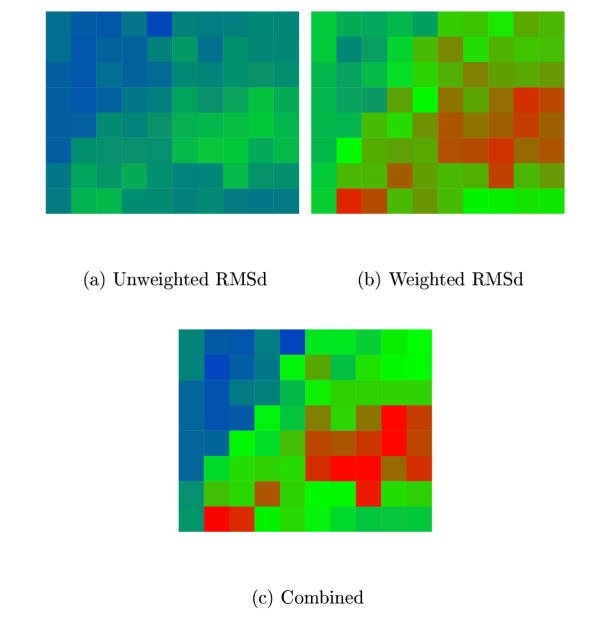
The MLV alignment with predicted secondary structure was matched in the MST program with the four capsid proteins of known structure along with their associated aligned sequences. This was done for each parameter combination specified in Methods Sectn. and the quality of the agreement among the four resulting models quantified using both the Fu and Fw measures defined in Methods Sectn. There was reasonable overall agreement between the two measures of the parameter combinations (Figure 2) and with otherwise no basis on which to prefer one measure over the other, the combined measure (Fv) was used to select the parameters on which the final models were calculated. These were S = 7, G = 30.
Figure 2.
MLV model agreement. The degree of similarity among the four MLV CA models is plotted for different parameter combinations of MST. Combinations resulting in better agreement have colours towards the red end of the spectrum. (a) Using the unweighted RMSd measure (Fu) and (b) Using the weighted RMSd measure (Fw). (See Methods Sectn. for details.) (c) Combined score (Fu·Fu). The MST parameters varied were X-axis: S = 0→9 (in steps ofone) and Y-axis: G = 10→90 (in steps of 10). (See Methods Sectn. for details.) The best combined score is at: S = 7, G = 30.
N-terminal domain
The multiple structure alignment of the four models showed good agreement. In the N-terminal domain there were two extensive regions over which all models were aligned in register, covering helices N2-N3 and N5 (including the preceding short helix in the loop region). Two alternate registers were observed for helices N1 and N4, with relative shifts of 3 and 4 residues, respectively. The models based on 1qrjA (HTLV-I) and 1d1dA (RSV) were in complete agreement and the summed Fw score indicated that 1d1dA provided the best consensus model.
Given that the alignment of the capsid protein sequences is ambiguous, the superposition of the models on the structures from which they were derived provides a better way to assess whether there is any significant sequence similarity that could be used as a basis on which any one model might be preferred over the others. The PSId values were: 5.6, 18.5, 10.5, 13.0 for 1d1dA, 1qrjA, 1eia and 1e6jP, respectively. (No differences were observed whether using the standard version or the sequence-biased version of SAP).
Although the 1d1dA model was the best consensus representative, it has only 5.6% sequence similarity with MLV and the higher sequence similarity of the 1qrjA based model with its template (18.5%) was considered sufficient to justify its adoption as the preferred model over the 1d1dA model. Structurally, both were very similar (1.4 Å wRMSd) with the only significant structural difference being a slight reorientation of the helical region in the loop preceeding the final helix. This structure for the 1qrjA based model is shown along with its comparison with the 1d1dA model in Figure 3.
Figure 3.
