Abstract
One limitation in electron cryo-microscopy (cryo-EM) is the inability to recover high-resolution signal from the image-recording media at the full-resolution limit of the transmission electron microscope. Direct electron detection using CMOS-based sensors for digitally recording images has the potential to alleviate this shortcoming. Here, we report a practical performance evaluation of a Direct Detection Device (DDD) for biological cryo-EM at two different microscope voltages: 200 and 300 kV. Our DDD images of amorphous and graphitized carbon show strong per-pixel contrast with image resolution near the theoretical sampling limit of the data. Single-particle reconstructions of two frozen-hydrated bacteriophages, P22 and ε15, establish that the DDD is capable of recording usable signal for 3-D reconstructions at about 4/5 of the Nyquist frequency, which is a vast improvement over the performance of conventional imaging media. We anticipate the unparalleled performance of this digital recording device will dramatically benefit cryo-EM for routine tomographic and single-particle structural determination of biological specimens.
Keywords: Cryo-EM, Electron cryo-microscopy, Direct detection device, Active pixel sensor, CMOS detector, Nyquist frequency
1. Introduction
Electron cryo-microscopy (cryo-EM) is a powerful technique for generating structural models of biological macromolecules and assemblies at near-atomic resolution (see recent reviews: Grigorieff and Harrison, 2011; Hryc et al., 2011). Imaging in electron microscopy is commonly facilitated by either photographic film or charge-coupled device (CCD) cameras. Digital imaging using CCD cameras offers several advantages for data collection, including excellent linearity and dynamic range (Brink and Chiu, 1994) and the ability for automated data collection (Nakamura et al., 2010; Zhang et al., 2009; Zhang et al., 2003; Carragher et al., 2000). Additionally, digital imaging is much more efficient than photographic film since it is not prone to mechanical problems like film jams, and it does not require stringent and time-consuming digital scanning. Immediate access to acquired images also enables on-the-fly image analysis and evaluation to ensure specimen quality and instrument stability, defocus and astigmatism settings, and coma-free alignment (Ishizuka, 1994) necessary for high-resolution imaging.
Despite the many advantages of CCD detectors, photographic film remains the most popular choice of electron detector for cryo-EM (Bammes et al., 2011). Although CCDs provide increased low-frequency contrast (Booth et al., 2006; Sander et al., 2005), their detective quantum efficiency (DQE) at medium and high spatial frequencies is significantly lower than for photographic film, resulting in lower resolution images (McMullan et al., 2009). In the best practical cases, a typical CCD camera with 15 µm pixel size can record usable signal up to ~2/3 Nyquist frequency (Zhang et al., 2010; Chen et al., 2008). Unfortunately, new CCD cameras with smaller pixel sizes have not resulted in improved resolution, suggesting that current CCD technology for electron detection may have reached its limit (Bammes et al., 2011).
Direct electron detection using a CMOS-based active pixel sensor has been proposed as a promising alternative to CCD cameras for digital imaging in electron microscopy (Deptuch et al., 2007; Milazzo et al., 2005; Xuong et al., 2004; Faruqi et al., 2003). Such detectors have the advantage of providing immediate access to digital images, while also having a significantly improved DQE, similar to photographic film (Milazzo et al., 2010; McMullan et al., 2009). After more than a decade of development by multiple groups, direct electron detection is now being realized as a viable alternative to film and CCD detectors for transmission electron microscopy (Guerrini et al., 2011; Mooney et al., 2011; Milazzo et al., 2010; Jin et al., 2008), and recently, the first single-particle three-dimensional (3D) reconstruction of a biological macromolecule from images collected on a direct electron detector was published (Milazzo et al., 2011). However, the limits of its performance for practical data collection in cryo-EM have not yet been fully evaluated.
Our laboratory has previously evaluated the practical utility of digital imaging devices for low dose cryo-EM, including a 16 megapixel CCD camera (Chen et al., 2008; Booth et al., 2006) and a 111 megapixel CCD camera (Bammes et al., 2011). Here, we present a similar practical performance evaluation of a CMOS-based direct electron detector. Specifically, we have assessed the performance of a prototype DE-12 camera from Direct Electron, LP (San Diego, CA), containing 3072 × 4096 pixels (12.6 megapixels) with 6 µm pixel size (Jin and Bilhorn, 2010). Our quantitative and practical evaluation is based on spectral signal-to-noise ratios (SNR) from images of thin carbon film collected under typical low-dose cryo-EM conditions, as well as reconstructions of two biological test specimens (P22 procapsid and ε15 bacteriophage) at two different microscope voltages (200 and 300 kV, respectively). Our work demonstrates that the DE-12 can record unambiguously detectable signal up to its sampling limit (Nyquist frequency) and, for the first time since its development, the feasibility of its use for three-dimensional (3D) reconstructions of frozen, hydrated specimens to resolutions beyond 3/4 Nyquist frequency.
2. Materials and methods
2.1. Direct detection camera
We tested a prototype DE-12 camera system, which was based on a 12.6 megapixel (3072 × 4096) Direct Detection Device (DDD) sensor (Direct Electron LP, San Diego, CA). The detector had 6 µm pixel size resulting in a physical size of 18.4 × 24.6 mm and a superior DQE relative to a conventional CCD camera (Milazzo et al., 2010). The prototype camera was capable of capturing images at 5, 10, 15, 20, or 25 frames per second (fps). During image acquisition, individual frames could be saved and/or integrated by software into a single summed image. Camera control and image acquisition were facilitated by μ Manager (Edelstein et al., 2010).
We installed the DE-12 in the film drawer of two different JEOL (Tokyo, Japan) electron microscopes operated at 200 and 300 kV accelerating voltages, respectively. The camera was cooled to −30° C during operation.
2.2. Electron cryo-microscopy
For data collection, we set microscope conditions according to our routine procedure for high-resolution imaging of biological specimens. Images were collected at liquid nitrogen temperature (~100 K) on two different transmission electron cryo-microscopes equipped with field emission guns (FEG): (1) a JEM-2010F (JEOL Inc, Tokyo, Japan) operating at 200 kV and equipped with a Gatan cryo-holder (model 626), and (2) a JEM-3200FSC (JEOL Inc, Tokyo, Japan) operating at 300 kV with an in-column energy filter and equipped with a JEOL cryo-stage. Prior to imaging, astigmatism correction and evaluation of the stage stability were completed using images of the specimen-supporting amorphous carbon film. In accordance with our experiments on CCD detectors (Bammes et al., 2011), we used a total specimen exposure for each image of ~20 e−/Å2 over 1 second, unless otherwise specified. Specimen exposure rates were estimated using the microscope screen current, which was calibrated previously using a Faraday cage.
2.3. Dark and gain correction
All digital detectors are subject to several sources of noise. First, read noise artifacts arise due to detected signal in the absence of any true signal (Howell, 2000; Gilliland, 1992). To correct for these artifacts (dark correction), a dark reference image (without any electron exposure) is subtracted from the raw image. For all our experiments, a dark reference image was collected automatically by the μ Manager acquisition software at the beginning of each imaging session, and any time the image exposure time was changed. Accordingly, all collected images were then automatically dark corrected by subtracting the dark reference from the raw image.
Additionally, digital detectors are subject to noise arising from variations in the gain (sensitivity) of each pixel in the detector (Howell, 2000; Gilliland, 1992). Correcting for these artifacts is called gain correction, and must be done after dark correction. Gain correction is realized by acquiring a dark-corrected flood-beam image (without specimen) as a gain reference. Each dark-corrected specimen image is then divided by the gain reference to produce the final, corrected specimen image. The μ Manager data acquisition software for the prototype DE-12 camera we tested was not initially capable of gain-correcting the individual specimen images, but was subsequently modified to permit this operation. Therefore, gain correction was not performed for any initial 300 kV image data while all the subsequent 200 kV data were gain corrected with the updated data acquisition software.
