Abstract
The proteins responsible for the initiation of DNA replication are thought to be essentially unrelated in bacteria and archaea/eukaryotes. Here we show that RepA, the initiator from the Pseudomonas plasmid pPS10, and the C-terminal domain of ScOrc4p, a subunit of Saccharomyces cerevisiae (Sc) origin recognition complex (ORC), share sequence similarities. Based on biochemical and spectroscopic evidence, these similarities include common structural elements, such as a winged-helix domain and a leucine-zipper dimerization motif. We have also found that ScOrc4p, as previously described for RepA-type initiators, interacts with chaperones of the Hsp70 family both in vitro and in vivo, most probably to regulate the assembly of active ORC. In evolutionary terms, our results are compatible with the recruitment of the same protein module for initiation of DNA replication by the ancestors of present-day Gram-negative bacteria plasmids, archaea, and eukaryotes.
The universality of the processes involved in the transmission of genetic information has been interpreted to reflect a common evolutionary history. Thus it is accepted that life originated from self-replicating RNA molecules capable of directing protein synthesis, to be replaced later by DNA as the genetic material (1). However, this view has been challenged by modern genomics, inasmuch as protein sequence comparisons conclude that the set of genes for replication, transcription, and translation in bacteria differs from that found in archaea and eukarya (2). Divergence is noteworthy for proteins that initiate chromosomal DNA replication in bacteria (DnaA) (3) and eukarya (the six subunits of the origin recognition complex, ORC) (4, 5); despite their common function in binding to DNA replicators, they lack significant sequence similarity, other than an AAT+ module for ATP binding (6–8). However, structural similarities have been found among some of the accessory factors of prokaryotic and eukaryotic replicative DNA polymerases (9–11).
We had previously found in RepA, the Pseudomonas plasmid pPS10 DNA replication initiator protein (12), the leucine-zipper (LZ) (13, 14) and helix-turn-helix (15) sequence motifs that function in protein dimerization and protein–DNA interaction, respectively. These motifs are part of two repeated winged-helix (WH) domains (16). In common with other homologous plasmid initiators (17, 18), RepA molecules exist as monomers and dimers in equilibrium (14). The monomers activate initiation of replication by binding to directly repeated DNA sequences (14, 16), whereas the dimers repress repA transcription by binding to an inversely repeated DNA operator (19, 16). Dissociation of RepA dimers can either occur spontaneously (14, 20) or be mediated by DnaK/Hsp70 chaperones (21). Monomerization is coupled to a conformational change (22, 23) in the N-terminal domain in RepA. As a result both the N- and C-terminal domains of the monomer show DNA binding, whereas in the dimer only the C-terminal domain is involved in DNA binding (16). This model was recently confirmed in the crystal structure of monomeric RepE54, a homologue of RepA (24).
This article describes our findings on the similarities shared by RepA and the C-terminal domain of ScOrc4p, a subunit of Saccharomyces cerevisiae (Sc) ORC (25), in amino acid sequences, secondary structures, three-dimensional folds, and association states. We also describe how Hsp70 chaperones bind and dissociate oligomers of the N-terminal domain of ScOrc4p.
Materials and Methods
Cloning and Expression of Proteins.
The NcoI-BamHI fragment from pRG-recA-NHis-repA (16) was recloned in pET3d (Novagen) and then used to clone ScORC4s (25), obtained by PCR on genomic DNA (26) from the S. cerevisiae strain W303. Escherichia coli BL21(DE3)/pLysS cells exponentially growing at 28°C were induced by adding isopropyl-β-d-thiogalactoside (0.1 mM) for 4 h. Cloning in the yeast expression vector pYeF2 (27) was performed by PCR on the pET3d recombinants, with oligonucleotides coding for (i) the ClaI site, six histidines (His6) and forward repA/ScORC4 5′sequences and (ii) NotI, complementary simian virus 40 nuclear localization signal and repA/ScORC4 3′ sequences. The vector-encoded hemagglutinin (HA) epitope remains as a C-terminal fusion (27). All recombinants were sequenced. pYeF2 derivatives were transformed by the Li-acetate procedure (26) into the haploid S. cerevisiae strain W303-1Bα and selected in complete medium dropout (-uracil) agar plates (26). Overexpression was achieved by growing yeast in 0.2 liters of the same medium at 27°C to OD600 ≈ 0.5. Then cells were washed and left to grow to OD600 ≈ 2.0 in complete medium, but with galactose replacing glucose.
Protein Purification.