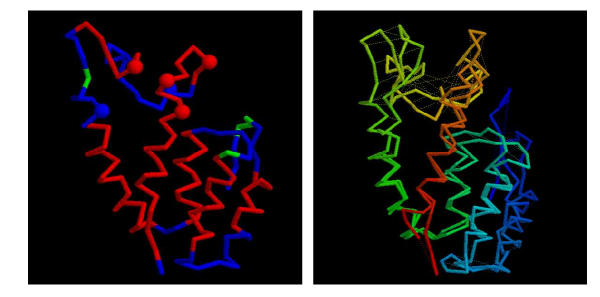
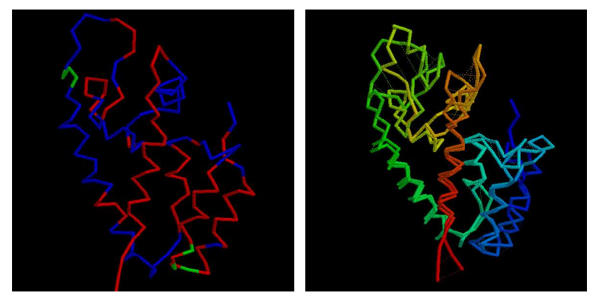
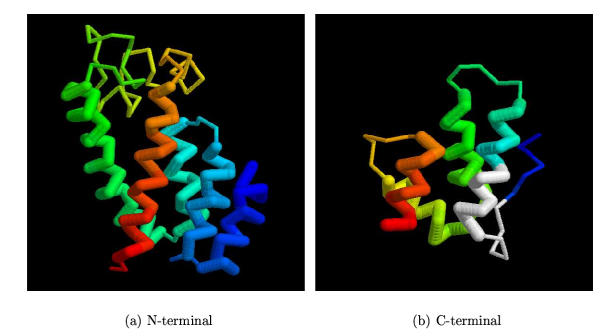
Consensus model for the MLV N-domain. (a) The model for the MLV CA N-domain based on 1qrjA shown as a α-carbon trace with predicted α-helices coloured red (some fragments of predicted β-structure are coloured green). The molecule is in approximately the same orientation as in Figure 8(a) and residues identified by Stevens et al. (2003) are marked as small spheres. (b) The models based on 1qrjA and 1d1dA are shown superposed and coloured from blue (amino) to red (carboxy). Feint dashed lines connect identical residues. The wRMSd = 1.4 (uRMSd = 5.8) over 123 residues.
It can be seen from Figure 3(a) that there is good location of the predicted and model helices with deviations occurring only at the ends of some helices and into the loop regions at the 'top' of the molecule. The ends of helices N4 and N5 and the loops at this end of the molecule also encompass the mutations identified as affecting the sensitivity of the virus to Fv1 [29].
C-terminal domain
With its relatively unambiguous MHR, all the models of the C-terminal domain were in complete agreement over the first half of the domain. The more C-terminal half, however, was less consistent due to a combination of its generally less conserved nature combined with uncertainty in the location of the terminus in some of the sequences.
As the C-terminal domain has been shown to be less important in determining the property of virus susceptibility, further effort was not expended to try and refine the alignment at the carboxy terminus of the molecule, especially in the more difficult alignment of the FV1 sequence described below.
FV1 Modelling
The databank search using the FV1 sequence as a probe provided a less useful collection of only two distinct sequences (Table 1(b)). Although other sequences were found these were rejected by QUEST as being too similar to those retained.
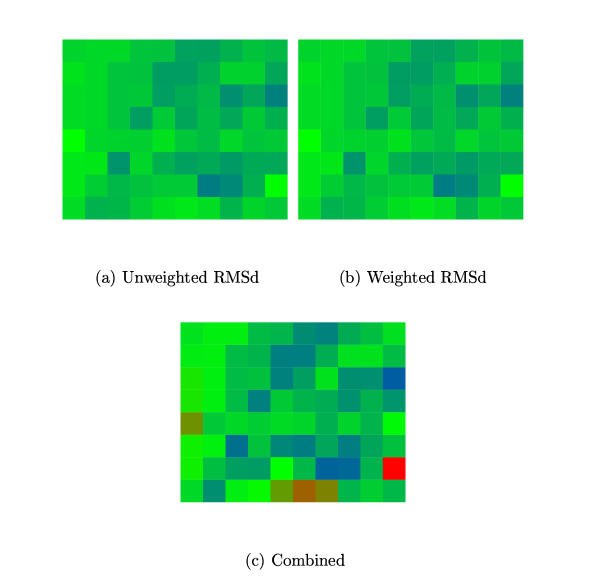
The FV1 alignment with predicted secondary structure was matched to the four capsid proteins of known structure as described above for each MST parameter combination and agreement among the four models monitored using the F scores (Methods Sectn.). These were 50.3 and 131.874 for Fu and Fw while the corresponding score obtained for the MLV models were 78.7 and 188.0, respectively. The greater variation among the FV1 models was undoubtedly due to uncertainty in the placement of the large 'unstructured' region. Nevertheless, there was reasonable overall agreement between the two measures and both found maximal model agreement when S = 9, G = 20 (Figure 4).
Figure 4.
FV1 model agreement. The degree of similarity among the four FV1 CA models is plotted for different parameter combinations of MST. (a) Using the unweighted RMSd measure (Fu) and (b) Using the weighted RMSd measure (Fw). (See Methods Sectn. for details.) (c) Combined score (Fu·Fu). The MST parameters varied were X-axis: S = 0→9 (in steps of one) and Y-axis: G = 10→90 (in steps of 10). (See Methods Sectn. for details.) The best combined score is at: S = 9, G = 20.
