Abstract
NphB is an aromatic prenyltransferase that catalyzes the attachment of a 10-carbon geranyl group to aromatic substrates. Importantly, NphB exhibits a rich substrate selectivity and product regioselectivity. A systematic computational study has been conducted in order to address several question associated with NphB catalyzed geranylation. The reaction mechanism of the prenylation step has been characterized as a SN1 type dissociative mechanism with a weakly stable carbocation intermediate. A novel π-chamber composed of Tyr121, Tyr216 and 1,6-DHN is found to be important in stabilizing the carbocation. The observed difference in the rates of product formation from 5 and 2-prenylation arises from the differing orientations of the aromatic substrate in the resting state. 4-prenylation shares the same resting state with 5-prenylation, but the lower free energy barrier for carbocation formation makes the latter reaction more facile. The high free energy barrier associated with 7-prenylation is caused by the unfavorable orientation of 1,6-DHN in active site pocket, along with the difficulty of proton elimination after the prenylation step. A water mediated proton transfer facilitates the loss of hydrogen at the prenylation site to form the final prenylated product. Interestingly, the same crystallographically observed water molecule has been found to be responsible for proton loss in all three experimentally identified products. After proton transfer, the relaxation of the final product from a sp3 carbon center to a sp2 center triggers a “spring-loaded” product release mechanism which pushes the final product out of the binding pocket towards the edge of the active site. The hydrogen bond interactions between the two hydroxyl groups of the aromatic product and the sidechains of Ser214 and Tyr288 help to “steer” the movement of the product. In addition, mutagenesis studies identify these same two sidechains as being responsible for the observed regioselectivity, particularly 2-prenylation. These observations provide valuable insights into NphB chemistry, offering an opportunity to better engineer the active site and to control the reactivity in order to obtain high yields of the desired product(s). Furthermore, the SN1 reaction mechanism observed for NphB differs from the prenylation reaction found in, for example, the farnesyltransferase, which proceeds via an SN2-like reaction pathway. The spring-loaded release mechanism highlighted herein also offers novel insights into how enzymes facilitate product release.
Prenyltransferases catalyze the attachment of prenyl groups to various substrates1. Based on substrate preferences, this family of enzymes can be further categorized as protein prenyltransferase1-4 (PPTase) and aromatic prenyltransferase5-7 (APTase). PPTases catalyze the posttranslational attachment of either a 15-carbon farnesyl or a 20-carbon geranylgeranyl group to cysteine residue(s) contained in specific amino acid sequences (e.g., Ca1a2X farnesyltransferase) including G-proteins that are well known for their roles in signaling cell growth and differentiation. APTases usually transfer smaller prenyl groups like the 5-carbon dimethylallyl or 10-carbon geranyl moieties to a variety of aromatic substrates. Known APTases include, but are not limited to, NphB5,8 (formerly Orf2) purified from Streptomyces sp. Strain CL190 and the fungal indole prenyltransferase FtmPT17,9 extracted from Aspergillus fumigatus.
APTases are involved in the biosynthesis of terpenoids, in combination with a variety of terpenoid synthases10. Terpenoids are widely distributed natural products, which are found in fungi, animals and plants11-13. Plant isoprenoids are well known for their application in traditional herbal remedies due to their aromatic qualities14. Many isoprenoids and their derivatives possess pharmaceutically desirable properties such as antiviral, anti-inflammatory, antioxidant and anticancer characteristics15-24. In addition, the low cellular toxicity and excellent penetrability into cell membranes have made this class of compounds desirable as drug templates25. Therefore, the biosynthesis of these terpenoids is of great interest and importance to chemists, biochemists and medicinal chemists. As a result, APTases have drawn considerable attention from both experimental and computational labs interested in biosynthesis of terpenoids5-8,26-30.
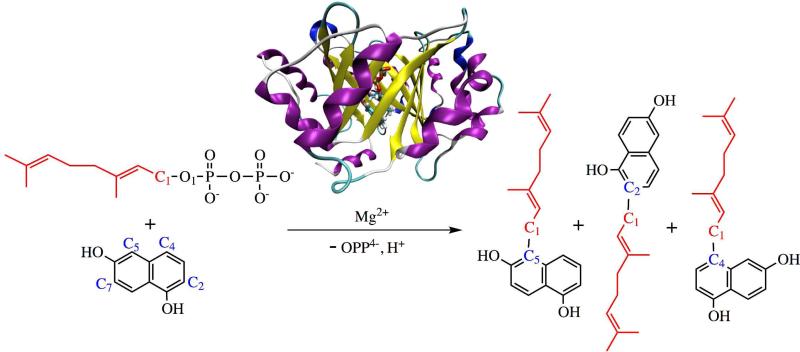
NphB, identified from Streptomyces, catalyzes the addition of a 10-carbon geranyl group to a number of small organic aromatic substrates (see Figure 1), and is involved in the biosynthetic pathway of the antioxidant naphterpin5. The 3-D structure of NphB features a novel α/β barrel fold termed PT-barrel (prenyltransferase barrel, see Figure 1)5,31, composed of 10 anti-parallel β-strands surround by 10-α helices. Inside the barrel, a spacious and solvent accessible binding pocket is provided and two substrates molecules, namely geranyl diphosphate (GPP) and 1,6-dihydroxynaphthalene (1,6-DHN), are bound. GPP is stabilized via interactions between its negatively charged diphosphate moiety and several amino acid sidechains, including Lys119, Thr171, Arg228, Tyr216 and Lys284, in addition to Mg2+. A Mg2+ cofactor is required for the activity of NphB, despite the lack of a (N/D)DXDX motif usually found in Mg2+ containing enzymes32,33. The metal ion is coordinated to a carboxylate oxygen atom from Asp62, an α-diphosphate oxygen atom and four water molecules. The divalent metal cation has been proposed to stabilize the developing negative charge on the diphosphate leaving group in the chemical step. For instance, in FTase catalysis, a magnesium ion reduces the free energy barrier height by approximately 4kcal/mol, despite the fact that Mg2+ is only required for optimal activity of FTase34.
Figure 1.
Schematic representation of geranylation catalyzed by NphB complexed with GPP and 1,6-DHN.
NphB chemistry features diverse substrate selectivity and interesting product regioselectivity5,8,29. NphB catalysis of the prenylation of 1,6-DHN yields three different products, where the geranyl chain being attached to different carbon atoms of the aromatic unit (see Figure 1). The major product, a 5-geranyl-1,6-DHN, and the minor product, a 2-geranyl-1,6-DHN, were originally characterized, with a product ratio of 10:15. In subsequent work, another minor product - 4-geranyl-1,6-DHN, was identified, with a much smaller kcat than that observed for the other two products8. Earlier computational work identified the source of the product diversity as arising from the different orientations of 1,6-DHN found within the active site29. In the resultant free energy profile, the free energy minimum that leads to the major product and another local minimum stabilized by hydrogen bond (H-bond) interactions between the hydroxyl groups of 1,6-DHN and the sidechains of Ser214 and Tyr288 responsible for the minor product were found to be connected via an intermediate state. The free energy minimum is favored by 2.3kcal/mol over the second minimum. However, this work did not locate a binding orientation that could be responsible for 4-geranyl-1,6-DHN.
Similar to FTase catalysis, the reaction mechanism of for NphB prenylation is not certain. Proposed mechanisms include an associative pathway featuring a SN2-like nucleophilic attack and a dissociative pathway via a carbocation-mediated electrophilic capture5. The so-called associative mechanism with dissociative characteristics ascribed to FTase chemistry can't be excluded as well35-37. Despite the aromatic nature of the target substrate, most APTases also possess an aromatic-rich active site that PPTases usually do not have. Whether these aromatic residues stabilize a positively charged prenyl carbocation is as yet unknown. Structural analysis and currently available experimental evidence are insufficient to determine the preferred prenylation pathway. Furthermore, a proton transfer step is required after the prenylation step to re-aromatize the product. A water mediated proton transfer has been proposed for this step, but hasn't been unequivocally proven5.
Computational tools have proven useful in the mechanistic study of PPTases and other enzymes36-39. Combined QM/MM methods are effective in the study of enzyme catalysis, but given the sizes typically employed and sampling requirement most studies have been restricted to semiempirical models. Herein we use the self-consistent charge density-functional based tight-binding (SCC-DFTB) method40 as our QM tool of choice given its accuracy and computational performance. SCC-DFTB/MM method has been widely applied to simulate enzymatic reactions and reported to generate good accuracies41-48. Moreover, SCC-DFTBPR has proven effective in the study of phosphate complexes49,50. In our studies of FTase carried out using the SCC-DFTB(PR)/AMBER QM/MM level of theory coupled with molecular dynamics (MD) and umbrella sampling (US) techniques have yielded excellent results in comparison with experiment37. Hence, given this latter success we decided to extend our efforts to the fascinating case of NphB catalysis. The performance of SCC-DFTBPR for this prenylation reaction was also examined and is discussed in SI.
Methods
All QM/MM51 MD simulations in this study were performed using SANDER and PMEMD was utilized for long classical MD simulations. Both programs are part of the AMBER1152 suite of programs.. The particle mesh Ewald (PME) method53 was used to model the long-range electrostatic interactions. X-H bonds were constrained using the SHAKE algorithm54. Quantum calculations were conducted using the Gaussian’09 package55.
QM/MM equilibration of NphB complexed with GPP and 1,6-DHN
The QM/MM X-ray refined structures of the free energy minimum (leading to 5-prenylation, termed P5) and the second local minimum (leading to 2-prenylation, termed P2) were used to start all of our studies29. Each structure was first solvated in a truncated octahedral water box filled with TIP3P56 water molecules and then equilibrated using classical MD simulations for several ns. The protocol was similar to that described by Cui et al.29 and will not be discussed in detail here. For each system, the two substrates, GPP and 1,6-DHN, were incorporated into the QM region while the rest of the system remained in the MM part (see discussion in SI). This partitioning includes the important groups involved in the chemistry steps, while allowing for long umbrella sampling57,58 (US) simulations in the following potential of mean force (PMF) studies. The systems were a fully minimization using the hybrid QM/MM (SCC-DFTB/ff99SB) potential for 1000 steps the using steepest descent algorithm. These structures were then slowly heated to 300K over 50ps using a Langevin thermostat. The collision frequency was set to 5ps-1 while the time step was 0.5fs. Subsequently, the systems were equilibrated at 1atm in the NPT ensemble for 450ps with a 1fs time step. SHAKE was turned off for the QM region during all QM/MM simulations. A modified PME approach was used to treat the QM/MM long-range electrostatics59.