His6–RepA purification was performed as described (16). His6–ScOrc4 proteins were purified at 4°C by a similar protocol: Ni2+ affinity (linear gradient 0.02–0.2 M imidazole) plus ionic exchange chromatography. Full-length ScOrc4p and C-terminal deletion (ΔC367) fragment flow through SP-Sepharose and then bind to Q-Sepharose, both equilibrated in 0.02 M Hepes (pH 7.0), 5 mM βMeEtOH, 0.1 mM EDTA, 10% glycerol. They were eluted with a 0–0.6 M KCl gradient in the same buffer. N-terminal deletion (ΔN366) fragment binds to SP-Sepharose in 0.05 M K-acetate (pH 6.0), 5 mM βMeEtOH, 0.1 mM EDTA, 10% glycerol and was eluted with a gradient to 0.5 M KCl. For structural analysis, His6 tags were removed with thrombin after Ni2+ affinity (16). The ΔN402 fragment was refolded from inclusion bodies as described for the equivalent RepA construct ΔN37 (16) and used with the His6 tag, because of its higher solubility. Proteins were concentrated and stored as described (16).
Gel Filtration Analysis.
Proteins were diluted to 8 μM in 200 μl of gel filtration buffer (0.15 M KCl/0.02 M Hepes, pH 8.0/1 mM βMeEtOH/0.1 mM EDTA/0.01% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate/5% glycerol), injected into a Superose-12 (HR-10/30) FPLC column, and then run at 0.4 ml/min. For DnaK-ScOrc4p complexes, 85 μg of the Q-Sepharose peak fraction (≈0.29 M KCl) containing both proteins was supplied with 0.01 M MgCl2 and 0.1 mM ATP and then incubated for 2 h at 4°C or 37°C. Gel filtration was then carried out in 0.25 M KCl, 0.02 M Hepes (pH 7.5), 1 mM βMeEtOH, 0.01 M MgCl2, 5% glycerol. Fractions (0.5 ml) were dialyzed against 0.025 M NH4-acetate and dried out. After electrophoresis (see below), protein bands were quantified in a Molecular Dynamics 300A densitometer.
CD Spectroscopy.
Spectra and thermal denaturation profiles were acquired and analyzed as described for RepA (16).
Isolation of Whole-Cell Extracts (WCE) and Chromatin from Yeast.
Yeast, W303 or W303–1Bα/pYeF2s, was grown as described above. For WCE preparation (26), cell pellets were resuspended (1:1, wt:vol) in lysis buffer (1.0 M KCl/0.01 M imidazole/0.02 M Hepes, pH 8.0/5% glycerol/0.1% Nonidet P-40/1 mM pNH2-benzamidine) plus protease inhibitors (Roche Molecular Biochemicals) at 4°C. Then 1 volume of glass beads (Ø ≈ 0.45 mm) and 20 μg of lyticase (Sigma) were added. Lysis was achieved by vortexing. After the beads and cell debris were sedimented at 5,000 × g, the supernatant was centrifuged for 15 min at 15,000 × g. Protein concentration in the supernatant (WCE) was determined (Bio-Rad protein assay), and aliquots (20–30 mg/ml) were stored at −80°C. For chromatin isolation, yeast from 25-ml cultures was treated with lyticase to get spheroplasts, followed by centrifugation in Ficoll to extract nuclei (26), and finally lysed in 0.01 M NaCl, 0.01 M Hepes (pH 7.5), 1 mM EDTA, 5 mM βMeEtOH, 0.1% Triton X-100 containing protease inhibitors. Nuclear lysates were centrifuged (15,000 × g, 15 min), and pellets (chromatin fraction) and supernatants (low-salt nuclear extracts) were stored at −80°C.
Binding of WCE to Immobilized RepA/ScOrc4p.
Two hundred micrograms of purified His6–proteins were bound to 50 μl of Ni2+-activated Chelating Sepharose beads (Amersham Pharmacia) in WCE lysis buffer (see above) and gently mixed for 1 h at 4°C. The beads were then spun down and washed with 500 μl of the same buffer. One hundred fifty microliters of WCE (W303, ≈1.5 mg total protein) was incubated with the immobilized His6–proteins for 3 h. The beads were then washed twice (1 h) with 500 μl of 0.1 M KCl, 0.01 M imidazole, 0.02 M Hepes (pH 8.0), 5% glycerol, 0.1% Nonidet P-40, 1 mM pNH2-benzamidine and protease inhibitors and finally resuspended in SDS/PAGE load buffer.
Immunoprecipitation.
One hundred fifty microliters of WCE (W303) was mixed with 100 μg of purified proteins for 1 h at 4°C and then incubated for 2 h in the presence of anti-Hsp70 mAb (Affinity Bioreagents, Golden, CO; clone 5A5, dilution 1:100). Twenty-five microliters of protein-A agarose (Roche Molecular Biochemicals) was added and mixed for 2 h. The agarose beads were sedimented and washed twice (see above). For WCE from W303-1Bα/pYeF2s, mAb was added directly to the extracts, and then the procedure continued as indicated above.
Protein Identification.