When the predicted secondary structures were viewed on the aligned sequences, although several distinct α-helices were apparent, these had a less direct correspondence with those expected for a capsid protein (Figure 5). In particular, there is little predicted α-structure for helix N4 and the alignment around N5 is ambiguous. If the secondary structure prediction were to be believed, this might indicate the introduction of a large insertion, or given that there were less sequences on which to base the predictions, it is possible that the predictions are not as accurate as those obtained for the MLV sequences.
Figure 5.
FV1 sequence alignment with the sequence of 1e6jP and related sequences. The alignment is displayed twice in different colouring schemes. In the top two panels, the amino half is coloured by predicted secondary structure (red = α, green = β) and amino acid type (Taylor, 1997). See the legend to Figure 2 for further details. with the known helices (N1–N5) marked as red lines between (minor helices are orange). The carboxy half of the alignment is displayed in a similar way in the lower panels. The sequence identifiers are given below the alignment in the same order as they are aligned. The mid-line divides the 1e6jP sub-family from the FV1 sub-family.
N-terminal domain
The multiple structure alignment of the four models showed unexpectedly good agreement. As in the MLV models, two regions were aligned in register over all the models. These included most of helix N4 and the following loop region then, after only one shift, the full register was regained for helix N5. The sum-of-F scores for each model indicated that 1eia provided the best consensus, followed by 1e6jP (to which it was most similar). The PSId values were: 9.3, 7.8, 6.6, 15.9 for 1d1dA, 1qrjA, 1eia and 1e6jP, respectively. As the 1e6jP model aligns better on its target by almost 10 PSId, this was considered sufficient to justify its adoption as the preferred model over the 1eia based model. Both models are similar in the core region (1.3 Å wRMSd) but with the significant differences in the extensive loop regions at the 'top' of the molecules. This structure for the 1e6jA based model is shown along with its comparison with the 1eia model in Figure 6.
Figure 6.
Consensus model for the FV1 N-terminal domain. (a) The model for the FV1 N-domain based on 1e6jP shown as a α-carbon trace with predicted α-helices coloured red (some fragments of predicted β-structure are coloured green). The molecule is in approximately the same orientation as in Figure 8(a). (b) The models based on 1e6jP and 1eia are shown superposed and coloured from blue (amino) to red (carboxy). Feint dashed lines connect identical residues. The wRMSd = 1.3 (uRMSd = 5.4) over 124 residues. Note: the modelling program rescales the secondary structure prediction values so that there is the same proportion of predicted structure as measured secondary structure on the model.
It can be seen from Figure 5 and Figure 6(a) that there is reasonable location of the predicted and model helices except for helix N4 which is almost completely unpredicted. Otherwise deviations occurred at the ends of helices and in the loop regions at the 'top' of the molecule.
Table 2.
Target sequences selected for alignment. The Ψ-BLAST/QUEST search strategy (Methods Sectn.) when started with the two target sequences indicated by "SEED", selected the sequences indicated in each subtable: (a) MVL and (b) FV1. The sequences are identified as in Table 1. (* the C-terminal domain of this sequence appears to be replaced by an oncogene.)
Control Models
To help assess the accuracy of the models constructed above, the protocols described in the Methods sections were applied to each of the sequences of known structure. For each protein, this results in four models based on the other structures (and its own structure). Each of these models, was then compared to the known structure using the modified SAP program and the RMSd/PSId measures reported in Table 3.
Table 3.
Control model similarity. The similarity of the capsid protein domains are tabulated as: uRMSd/PSId, both as calculated by the SAP program. The proteins are identified as in Table 4. The column "params" gives the MST parameter values S, G at which the four models in each row had maximum agreement as measured by the combined agreement score Fv (Methods Sect n.).