2.4. Magnification calibration
Magnification calibration on the JEM-2010F was completed using commercially available graphitized carbon grids (Electron Microscopy Sciences, Hatfield, PA). We collected unbinned images of graphitized carbon particles from 25,000× to 100,000× nominal microscope magnification. Multiple images at 0–1 µm under-focus were collected for each magnification. The Fourier transforms of boxed areas of graphitized carbon with at least 512 × 512 pixels revealed a bright ring at 1/3.35 Å−1 (Houska and Warren, 1954). Based on the diameter in pixels of this bright ring, we calculated the Ångströms-per-pixel sampling and the effective detector magnification for each nominal microscope magnification. Nominal microscope magnifications lower than 25,000× were calibrated with line grating replica (Electron Microscopy Sciences).
Magnification calibration on the JEM-3200FSC was completed using negatively-stained catalase crystal grids (Electron Microscopy Sciences). We collected unbinned images of catalase crystals from 10,000× to 25,000× nominal microscope magnification. Images were processed and analyzed using 2dxh software (Gipson et al., 2007) to determine the lattice spacing, overall crystal tilt, and corresponding detector magnification from the Fourier transformed image regions using a 175 × 68.5 Å unit cell spacing based on the published commercial specification.
As expected, since the detector was situated near the film plane, the post-magnification factor was measured to be ~1.0(0.87 for the 200 kV microscope, and 0.98 for the 300 kV microscope).
2.5. Carbon film imaging and analysis
The carbon support film on the commercial graphitized carbon and catalase crystal grids was used for analysis of spectral SNR using a radiation insensitive specimen. We acquired dark-corrected images within the range of 2 – 6 µm under-focus at various magnifications and detector frame rates. Gain normalization was not performed on the raw data, since this feature had not yet been implemented in the software controlling the camera. We examined the Fourier transform of each image to check for drift and astigmatism before saving it for subsequent analysis. The noise profile was estimated by log-linear interpolation using the contrast transfer function (CTF) zeros in the circularly averaged power spectrum of each image. The spectral SNR for each image was calculated from the measured power spectrum and the estimated noise profile. To estimate the spectral SNR in the absence of defocus-induced CTF oscillations, we used log-linear interpolation based on the smoothed SNR values at each CTF peak and the square of the spatial frequency.
2.6. Evaluating linearity
To evaluate the linearity of the detector, we collected images with and without any specimen at magnifications from 20,000× to 80,000× on a 300 kV microscope. The use of different magnifications was designed to use the same specimen exposure with different detector exposures. The equivalent specimen exposure rate was 10 e−/Å2/s over a total time of 4 seconds. Dark-corrected images were collected at either 15, 20, or 25 fps, corresponding to 0.067, 0.050, and 0.040 seconds per frame, respectively. Rather than saving a single summed image representative of the entire exposure time, we set the camera software to save each individual frame. The frames from the first 0.2 seconds of exposure were discarded to ignore any possible effects of imprecise synchronization between shutter opening and imaging. Using a custom Python script based on the EMAN2 libraries (Tang et al., 2007), we generated a series of images representing the sum of the first n frames (not including the discarded frames from the first 0.2 seconds), resulting in images with incrementally increasing cumulative exposure. The resulting images were gain normalized by dividing with the maximum cumulative exposure at the corresponding magnification and frame rate. Empty flood-beam images were analyzed by calculating the mean pixel intensity over each resulting image. Images of thin carbon film were analyzed based on the Fourier transform of the central 1024 × 1024 region of each resulting image. The overall linearity was evaluated based on the coefficient of determination (R2) from linear regression analysis.
2.7. ε15 bacteriophage imaging and analysis
Frozen, hydrated ε15 bacteriophage cryo-EM grids were prepared as previously described (Bammes et al., 2011). Dark-corrected images of ε15 bacteriophage were collected on the JEM-3200FSC (300 kV) at a detector magnification of 20,000×, resulting in 3.00 Å/pixel sampling. The detector was set to a frame rate of 20 fps. As with carbon film imaging, we did not perform gain correction. The total specimen exposure per summed image was 20 e−/Å2. Particle boxing and automated CTF fitting were completed using EMAN2 (Tang et al., 2007), followed by manual refinement of CTF parameters using the EMAN program ctfit (Ludtke et al., 1999). An initial 3D model was generated from 500 single-particle images of ε15 bacteriophage, which were assigned random orientations. With this initial model, alignment parameters for each particle were determined by the Multi-Path Simulated Annealing algorithm (Liu et al., 2007), and were used to generate an icosahedral 3D reconstruction of ε15 bacteriophage using EMAN’s make3d program (Ludtke et al., 1999). The resulting reconstruction was sharpened using the experimental structure factor computed in EMAN2 and then masked to exclude the dsDNA contained within the capsid shell. Resolution was estimated by Fourier shell correlation (FSC) (Harauz and van Heel, 1986) by comparing the resulting 3D reconstruction with the previously published 4.5 Å model (Jiang et al., 2008). The overall resolution was determined based on a 0.5 FSC criterion (Böttcher et al., 1997; Conway et al., 1997), however, we also show the 0.143 FSC criterion (Rosenthal and Henderson, 2003) to facilitate comparison with other published results. Surface representations of density maps were generated using Chimera (Goddard et al., 2005).
In the case of oversampling, we used the “scale” and “clip” options in the EMAN program proc2d to increase the pixel dimensions of each particle image by 1.5× using real-space interpolation (Ludtke et al., 1999). For example, if the original particle image was 320 × 320 pixels with 3 Å/pixel sampling, the oversampled particle image would be 480 × 480 pixels with 2 Å/pixel sampling. After applying oversampling to the raw images, we repeated all other image processing steps from CTF parameter estimation to generating a 3D reconstruction.
2.8. P22 procapsid imaging and analysis
Biochemical and cryo-specimen preparations for P22 procapsid were carried out as previously described (Chen et al., 2011). Dark and gain-corrected images of P22 procapsid were collected with the latest data acquisition software on the JEM-2010F (200 kV) at a detector magnification of 17,200×, resulting in 3.48 Å/pixel sampling. The detector was set to a frame rate of 25 fps. The total specimen exposure per summed image was 17 e−/Å2. Particles were automatically boxed using ethan (Kivioja et al., 2000) and all particle boxes were oversampled by 1.5× using the method described above. Automated CTF fitting was done using fitctf.py (Yang et al., 2009), followed by manual refinement of the CTF parameters using the EMAN program ctfit (Ludtke et al., 1999). The initial model for the reconstruction was built from scratch using EMAN program starticos. Orientation and center parameters for each particle were determined by the Multi-Path Simulated Annealing algorithm (Liu et al., 2007), and were used to generate an icosahedral 3D reconstruction of P22 procapsid using EMAN’s make3d program (Ludtke et al., 1999). Resolution was estimated using the 0.5 and 0.143 FSC criterions, comparing the resulting 3D reconstruction with the previously published 3.8 Å model (Chen et al., 2011). Surface representations of density maps were generated using Chimera.
3. Results
3.1. Graphitized carbon imaging
The Fourier transform of an image of graphitized carbon at sufficient magnification contains a bright ring at 1/3.35 Å−1, which corresponds to the lattice spacing of graphite (Houska and Warren, 1954). Because of this clear peak at a defined spatial frequency, graphitized carbon is a popular specimen for TEM magnification calibration and resolution tests.