QM/MM PMF study of NphB prenylation
QM/MM equilibrated P2 and P5 structures were used to start the PMF studies. For each prenylation reaction, the reaction coordinate (RC) was defined as the distance between two reacting atoms - C1 of GPP and the corresponding prenylation center in 1,6-DHN; For example, C5 of 1,6-DHN for P5. For P2 and P5, it was straightforward to use the two existing systems adopted from a previous MM based PMF study29. For 4-prenylation (P4), we used the P5 minimum because it had the closest contact between the C1 of GPP and the C4 of 1,6-DHN making it appropriate as a starting point for the QM/MM PMF simulation of P4 prenylation. We also examined the possibility of 7-prenylation (P7). In this case, another snapshot from the equilibrated P5 system was chosen as the initial structure. Steered MD60-62 (SMD) scans were first conducted for all systems in order to propagate the trajectory along the RC. The force constant was 5000 kcal/mol•Å2 and the pulling speed was 0.02Å/ps. Starting structures for the subsequent US simulations were extracted from the corresponding SMD trajectory with an interval of 0.1Å between adjacent windows. Thus, based on dRC, there were 26, 31, 26, 31 windows generated for P2, P4, P5 and P7, respectively. These windows covered the RC from 1.5Å to 4.0Å for P2 and P5, and from 1.5Å to 4.5Å for P4 and P7. Each window was first equilibrated for 100ps followed by a 150ps production run during which RC data was collected. The applied harmonic force constant was 100-500 kcal/mol•Å2 for each window (with larger force constants applied to higher energy regions to ensure thorough sampling). In order to improve the overlap between adjacent windows in areas associated with bond breaking or bond forming, 14 additional windows were added, yielding over 10ns US simulations for each state. Grossfield's WHAM code63 was used to reconstruct the unbiased RC distribution for all US simulations reported in this work.
QM/MM PMF study of the effect of point mutations in NphB prenylation
In the prenylations step, the sidechain of Tyr121 was in close contact with the geranyl group throughout the course of the prenylation reaction and likely stabilizing the developing carbocation. In this regard, a computational mutation study was carried out to investigate the effect of the Y121L mutation. A snapshot from the P5 QM/MM equilibration was selected, modified and re-equilibrated for 100ps, connected by another set of SMD and US PMF studies following the same strategy previously described. In an previous study by Cui et al.29 Ser214 and Tyr288 were identified as stabilizing 1,6-DHN in an orientation required for 2-prenylation. Therefore, we carried out another two sets of QM/MM PMF studies to investigate the effect of S214A and Y288F mutation on 2-prenylation. A snapshot of the P2 QM/MM equilibration was extracted and followed by two series of QM/MM simulations adopting an identical procedure as previously described for the Y121L study.
QM/MM PMF study of the proton transfer step of NphB catalysis
NphB catalysis requires a proton transfer step to remove the extra hydrogen atom at the prenylation site in 1,6-DHN to generate the final product. In order to study this, each prenylation product was equilibrated for another 250ps and prepared for a second PMF study. The RC at this stage was defined as the distance between the hydrogen atom attached to the prenylation center in 1,6-DHN and the oxygen atom of the water molecule serving to “shuttle” the proton. A SMD simulation was performed with the same force constant and pulling speed as applied in the prenylation step for each state. For these studies, the “shuttle” water molecule was also included in the QM region. Starting configurations were then extracted from the SMD trajectory with an interval of 0.1Å for the US studies. In this way 17, 21 and 18 windows were prepared for the P2, P4 and P5 states. The P7 state was excluded from this series of US simulation (see discussion below). For each of the final product states, a snapshot was extracted from the final US simulations (for the P7 state this was taken from a SMD study) and equilibrated for another 100ps using the QM/MM model. In order to test the feasibility of a Tyr216 mediated proton transfer, another SMD was performed for the P5 state with the distance between the tyrosine hydroxyl oxygen atom (in these simulations the Tyr216 sidechain was placed into the QM the region) and the extra hydrogen atom connected to C5 of 1,6-DHN as the reaction coordinate.
QM calculation of the weakly stable carbocation intermediates in the prenylation step of NphB catalysis
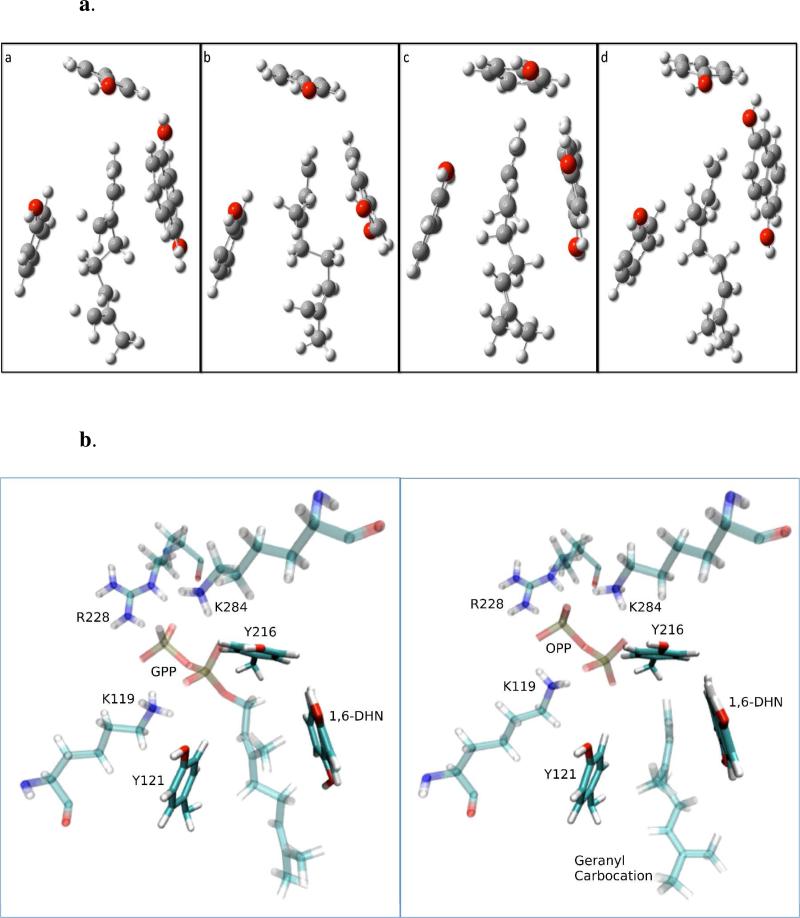
For each of three states that lead to experimentally observed products (P2, P4 and P5), along with the possible product P7, an interesting π-chamber was identified and appeared important to carbocation stabilization. Therefore, we decided to further investigate the weakly stable carbocation in the intermediate state of each prenylation reaction. The atomic coordinates of the geranyl carbocation, 1,6-DHN and the phenol sidechains of Tyr121 and Tyr216 were extracted from a snapshot close to the carbocation intermediate for each state and subjected to higher-level QM analysis. A hydrogen atom was added to each tyrosine sidechain to generate a phenol group. For each structure, an optimization at the M06-2X/6-31G** level64,65 was first performed to locate the potential energy minimum, followed by a set of single point calculations using basis set superposition error (BSSE) corrections to quantitatively study the interaction between the carbocation and each aromatic group66,67. All carbon atoms were restrained during the optimization. The water and the low dielectric calculations (see SI) were run using the Polarized Continuum Model (PCM) using the integral equation formalism variant (IEFPCM) for both optimizations and single point calculations68. The single point calculation were also carried out at the M06-2X/6-31G** level. In addition, we also carried out QM calculations at the same level of theory on the prenylated carbocation intermediates for each of four possible prenylation pathways in order to evaluate the inherent stability of such an intermediate state for each reaction channel. The atomic coordinates for the prenylated carbocation were obtained from a snapshot of the intermediate for each reaction pathway. Each intermediate was optimized at the M06-2X/6-31G** level with the PCM solvation model and all heavy atoms restrained.
MD simulation and point mutation study of the product release process from NphB catalyzed 5-prenylation
The structure of the QM/MM equilibrated P5 product was extracted with all water molecules except the Mg2+ bound water molecules removed in order to study the product release process. The resulting structure was modeled using the AMBER force field ff99SB69 for the protein and the generalized AMBER force field (GAFF)70 for the ligand. The atomic charges of diphosphate (OPP) and 5-prenylated-1,6-DHN (5P-1,6-DHN) were derived following a two-stage restrained electrostatic potential (RESP) fitting procedure71,72. In addition to this wildtype (WT) system, point mutations were made S214A (MUT1) and Y288F (MUT2) in order to study their influence on the product release process. Thus, a total of three systems were prepared for subsequent MD simulations. All three systems were solvated in a truncated octahedral TIP3P water box with an 8Å separate between the edge of the box and nearest solute atom. The resultant structures were first minimized with steepest descent and conjugated gradient methods for 2000 steps. Subsequently, a gradual heating procedure was conducted on each of the three systems to bring them to 300K over 100ps of simulation with a 1fs time step. A Langevin thermostat with a 5ps-1 collision frequency was applied. This was followed by a 500ps NTP equilibration and 100ns production runs, for each of the three systems.
Results and Discussions
It has been proposed that different orientations of 1,6-DHN in the starting state of NphB's catalytic cycle are responsible for the observed product regioselectivity8,29. However, to further understand the nature of NphB catalysis, we carried out QM/MM studies to elucidate the reaction mechanism for both the prenylation and the subsequent proton transfer steps. With an approach based on statistical mechanics principles and numerical simulation techniques, we were able to both qualitatively and quantitatively study the course of the catalytic reaction. Furthermore, unguided MD simulations at 10-100ns timescale were conducted to study the mechanism of product release, yielding molecular-level insights into protein-ligand, protein-solvent and ligand-solvent interactions during product release. Given the detailed understanding we have obtained with regards to NphB catalysis we have set the stage to engineer this system to synthesize compounds of interest with the desired stereochemistry.