| str \ seq | D1D | QRJ | E6J | EIA | params |
| D1D | 0.60/78.9 | 4.14/90.0 | 2.05/56.7 | 2.41/38.3 | 9,10 |
| QRJ | 5.58/100. | 0.32/100. | 3.58/88.4 | 5.51/100. | 3,10 |
| E6J | 1.88/95.2 | 1.70/84.7 | 0.31/100. | 1.39/100. | 7,10 |
| EIA | 2.00/93.3 | 2.00/100. | 1.28/100. | 0.35/100. | 8,20 |
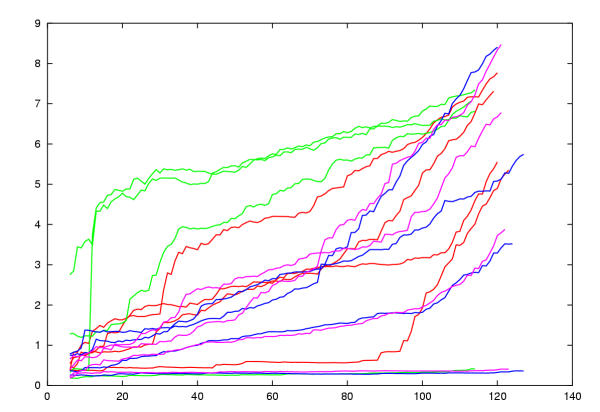
With few exceptions, the wRMSd/PSId values indicated generally good models and by visual inspection, all had the correct fold. The models based on their own structures were almost identical to their templates with the exception of 1d1dA. When the uWRMSd values of the models against the known structure are plotted against number of residues ranked by local score (Figure 7) it can be seen that for 1d1dA the error rises sharply after 85 positions. Visual inspection revealed that this was due to a misaligned segment in the large loop region. The majority of models exhibited a gradual increase in wRMSd with increased error mounting as the loop regions were introduced. Two models that ran markedly below this trend were those based on 1eia and 1e6jP (22% identity), the two most similar proteins. A few models were also markedly worse. Two of these were derived from the 1qrj alignment based models which had a distinctly lower weight on the structure component (S weight) in the MST alignment.
Figure 7.
Control model uRMSd values. Each model was compared with its known structure and the local residue matches calculated by SAP were ranked. The uRMSd was calculated for increasingly larger sets of ranked residues and plotted against set size. This means that the best fitted residues in each comparison are found to the left of the plot with increasing divergence towards the right. Four models are plotted for each of four CA proteins of known structure: 1d1dA = red, 1qrjA = green, 1eia = purple, 1e6jP = blue. The best models are those of sequences built on their own structure. Three of these remain low throughout their length while one has poor loops and rises towards the right. Above these are two models for the most similar sequences of 1e6jP and 1eia built on each others structure (22% identity). Most models lie in the mid range with a few accumulating early errors due to shifts in the core regions.
Conclusions
Based on relative degrees of sequence similarity among the control models (Table 3(b)) and the MLV and FV1 sequences (MLV:1qrj = 18.5%, FV1:1e6jP = 15.9%), it would be expected that the models constructed for the MLV sequence would fall in the mid-range of the spread in quality observed in Figure 7 – typically, a good core model (3 Å RMS over 80 residues) with increasing divergence in the more variable loop regions. This is where the majority of the control models lie which were all constructed from sequence similarities that are generally lower than either of the above relationships used to model the MLV or FV1 sequences.
While a similar confidence might be hoped for the FV1 model, given its overall lower sequence similarity to the proteins of known structure and the less consistent nature of its secondary structure prediction, it is more likely that it will be of lesser quality – corresponding more to the poorer models constructed among the control proteins (Figure 7). Typically, this would include shifts in the core helices (giving the characteristic immediate rise in the traces in Figure 7) with further shifts in the loop regions. Despite this, as with all of the control models, it is likely that the core fold of the protein should remain unaltered.
This study has shown that reasonable models can be constructed for both the FV1 and its target MLV protein based on other retroviral capsid proteins. Although this result was suggested by the existence of the MHR in both sequences, the fluid nature of retroviral genomes does not necessarily constrain the preceding domain to remain constant in structure or even remain at all. Despite only weak sequence similarities in this region, the addition of multiple sequences with predicted secondary structure has allowed plausible models to be constructed.
These models can now be used in the interpretation of experimental studies on the mode of action of retroviral susceptibility. As will be reported in more detail elsewhere [29], a series of amino acids in CA affecting the CA – FV1 interaction have been identified in the loops at the 'top' of the N-terminal domain (Figure 3). Based on the model, they suggest a potential FV1 binding domain in the MLV CA. Experiments are currently under way to test this prediction by crystallographic studies.
For many years, the Fv1 gene has been the only known intracellular non-immune natural defense against a retroviral infection. Recently, two additional genes, Ref1 and Lv1, with antiviral activity have been described in human cells [30,31]. Phenotypically, they resemble Fv1 [32] but the genes themselves remain to be characterised. Understanding the mechanism of Fv1 action will provide insights into how natural defences to retroviral infection might be deployed against HIV.
Methods and Data
Data
Sequence Data
All sequences were extracted from (and searches were made over) the non-redundant protein sequence databank (NRDB) at the National Centre for Biotechnology Information (NCBI) as it was found on 28th of January 2003.
The sequence of the MLV used was the gag protein AAD55051 (GI:5881091) [10] and a region was extracted from residues 215–432, corresponding to the CA protein.
The sequence of the Fv1 gene [7] was taken from FV1_MOUSE (GI:3913713). The region corresponding to the CA was identified as residues 100/120–340/360 where the inner numbers represent the probable core of the protein. This range corresponds to the region of highest similarity to the MuERV-L sequence [8]. The leading 100 residues of the polyprotein may correspond with a relic matrix protein. Perhaps because of this, there is no obvious protease cleavage motif [11] to give any indication of the true terminus. However, in other situations this has not always been an accurate guide [12].