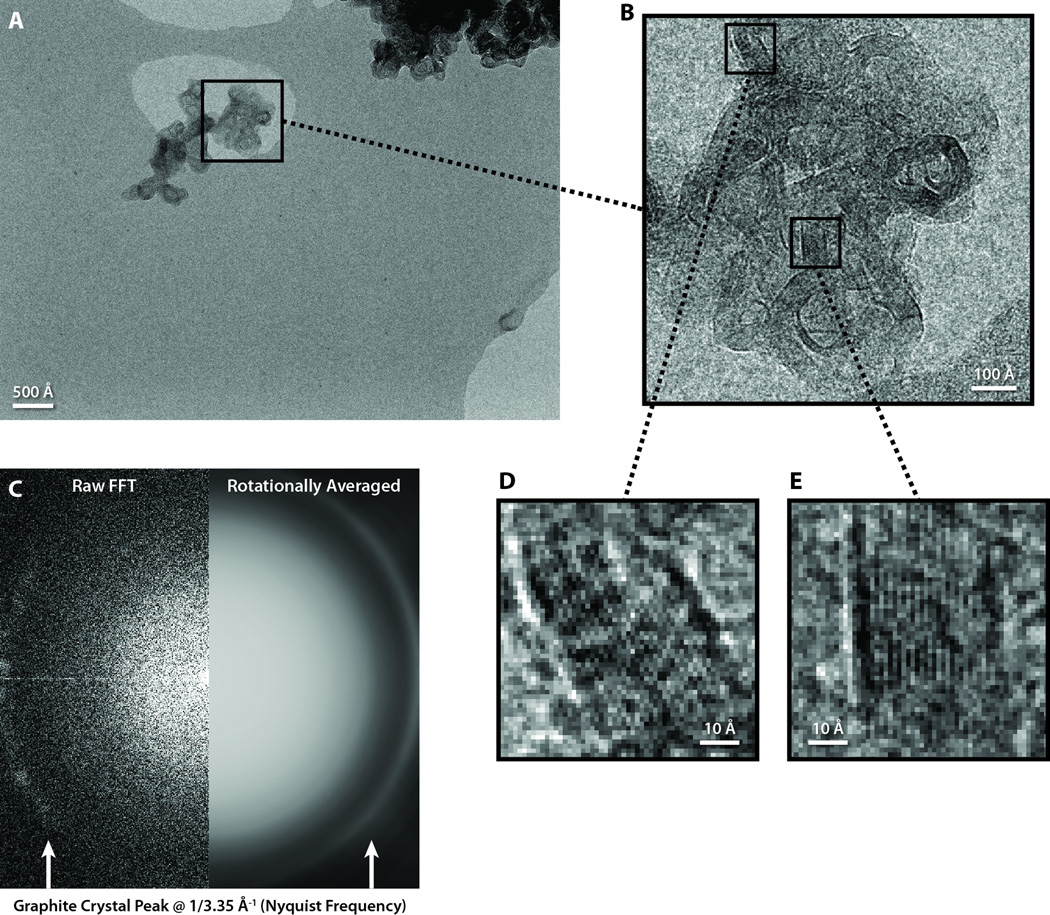
Figure 1 shows an example of an image collected at 35,700× detector magnification on a 200 kV microscope at room temperature. The sampling at this magnification is 1.68 Å/pixel, yielding a Nyquist frequency of 1/3.36 Å−1, which is just shy of the spatial frequency of the graphite’s reciprocal lattice peak.
Figure 1. Graphitized carbon performance test on a JEM-2010F microscope operated at 200 kV.
(A) A full frame of a graphitized carbon specimen collected at 35,700× magnification. (B) A zoomed-in view of a graphite crystal from A. (C,D) Zoomed-in views of two regions of graphite crystal from B, with lattice spacing approximately equal to the size of one pixel. (E) The raw (left) and processed (right) Fourier transform of the crystal image in B, with the characteristic graphite crystal peak visible at Nyquist frequency.
Since the graphitized carbon particle is relatively small compared to the entire frame (Fig. 1A), we extracted a 512 × 512 box containing some of the graphitized carbon particle (Fig. 1B). The Fourier transform of this boxed region revealed a bright peak at the edge of the transform (Fig. 1C), indicating strong SNR even at Nyquist frequency. Indeed, the SNR at Nyquist frequency is high enough that differences in specimen signal between individual pixels are clearly visible in the image. Figures 1D and 1E are highly zoomed-in views of two 58 × 58 pixel areas from Figure 1B, so that individual pixels can be distinguished. These zoomed views clearly show a pixel-to-pixel oscillating pattern of single dark and light intensity pixels, which is especially obvious when the graphite lattice is oriented vertically in the boxed image (Fig. 1E).
3.2. Carbon film imaging
While graphitized carbon can demonstrate the potential resolution of a detector, the spectral SNR from images of amorphous carbon film provides a much more robust metric for assessing the overall performance of electron detectors mimicking the conditions for single-particle cryo-EM imaging (Booth et al., 2006; Zhang et al., 2003). Since the SNR of each image depends on the total exposure, the total exposure must be carefully chosen. For subnanometer target resolutions, radiation damage considerations limit cryo-EM exposures of biological specimens to ~20–30 e−/Å2 (Baker et al., 2010; Bammes et al., 2010). As with our previous evaluations of CCD detectors (Bammes et al., 2011), we have conservatively used 20 e−/Å2 for our practical SNR performance tests.
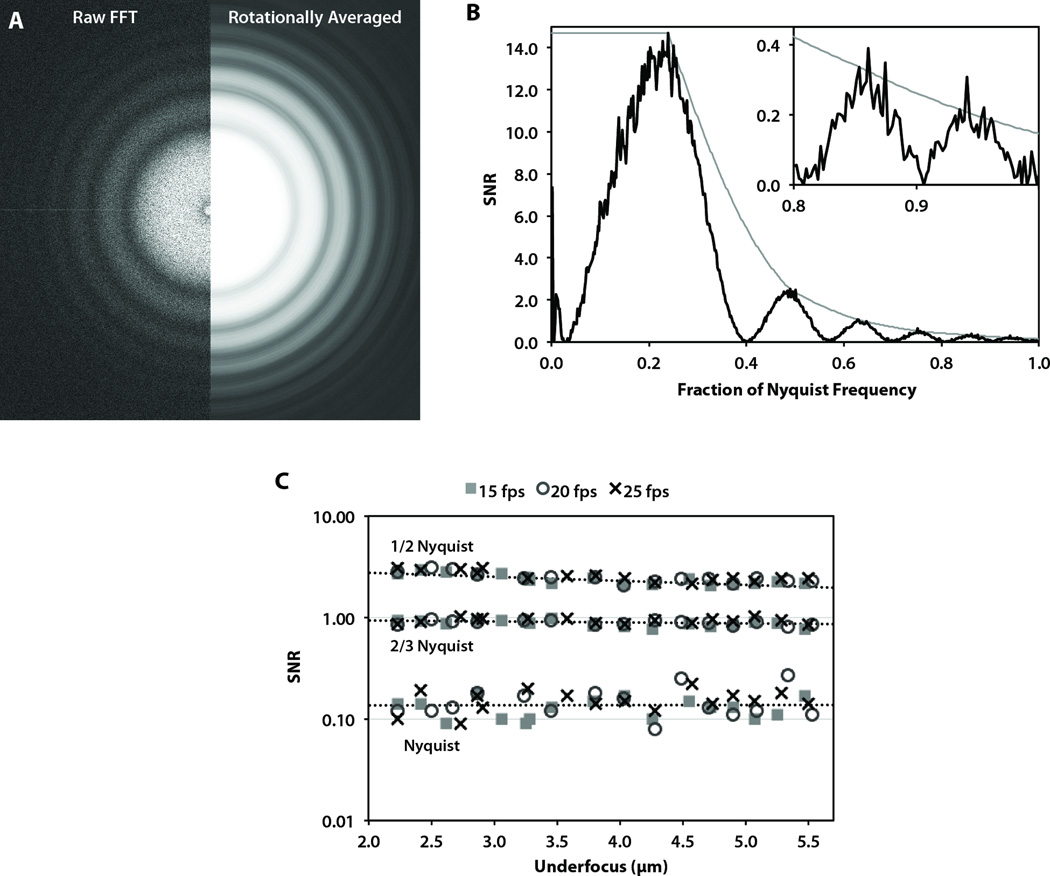
Figure 2A shows the entire Fourier transform from an image of amorphous carbon film at 10,800× detector magnification on a 300 kV microscope. The edges of the Fourier transform frame correspond to Nyquist frequency. In agreement with the graphitized carbon results, CTF rings are clearly distinguishable up to and even past (in the corners) Nyquist frequency. Based on the rotational average of this Fourier transform, the spectral SNR again shows significant information content up to Nyquist frequency (Fig. 2B). In fact, the SNR near Nyquist frequency is 0.20, which is much higher than the threshold SNR value of ~0.05 typically used to estimate the practically useful signal (Booth et al., 2006).