QM/MM MD simulation and PMF study of prenylation catalyzed by NphB complexed with GPP and 1,6-DHN
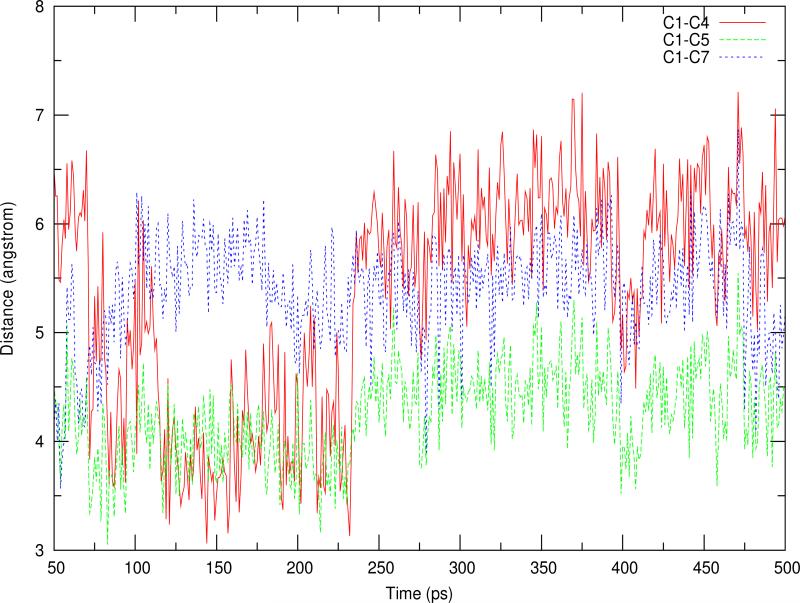
Two separate 500ps QM/MM simulations were performed for P2 and P5 and in both cases the backbone conformation of the protein and the orientation of both GPP and 1,6-DHN were well preserved throughout. Mg2+ maintained its octahedral coordination consisting of the α-diphosphate of GPP, Asp62 and four crystal water molecules. The diphosphate group was anchored throughout by H-bond interactions between itself and the sidechains of residues Lys119, Asn173, Tyr216, Arg228 and Lys284. It has been previously observed that the P2 and P5 states lead to two different experimentally observed products. However, no stable resting state has been found for P4 prenylation, which was experimentally characterized more recently. Moreover, the intermediate binding state found in a previous study by Cui et al.29 could lead to 7-prenylation, because the C7 of 1,6-DHN was the closest aromatic carbon atom to C1 of GPP. In addition, both the C2 and C5 atoms of 1,6-DHN are ortho to one of the hydroxyl groups while C4 is para to a hydroxyl group, but C7 - another ortho position, has not been reported to be prenylated. Furthermore, the average Mulliken charges of C2, C4, C5 and C7 are -0.24, -0.16, -0.24 and -0.18q respectively (averaged over the last 25ps of simulation), and the RESP charges of such four carbon atoms are -0.14, -0.17, -0.15 and -0.17q separately. Combining the above points, it appears that C7 atom should be able to carry prenylation also. The observed distance variations from the last 45ps of the P5 simulation reveal that both C4 and C7 of 1,6-DHN were accessible to the C1 atom of GPP (dC1-C4 and dC1-C7 ~3.5Å in spots; see Figure 2). Therefore, in the subsequent PMF studies, both P4 and P7 were examined starting from a snapshot extracted from the P5 simulation.
Figure 2.
Distance variations observed during last 450ps of QM/MM MD simulation of the P5 reaction channel.
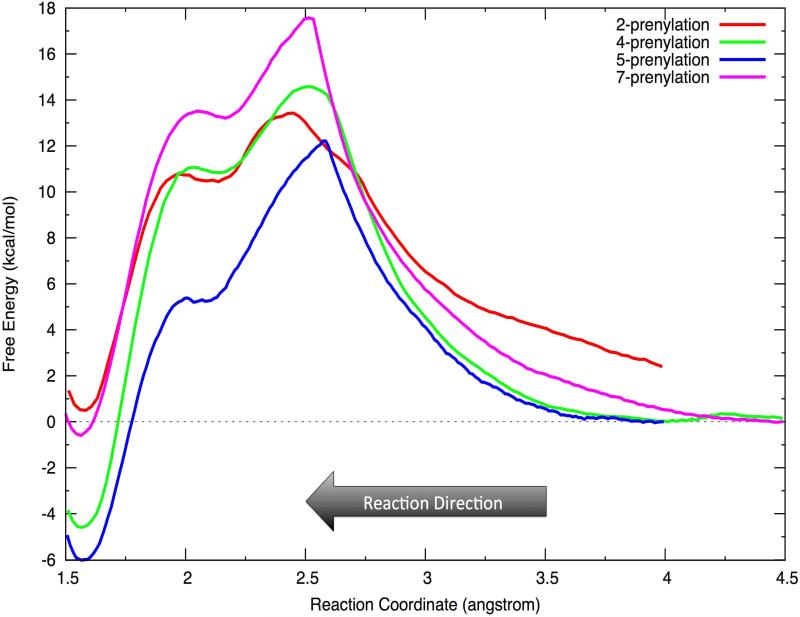
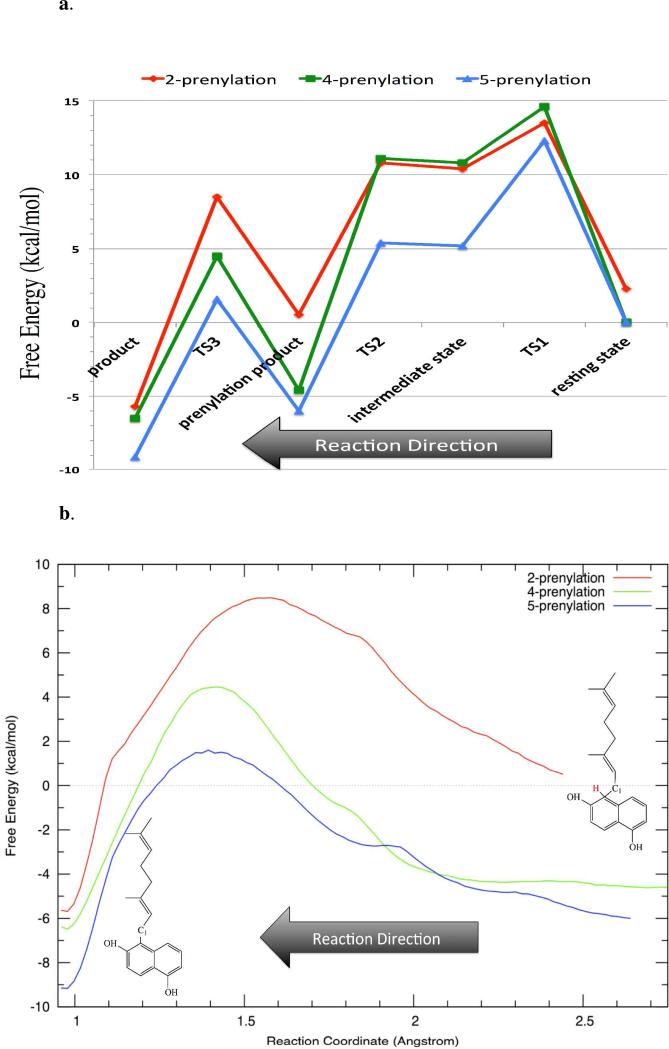
In the subsequent PMF studies of the prenylation step, over 40ns of US simulations were performed for the four systems examined. The weighted histogram analysis method (WHAM) was utilized to unbias the distribution of the corresponding RCs and to construct the free energy profiles (FEPs) (see Figure 3). The RC was defined as dC1-C2, dC1-C4, dC1-C5, and dC1-C7 for P2, P4, P5 and P7, respectively. Alternate RCs choices were tried (see SI), but the model chosen here performed the best overall. Figure 3 shows the computed free energy profiles for each of the four reaction pathways. In each case the statistical errors estimated via the Monte Carlo bootstrap analyses were typically less than 0.2kcal/mol across the entire profile.
Figure 3.
Free energy profiles for prenylation at four sites in 1,6-DHN. P2 starts at 2.3 kcal/mol, which is the relative free energy difference of the P2 state to that of P5.