Structural Data
The structures of capsid proteins were extracted from the PDB [13] with the aid of the FSSP structure comparison database [14]http://www.ebi.ac.uk/dali/fssp/fssp.html. Of the six structures in the FSSP alignment, only four extended over the full length of the two structural domains. There were as follows, with their PDB code (and chain delimiter, if any) shown in brackets: Rous Sarcoma Virus (RSV) [1d1dA] [15], Human T-cell Leukemia Virus (HTLV-I) [1qrjA] [16,17], Equine Infectious Anemia Virus (EIAV) p26 [1eia] [18] and Human Immunodeficiency Virus (HIV-I) p24 [1e6jP] [19].
The common core of the N-terminal domains of these proteins (in the numbering of the PDB structure) was defined as: 1d1dA 15–148, 1qrjA 16–129, 1eia 16–145 and 1e6jP 16–146. These fragments will be distinguished below as: 1d1dAn, 1qrjAn, 1eia-n and 1e6jPn and each terminates 8 or 9 residues before the conserved glutamine of the MHR. The N-terminal domain can be described as having five α-helices (N1...N5) with a long 'disordered', partly helical, loop between helices N4 and N5. For ease of reference below, this region will be called the 'top' of the molecule and its representation in the Figures will preserve this orientation.
The C-terminal domains were defined as: 1d1dA 152–224, 1qrjA 132–204, 1eia 149–220, 1e6jP 149–220 and were distinguished by the suffix "c". The common core of this domain consists of an extended strand leading into the MHR region followed by four helices designated C1...C4.
Despite their different sizes, both the N and the C domains have the same fold, perhaps suggesting an ancient gene duplication. This is most obvious in the HIV structure [1e6jP] where the domains can be superposed with 4.6 (2.0) unweighted (weighted) RMSd over 68 residues.
The similarity of sequence and structure over these domains was calculated using the SAP structure comparison program [20] for each pair of like domains (Table 4). The N-terminal domains have clearly related structures (3–6 Å RMSd) but have no significant sequence similarity (over 20%) except for the EIAV and HIV-1 structures. The smaller C-terminal domain (which contains the MHR) has greater overall sequence and structural similarity compared to the N-terminal domain (mostly over 20% with 2–3 Å RMSd) and over 40% for the EIAV/HIV-1 pair.
Table 4.
Capsid protein similarity. The similarity of the capsid protein domains are tabulated as: (a) RMSd (unweighted) and (b) PSId, both as calculated by the SAP program. Upper-right triangle: over the common core of the C-terminal domain and Lower-left triangle: over the common core of the N-terminal domain. The proteins are identified as: D1D = RSV [1d1dA], QRJ = HTLV-I [1qrjA], E6J = HIV-I [1e6jP], EIA = EIAV [1eia ].
| (a) structure | (b) sequence | ||||||||
| N \ C | D1D | QRJ | E6J | EIA | N \ C | D1D | QRJ | E6J | EIA |
| D1D | 2.88 | 2.64 | 2.46 | D1D | 21.9 | 22.2 | 14.7 | ||
| QRJ | 3.83 | 2.11 | 2.39 | QRJ | 12.5 | 30.6 | 26.4 | ||
| E6J | 6.21 | 4.76 | 1.40 | E6J | 8.6 | 10.8 | 41.7 | ||
| EIA | 5.75 | 4.31 | 3.22 | EIA | 11.6 | 12.5 | 22.0 | ||
Sequence Databank Searches
Initially, each probe sequence was compared against the sequence databank using the Ψ-BLAST program [21] with a significance level set at 0.001 and 5 cycles of iteration. When the probe is a retroviral sequence, the number of hits found by Ψ-BLAST can be large (typically over 1000). These were reduced to manageable numbers by the use of the search program QUEST which is similar to Ψ-BLAST but incorporates a multiple sequence alignment stage in its iterations to exclude redundant sequences as well as excluding poorly related or incomplete sequences [22]. The alignments produced by QUEST typically contain between 6–12 sequences (including the probe sequences), none of which have more than 60% sequence identity (PSId) with each other.
The sequences retained by QUEST are selected on the basis of associated biological information, with those including useful annotation and structural data being given preference over those with no annotation or keywords such as "hypothetical". The filters are part of the MULTAL sequence alignment program [23] which are fully described in Ref. [24].
Secondary Structure Prediction
The multiple alignments resulting from the Ψ-BLAST/QUEST search protocol were passed to the program PsiPred http://bioinf.cs.ucl.ac.uk/psipred/, Version 2.3) [25]. This program normally performs its own databank searches using Ψ-BLAST to build-up an alignment. Given the problems described above that arise when searching with retroviral sequences, the PsiPred program was used locally to search only a database consisting of the sequences that had already been selected by QUEST.