Proteins were analyzed by SDS/PAGE (12.5% polyacrylamide) and Coomassie blue staining. When required, duplicated gels were Western-blotted to a nitrocellulose membrane (Bio-Rad semidry transfer cell) at 10 V for 2 h, in 0.025 M Tris, 0.192 M Gly, 0.1% SDS, 20% MetOH. Membranes were then blocked with 5% skimmed powder milk in 0.137 M NaCl, 0.02 M Tris (pH 7.5), 0.05% Tween-20 at 4°C overnight. Immunodetection was performed in this buffer with mAbs: anti-HA (Roche Molecular Biochemicals; 12CA5, 1:1,000), anti-Hsp70 (1:1,000), or horseradish peroxidase-conjugated anti-His (Sigma; HIS-1, 1:5,000). For the first two, a horseradish peroxidase-conjugated secondary IgG (Amersham Pharmacia; 1:10,000) was required before enhanced chemiluminescence detection (Amersham Pharmacia). Membranes were stripped for reprobing according to the enhanced chemiluminescence protocol. The DnaK N terminus was sequenced (Applied Biosystems Procise-494 sequencer) from ≈100 pmol of protein bound to a poly(vinylidene difluoride) membrane, identified at the E. coli proteome (http://sun1.bham.ac.uk/bcm4ght6/res.html) and confirmed by Western blotting with a rabbit anti-DnaK antiserum (1:1,000).
Results
Sequence and Structural Similarities between RepA and ScOrc4p C-Terminal Domain.
We surveyed for common protein motifs in the DNA replication initiators of prokarya and eukarya, by aligning the Pseudomonas plasmid pPS10 RepA protein sequence (12) against the ORC subunits from the budding yeast S. cerevisiae (4). We found that a C-terminal fragment of ScOrc4p (25) (ΔN366, residues 366–529) shares sequence similarity with RepA (19% identical residues plus 25% conservative changes) (Fig. 1a). Although this level of sequence identity is not high enough to become obvious in standard database searches, it could conceal a true structural similarity. For example, the DNApol processivity factors β-clamp and proliferating cell nuclear antigen share only a 6% identity, yet have similar three-dimensional structures (10). It is significant that the isoelectric points calculated (http://www.expasy.ch/tools/pi-tool.html) for RepA and ScOrc4p-ΔN366 are nearly identical (9.42 and 9.45, respectively). Similarities, albeit weaker, can also be found in alignments that include the C termini of Orc4p homologues from other eukaryotes (not shown).
Figure 1.
(a) Sequence alignment between pPS10 RepA, ScOrc4p, and PaCdc6p shows similarities between these DNA replication initiators. Identical residues (*) and conservative changes (+) in RepA vs. ScOrc4p alignment are shown. Residues in PaCdc6p found to be identical (24%) in RepA and/or ScOrc4p (black squares). Secondary structural elements are labeled according to the crystal structures of a Rep-type monomer (24) and PaCdc6p (28): the two WH domains in RepA are colored red (WH1) and blue (WH2), whereas that in PaCdc6p is in green. The conserved hydrophobic heptads in the two N-terminal α-helices and Trp-94 (RepA)/Trp-451 (ScOrc4p) are boxed in orange. The alignment between RepA and ScOrc4p was generated with clustalw (29), with minor manual adjustments. After the yeast sequence was shuffled 1,000 times (http://www.ch.embnet.org/software/PRSS_form.html), the probability of a better alignment (score ≥ 294) between RepA and a sequence of the randomized population was 0.0108. Once the sequence of PaCdc6p was available (28), it was included using the alignment as a profile (29). European Molecular Biology Laboratory database accession numbers: RepA, X58896; ScOrc4p, SC34862. (b) Least-squares superposition of the peptide backbones of the WH domains from the RepA homologue RepE54 (24) (Protein Data Bank entry 1REP) and PaCdc6p (28) (1FNN). Forty-one Cα atoms from both three-helix bundle cores (α2-α4 and α16-α18, respectively) were fit with a rms deviation of 2.14 Å. Coordinate transformation was performed with o (30) and displayed with molscript (31).
While this article was under review, the crystal structure of Cdc6p from the archaea Pyrobaculum aerophilum (Pa) was solved and found to include a C-terminal WH domain (28). We have noticed that its sequence also aligns with the first domain of RepA (WH1) (Fig. 1a). The underlying structural similarity becomes evident when the three-dimensional folds of a RepA homologue (RepE54) and PaCdc6p are superimposed (Fig. 1b). Because Cdc6p is an ortholog of Orc4p (32, 33) the sequence alignment in Fig. 1a reflects a true structural similarity between all three initiators.
The structural similarity between RepA and ScOrc4p points to the presence of a potential LZ motif, previously characterized in RepA and in other plasmid initiators (12–14, 16), between residues 373 and 400 in ScOrc4p. Some functionally related residues are also conserved. Thus Trp-94 in RepA aligns with Trp-451 in ScOrc4p (Fig. 1a). In RepE54 monomers, this tryptophan establishes hydrophobic interactions with two leucines in the LZ that keep this motif buried in the WH1 domain (24). Trp-94 is also involved in a conformational switch that would transform α1 and α2 helices into a LZ upon dimerization (16, 24). Similar conformational changes could also occur with the ScOrc4p C-terminal domain. Several residues in the RepA C-terminal domain (WH2) are also conserved in ScOrc4p. However, this alignment shows some gaps (Fig. 1a): the helix-turn-helix motif in RepA (α-helices 3′ and 4′) (15, 16, 24) is not complete in ScOrc4p.