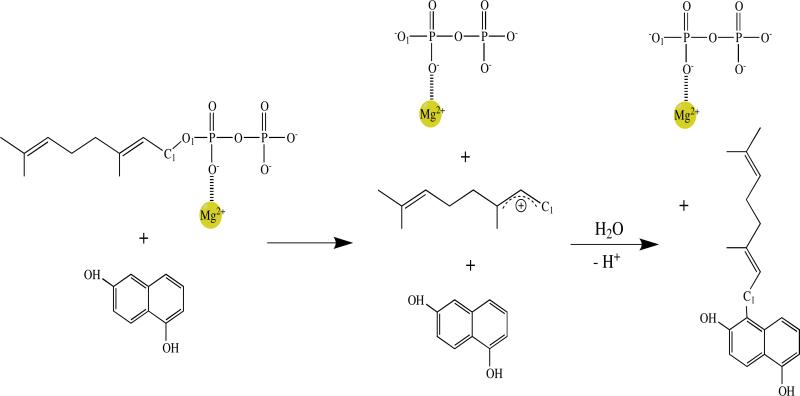
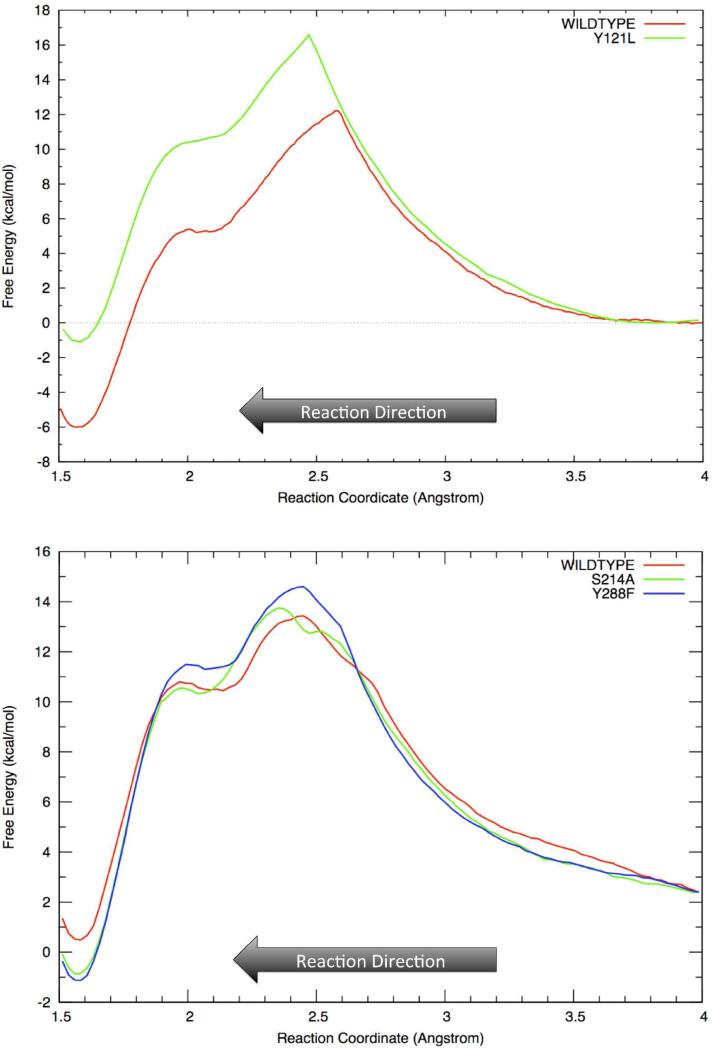
The free energy profile indicates a SN1-like dissociative reaction mechanism for prenylation of the aromatic substrate 1,6-DHN. Two free energy barriers are observed separated by an intermediate state (see Figure 4). The first barrier, which corresponds to the cleavage of the C1-O1 bond of GPP and the formation of a carbocation, is the rate-limiting step for the prenylation reaction. In each case, the geranyl carbocation is beautifully wedged between two aromatic groups – Tyr121 and 1,6-DHN, and capped by another aromatic group – Tyr216, leading to an interesting cation-π “π-chamber”73-75 scheme (see Figure 5a and 5b). More interestingly, visual examination of the reaction course clearly shows that the carbocation repositions to better center itself between two aromatic units after formation. The truncated model of this cation-π interaction scheme (see Figure 5a) were optimized at the M06-2X/6-31G** level with the IEFPCM water model. The BSSE corrected interaction energies between the carbocation and each aromatic unit were computed using the same level of theory to investigate their magnitudes. For the three experimentally characterized prenylations, the resultant energies were ~-1kcal/mol and ~-3 to -6kcal/mol for the Tyr121 and Tyr216 carbocation interactions, while for the carbocation-1,6-DHN interaction values of ~-7 to -20kcal/mol were computed (see Table 1). Moreover, a Y121L computational point mutation study revealed that the loss of the cation-π interaction increases the free energy barrier associated with carbocation formation by ~4.5kcal/mol and destabilizes the intermediate state (see Figure 6). Hence, it is not surprising that a weakly stable carbocation is observed in the NphB prenylation reaction. The same trend was observed in the IEFPCM low dielectric water (see SI). Furthermore, with a strong interaction between the geranyl group and the aromatic substrate, it is not surprising to observe a low second barrier height (corresponding to the Cgeranyl-Caromatic bond formation) of ~1kcal/mol relative to the intermediate state.
Figure 4.
Dissociative mechanism of NphB catalyzed geranylation (P5 channel).
Figure 5.
Schematic representation of π-chamber. (a) Truncated representation of the π-chamber found at the intermediate state of (a) 2-prenylation, (b) 4-prenylation, (c) 5-prenylation and (d) 7-prenylation. (b) π-chamber in the NphB binding pocket consisting of Tyr121, Tyr216 and 1,6-DHN found at the resting state (left) and intermediate state (right) of 5-prenylation.
Table 1.
Cation-π interactions in “π-chamber” of each product. (Unit: kcal/mol. GCC = geranyl carbocation, negative value means attractive and positive value suggests repulsive). (See also Table S1.)
| 2-prenylation | 4-prenylation | 5-prenylation | 7-prenylation | |
|---|---|---|---|---|
| GCC – Y121 | -2.9 | -4.4 | -5.8 | -2.7 |
| GCC – Y216 | -0.7 | -1.3 | -0.7 | -0.5 |
| GCC – 1,6-DHN | -20.6 | -6.8 | -13.4 | +1.0 |
Figure 6.
Schematic representation of point mutation effects of Y121L on 5-prenylation (top), and S214A & Y288F on 2-prenylation (bottom). Note: reaction direction is from LEFT to RIGHT.
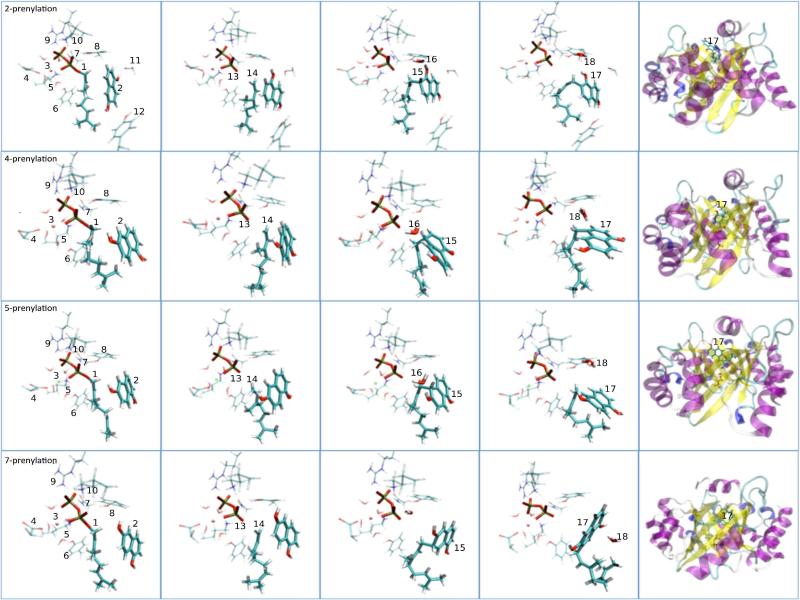
The intrinsic stabilities of the prenylated intermediate states also influences the different reaction channels. Hence, the potential energies of all prenylated 1,6-DHN intermediates arising from the various reaction channels were computed and compared to P5 channel intermediate (see Table 2). In our M06-2X/6-31G** (with IEFPCM) optimizations of the four prenylated intermediate state structures, the major product channel, P5, has the lowest energy, while P2, P4 and P7 are ~7, ~10 and ~16 kcal/mol higher, respectively. The second barrier (around 2 Å) from Figure 3 also indicates the ease of formation of the carbocation intermediate. Importantly, both the trend and the energy difference are in agreement with our QM calculations. On the other hand, the negatively (-4) charged diphosphate leaving group is stabilized by Mg2+ coordination and H-bond interactions with the sidechains of Lys119, Asn173, Tyr216, Arg228 and Lys284. Tyr216 situates itself between diphosphate and prenylated-1,6-DHN, suggesting it might be involved in shuttling out the excess proton (see Figure 7). Alternatively, a crystallographically observed water molecule that remains in the NphB active site throughout our simulations could serve as a proton shuttling group as well. Indeed, the short distance between the excess proton and this water molecule (further localized via a H-bond with a hydroxyl group of 1,6-DHN) makes it the most likely candidate to facilitate the proton transfer step (see Figure 7).
Table 2.
Inherent stability of prenylated carbocation intermediates. (Unit: kcal/mol)
| 2-prenylation | 4-prenylation | 5-prenylation | 7-prenylation | |
|---|---|---|---|---|
| GCC – Y121 | 7.5 | 9.9 | 0.0 | 15.5 |
Figure 7.
Key snapshots from NphB catalysis taken during the reaction course of (From top to bottom) 2-, 4-, 5- and 7-prenylation. From left to right: resting state, intermediate state, prenylation intermediate state, final product state and product released state. Note: (1) GPP; (2) 1,6-DHN; (3) Mg; (4) D62; (5) K119; (6) Y121; (7) T171; (8) Y216; (9) R228; (10) K284; (11) S214; (12) Y288; (13) OPP; (14) Geranyl carbocation; (15) Prenylated-1,6-DHN; (16) Crystal water (shuttle); (17) Final product; (18) Hydronium.
For P2, a rotation of 1,6-DHN was observed in our study, possibly in order to reduce the steric repulsion between itself and the geranyl group due to their original orientation. In the resting state of P2, 1,6-DHN is stabilized by H-bond interactions with the sidechains of Ser214 and Tyr288. However, in the intermediate state, the aromatic substrate starts to rotate slightly, reorienting itself relative to the geranyl carbocation. While in the product state, the 1,6-DHN rotates ~90° counterclockwise (see Figure 7), reducing the steric repulsion in the complex thereby generating a configuration akin to what is observed for P4 and P5. In the free energy profile, the “zero” point for P2 starts at 2.3kcal/mol, which is the relative free energy difference between the resting states for P2 and P5. Indeed, in the rate-determining step the computed energy difference (~1kcal/mol) suggests that the less stable starting state for P2 prenylation is solely responsible for the observed product distribution. This is consistent with the QM calculations of the interaction energy found in the “π-chamber” since in the intermediate state the carbocation and 1,6-DHN were more negative for P2 than P5. Moreover, the free energies of the intermediate and product states for P2 are higher than for P5, possibly due to conformational effects. In the S214A and Y288F computational point mutation studies, H-bond interactions between the sidechains of Ser214 and Tyr288 and the two hydroxyl groups of 1,6-DHN were lost. As a result, the barrier for carbocation formation increased by 0.7 and 1.3kcal/mol (see Figure 6), respectively, further confirming that these two residues help stabilize the required 1,6-DHN orientation via H-bond interactions over the course of P2 prenylation. Moreover, the free energies associated with the prenylation products of S214A and Y288F were both lower than the wildtype, implying that the H-bonds between these two residues and 1,6-DHN destabilize the initial product state and thereby facilitate the formation of the final product via proton transfer.