Figure 2. Carbon film performance test on a JEM-3200FSC microscope operated at 300 kV.
(A) The raw (left) and processed (right) Fourier transform of an image of carbon film acquired at 10,800× detector magnification. (B) The corresponding spectral SNR curve (black) and computed SNR envelope (gray). (C) The SNR values at 1/2 Nyquist, 2/3 Nyquist, and Nyquist frequencies for carbon film images collected at 10,800× magnification at three different frame rates.
The light gray curve shows the estimated spectral SNR in the absence of defocus-induced CTF oscillations. We used this interpolated curve to estimate the SNR values at specific spatial frequencies. Figure 2C shows the SNR values at several specific spatial frequencies for images of amorphous carbon film at the same detector magnification (10,800×) across a range of defocus values on a 300 kV microscope. Not surprising, we observed a slight negative correlation between SNR and defocus due to the effect of defocus on spatial coherence (Glaeser et al., 2007). Table 1 summarizes the observed SNR values at each spatial frequency examined. Indeed, the minimum SNR value we observed at Nyquist frequency was higher than 0.08 under a broad range of defocus values.
Table 1.
Summary of observed SNR values of carbon film images at 10,800× detector magnification across a defocus range of 2.2 – 5.5 µm with images recorded at 15, 20, or 25 fps on a JEM-3200FSC microscope operated at 300 kV.
| Spatial Frequency | Mean | Minimum | Maximum |
|---|---|---|---|
| 1/2 Nyquist | 2.48 | 2.08 | 3.13 |
| 2/3 Nyquist | 0.90 | 0.76 | 1.01 |
| Nyquist | 0.14 | 0.08 | 0.27 |
3.3 Detector linearity
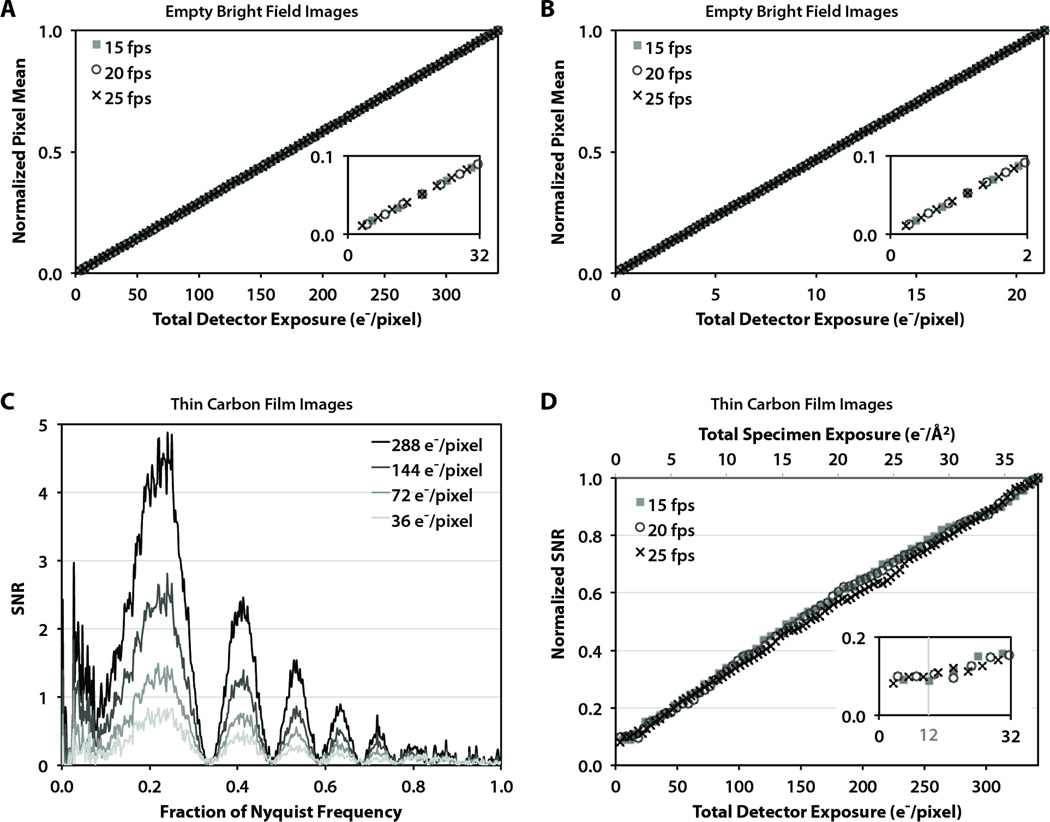
To test the linearity of the detector, we analyzed the mean pixel intensity from empty flood-beam images (without specimen) on a 300 kV microscope over a range of total exposures (Fig. 3A and B). In order to cover a wide range of per-pixel exposures, we used a constant specimen exposure rate of 10 e−/Å2/s at different frame rates and magnifications. Specifically, this yielded incremental exposures per frame in the range of 3.6 – 6.0 e−/pixel at low magnification (~20,000×, Fig. 3A) and 0.225 – 0.375 e−/pixel at high magnification (~80,000×, Fig. 3B). In both cases, there were no detectable differences between the three different frame rates and the detector linearity was nearly perfect (R2>0.9999).
Figure 3. Evaluation of detector linearity at various detector frame rates.
(A) The normalized mean intensity of flood-beam images for a wide range of total image exposures, with a zoomed view (inset) to distinguish individual data points. (B) The normalized mean intensity of flood-beam images for very low total image exposures, with a zoomed view (inset) to distinguish individual data points. (C) Spectral SNR of images of carbon film collected at 25 fps with various total image exposures. (D) The change in the spectral SNR of images of carbon film under a wide range of total image exposures.
We also evaluated the effect of total cumulative exposure on the spectral SNR of amorphous carbon film images. Figure 3C shows four spectral SNR curves up to Nyquist frequency for images with various total exposures collected at ~20,000× detector magnification and 25 fps. At this magnification, the lowest total exposure of 36 e−/pixel corresponds to 4 e−/Å2 at the specimen plane. Even with this low exposure, the CTF oscillations in the spectral SNR are clearly distinguishable, and SNR peaks are visible to near Nyquist frequency. As expected, at 2×, 4×, and 8× the total exposures, the SNR values are increased by about the same factor across all spatial frequencies, indicating a high degree of linearity.