In summary, our sequence analysis suggests that DNA replication initiators from Gram-negative bacteria plasmids, archaea, and yeast share a common WH domain.
Experimental Evidence for the Structural Similarity Between RepA and ScOrc4p C-Terminal Domain.
We have expressed in E. coli and purified RepA (16) full-length ScOrc4p and three fragments N- or C-terminal to its proposed LZ (Fig. 2a). We have then compared their association state (Fig. 2b), overall secondary structure (Fig. 2c), and structural stability (Fig. 2d).
Figure 2.
Structural similarities between RepA and ScOrc4p. (a) Scheme of cloned ScOrc4p (full length and fragments) and SDS/PAGE of the purified proteins (4 μg per lane). (b) FPLC gel filtration profiles for proteins shown in a. Molecular weights and association states assigned to the peaks are indicated: m, monomer; d, dimer; t, tetramer; o, oligomer. Standards (kDa): alcohol dehydrogenase (ADH), 150; BSA, 66; ovalbumin, 45; carbonic anhydrase, 29; cytochrome c, 12. (c) CD spectra of RepA (16) and ScOrc4p-ΔN366. (d) CD thermal denaturation profiles for ScOrc4 proteins: WT, 10 μM; ΔC367, 14 μM; ΔN366, 30 μM; ΔN402, 22 μM. Calculated Tm are indicated. To be compared with the others, the ΔN402 curve is down-shifted by 4.2 × 103 θMR units.
Gel filtration analysis (Fig. 2b) shows that full-length ScOrc4p and its RepA-like fragment (ΔN366) are mainly dimers (confirmed by sedimentation equilibrium analysis; unpublished data), as previously found for RepA (14, 16). Deletion of the putative LZ in ΔN366 generates a fragment (ΔN402) that behaves like a monomer. This result supports the conclusion that the LZ is responsible for ScOrc4p dimerization. ScOrc2p, Orc4p, and Orc5p bind two sequence patches in autonomously replicating sequence DNA (34); thus dimers of these subunits could be relevant in some functional state of ORC. The N-terminal portion of ScOrc4p (ΔC367) mainly forms soluble oligomers.
The similar shapes of the CD spectra of RepA (16) and ScOrc4p-ΔN366 (Fig. 2c) indicate that their average secondary structures must also be similar, including an α-helical content of about 35%. This conclusion is compatible with the values estimated for the WH domains from the structures of RepE54 and PaCdc6p (Fig. 1).
In CD thermal profiles (Fig. 2d), the temperatures for 50% denaturation (Tm) of the ScOrc4p fragments indicate that they are stable, except for ΔN402 (probably because of the deletion of the LZ). Full-length ScOrc4p shows two thermal transitions that can be assigned to its two domains. ΔN366 exhibits the same stability either isolated or as a part of whole ScOrc4p (first thermal transition). Thus it is a truly independent structural unit. ΔC367 remains folded at 90°C, probably because of its oligomeric state (Fig. 2b) but could cause the second transition when part of full-length ScOrc4p.
Thus the biochemical and biophysical studies reinforce our proposal that RepA and ScOrc4p share a domain with structural homology.
E. coli DnaK Chaperone Binds and Disassembles Oligomers of the N-Terminal Domain of ScOrc4p.
DnaK dissociates RepA dimers into functional monomers (20–23) and disassembles oligomers of DnaA into an active form (35). Hsp70, the eukaryotic homologue of DnaK, binds to the human papilloma virus initiator E1, stimulating origin recognition and replication (36).
We found that bacterially expressed ScOrc4p copurifies in Ni2+ affinity chromatography with a protein of 70 kDa, which also coelutes with ScOrc4p-ΔC367 and, to a lesser degree, with RepA, but not with ScOrc4p-ΔN366. N-terminal sequencing of the 70-kDa band (NH2-GKIIGIDL) identified it as DnaK, which was further confirmed by Western blotting (Fig. 3a). Copurifying ScOrc4p (either full length or ΔC367) and DnaK proteins yield three separate Q-Sepharose fractions: (i) ScOrc4p, (ii) ScOrc4p plus DnaK, and (iii) DnaK (not shown). Fraction ii implies that the proteins form a complex (see below, Fig. 3b). ScOrc4p in fraction i is both soluble and stable (Fig. 2d), and no significant overexpression of DnaK was observed (induction was carried out at low temperature). Thus it is unlikely that DnaK-ScOrc4p complexes arise from the interaction of the chaperone with a misfolded protein, but they could be a means of controlling a biological activity, e.g.,: disassembling oligomers (37).