The rates of product formation for the three experimentally characterized products are (1.1 ± 0.1) × 103s-1, (0.32 ± 0.02) × 103s-1 and (4.2 ± 0.2) × 103s-1 for P2, P4 and P58, respectively. These correspond to free energy barrier heights for these three systems of 13.4kcal/mol, 14.1kcal/mol and 12.6kcal/mol, respectively. Our predicted free energy barrier heights for the three observed prenylation reactions are 13.5kcal/mol, 14.6kcal/mol and 12.3kcal/mol, all in good agreement with experimental measurements. On the other hand, we predict a free energy barrier height of ~17.6kcal/mol for P7 prenylation, which prohibits the formation of this product relative to the others. The much higher free energy barrier for P7 might be associated with the unfavorable arrangement of the two substrates (see Figure 7). Indeed, QM calculation on the intermediate state of P7 reveals repulsive interactions between the carbocation and aromatic substrate (see Table 1), which suggests an unfavorable arrangement in the transition state and intermediate state. In fact, based on the relative orientation of 1,6-DHN and GPP in the so-called S2 state reported by Cui et al.29, could be a better starting state, but is was unstable relative to the P5 and P2 starting states in our MD simulations suggesting that it is a poor starting state for P7 prenylation.
The trend in free energy barriers for the four prenylation reactions is largely consistent with the results obtained from the M06-2X/6-31G** calculations of the π-chamber stabilization energy of the corresponding intermediate states (see Table 1). The main exception is between P5 and P2 prenylation where the former is the favored product, but the latter is predicted to have the lowest barrier for prenylation. In this case the observed product distribution reflects the relative stability of the starting state (P5 more stable than P2) from which the product is formed. Hence, P2 possesses the most attractive interaction between the geranyl carbocation and 1,6-DHN, consistent with the fact that the actual free energy barrier of carbocation formation for P2 is lower than P5. We propose that the binding pose of 1,6-DHN in P2 provides the largest contact “surface” for cation-π interaction with the carbocation resulting in a greater interaction energy than for P5 (see Figure 7, resting and intermediate state).
Based on our predicted free energy barriers, the C1-O1 bond breaking/carbocation formation step is less facile than attack of the carbocation on the aromatic substrate. In order to identify the rate-determining step in the overall prenylation reaction we next needed to explore the proton transfer step. However, prior to doing this it is instructive to compare NphB prenylation to that of farnesyltransferase (FTase).
Comparison of the prenylation mechanism of NphB with that of FTase
The FTase catalytic mechanism has also been hotly debate for a number of years with evidence supporting both an associative mechanism and a dissociative mechanism being reported76. It is not until recently that the so-called associative mechanism with dissociative character was experimentally delineated for FTase chemistry, and subsequently seen computationally35-37. At this point it is instructive to compare the prenylation mechanism of NphB relative to FTase.
The major difference in these two systems is the target substrates: FTase prenylates the S atom in a cysteine residue contained in a CaaX motif while NphB prenylates aromatic compounds. NphB possess an aromatic rich binding pocket, with Tyr121 and Tyr216 situated in such a way to form a π-chamber along with 1,6-DHN (see Figure 5b). Such an environment stabilizes the developing positive charge on prenyl group or carbocation via cation-π interactions (see above), thus favoring a SN1-like mechanism. On the other hand, FTase does not exhibit an aromatic rich active site and prenylates a S atom which is bound to a zinc ion. The lack of an aromatic rich active site may be the major deciding factor disfavoring carbocation formation in FTase catalysis. Fungal prenyltransferase FtmPT1 is another good example of an APTase. The target substrate is aromatic and, in addition, has a unique “tyrosine shield” composed of 4 tyrosine residues surrounding the dimethylallyl group. This environment appears to stabilize the developing carbocation and experimental evidence has been obtained supporting the generation of a weakly stable carbocation7.
In summary, we propose that the aromatic nature of the target substrate as well as the active site favors a dissociative mechanism for the APTases. In contrast, the lack of aromatic character in both the target substrate and the binding pocket in PPTases makes it more difficult to stabilize a carbocation, thereby, favoring mechanisms with more SN2 character.
QM/MM MD simulation and PMF study of the proton transfer step in NphB catalysis
A proton at the prenylation center of 1,6-DHN must be removed in order to generate the final re-aromatized product. To obtain the full picture of the NphB catalytic mechanism, QM/MM MD simulations were carried out for all four systems and three PMF simulations were carried out map out the free energy profiles for P2, P4 and P5.
During preliminary 250ps QM/MM MD simulations for each prenylated intermediate, the same crystal water molecule was observed less than 3Å away from 1,6-DHN during the simulation timescale. For most of these simulations, this water molecule forms a H-bond with a hydroxyl group from 1,6-DHN nearest to the prenylation center, ensuring that it remains proximal to the extra hydrogen atom. Tyr216 also forms a stable H-bond with an α-phosphate oxygen atom of the OPP leaving group and situates itself between OPP and prenylated-1,6-DHN. Hence, Tyr216 mediated proton transfer is also possible.
In order to elucidate the favored reaction path for proton transfer, SMD simulation and subsequent US PMF study were initially performed for P5, which yields the major product. The direct proton abstraction from prenylated 1,6-DHN by the hydroxyl group of Tyr216 generates an unreasonably high free energy barrier of ~45kcal/mol. Therefore, proton transfer via such a pathway is highly unlikely. The water mediated proton transfer is much more energetically feasible, with a free energy barrier of ~7.7kcal/mol higher than the prenylated intermediate, which is ~1-2kcal/mol higher than the resting state (see Figure 8). Hence, we conclude that the formation of carbocation in the prenylation step is the rate-limiting chemical step in NphB catalysis.
Figure 8.
(a) Computed free energy chart for the entire reaction course and (b) computed free energy profile for the proton transfer step for the three experimentally observed products (Note: reaction direction is from LEFT to RIGHT.). Note: (a) TS1 – carbocation formation step; TS2 – prenylation reaction step; TS3 – proton transfer step. (b) the starting points are chosen based on their corresponding prenylation product states and free energies shifted accordingly.
The computed PMF surfaces for the P2, P4 and P5 proton transfer step are shown in Figure 8. P7 was not examined since its barrier was significantly higher than the other cases for the prenylation step. The P2 and P4 proton transfer reactions also have lower free energy barriers than their corresponding prenylation steps. Therefore, we propose that the C1-O1 bond cleavage/carbocation formation step is the rate-limiting product formation step for each product observed in NphB catalysis.
The four final product states were further equilibrated at the QM/MM level for an additional 100ps (initial structure for P7 was generated from a SMD simulation). Interestingly, for P2, P4 and P5, the same trend was observed: accompanying internal geometric relaxation, the corresponding XP-1,6-DHN (X=2, 4 or 5) product pushes itself out of the binding pocket toward one edge of the PT-barrel. In fact, all three simulations end with XP-1,6-DHN product localized in a similar location at the edge of the PT-barrel (see Figure 7). However, the story for P7 is different: the H3O+ ion is situated on the opposite side of 7P-1,6-DHN from OPP and this arrangement seems to hinder the release of this the product (see Figure 7). Clearly P7 prenylation is disfavored throughout all phases of NphB catalysis.
Classical MD simulation and mutational study of NphB's product release process
During the final QM/MM equilibration of the NphB product states, the prenylated products begin to release from the binding pocket largely driven by geometric relaxation after the loss of the extra proton (see Figure 9). After proton transfer, the relaxation of the final product from a sp3 carbon center to a sp2 center triggers a “spring-loaded” product release mechanism which pushes the final product out of the binding pocket towards the edge of the active site. To further study this process, classical MD simulations were carried out on the wildtype P5 system (WT) along with S214A (MUT1) and Y288F (MUT2) point mutated models. The goal of these studies were to qualitatively understand what key residues, etc. that might be important in the product release process rather than obtaining a free energy profile, which would require much more sophisticated and time consuming simulations.
Figure 9.
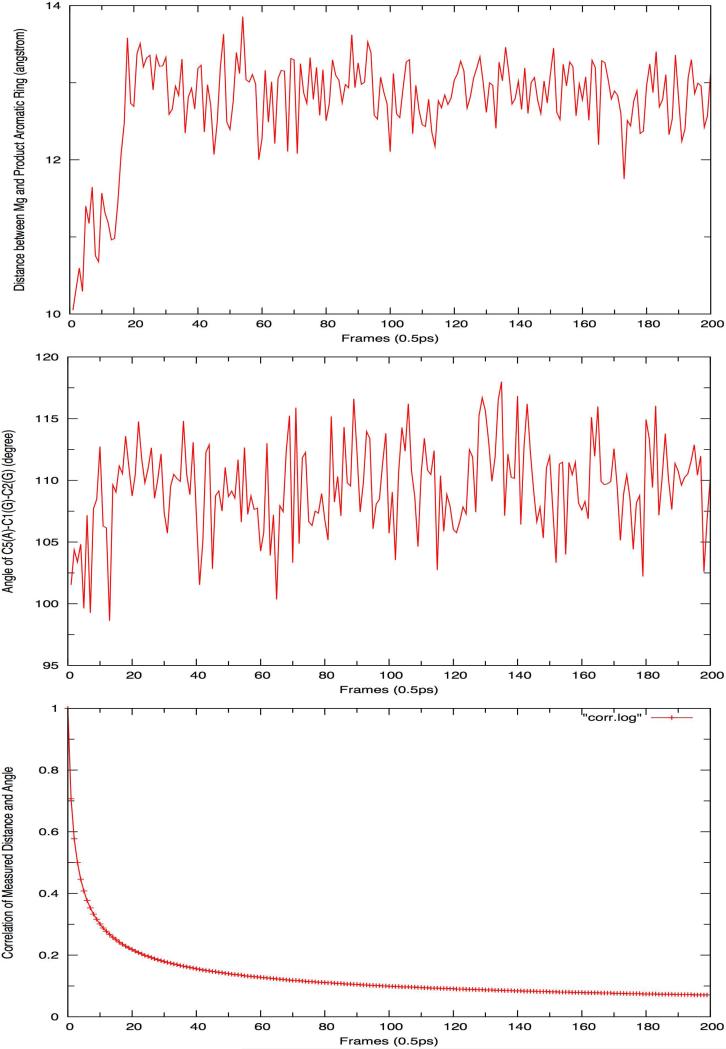
Schematic representation of the correlation between the 5P-1,6-DHN geometry relaxation and product release: (1) variation of distance (between Mg(II) and the aromatic ring of the product, (top, showing product release), (2) angle of C5(1,6-DHN)-C1(GPP)-C2(GPP) (medium, showing geometry relaxation), and (3) correlation between them over 100ps QM/MM equilibration of P5 final product (bottom, showing correlation).