We quantitatively analyzed the SNR response to increasing total exposure by examining the relative scaling the SNR peaks up to 2/3 Nyquist. Again the detector magnification was ~20,000×, resulting in an incremental exposure per frame of 3.6, 4.5, and 6.0 e−/pixel for images collected at 25, 20, and 15 fps, respectively. Figure 3D shows the normalized SNR response to increasing total exposure, shown in terms of both the per-pixel exposure on the detector (bottom axis) and the per-area exposure on the specimen (top axis) for the magnification we used. Although the results are noisier than the case of empty flood-beam images, again we observed similar responses for the three different frame rates and a high degree of linearity (R2> 0.996). However, at very low exposures of ~12 e−/pixel or less (corresponding to <1.33 e−/Å2 on the specimen at 20,000× magnification), the normalized SNR values appear nearly flat due to the noise in the spectral SNR curves overwhelming the small SNR peaks due to insufficient signal (inset).
3.4. ε15 bacteriophage reconstructions
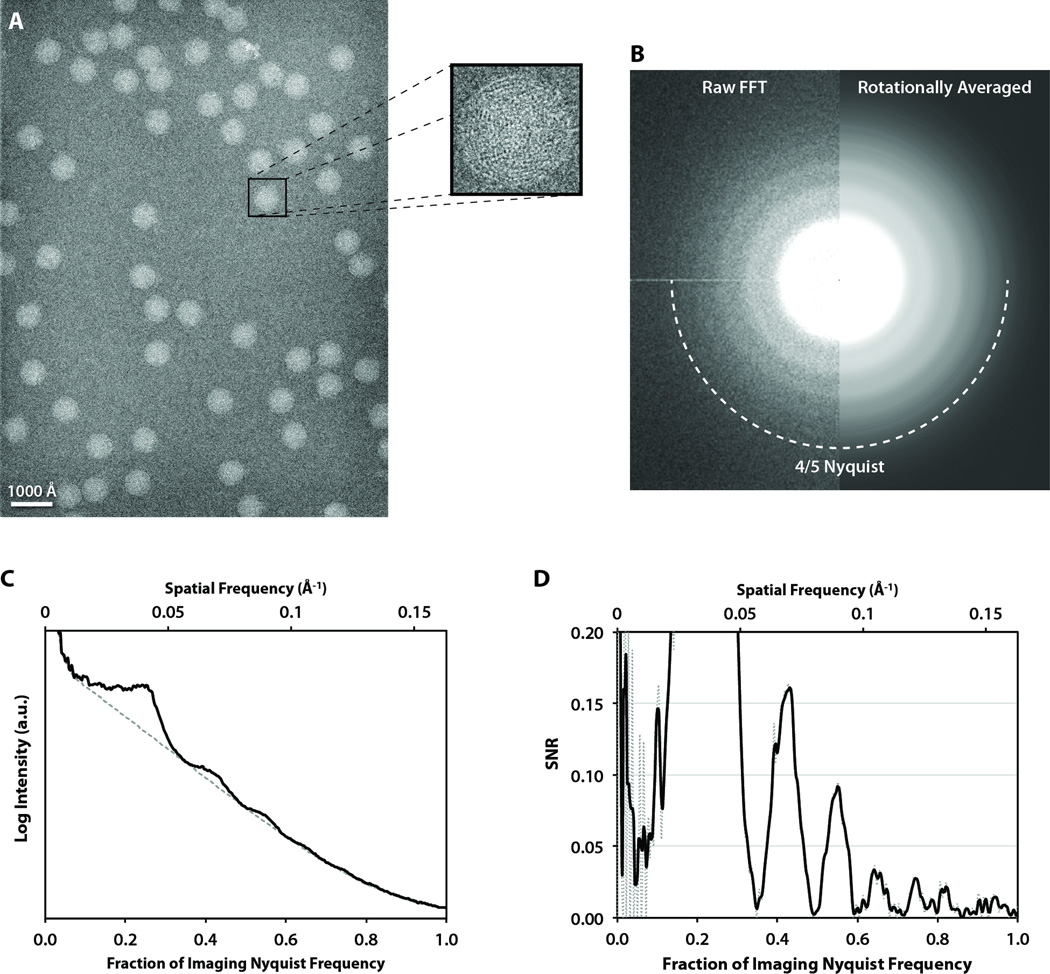
The structure of ε15 bacteriophage has been solved by cryo-EM to 4.5 Å resolution (Jiang et al., 2008), making it an ideal biological specimen for evaluating the practical performance of new technology. We have previously used ε15 bacteriophage to assess the performance of a variety of CCD detectors (Bammes et al., 2011; Chen et al., 2008; Booth et al., 2006). We acquired 75 frames of frozen, hydrated ε15 bacteriophage at 20,000× detector magnification, yielding sampling of 3.00 Å/pixel. Figure 4A shows a representative frame acquired at 1.55 µm under-focus, containing 55 ε15 bacteriophage particle images. This particle distribution was typical for our particular sample preparation, which yielded an average of 48 particle images per frame. Even at this low magnification, the dsDNA fingerprint inside each particle is clearly visible (Fig. 4A, inset). The separation of the dsDNA strands inside the ε15 bacteriophage capsid is ~25 Å (Jiang et al., 2006), corresponding to 0.24 Nyquist frequency in our images.
Figure 4. ε15 bacteriophage imaging.
(A) A full frame collected at 20,000× detector magnification, on a JEM-3200FSC microscope operated at 300 kV, with a zoomed-in view of a single ε15 capsid. (B) The raw (left) and processed (right) Fourier transform sum of all the particles in A with box size of 320×320 pixels. (C) The circularly averaged power spectrum computed from the Fourier transform in B. (D) The corresponding smoothed spectral SNR curve (dotted line is unsmoothed) showing distinguishable CTF oscillations close to Nyquist frequency.
The sum of the Fourier transforms of the 55 particle images in the single DDD frame showed clear CTF rings up to ~4/5 Nyquist (Fig. 4B). These CTF oscillations were also visible in the rotationally averaged power spectrum (Fig. 4C), and the corresponding spectral SNR (Fig. 4D). The SNR curve shows CTF oscillations corresponding to the image defocus to near Nyquist frequency, with three clear SNR peaks (with SNR values from 0.034 to 0.021) visible between 0.10 and 0.15 Å−1, corresponding to 60% and 90% of Nyquist frequency, respectively. Note that a continuous thin carbon film was used on the holey grid before freezing, which may enhance the signal compared to imaging with no continuous carbon film.