Each sequence in the alignment was taken in turn and used as a probe against this small local database. As the Ψ-BLAST parameters used by PsiPred were more restrictive than those used in the full search (only 3 cycles) and there are fewer sequences in the databank, each sequence may only find those to which it is more closely related. This introduces some variation into the predictions which provides a useful indication of the confidence of each predicted secondary structure element (SSE).
Multiple Sequence Threading
The alignment gathered on the probe sequence was then aligned with a protein structure using the multiple sequence threading MST program [26]. This program uses multiple sequence and structural information to automatically construct an α-carbon molecular model for the probe sequence with some limited remodelling in regions of insertion and deletion.
Template Sequence Alignments
The MST program can incorporate multiple aligned sequences along with both the probe sequence and the template structure. The latter were gathered in an identical manner to the probe sequence using the Ψ-BLAST/QUEST search protocol described above. Each search against the NRDB was started with the sequence of the protein of known structure and the resulting multiple alignments examined 'by-eye' in the light of the known secondary structures. If any large insert had been made in a secondary structure element (SSE) then it was assessed whether the gap could be shifted outside the SSE without significant loss of residue matches. Similarly, if a large insert (more than 6 residues) was made by any sequence other than the probe sequence (of known structure) then the insert was reduced to six residues by removing the positions with most gaps.
Parameter Choice
The MST program has parameters that allow different weights to be attached to the matching contribution of the sequences, their secondary structures, residue exposure and the residue packing in the resulting model. There is also a gap-penalty. The best values for these weights depends on the number and degree of relatedness among both the probe and the template sequences [26]. Rather than vary all these parameters individually, the weights on the structural components (secondary structure, exposure and packing) were 'ganged' together into a single parameter reflecting the contribution of structural terms relative to the sequence matching component. This gave two parameters: S (for structure) and G (the gap-penalty). Previously, the structural parameters had all been scaled into the same range so a value of S = 3 corresponds to a value of 3 for each individual weight. Although the gap-penalty is correlated with S, it cannot be linked in the same way without the risk of missing good alignments.
In the current application, there was more than one available template structure and advantage was taken of this by constructing models based on all available templates and choosing the MST parameters such that the agreement among the models was greatest. The parameters were varied over the ranges: S = 0→9 (in steps of one) and G = 10→90 (in steps of 10).
Measuring Model Agreement
Whatever the parameters for MST, all the models constructed from the same probe have an identical sequence. These might therefore be compared using the α-carbon RMSd based on a one-to-one (100 PSId) sequence equivalence. However, using this simple measure, a 'trivial' shift in space in which, say, an α-helix shifts by one turn relative to another α-helix might result in a large RMSd between what are, topologically, similar models. It is better to allow a local relative shift in sequence of four residues to restore the spatial equivalence at the expense of residue identity.
To implement this trade-off between RMSd and PSId, the models constructed for each parameter combination were compared against each other using the program SAP [20]http://mathbio.nimr.mrc.ac.uk/ftp/wtaylor/sap/. This program calculates both a weighted (Rw) and unweghted (Ru) RMSd for the two structures being compared and reports the percentage sequence identity of the alignment. The weighted RMSd down-weights regions of weak similarity which are mainly loop regions that can have large relative displacements. Despite its origins [27], in its current implementation the SAP program http://mathbio.nimr.mrc.ac.uk/ftp/wtaylor/sapid/ does not include a sequence matching component and this was restored (for sequence identity only) by doubling the local residue pair score for identical residue types and otherwise halving all other residue match scores in the initial score matrix.
A score reflecting match quality (f) was calculated as: f = M/(1+R), where M is the PSId measured over the positions aligned by SAP and R is one of the RMSd measures. Identical structures would score 100. For a set of N models, a sum was calculated over the (N2 - N)/2 pair combinations giving an overall measure of agreement (F) among the set. For a set of four models that align perfectly (100 PSId) with 2 Å RMSd, the overall score obtained would be 200. This score was calculated for both the wRMSd and uRMSd values (giving Fw and Fu, respectively) and a combined score (Fv) as the product of Fw and Fu.
While this procedure provides a general method for choosing parameter values, in the current application to a multi-domain protein it was not meaningful to calculate the RMSd over the full atomic model (because of relative domain movements). Instead, the agreement was calculated over the more distantly related N-terminal domain.