Figure 3.
E. coli DnaK binds to ScOrc4p and disassembles oligomers. (a) SDS/PAGE of soluble lysates and Ni2+ affinity chromatography peak fractions for proteins in Fig. 2a and RepA. The arrow points to the 70-kDa copurifying band. (Top) Western blot with anti-DnaK polyclonal antiserum. (b) (Top) FPLC gel filtration analysis of purified DnaK-ScOrc4p (ΔC367) complexes (lane L) after incubation with ATP, at 4°C or 37°C. Elution positions of standards (Fig. 2b) and unbound ATP/ADP are marked on the absorption profiles. (Bottom) SDS/PAGE and densitometric ratios for significant fractions.
To test this hypothesis, gel filtration was carried out on the Q-Sepharose fractions, including the complex between ΔC367 (42 kDa) and DnaK (Fig. 3b): because it survives two chromatographic steps plus an intermediate dialysis, it could correspond to the high-affinity/slow-exchange chaperone-ADP-substrate intermediate in the DnaK cycle (37). Thus, ATP and Mg2+ were supplied to samples and incubated, at either 4°C or 37°C, before chromatography. At low temperature, a large ΔC367-DnaK complex elutes in the void volume (Fig. 3b). According to densitometric measurements after gel electrophoresis, its molar ratio is 4:1, implying a mass of n × 240 kDa (n = 1, 2, 3… ) The same was observed in the absence of ATP (not shown). However, after incubation with ATP at physiological temperature, the complex elutes in a volume that fits to a 2:1 ΔC367/DnaK molar ratio (≈150 kDa) (Fig. 3b). Therefore, DnaK disassembles oligomers of ScOrc4p (established through its N-terminal domain) into dimers. This dimer form is the association state found for both full-length ScOrc4p and its C-terminal domain (Fig. 2b). Whether ScOrc4p would be assembled as a monomer in the presence of the other ORC subunits remains to be explored. We then attempted to determine whether the yeast homologue of DnaK (Hsp70) also binds to ScOrc4p and RepA, because the existence of common partners for both initiators would imply functional similarities.
RepA and ScOrc4p Interact with Yeast Hsp70 Chaperone in Vitro.
We carried out two in vitro procedures to look for protein–protein interactions in yeast WCE (Fig. 4). In the first approach, purified His6-tagged ScOrc4p proteins and RepA or BSA (as a control) were incubated with Ni2+-activated Sepharose before WCE was added. Western blotting was then performed on the immobilized proteins with anti-Hsp70 mAb (Fig. 4a). In the second approach, mixtures of WCE and recombinant proteins were immunoprecipitated with the anti-Hsp70 mAb, and then the presence or absence of ScOrc4p or RepA bound to the chaperone was revealed with anti-His tag mAb (Fig. 4b). In both procedures ScOrc4p, either full length or ΔC367, and RepA bind to Hsp70, whereas ScOrc4p-ΔN366 does not, as observed in copurifications (Fig. 3). The low level of ScOrc4p-ΔN402 coimmunoprecipitated (Fig. 4b) could reflect binding of the chaperone to a loosely folded protein, as suggested by its low Tm and slope in Fig. 2d, which resemble those reported for an equivalent RepA fragment (16).
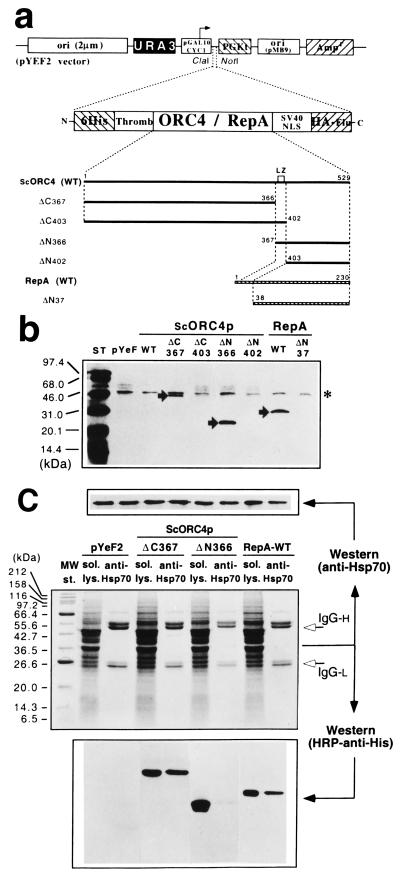
Figure 4.