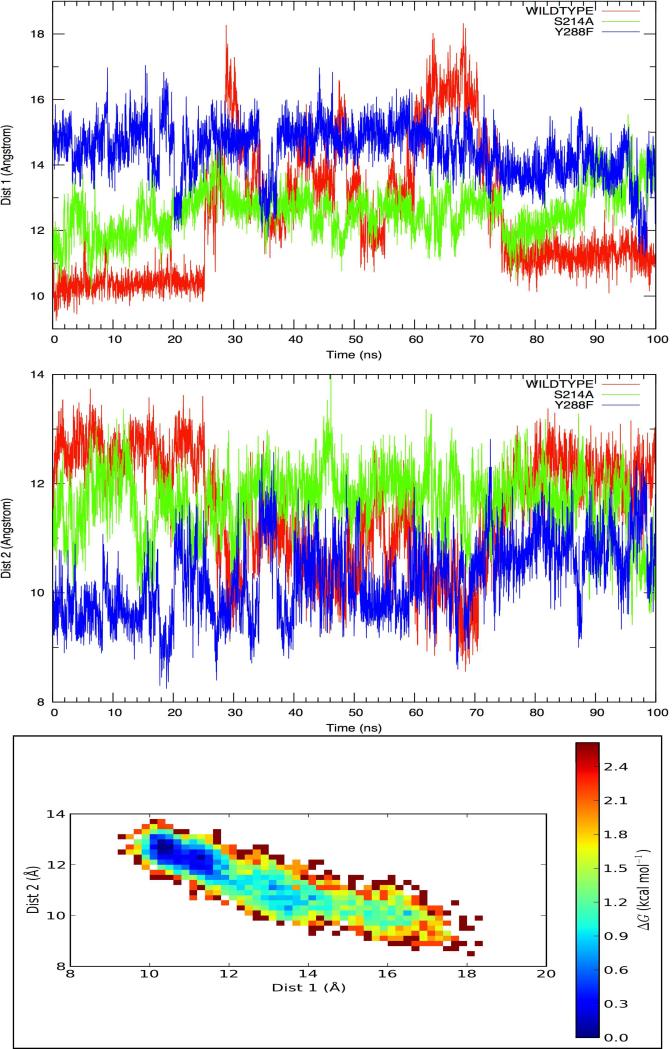
The 100ns MD simulation of WT aimed to study the process of 5P-1,6-DHN release and to study the protein-ligand and ligand-solvent interactions involved. During the simulation, movement towards the edge of the PT-barrel was observed after ~20ns, as the distance between the magnesium ion and the center of mass of the aromatic carbon atoms 5P-1,6-DHN increases significantly, (see Figure 10) indicating that the product is departing the active site. Once 5P-1,6-DHN reaches the lip of the active site it loosely associates with protein surface for the remainder of the simulation. This observation might be related to the hydrophobic nature of the product that inhibits its ready passage into aqueous solvent. By monitoring the variation of two different distances during the 100ns product release process of the major product (5-prenylation), we are able to generate a heat map (see Figure 10), which displays an analysis of the distribution of these distances during the simulation and estimates the free energy cost of product loss to the protein surface. Therefore, quantitatively, we predict that the free energy barrier of this product release process is ~2kcal/mol for the major product. Hence, we further predict that the carbocation formation step is the rate-determining step in NphB catalysis.
Figure 10.
Top: (Dist 1) distance variation between the magnesium site and the aromatic ring of 5P-1,6-DHN. Middle: (Dist 2) distance variation between the C-terminus capping α-helix and the aromatic ring of 5P-1,6-DHN. Bottom: heat map based on Dist 1 and Dist 2.
The motion of 5P-1,6-DHN out of the active site pocket appears to be steered by H-bond interactions between the hydroxyl groups of the product and the sidechains of several protein amino acid residues; notably, Gln161, Ser214 and Tyr288. Among these, both Ser214 and Tyr288 have been previously determined as anchor residues for the less stable P2 resting state. The importance of these interactions was further confirmed in the MUT1 and MUT2 simulations; the former mutation (S214A) inhibits the movement of the final product toward the edge of the enzyme, while the latter case (Y288F) appears to facilitate product release. Based on these observations, we propose a H-bond driven release mechanism: the sidechains of Ser214 and Tyr288 help localize 5P-1,6-DHN in the active site after geometry relaxation, while the former residue, along with sidechain of Gln161, also facilitates the movement of the product towards the edge of the barrel via H-bonds with the hydroxyl groups found in 5P-1,6-DHN. Additionally, cleavage of the H-bond interaction between Tyr288 and the product appears to be required for facile release of 5P-1,6-DHN from the PT-barrel (see Figure 10). Qualitatively we suggest that these three residues control access and egress from the active site of NphB.
Rate-limiting step in NphB catalysis
Although we observed excellent agreement between the computed free energy barriers of carbocation formation in the three experimentally observed reaction channels and the experimental free energy barriers, its has not been demonstrated via experiment that either the proton transfer step or the product release step are not the rate limiting steps. A case in point is the catalytic mechanism of protein farnesyltransferase or protein geranylgeranyltransferase, where the rate-limiting step is product release instead of the chemical step. However, because we have computationally examined all steps in NphB catalysis we propose that the chemical step, or more accurately, the carbocation formation during the prenylation step, is the rate-limiting step in NphB catalysis.
Conclusions
Unlike PPTases that prenylate sulfur in specific cysteine residue(s), the APTases and NphB, in particular, exhibit diverse substrate selectivity and product regioselectivity. Hence, NphB is essentially a reaction vessel for the prenylation of a great variety of small aromatic organic compounds. In this regard, NphB can be readily tuned to synthesize products with the desired functionality based on therapeutic needs as long as the details of its catalytic mechanism are well understood. The present work goes a long way to fulfilling this need.
In this study, a systematic study consisting of QM, QM/MM and MM calculations was carried out to shed light on NphB catalyzed prenylation of 1,6-DHN by GPP. First, QM/MM PMF studies found that NphB follows a SN1-like dissociative mechanism featuring a weakly stable carbocation that is electrophilically captured in the prenylation step by 1,6-DHN. The prenylated product is then re-aromatized via a water-mediated proton transfer pathway and the resultant hybridization change (sp3 -> sp2) triggers the start of the product release. The aromatic character of the active site of NphB has a strong impact in catalysis by stabilizing the developing carbocation. A novel π-chamber composed by Tyr121, Tyr216 and 1,6-DHN was characterized in the aromatic-rich binding pocket of NphB and proved to help stabilize the developing carbocation via cation-π interactions. Different orientations of 1,6-DHN results in different interaction energies between itself and the geranyl carbocation, thereby governing the ease by which a specific atom in 1,6-DHN is prenylated. The computed free energy barriers for the three observed prenylation reactions (P2, P4 and P5) are 13.5kcal/mol, 14.6kcal/mol and 12.3kcal/mol and are all in good agreement with experimental measurements (13.4kcal/mol, 14.1kcal/mol and 12.6kcal/mol, respectively). Moreover, we predict a higher free energy barrier for 7-prenylation (~17kcal/mol), thereby, explaining why this product is not experimentally observed. We also find that the water-mediated proton transfer is quite facile and is not rate limiting for the chemistry part of the catalytic mechanism. Furthermore, Ser214 and Tyr288 have been found to affect the release of the aromatic product in addition to facilitating 2-prenylation. Finally, we estimate a ~2kcal/mol free energy cost for 5P-1,6-DHN release to the protein surface and, thereby, predict that the product release step is not rate limiting. However, we have not explored its full release into aqueous solution, which will increase the free energy barrier for product release.
Supplementary Material
Acknowledgments
We thank the NIH via grant GM044974 for supporting this research. We thank Professor Qiang Cui for helpful comments.
Abbreviation
- PPTase
protein prenyltransferase
- APTase
aromatic prenyltransferase
- G-proteins
guanine nucleotide-binding protein
- PT-barrel
prenyltransferase barrel
- GPP
geranyl diphosphate
- 1,6-DHN
1,6-dihydroxynaphthalene
- H-bond
hydrogen bond
- QM/MM
quantum mechanical molecular mechanical
- SCC-DFTB
self-consistent charge density-functional tight-binding
- MD
molecular dynamics
- US
umbrella sampling
- PME
particle mesh Ewald
- PMF
potential of mean force
- RC
reaction coordinate
- SMD
steered molecular dynamics
- BSSE
basis set superposition error
- PCM
polarized continuum model
- OPP
diphosphate leaving group
- WT
wildtype
- WHAM
weighted histogram analysis method
- FEP
free energy profile
Footnotes
Supporting Information Available: Additional method and computational details, additional figures including transition state active site snapshots, additional tables and Gaussian input structures. This material is available free online via the internet at http://pubs.acs.org.