Selecting a consensus model
Although any model in the set could be taken as a representative, it is best to try and select one that, by some criteria, can be considered to be the most representative. To do this, we compared each pair of models usuing the structure comparison protocol described in the previous section. This provides a pairwise alignment based on structure, and even though each model has an identical sequence, the structural alignment may not match-up identical residues. The pairwise alignment were then combined into a multiple structure alignment [28] and as the models all have an identical sequence, their relative shifts can be seen easily. Rather than use a pure structure or sequence based measure of similarity between the proteins, the score F was devised in the previous section (Methods Sectn.) that combines both a sequence and a structural component. This was used to find the model with the greatest sum-of-scores to the others.
An alternative selection test was also considered of selecting the model that had greatest sequence similarity when superposed with the template structure from which it was derived. As most of the sequence similarities considered below lie in the 'twilight-zone', the latter option was only used when one model was clearly better than the others. For this, we choose the criterion that it had to be 10 PSId points clear of its 'rivals'.
Abbreviations
Fv1/FV1, gene/gene-product of Friend Virus susceptibility locus-1;
MLV, Murine Leukaemia Virus;
CA, CApsid protein;
HERV-L, Human Endogenous RetroVirus (L family);
MuERV-L, Murine Endogenous RetroVirus (L family);
MHR, Major Homology Region;
NCBI, National Centre for Biotechnology Information;
NRDB, Non-Redundant DataBank;
MST, Multiple Sequence Threading (program);
SSE, Secondary Structure Element;
PSId, Percent Sequence Identity;
PDB, Protein DataBank;
RMSd, Root-Mean Square deviation;
wRMSd, weighted Root-Mean Square deviation;
uRMSd, unweighted Root-Mean Square deviation;
Figure 8.
Capsid protein domains. The structures of the common core of the capsid proteins are shown using the RSV protein [1d1dA] as a representative. (a) The N-terminal domain, coloured blue→red from amino→carboxy termini with the five major helices represented by thickened lines. (b) The C-terminal domain, represented as in part (a) but with the MHR region marked in white.
Contributor Information
William R Taylor, Email: wtaylor@nimr.mrc.ac.uk.
Jonathan P Stoye, Email: jstoye@nimr.mrc.ac.uk.
References
- Lilly F. Fv-2: identification and localation of a second gene governing the spleen focus response of friend leukemia virus in mice. J Natl Cancer Inst. 1970;45:136–169. [PubMed] [Google Scholar]
- Lilly F, Pincus T. genetic control of muring viral leukemogenesis. Adv Cancer Res. 1973;17:231–277. [Google Scholar]
- Hartley JW, Rowe WP, Huebner RJ. Host-range restrictions of murine leukemia virus in mouse embryo cell cultures. J Virol. 1970;5:221–225. doi: 10.1128/jvi.5.2.221-225.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg N, Jolicoeur P. Retroviral pathenogenisis. In: Coffin JM, Hughes SH, Varmus HE, editor. In Retroviruses. Cold Spring Harbor Press, Cold Spring Harbor, N.Y; 1997. pp. 475–585. [PubMed] [Google Scholar]
- Hopkins N, Schindler J, Hynes R. Six NB-tropic leukemia viruses derived from a b-tropic virus of BALB/c have altered p30. J Virol. 1977;21:309–318. doi: 10.1128/jvi.21.1.309-318.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rommelaere J, Donis-Keller H, Hopkins N. RNA sequencing provides evidence for allelism of determinants of the N-, B-, or NB-tropism of murine leukemia viruses. Cell. 1979;16:43–50. doi: 10.1016/0092-8674(79)90186-7. [DOI] [PubMed] [Google Scholar]
- Best S, Le Tissier P, Towers G, Stoye JP. Positional cloning of the mouse retrovirus restriction gene fv1. Nature. 1996;382:826–829. doi: 10.1038/382826a0. [DOI] [PubMed] [Google Scholar]
- Béenit L, de Parseval J-F, Callebaut I, Cordonnier A, Heidmann T. Cloning of a new nurine endogenous retrovirus MuERV-L with strong similarity to the human HERV-L element and a gag coding sequence closely related to the fv1 restriction gene. J Virol. 5657;71:5652–1997. doi: 10.1128/jvi.71.7.5652-5657.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff SP. Operating under a gag order: a block against incoming virus by the fv1 gene. Cell. 1996;86:691–693. doi: 10.1016/s0092-8674(00)80141-5. [DOI] [PubMed] [Google Scholar]