Yeast Hsp70 binds to ScOrc4p and RepA in vitro. (a) SDS/PAGE of proteins from yeast WCE bound to Ni2+-Sepharose saturated with His6-tagged ScOrc4p or RepA (major bands). (Top) Western blot with anti-Hsp70 mAb. (b) Immunoprecipitation with anti-Hsp70 of mixtures of WCE and proteins shown in a. (Center) SDS/PAGE. Unfilled arrows point to IgG heavy (H) and light (L) chains; full-length ScOrc4p, ΔN366, and RepA are hidden under them (see next). (Bottom) Western blot with horseradish peroxidase–anti-His mAb. (Top) Membrane was reprobed with anti-Hsp70 mAb.
Evidence for the Interaction of RepA and ScOrc4p with Yeast Hsp70 in Vivo.
ScORC4, repA, or combinations of their protein domains, including His6 and HA tags, were cloned in S. cerevisiae and expressed under a galactose-inducible promoter (Fig. 5a). Western blotting with anti-HA mAb on cytosolic and nuclear fractions, either low-salt soluble (not shown) or chromatin-bound (Fig. 5b), indicates that ScOrc4p-ΔC367, ΔN366, and RepA are associated with chromatin in yeast nuclei. Full-length ScOrc4p is expressed at very low levels, possibly because of functional interference with endogenous ORC (data not shown). The ScOrc4p fragments detected (ΔC367 and ΔN366) are stable protein domains (Fig. 2). The other fragments are probably degraded in vivo, including species with the LZ motif deleted (ΔN402 and RepA-ΔN37) (16) or a fragment with the LZ misplaced at the C terminus of the N-terminal domain (ΔC403). This possibility supports the conclusion that the LZ is a constituent element of the C-terminal domain of ScOrc4p.
Figure 5.
ScOrc4p and RepA interact with yeast Hsp70 in vivo. (a) Constructs for the expression of ScOrc4p and RepA in S. cerevisiae. (b) Western blot with anti-HA mAb of chromatin fractions from induced yeasts carrying plasmids shown in a. The asterisk marks a nonspecific band that provides a control for loading. Arrows point to ScOrc4p/RepA products. (c) SDS/PAGE of proteins immunoprecipitated by anti-Hsp70 in WCE from yeast found in b to have ScOrc4 proteins (or RepA) associated with chromatin. (Bottom) Western blot with horseradish peroxidase–anti-His mAb. (Top) The membrane reprobed with anti-Hsp70 mAb.
Immunoprecipitation with anti-Hsp70 mAb of WCE from yeast expressing either RepA or the ScOrc4p fragments yielded ScOrc4p-ΔC367 and RepA, but not ScOrc4p-ΔN366 (Fig. 5c), in agreement with the results shown in Fig. 4. Thus the interaction between Hsp70 and ScOrc4p is likely to be significant.
Discussion
Structural and Functional Similarities Between RepA and ScOrc4p Initiators.
Our results show that ScOrc4p contains at least two domains (residues 1–365 and 366–529). The C-terminal domain, a WH as in its ortholog Cdc6p (28), shares structural similarity with the N-terminal WH domain of RepA, a DNA replication initiator common to plasmids in Gram-negative bacteria (16, 24). The C-terminal domain of ScOrc4p includes a LZ motif that, as in RepA, has a role in dimerization in vitro (Figs. 1 and 2). The N-terminal domain of ScOrc4p shows a functional feature of RepA-type prokaryotic initiators—binding to Hsp70 chaperones (Figs. 3–5). We have found that they modulate the association state of ScOrc4p by dissociating oligomers into dimers (Fig. 3). We speculate that it could be a step in the assembly of the multisubunit ORC. The conformation of some ORC subunits seems to be different when the complex is assembled free or associated with autonomously replicating sequence DNA, a process in which ATP is the allosteric effector (8, 34). We propose that, by analogy with DnaK effects on RepA (16, 20–23), Hsp70 could also switch ScOrc4p between different functional states in the replication complex. Further experiments are required to test this hypothesis.
At the bacterial chromosomal replicator oriC, the initiator DnaA binds ATP and establishes homooligomeric assemblies (3). Plasmid initiators also form homooligomers at the replicators, but because they do not bind ATP, they still require DnaA (17, 18). The eukaryotic ORC is a heterooligomer (4, 8, 34) in which several subunits [Orc1p and Orc5p in S. cerevisiae (5) and the N terminus of Orc4p in higher eukaryotes (32, 33)] bind ATP. It has been found that Orc1p and Orc5p share sequence similarity with Orc4p and eukaryotic and archaeal Cdc6p (28, 33). Thus, if the RepA-like domain in Orc4p were also found in its homologues (8, 34), the prokaryotic and eukaryotic initiation complexes would be “variations on the same theme.”
Implications for the Phylogeny of DNA Replication Initiators.
The findings presented in this paper establish a molecular link between the DNA replication initiators in bacterial plasmids with those in eukaryotes/archaea, which were previously thought to be unrelated (6–8), and argue in favor of a universal, DNA-based, mechanism for the transmission of genetic information going back to the last common ancestor of all living organisms. We cannot exclude the possibility that the WH domains found in Rep and Orc4p/Cdc6p initiators are a case of convergent evolution, although alternative scenarios are also feasible.