References
- 1.Zhang FL, Casey PJ. Protein Prenylation: Molecular Mechanisms and Functional Consequences. Annu. Rev. Biochem. 1996;65:241–269. doi: 10.1146/annurev.bi.65.070196.001325. [DOI] [PubMed] [Google Scholar]
- 2.Park HW, Boduluri SR, Moomaw JF, Casey PJ, Beese LS. Crystal structure of protein farnesyltransferase at 2.25 angstrom resolution. Science. 1997;275:1800–1804. doi: 10.1126/science.275.5307.1800. [DOI] [PubMed] [Google Scholar]
- 3.Long SB, Casey PJ, Beese LS. Reaction path of protein farnesyltransferase at atomic resolution. Nature. 2002;419:645–650. doi: 10.1038/nature00986. [DOI] [PubMed] [Google Scholar]
- 4.Taylor JS, Reid TS, Terry KL, Casey PJ, Beese LS. Structure of mammalian protein geranylgeranyltransferase type-I. EMBO J. 2003;22:5963–5974. doi: 10.1093/emboj/cdg571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuzuyama T, Noel JP, Richard SB. Structural basis for the promiscuous biosynthetic prenylation of aromatic natural products. Nature. 2005;435:983–987. doi: 10.1038/nature03668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heide L. Prenyl transfer to aromatic substrates: genetics and enzymology. Curr. Opin. Chem. Biol. 2009;13:171–179. doi: 10.1016/j.cbpa.2009.02.020. [DOI] [PubMed] [Google Scholar]
- 7.Jost M, Zocher G, Tarcz S, Matuschek M, Xie X, Li S-M, Stehle T. Structure-Function Analysis of an Enzymatic Prenyl Transfer Reaction Identifies a Reaction Chamber with Modifiable Specificity. J. Am. Chem. Soc. 2010;132:17849–17858. doi: 10.1021/ja106817c. [DOI] [PubMed] [Google Scholar]
- 8.Kumano T, Richard SB, Noel JP, Nishiyama M, Kuzuyama T. Chemoenzymatic syntheses of prenylated aromatic small molecules using Streptomyces prenyltransferases with relaxed substrate specificities. Bioorg. Med. Chem. 2008;16:8117–8126. doi: 10.1016/j.bmc.2008.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grundmann A, Li SM. Overproduction, purification and characterization of FtmPT1, a brevianamide F prenyltransferase from Aspergillus fumigatus. Microbiology-Sgm. 2005;151:2199–2207. doi: 10.1099/mic.0.27962-0. [DOI] [PubMed] [Google Scholar]
- 10.Christianson DW. Structural biology and chemistry of the terpenoid cyclases. Chem. Rev. (Washington, DC, U. S.) 2006;106:3412–3442. doi: 10.1021/cr050286w. [DOI] [PubMed] [Google Scholar]
- 11.Spurgeon SL, Porter JW. In: Biosynthesis of Isoprenoid Compounds. Porter JW, Spurgeon SL, editors. John Wiley and Sons; New York: 1981. p. 1. [Google Scholar]
- 12.Qureshi N, Spurgeon SL. In: Biosynthesis of Isoprenoid Compounds. Porter JW, Spurgeon SL, editors. John Wiley and Sons; New York: 1981. p. 47. [Google Scholar]
- 13.Rohmer M. The discovery of a mevalonate-independent pathway for isoprenoid biosynthesis in bacteria, algae and higher plants. Nat. Prod. Rep. 1999;16:565–574. doi: 10.1039/a709175c. [DOI] [PubMed] [Google Scholar]
- 14.Piironen V, Lindsay DG, Miettinen TA, Toivo J, Lampi AM. Plant sterols: biosynthesis, biological function and their importance to human nutrition. J. Sci. Food Agric. 2000;80:939–966. [Google Scholar]
- 15.Santos FA, Rao VSN. Inflammatory edema induced by 1,8-cineole in the hindpaw of rats: a model for screening antiallergic and anti-inflammatory compounds. Phytomedicine. 1998;5:115–119. doi: 10.1016/S0944-7113(98)80007-X. [DOI] [PubMed] [Google Scholar]
- 16.Egido J, Blanco-Colio LM, Tunon J, Martin-Ventura JL. Anti-inflammatory and immunomodulatory effects of statins. Kidney Int. 2003;63:12–23. doi: 10.1046/j.1523-1755.2003.00744.x. [DOI] [PubMed] [Google Scholar]
- 17.Schroder H, Grosser N, Hemmerle A, Berndt G, Erdmann K, Hinkelmann U, Schurger S, Wijayanti N, Immenschuh S. The antioxidant defense protein heme oxygenase 1 is a novel target for statins in endothelial cells. Free Radical Biol. Med. 2004;37:2064–2071. doi: 10.1016/j.freeradbiomed.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 18.Sakagami H, Chowdhury SA, Kishino K, Satoh R, Hashimoto K, Kikuchi H, Nishikawa H, Shirataka Y. Tumor-specificity and apoptosis-inducing activity of stilbenes and flavonoids. Anticancer Res. 2005;25:2055–2063. [PubMed] [Google Scholar]
- 19.Jahangir T, Khan TH, Prasad L, Sultana S. Alleviation of free radical mediated oxidative and genotoxic effects of cadmium by farnesol in Swiss albino mice. Redox Rep. 2005;10:303–310. doi: 10.1179/135100005X83671. [DOI] [PubMed] [Google Scholar]
- 20.Soria-Mercado IE, Prieto-Davo A, Jensen PR, Fenical W. Antibiotic terpenoid chloro-dihydroquinones from a new marine actinomycete. J. Nat. Prod. 2005;68:904–910. doi: 10.1021/np058011z. [DOI] [PubMed] [Google Scholar]
- 21.Zhou YD, Kim YP, Mohammed KA, Jones DK, Muhammad I, Dunbar DC, Nagle DG. Terpenoid tetrahydroisoquinoline alkaloids emetine, klugine, and isocephaeline inhibit the activation of hypoxia-inducible factor-1 in breast tumor cells. J. Nat. Prod. 2005;68:947–950. doi: 10.1021/np050029m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boucher K, Chad SS, Sharma P, Hauschka PV, Solomon KR. HMG-CoA reductase inhibitors induce apoptosis in pericytes. Microvasc Res. 2006;71:91–102. doi: 10.1016/j.mvr.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 23.Hwang DR, Chang CW, Lien TW, Chen WC, Tan UK, Hsu JTA, Hsieh HP. Synthesis and anti-viral activity of a series of sesquiterpene lactones and analogues in the subgenomic HCV replicon system. Bioorg. Med. Chem. 2006;14:83–91. doi: 10.1016/j.bmc.2005.07.055. [DOI] [PubMed] [Google Scholar]
- 24.Sultana S, Jahangir T, Khan TH, Prasad L. Farnesol prevents Fe-NTA-mediated renal oxidative stress and early tumour promotion markers in rats. Hum Exp Toxicol. 2006;25:235–242. doi: 10.1191/0960327106ht616oa. [DOI] [PubMed] [Google Scholar]
- 25.Kirby J, Keasling JD. Biosynthesis of Plant Isoprenoids: Perspectives for Microbial Engineering. Annu. Rev. Plant Biol. 2009;60:335–355. doi: 10.1146/annurev.arplant.043008.091955. [DOI] [PubMed] [Google Scholar]
- 26.Sacchettini JC, Poulter CD. Biochemistry - Creating isoprenoid diversity. Science. 1997;277:1788–1789. doi: 10.1126/science.277.5333.1788. [DOI] [PubMed] [Google Scholar]
- 27.Croteau R, Bohlmann J, Meyer-Gauen G. Plant terpenoid synthases: Molecular biology and phylogenetic analysis. Proc. Natl. Acad. Sci. U. S. A. 1998;95:4126–4133. doi: 10.1073/pnas.95.8.4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heide L, Pojer F, Wemakor E, Kammerer B, Chen HW, Walsh CT, Li SM. CloQ, a prenyltransferase involved in clorobiocin biosynthesis. Proc. Natl. Acad. Sci. U. S. A. 2003;100:2316–2321. doi: 10.1073/pnas.0337708100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cui G, Li X, Merz KM., Jr. Understanding the Substrate Selectivity and the Product Regioselectivity of Orf2-Catalyzed Aromatic Prenylations. Biochemistry. 2006;46:1303–1311. doi: 10.1021/bi062076z. [DOI] [PubMed] [Google Scholar]
- 30.Lawson DM, Metzger U, Keller S, Stevenson CEM, Heide L. Structure and Mechanism of the Magnesium-Independent Aromatic Prenyltransferase CloQ from the Clorobiocin Biosynthetic Pathway. J. Mol. Biol. 2010;404:611–626. doi: 10.1016/j.jmb.2010.09.067. [DOI] [PubMed] [Google Scholar]
- 31.Heide L, Metzger U, Schall C, Zocher G, Unsold I, Stec E, Li SM, Stehle T. The structure of dimethylallyl tryptophan synthase reveals a common architecture of aromatic prenyltransferases in fungi and bacteria. Proc. Natl. Acad. Sci. U. S. A. 2009;106:14309–14314. doi: 10.1073/pnas.0904897106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Starks CM, Back KW, Chappell J, Noel JP. Structural basis for cyclic terpene biosynthesis by tobacco 5-epi-aristolochene synthase. Science. 1997;277:1815–1820. doi: 10.1126/science.277.5333.1815. [DOI] [PubMed] [Google Scholar]
- 33.Lesburg CA, Zhai GZ, Cane DE, Christianson DW. Crystal structure of pentalenene synthase: Mechanistic insights on terpenoid cyclization reactions in biology. Science. 1997;277:1820–1824. doi: 10.1126/science.277.5333.1820. [DOI] [PubMed] [Google Scholar]
- 34.Pickett JS, Bowers KE, Fierke CA. Mutagenesis studies of protein farnesyltransferase implicate aspartate beta 352 as a magnesium ligand. J. Biol. Chem. 2003;278:51243–51250. doi: 10.1074/jbc.M309226200. [DOI] [PubMed] [Google Scholar]
- 35.Pais JE, Bowers KE, Fierke CA. Measurement of the alpha-secondary kinetic isotope effect for the reaction catalyzed by mammalian protein farnesyltransferase. J. Am. Chem. Soc. 2006;128:15086–15087. doi: 10.1021/ja065838m. [DOI] [PubMed] [Google Scholar]
- 36.Ho M-H, Vivo MD, Peraro MD, Klein ML. Unraveling the Catalytic Pathway of Metalloenzyme Farnesyltransferase through QM/MM Computation. J. Chem. Theory Comput. 2009;5:1657–1666. doi: 10.1021/ct8004722. [DOI] [PubMed] [Google Scholar]