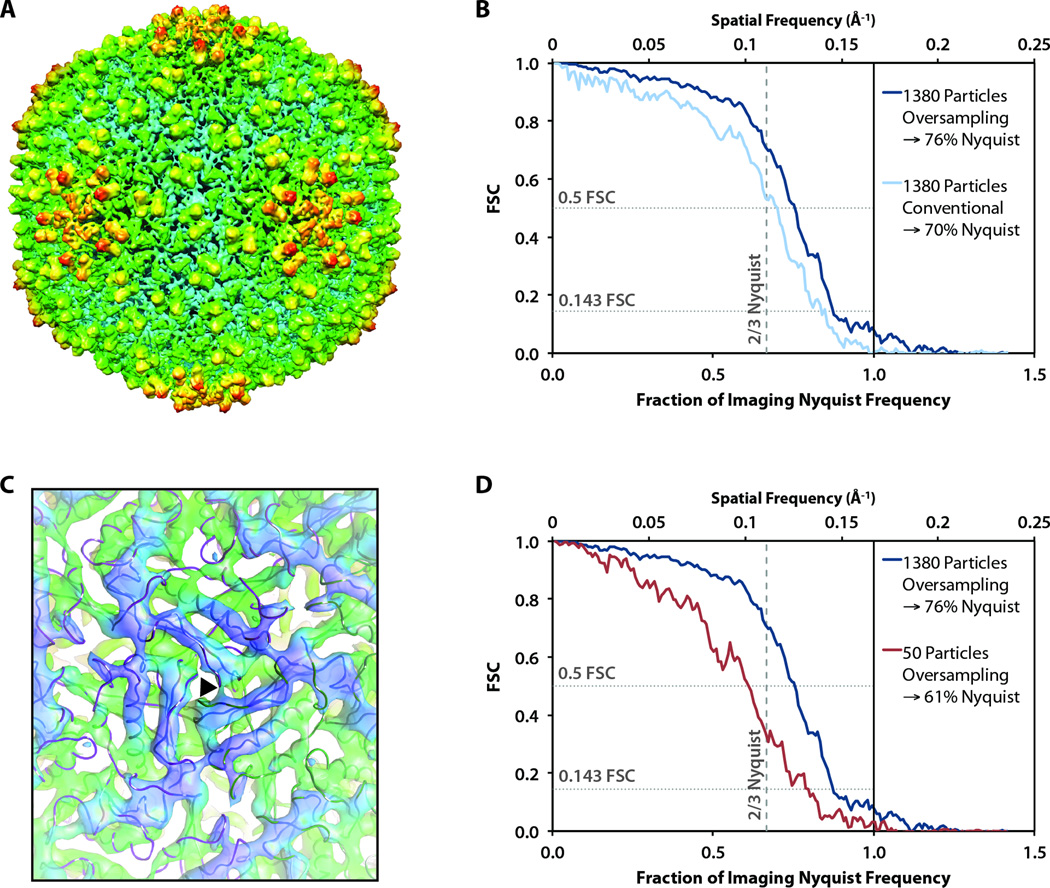
In total, we boxed ~3,500 particle images (210,000 asymmetric units based on icosahedral symmetry) from 75 frames, which should be more than enough asymmetric units to reach subnanometer resolution (Liu et al., 2007). 500 of these particle images were randomly selected and assigned random orientations to produce an initial model. From this data set, we selected the best 1380 particles to generate an icosahedral 3D reconstruction using the MPSA protocol (Liu et al., 2007). According to the 0.5 FSC criterion, the resolution of this reconstruction was 8.6 Å, corresponding to 70% Nyquist frequency (Fig. 5B). Since we observed CTF oscillations in individual frames up to 80% Nyquist frequency, we attempted to recover additional signal by oversampling each of our original 1380 particle images by 1.5× using real-space linear interpolation. Using these oversampled particle images, we performed CTF correction and generated an icosahedral reconstruction (Fig. 5A). The FSC from the oversampling reconstruction (from 1380 particles) crosses 0.5 at 7.9 Å (corresponding to 76% Nyquist frequency) and 0.143 at 6.8 Å (corresponding to 88% Nyquist frequency) (Fig. 5B). At this resolution, we could unambiguously visualize α-helices (Fig. 5C).
Figure 5. ε15 bacteriophage reconstructions.
(A) The entire capsid reconstructed from 1380 particles, oversampled by 1.5×. (B) FSC curve of the reconstruction in A compared to a 4.5 Å reference model of ε15 (Jiang et al., 2008) for reconstructions from the original and oversampled particle images. (C) A view of the three-fold symmetry axis (black triangle) viewed from inside the reconstructed capsid with α-helices clearly visible. The backbone trace from the 4.5 Å reference model of ε15 is overlaid on our density map. (D) The FSC of reconstructions of oversampled images from 1380 and 50 particle images.
We also generated a 3D reconstruction of ε15 bacteriophage using only the best 50 particle images (3,000 asymmetric units based on icosahedral symmetry) chosen from 8 of the 75 frames. To facilitate a direct comparison with the reconstruction using 1,380 particle images (82,800 asymmetric units based on icosahedral symmetry), we applied the same software oversampling in this case. The FSC from the resulting reconstruction crosses 0.5 at 9.8 Å (corresponding to 61% Nyquist frequency) and 0.143 at 7.7 Å (corresponding to 78% Nyquist frequency) (Fig. 5D).
3.5. P22 procapsid reconstructions
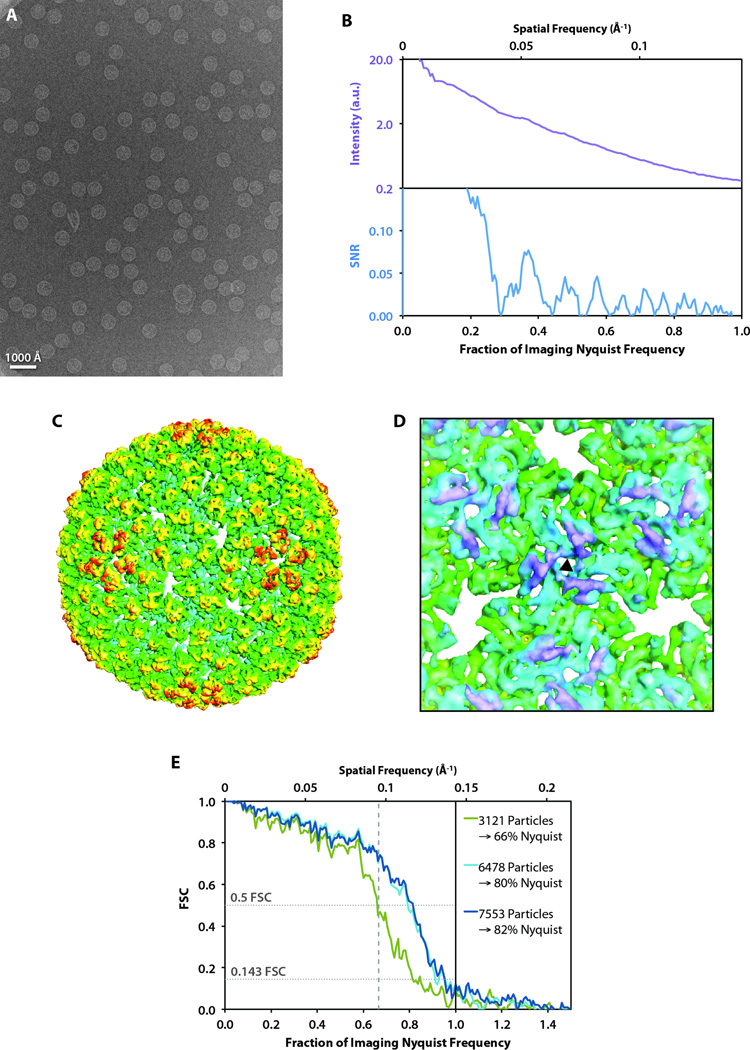
As a second biological test specimen, we used frozen, hydrated bacteriophage P22 procapsid, whose structure has been solved by cryo-EM to 3.8 Å resolution (Chen et al., 2011). We acquired 217 frames of P22 procapsid (Fig. 6A) at ~17,200× detector magnification, yielding sampling of 3.48 Å/pixel. Similar to the case of ε15 bacteriophage, the sum of the Fourier transforms of the particle images from the best individual frame showed clear CTF oscillations up to at least ~4/5 Nyquist frequency (Fig. 6B).
Figure 6. P22 procapsid imaging and reconstructions.
(A) A full frame collected at 17,200× detector magnification acquired on a JEM-2010F microscope operating at 200 kV. (B) The circularly averaged power spectrum (top) and spectral SNR (bottom) computed from the sum of the Fourier transforms of the particle images in A. (C) 3D reconstruction of the P22 procapsid. Images were software oversampled (1.5×) prior to data processing. (D) A view of the three-fold symmetry axis (black triangle) viewed from inside the reconstructed capsid. (E) FSC curves comparing our structures using varying numbers of particle images with the previously published 3.8 Å model (Chen et al., 2011).