- Lund AH, Pedersen FS. The nucleotide sequence of the high-leukemogenic murine retrovirus SL3-3 reveals a patch of mink cell focus forming-like sequences upstream of the ecotropic envelope gene. Arch Virol. 1999;144:2207–2212. doi: 10.1007/s007050050634. [DOI] [PubMed] [Google Scholar]
- Pearl LH, Taylor WR. Sequence specificity of retroviral proteases. Nature. 1987;328:482. doi: 10.1038/328482b0. (Communication) [DOI] [PubMed] [Google Scholar]
- Pearl LH, Taylor WR. A structural model for the retroviral proteases. Nature. 1987;329:351–354. doi: 10.1038/329351a0. [DOI] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. Each PDB code specifies a protein that can be viewed or downloaded from: http://www.rcsb.org/pdb/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Sander C. Dali/FSSP classification of three-dimensional protein folds. Nucleic acids research. 1997;25:231–234. doi: 10.1093/nar/25.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos-Olivas R, Newman JL, Summers MF. Solution structure and dynamics of the rous sarcoma virus capsid protein and comparison with capsid proteins of the other retroviruses. J Molec Biol. 2000;296:633–649. doi: 10.1006/jmbi.1999.3475. [DOI] [PubMed] [Google Scholar]
- Khorasanizadeh S, Campos-Olivas R, Summers MF. Solution structure of the capsid protein from human T-cell leukemia virus type-1. J Molec Biol. 1999;291:491–505. doi: 10.1006/jmbi.1999.2986. [DOI] [PubMed] [Google Scholar]
- Khorasanizadeh S, Campos-Olivas R, Clark CA, Summers MF. Sequence-specific 1H, 13C and 15N chemical shift assignment and secondary structure of the HTLV-I capsid protein. J Biomol NMR. 1999;14:199–209. doi: 10.1023/A:1008307507462. [DOI] [PubMed] [Google Scholar]
- Jin Z, Jin L, Peterson DL, Lawson CL. Model for lentivirus capsid core assembly based on crystal dimers of EIAV p26. J Molec Biol. 1999;286:83–93. doi: 10.1006/jmbi.1998.2443. [DOI] [PubMed] [Google Scholar]
- Monaco-Malbet S, Berthet-Colominas C, Novelli A, Battai N, Piga N, Cheynet V, Mallet F, Cusack S. Mutual conformational adaptations in antigen and antibody upon complex formationr between an fab and HIV-1 capsid protein p24. Structure Fold Des. 1079;8:1069–2000. doi: 10.1016/S0969-2126(00)00507-4. [DOI] [PubMed] [Google Scholar]
- Taylor WR. Protein structure alignment using iterated double dynamic programming. Prot Sci. 1999;8:654–665. doi: 10.1110/ps.8.3.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nuc Acid Res. 3402;25:3389–1997. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor WR. Dynamic databank searching with templates and multiple alignment. J Molec Biol. 1998;280:375–406. doi: 10.1006/jmbi.1998.1853. [DOI] [PubMed] [Google Scholar]
- Taylor WR. A flexible method to align large numbers of biological sequences. J Molec Evol. 1988;28:161–169. doi: 10.1007/BF02143508. [DOI] [PubMed] [Google Scholar]
- Higgins DG, Taylor WR. Multiple sequence alignment. In: Webster DM, Walker JM, editor. In Protein structure prediction, Methods in Molecular Biology. Vol. 143. Humana Press, Totowa, New Jersey, USA; 2000. pp. 1–18. [DOI] [PubMed] [Google Scholar]
- Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- Taylor WR. Multiple sequence threading: an analysis of alignment quality and stability. J Molec Biol. 1997;269:902–943. doi: 10.1006/jmbi.1997.1008. [DOI] [PubMed] [Google Scholar]
- Taylor WR, Orengo CA. Protein structure alignment. J Molec Biol. 1989;208:1–22. doi: 10.1016/0022-2836(89)90084-3. [DOI] [PubMed] [Google Scholar]
- Taylor WR, Sælensminde G, Eidhammer I. Multiple protein structure alignment using double-dynamic programming. Comp Chem. 2000;24:3–12. doi: 10.1016/S0097-8485(99)00043-1. [DOI] [PubMed] [Google Scholar]
- Stevens A, Bock M, Ellis S, Le Tissier P, Bishop KN, Taylor WR, Stoye JP. Retroviral capsid determinants of the Fv1 NB- and NR-tropism. 2003. [DOI] [PMC free article] [PubMed]
- Towers GJ, Bock M, Martin S, Takeuchi Y, Stoye JP, Danos O. A conserved mechanism of retrovirus restriction in mammals. Proc Natl Acad Sci USA. 2000;97:12295–12299. doi: 10.1073/pnas.200286297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan S, Hatziioannou T, Cunningham T, Muesing MA, Gottlinger HG, Bieniasz PD. Rcellular inhibitors with fv1-like activity restrict human and simian immunodeficiency virus tropism. Proc Natl Acad Sci USA. 2002;99:11914–11919. doi: 10.1073/pnas.162299499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoye JP. An intracellular block to primate lentivirus replication. Proc Natl Acad Sci USA. 2002;99:11549–11551. doi: 10.1073/pnas.192449399. [DOI] [PMC free article] [PubMed] [Google Scholar]