Horizontal gene transfer between members of distinct domains of life has been recently proposed to be a major source of the evolution of microorganisms (38, 39) but was thought to be restricted to operational (metabolic) genes (40). Our results suggest that it might also involve informational genes, in particular those with a central role in DNA replication. The gene coding for an ancestral chromosomal initiator could have fused, in the lineage leading to archaea and eukarya, with a gene of plasmid origin, to constitute independent domains in a chimeric initiator. This initiator would then duplicate and diverge into the multisubunit ORC. The success of the resulting chimera would have been initially facilitated by the common Hsp70-dependent mechanism to modulate the assembly of both protein partners, which in modern Orc4p remains just for its N-terminal domain.
On the other hand, if the last common ancestor genome “more resembled mobile genetic elements than typical modern chromosomes” (41), plasmid and archaeal/eukaryal replicons might be ancestral. The distinct DnaA chromosomal initiator could have been acquired, soon after divergence of bacteria from the last common ancestor, because of nonorthologous gene displacement (42) of the original gene by another that included the AAA+ module required for initiation (7, 8).
Acknowledgments
We thank the members of our laboratory for encouragement. Special thanks are due to the staff of the oligonucleotide synthesis, DNA and protein sequencing, and analytical ultracentrifugation facilities at the Centro de Investigaciones Biológicas. We are grateful to Drs. C. Cullin for the pYeF2 vector, M. Kohiyama for the anti-DnaK antiserum, and J. M. Andreu for access to his FPLC equipment. We are indebted to Drs. D. Rhodes, L. Giraldo, M. Lemonnier, and S. Padmanabhan for their critical reading of the manuscript. This work was financed by the Spanish Comisión Interministerial de Ciencia y Tecnología (Grants PB96-0917 and PM99-0096).
Abbreviations
- Sc
Saccharomyces cerevisiae
- ORC
origin recognition complex
- WH
winged-helix domain
- LZ
leucine-zipper motif
- HA
hemagglutinin
- Pa
Pyrobaculum aerophilum
- ΔN
N-terminal deletion
- ΔC
C-terminal deletion
- WCE
whole-cell extract
References
- 1.Orgel L E. Trends Biochem Sci. 1998;23:491–495. doi: 10.1016/s0968-0004(98)01300-0. [DOI] [PubMed] [Google Scholar]
- 2.Brown J R, Doolittle W F. Microbiol Mol Biol Rev. 1997;61:456–502. doi: 10.1128/mmbr.61.4.456-502.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Messer W, Weigel C. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt F C, Curtiss R III, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 1579–1601. [Google Scholar]
- 4.Bell S P, Stillman B. Nature (London) 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- 5.Klemm R D, Austin R J, Bell S P. Cell. 1997;88:493–502. doi: 10.1016/s0092-8674(00)81889-9. [DOI] [PubMed] [Google Scholar]
- 6.Edgell D R, Doolittle W F. Cell. 1997;89:995–998. doi: 10.1016/s0092-8674(00)80285-8. [DOI] [PubMed] [Google Scholar]
- 7.Leipe D D, Aravind L, Koonin E V. Nucleic Acids Res. 1999;27:3389–3401. doi: 10.1093/nar/27.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee D G, Bell S P. Curr Opin Cell Biol. 2000;12:280–285. doi: 10.1016/s0955-0674(00)00089-2. [DOI] [PubMed] [Google Scholar]
- 9.Baker T A, Bell S P. Cell. 1998;92:295–305. doi: 10.1016/s0092-8674(00)80923-x. [DOI] [PubMed] [Google Scholar]
- 10.Krishna T S R, Kong X -P, Gary S, Burgess P M, Kuriyan J. Cell. 1994;79:1233–1243. doi: 10.1016/0092-8674(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 11.Guenther B, Onrust R, Sali A, O'Donnell M, Kuriyan J. Cell. 1997;91:335–345. doi: 10.1016/s0092-8674(00)80417-1. [DOI] [PubMed] [Google Scholar]
- 12.Nieto C, Giraldo R, Fernández-Tresguerres E, Díaz R. J Mol Biol. 1992;223:415–426. doi: 10.1016/0022-2836(92)90661-3. [DOI] [PubMed] [Google Scholar]
- 13.Giraldo R, Nieto C, Fernández-Tresguerres M E, Díaz R. Nature (London) 1989;342:866. doi: 10.1038/342866a0. [DOI] [PubMed] [Google Scholar]
- 14.García de Viedma D, Giraldo R, Rivas G, Fernández-Tresguerres E, Díaz-Orejas R. EMBO J. 1996;15:925–934. [PMC free article] [PubMed] [Google Scholar]