In total, we boxed ~21,100 particle images (1,266,000 asymmetric units based on icosahedral symmetry) from 217 frames. Since software oversampling yielded a higher resolution in the case of ε15 bacteriophage, we applied the same strategy to our P22 procapsid particle images. Using~7,500 particle images (450,000 asymmetric units based on icosahedral symmetry), we generated a 3D reconstruction of P22 procapsid (Fig. 6C). We observed rod densities corresponding to α-helices in the resulting 3D density map (Fig. 6D), indicating that the resolution is subnanometer. The FSC revealed the resolution of our resulting density map to be 8.5 Å (corresponding to 82% Nyquist frequency) by the 0.5 FSC criterion, and 7.3 Å (corresponding to 95% Nyquist frequency) by the 0.143 FSC criterion (Fig. 6E).
4. Discussion
4.1 Significance
The achievable resolution of single-particle cryo-EM reconstructions is limited by numerous factors, including the total number of single-particle images, particle conformation uniformity, specimen preservation, detector, radiation damage, imaging exposure, defocus, spatial and temporal coherence, beam alignment, errors in image reconstruction, etc. (Glaeser et al. 2007; Rosenthal and Henderson, 2003; Zhou and Chiu 2003). All of these factors except the electron detector are largely insensitive to the imaging magnification. Imaging at higher magnification has been the choice for achieving high-resolution structures because it results in finerÅngströms-per-pixel sampling to overcome the effective resolution of the detector (Bammes et al., 2011). However, imaging at higher magnification also reduces the total field of view since the detector has a limited size. Choosing the electron detector and the optimal imaging conditions for that detector is critical to assuring cryo-EM images with maximum resolution.
Digital imaging using a CCD camera is an indirect technique for detecting electrons, since it relies on a scintillator to convert incident electrons to a low-light image that can be effectively captured by the CCD detector (Faruqi and Subramaniam, 2000). The conversion process imposes a finite point spread function (PSF) on the image, which degrades the overall image resolution (Downing and Hendrickson, 1999; Daberkow et al., 1991).
In contrast, CMOS-based direct electron detectors do not require electron-to-photon conversion, and thus the image-degrading scintillator is unnecessary. Physics-based performance metrics such as the DQE/MTF have already demonstrated that this direct detection results in significantly improved performance compared to existing CCD technology (Milazzo et al., 2010; McMullan et al., 2009). However, these performance metrics are difficult to compare and interpret since they depend on the calculation method and they do not account for all the sources of noise that are present in low-dose cryo-EM imaging under practical conditions (Mooney, 2007). Since the performance of an electron detector depends on its acquisition of signal and noise, the spectral SNR under typical experimental conditions is the most pragmatic method for performance evaluation. Such practical characterizations have been vital to the development and acceptance of CCD cameras for cryo-EM imaging (Bammes et al., 2011; Chen et al., 2008; Booth et al., 2006; Sander et al., 2005; Booth et al., 2004; Zhang et al., 2003; Downing and Hendrickson, 1999).
Here, we have presented a similar practical performance evaluation of a recently commercialized direct electron detector (Jin and Bilhorn, 2010). Our evaluation is based on quantitative analysis of images, with particular relevance to low-dose cryo-EM imaging. We have used images of graphitized carbon to demonstrate its resolution limits (Fig. 1), and images of amorphous carbon film to evaluate its spectral SNR under a variety of conditions (Figs. 2 and 3). Finally, we completed reconstructions of frozen-hydrated ε15 bacteriophage (Figs. 4 and 5) imaged from a 300 kV microscope and P22 procapsid (Fig. 6) imaged from a 200 kV microscope as practical examples of real cryo-EM experiments using radiation-sensitive biological specimens.
4.2. Practical performance of direct detection relative to CCD
Images of graphitized carbon from the DE-12 direct detection device showed significant contrast between individual pixels (Figs. 1C and D). The Fourier transform of these low-magnification images clearly showed the 1/3.35 Å−1 graphite ring at 100% Nyquist frequency (Fig. 1E), indicating that the achievable resolution of this detector is limited by its pixel size. In contrast, the reciprocal of the lattice line spacing in graphitized carbon was detectable up to only 90% Nyquist frequency on a 4k × 4k CCD (Zhang et al., 2003) with more than 5× the total exposure used in this study. Therefore, it appears that one can use lower exposures and still recover information closer to the sampling limit (Nyquist frequency) with the DE-12 direct detector compared to the current generation of CCD cameras.
Analysis of the spectral SNRs from images of amorphous carbon film further substantiates the improved performance of direct detection compared to CCD technology. Under low-dose cryo-EM imaging conditions, we previously observed that CCD cameras can record detectable SNR for cryo-EM experiments up to ~22.5 µm spacing at the detector plane (Bammes et al., 2011). Specifically, we found that a CCD with 15 µm pixel size can acquire usable SNR to about 15 µm ÷ 22.5 µm = 2/3 Nyquist frequency (Chen et al., 2008), whereas a CCD with pixel size of 9 µm pixel size can acquire usable SNR to about 9 µm ÷ 22.5 µm = 2/5 Nyquist frequency (Bammes et al., 2011). Therefore, decreasing the pixel size on a CCD camera does not appear to improve the effective resolution of images at a given magnification. This empirically derived limit may be attributable to the scintillator and/or fiber optics necessary for electron-to-photon conversion and photon transmission to the CCD detector (Bammes et al., 2011).
In contrast, we observed much better performance from the DE-12 (which does not use a scintillator), with significant SNR values (>0.1) at 100% Nyquist (Fig. 2), meaning that this camera can record detectable SNR up to its pixel size of 6 µm spacing at the detector plane. This ability for much finer sampling represents a substantial improvement in achievable resolution at any given magnification compared to existing CCD cameras. Furthermore, the SNR values across all spatial frequencies are significantly increased. For example, images of thin carbon film under single-particle cryo-EM conditions collected on a CCD with 15 µm pixel size has no detectable SNR at 2/3 Nyquist, while the DE-12 provides SNR values of ~1.0 at 2/3 Nyquist (Fig. 2C).
We have also shown that the DE-12 direct electron detector provides excellent linearity (Fig. 3), similar to CCD cameras (Brink and Chiu, 1994). In the process, we also observed a high dynamic range for imaging, with linear response from our minimum tested exposure of 0.225 e−/pixel to more than 1500× this value at 342 e−/pixel. Indeed, the dynamic range of direct electron detection for a typical cryo-EM exposure should be even higher than for a CCD, since the signal from the direct detector is continually read and integrated in software, rather than being hardware integrated in pixels wells over the entire exposure time (Fan et al., 1998).
4.3. Single-particle reconstructions to resolutions beyond 2/3 Nyquist frequency
Our graphite and carbon film performance tests demonstrate that the DE-12 provides significant SNR under low-dose conditions even at its Nyquist frequency (Figs. 1, 2, and 3). We further demonstrated the practical benefits of direct electron detection by performing single-particle reconstructions of two biological test specimens, ε15 bacteriophage imaged at 300 kV and P22 procapsid imaged at 200 kV. Even though the data acquisition and data processing protocols were different for both sets of data, both cases yielded 3D reconstructions with resolution beyond 3/4 Nyquist frequency (Figs. 5 and 6).
Realization of this high resolution ultimately required software oversampling of particle images prior to CTF estimation, image processing, and reconstruction. Although we observed CTF oscillations in the spectral SNR of our images from the DE-12 up to ~4/5 (80%) Nyquist frequency (Fig. 4), our initial reconstruction of ε15 only reached 70% of Nyquist frequency (Fig. 5B). Others have previously noted that spatial frequencies above 2/3 Nyquist increasingly suffer from interpolation and aliasing artifacts introduced during the image alignment and reconstruction algorithms (Frank,2006; Orlova et al., 1997). As a practical way to quickly reduce these deleterious artifacts past 2/3 Nyquist frequency and potentially push the resolution of our reconstructions slightly higher, we oversampled our data by 1.5× using linear real-space interpolation. Using oversampled particle images, we attained resolutions of ~4/5 (80%) Nyquist frequency (Figs. 5 and 6).