- 37.Yang Y, Wang B, Ucisik MN, Cui G, Fierke CA, Merz KM., Jr Insights into the Mechanistic Dichotomy of the Protein Farne-syltransferase Peptide Substrates CVIM and CVLS. J. Am. Chem. Soc. 2012;134:820–823. doi: 10.1021/ja209650h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cui G, Wang B, Merz KM., Jr. Computational Studies of the Farnesyltransferase Ternary Complex Part I: Substrate Binding. Biochemistry. 2005;44:16513. doi: 10.1021/bi051020m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cui G, Merz KM., Jr. Computational studies of the farnesyltransferase ternary complex part II: The conformational activation of farnesyldiphosphate. Biochemistry. 2007;46:12375–12381. doi: 10.1021/bi701324t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elstner M, Porezag D, Jungnickel G, Elsner J, Haugk M, Frauenheim T, Suhai S, Seifert G. Self-consistent-charge density-functional tight-binding method for simulations of complex materials properties. Physical Review B. 1998;58:7260–7268. [Google Scholar]
- 41.Cui Q, Elstner M, Kaxiras E, Frauenheim T, Karplus M. A QM/MM implementation of the self-consistent charge density functional tight binding (SCC-DFTB) method. J. Phys. Chem. B. 2001;105:569–585. [Google Scholar]
- 42.Elstner M, Cui Q, Munih P, Kaxiras E, Frauenheim T, Karplus M. Modeling zinc in biomolecules with the self consistent charge-density functional tight binding (SCC-DFTB) method: Applications to structural and energetic analysis. J. Comput. Chem. 2003;24:565–581. doi: 10.1002/jcc.10201. [DOI] [PubMed] [Google Scholar]
- 43.Guo H, Xu D, Cui G. Antibiotic deactivation by a dizinc beta-lactamase: Mechanistic insights from QM/MM and DFT studies. J. Am. Chem. Soc. 2007;129:10814–10822. doi: 10.1021/ja072532m. [DOI] [PubMed] [Google Scholar]
- 44.Seabra GD, Walker RC, Elstner M, Case DA, Roitberg AE. Implementation of the SCC-DFTB method for hybrid QM/MM simulations within the amber molecular dynamics package. J. Phys. Chem. A. 2007;111:5655–5664. doi: 10.1021/jp070071l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu DG, Guo H. Quantum Mechanical/Molecular Mechanical and Density Functional Theory Studies of a Prototypical Zinc Peptidase (Carboxypeptidase A) Suggest a General Acid-General Base Mechanism. J. Am. Chem. Soc. 2009;131:9780–9788. doi: 10.1021/ja9027988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang YH, Cao R, Yin F, Hudock MP, Guo RT, Krysiak K, Mukherjee S, Gao YG, Robinson H, Song Y, No JH, Bergan K, Leon A, Cass L, Goddard A, Chang TK, Lin FY, Van Beek E, Papapoulos S, Wang AHJ, Kubo T, Ochi M, Mukkamala D, Oldfield E. Lipophilic Bisphosphonates as Dual Farnesyl/Geranylgeranyl Diphosphate Synthase Inhibitors: An X-ray and NMR Investigation. J. Am. Chem. Soc. 2009;131:5153–5162. doi: 10.1021/ja808285e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu DG, Wu SS, Guo H. QM/MM Studies of Monozinc beta-Lactamase CphA Suggest That the Crystal Structure of an Enzyme-Intermediate Complex Represents a Minor Pathway. J. Am. Chem. Soc. 2010;132:17986–17988. doi: 10.1021/ja104241g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pierdominici-Sottile G, Horenstein NA, Roitberg AE. Free Energy Study of the Catalytic Mechanism of Trypanosoma cruzi trans-Sialidase. From the Michaelis Complex to the Covalent Intermediate. Biochemistry. 2011;50:10150–10158. doi: 10.1021/bi2009618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang Y, Yu HB, York D, Elstner M, Cui Q. Description of Phosphate Hydrolysis Reactions with the Self-Consistent-Charge Density-Functional-Tight-Binding (SCC-DFTB) Theory. 1. Parameterization. J. Chem. Theory Comput. 2008;4:2067–2084. doi: 10.1021/ct800330d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang Y, Cui Q. Does Water Relay Play an Important Role in Phosphoryl Transfer Reactions? Insights from Theoretical Study of a Model Reaction in Water and tert-Butanol. J. Phys. Chem. B. 2009;113:4930–4939. doi: 10.1021/jp810755p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Warshel A, Levitt M. Theoretical Studies of Enzymic Reactions - Dielectric, Electrostatic and Steric Stabilization of Carbonium-Ion in Reaction of Lysozyme. J. Mol. Biol. 1976;103:227–249. doi: 10.1016/0022-2836(76)90311-9. [DOI] [PubMed] [Google Scholar]
- 52.Case DA, T. A. D., Cheatham TE, III, Simmerling CL, Wang J, Duke RE, Luo R, Walker RC, Zhang W, Merz KM, Roberts BP, Wang B, Hayik S, Roitberg A, Seabra G, Kolossváry I, Wong KF, Paesani F, Vanicek J, Wu X, Brozell SR, Steinbrecher T, Gohlke H, Cai Q, Ye X, Wang J, Hsieh M-J, Cui G, Roe DR, Mathews DH, Seetin MG, Sagui C, Babin V, Luchko T, Gusarov S, Kovalenko A, Kollman PA. University of California; San Francisco: 2010. [Google Scholar]
- 53.Darden T, York D, Pedersen L. Particle Mesh Ewald - an N.Log(N) Method for Ewald Sums in Large Systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 54.Allen MP, Tildesley DJ. Computer Simulation of Liquids. Clarendon Press; Oxford, U.K.: 1987. [Google Scholar]
- 55.Frisch MJT, G. W., Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Mennucci B, Petersson GA, Nakatsuji H, Caricato M, Li X, Hratchian HP, Izmaylov AF, Bloino J, Zheng G, Sonnenberg JL, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Vreven T, Montgomery JA, Jr., Peralta JE, Ogliaro F, Bearpark M, Heyd JJ, Brothers E, Kudin KN, Staroverov VN, Kobayashi R, Normand J, Raghavachari K, Rendell A, Burant JC, Iyengar SS, Tomasi J, Cossi M, Rega N, Millam NJ, Klene M, Knox JE, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Martin RL, Morokuma K, Zakrzewski VG, Voth GA, Salvador P, Dannenberg JJ, Dapprich S, Daniels AD, Farkas Ö, Foresman JB, Ortiz JV, Cioslowski J, Fox DJ. Gaussian, Inc.; Wallingford CT: 2009. [Google Scholar]
- 56.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 57.Kumar S, Bouzida D, Swendsen RH, Kollman PA, Rosenberg JM. The Weighted Histogram Analysis Method for Free-Energy Calculations on Biomolecules .1. The Method. J. Comput. Chem. 1992;13:1011–1021. [Google Scholar]
- 58.Roux B. The calculation of the potential of mean force using computer simulations. Comput. Phys. Commun. 1995;91:275–282. [Google Scholar]
- 59.Case DA, Walker RC, Crowley MF. The implementation of a fast and accurate QM/MM potential method in Amber. J. Comput. Chem. 2008;29:1019–1031. doi: 10.1002/jcc.20857. [DOI] [PubMed] [Google Scholar]
- 60.Jarzynski C. Nonequilibrium equality for free energy differences. Phys. Rev. Lett. 1997;78:2690–2693. [Google Scholar]
- 61.Schulten K, Jensen MO, Park S, Tajkhorshid E. Energetics of glycerol conduction through aquaglyceroporin GlpF. Proc. Natl. Acad. Sci. U. S. A. 2002;99:6731–6736. doi: 10.1073/pnas.102649299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Estrin DA, Crespo A, Marti MA, Roitberg AE. Multiple-steering QM-MM calculation of the free energy profile in chorismate mutase. J. Am. Chem. Soc. 2005;127:6940–6941. doi: 10.1021/ja0452830. [DOI] [PubMed] [Google Scholar]
- 63.Grossfield A. 1.6d ed.
- 64.Truhlar DG, Zhao Y. The M06 suite of density functionals for main group thermochemistry, thermochemical kinetics, noncovalent interactions, excited states, and transition elements: two new functionals and systematic testing of four M06-class functionals and 12 other functionals. Theor. Chem. Acc. 2008;120:215–241. [Google Scholar]
- 65.Truhlar DG, Zhao Y. Density functionals with broad applicability in chemistry. Acc. Chem. Res. 2008;41:157–167. doi: 10.1021/ar700111a. [DOI] [PubMed] [Google Scholar]
- 66.Boys SF, Bernardi F. Calculation of Small Molecular Interactions by Differences of Separate Total Energies - Some Procedures with Reduced Errors. Mol. Phys. 1970;19:553–566. [Google Scholar]
- 67.Simon S, Duran M, Dannenberg JJ. How does basis set superposition error change the potential surfaces for hydrogen bonded dimers? J. Chem. Phys. 1996;105:11024–11031. [Google Scholar]
- 68.Tomasi J, Mennucci B, Cammi R. Quantum mechanical continuum solvation models. Chem. Rev. (Washington, DC, U. S.) 2005;105:2999–3093. doi: 10.1021/cr9904009. [DOI] [PubMed] [Google Scholar]
- 69.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Comparison of multiple amber force fields and development of improved protein backbone parameters. Proteins-Structure Function and Bioinformatics. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang JM, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. J. Comput. Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 71.Bayly CI, Cieplak P, Cornell W, Kollman PA. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: the RESP model. J. Phys. Chem. 1993;97:10269–10280. [Google Scholar]
- 72.Cieplak P, Cornell WD, Bayly C, Kollman PA. Application of the multimolecule and multiconformational RESP methodology to biopolymers: charge derivation for DNA, RNA, and proteins. J. Comput. Chem. 1995;16:1357–1377. [Google Scholar]
- 73.Dougherty DA. Cation-pi interactions in chemistry and biology: A new view of benzene, Phe, Tyr, and Trp. Science. 1996;271:163–168. doi: 10.1126/science.271.5246.163. [DOI] [PubMed] [Google Scholar]
- 74.Dougherty DA. Cation-pi interactions involving aromatic amino acids. J. Nutr. 2007;137:1504S–1508S. doi: 10.1093/jn/137.6.1504S. [DOI] [PubMed] [Google Scholar]
- 75.Jenson C, Jorgensen WL. Computational investigations of carbenium ion reactions relevant to sterol biosynthesis. J. Am. Chem. Soc. 1997;119:10846–10854. [Google Scholar]
- 76.Huang CC, Hightower KE, Fierke CA. Mechanistic studies of rat protein farnesyltransferase indicate an associative transition state. Biochemistry. 2000;39:2593–2602. doi: 10.1021/bi992356x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.