- 15.García de Viedma D, Serrano-López A, Díaz-Orejas R. Nucleic Acids Res. 1995;23:5048–5054. doi: 10.1093/nar/23.24.5048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giraldo R, Andreu J M, Díaz-Orejas R. EMBO J. 1998;17:4511–4526. doi: 10.1093/emboj/17.15.4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.del Solar G, Giraldo R, Ruíz-Echevarría M J, Espinosa M, Díaz R. Microbiol Mol Biol Rev. 1998;62:434–464. doi: 10.1128/mmbr.62.2.434-464.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Helinski D R, Toukdarian A E, Novick R P. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt F C, Curtiss R III, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 2295–2324. [Google Scholar]
- 19.García de Viedma D, Giraldo R, Ruíz-Echevarría M J, Lurz R, Díaz-Orejas R. J Mol Biol. 1995;247:211–223. doi: 10.1006/jmbi.1994.0134. [DOI] [PubMed] [Google Scholar]
- 20.DasGupta S, Mukhopadhyay G, Papp P P, Lewis M S, Chattoraj D K. J Mol Biol. 1993;232:23–34. doi: 10.1006/jmbi.1993.1367. [DOI] [PubMed] [Google Scholar]
- 21.Wickner S, Hoskins J, McKenney K. Proc Natl Acad Sci USA. 1991;88:7903–7907. doi: 10.1073/pnas.88.18.7903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pak M, Wickner S. Proc Natl Acad Sci USA. 1997;94:4901–4906. doi: 10.1073/pnas.94.10.4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dibbens J A, Muraiso K, Chattoraj D K. Mol Microbiol. 1997;26:185–195. doi: 10.1046/j.1365-2958.1997.5691920.x. [DOI] [PubMed] [Google Scholar]
- 24.Komori H, Matsunaga F, Higuchi Y, Ishiai M, Wada C, Miki K. EMBO J. 1999;18:4597–4607. doi: 10.1093/emboj/18.17.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bell S P, Mitchell J, Leber J, Kobayashi R, Stillman B. Cell. 1995;83:563–568. doi: 10.1016/0092-8674(95)90096-9. [DOI] [PubMed] [Google Scholar]
- 26.Lundblad V, Treco D A, Winston F, Reynolds A, Becker D M, Hoffman C S, Collart M A, Oliviero S, Dunn B, Wobbe C R, et al. In: Current Protocols in Molecular Biology. Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J R, Struhl K, editors. New York: Wiley; 1994. , Chap. 13. [Google Scholar]
- 27.Cullin C, Minvielle-Sebastia L. Yeast. 1994;10:105–112. doi: 10.1002/yea.320100110. [DOI] [PubMed] [Google Scholar]
- 28.Liu J, Smith C L, DeRyckere D, DeAngelis K, Martin G S, Berger J M. Mol Cell. 2000;6:637–648. doi: 10.1016/s1097-2765(00)00062-9. [DOI] [PubMed] [Google Scholar]
- 29.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones T A, Zou J-Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 31.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 32.Quintana D G, Hou Z -H, Thome K C, Hendricks M, Saha P, Dutta A. J Biol Chem. 1997;272:28247–28251. doi: 10.1074/jbc.272.45.28247. [DOI] [PubMed] [Google Scholar]
- 33.Tugal T, Zou-Yang X H, Gavin K, Pappin D, Canas B, Kobayashi R, Hunt T, Stillman B. J Biol Chem. 1998;273:32421–32429. doi: 10.1074/jbc.273.49.32421. [DOI] [PubMed] [Google Scholar]
- 34.Lee D G, Bell S P. Mol Cell Biol. 1997;17:7159–7168. doi: 10.1128/mcb.17.12.7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hwang D S, Crooke E, Kornberg A. J Biol Chem. 1990;265:19244–19248. [PubMed] [Google Scholar]
- 36.Liu J-S, Kuo S-R, Makhov A M, Cyr D M, Griffith J D, Broker T R, Chow L T. J Biol Chem. 1998;273:30704–30712. doi: 10.1074/jbc.273.46.30704. [DOI] [PubMed] [Google Scholar]
- 37.Bukau B, Horwich A L. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- 38.Aravind L, Tatusov R L, Wolf Y I, Walker D R, Koonin E V. Trends Genet. 1998;14:442–444. doi: 10.1016/s0168-9525(98)01553-4. [DOI] [PubMed] [Google Scholar]
- 39.Lake J A, Jain R, Rivera M C. Science. 1999;283:2027–2028. doi: 10.1126/science.283.5410.2027. [DOI] [PubMed] [Google Scholar]
- 40.Jain R, Rivera M C, Lake J A. Proc Natl Acad Sci USA. 1999;96:3801–3806. doi: 10.1073/pnas.96.7.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Woese C R. Proc Natl Acad Sci USA. 1998;95:6854–6859. doi: 10.1073/pnas.95.12.6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Forterre P. Mol Microbiol. 1999;33:457–465. doi: 10.1046/j.1365-2958.1999.01497.x. [DOI] [PubMed] [Google Scholar]