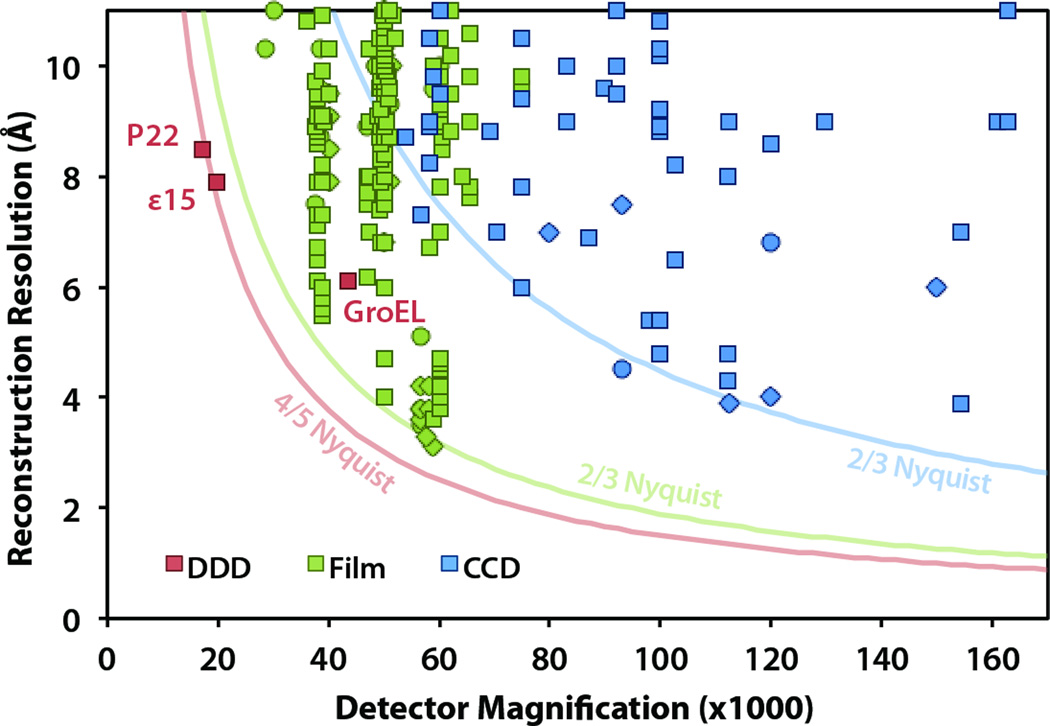
Based on our survey of the EM Databank (emdatabank.org) (Lawson et al., 2011), the structures shown here (Figs. 5 and 6) represent the first single-particle reconstructions from any detector with resolution significantly beyond 2/3 Nyquist frequency (based on the digitized image pixel size)(Fig. 7). In fact, both of our reconstructions from the DE-12 reached subnanometer resolution using images attained at less than 20,000× detector magnification. This is a significant improvement over previous results, which used at least 40,000× to reach subnanometer resolution using photographic film, and at least 80,000× to reach subnanometer resolution using a CCD detector (Fig. 7).
Figure 7.
Single-particle reconstruction results from the EMDB (emdatabank.org; accessed 11/30/2011), classified by the detector type used for data collection. The three published DDD reconstructions are specifically labeled: ε15 (Fig. 5), P22 (Fig. 6), and GroEL (Milazzo et al., 2011). Note that the method for estimating resolution varies, including 0.5 FSC (square), 0.143 FSC (diamond), and other methods (circle). The solid curves show approximate resolution limits for each detector type: CCD (15 µm pixel size limited to 2/3 Nyquist frequency), film (6.35 µm limited to 2/3 Nyquist frequency), and DDD (6 µm pixel size limited to 4/5 Nyquist frequency).
In addition to providing improved resolution, the high spectral SNR provides a significant benefit in reducing the total number of particles needed to reach a target reconstruction resolution. We generated a reconstruction of ε15 bacteriophage to 9.8 Å resolution (corresponding to 0.5 FSC) using only 50 particle images (3,000 asymmetric units) (Fig. 5D). Based on the 3.00 Å/pixel sampling at the magnification we used, this resolution corresponds to 61% Nyquist frequency. Using the same reconstruction algorithm (MPSA) with an icosahedral specimen, Liu and colleagues investigated the minimum number of particle images necessary to reach various resolutions using images collected on photographic film (Liu et al., 2007). Using a sampling of 2.4 Å/pixel, they generated a 9.6 Å resolution map (50% Nyquist frequency) from 62 particle images (3,720 asymmetric units). In terms of absolute spatial resolution, this is nearly identical to the resolution we obtained from 50 particle images (3,000 asymmetric units) using the DE-12 (Fig. 5D). However, in terms of detector performance, our resolution of 61% Nyquist frequency from 50 particle images outperforms the demonstrated results from photographic film.
Overall, the improved SNR allows for reconstructions that reach beyond 2/3 Nyquist frequency, while also reducing the number of particle images necessary to reach a target resolution. It is also likely that the improved SNR may improve image alignment accuracy, causing 3D reconstructions to converge faster and allowing for better separation of heterogeneous specimens.
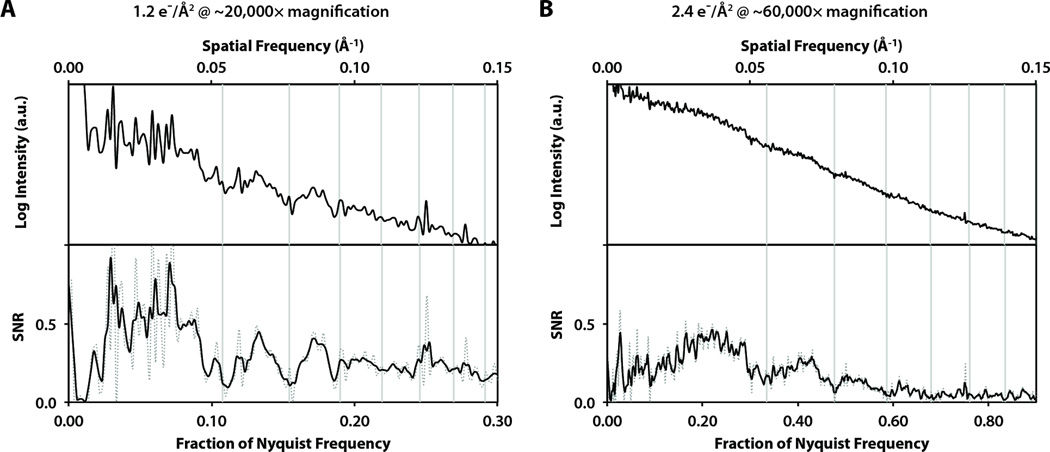
Besides these benefits for single-particle cryo-EM imaging, the high sensitivity and spectral SNR of direct detection also may provide significant advantages for ultra low-dose images, such as the dose-fractionated images collected in a tomographic tilt series. Figure 8 shows two example power spectra and spectral SNR curves from images of amorphous carbon film at low total exposures, similar to the exposure per image in cryo-tomography. In both cases, the CTF oscillations are clearly distinguishable, allowing for accurate determination of defocus. Thus, direct electron detection may benefit tomography both by improved resolution as well as allowing for CTF correction due to a more accurate determination of defocus in each very low-dose image.
Figure 8.
Circularly-averaged power spectra (top) and smoothed spectral SNR curves (bottom; dotted line is unsmoothed) from low-exposure images collected at 200 kV with the following conditions: (A) Detector exposure of 10.8 e−/pixel at ~20,000× magnification, corresponding to 1.2 e−/Å2 on the specimen. (B) Detector exposure of 2.4 e−/pixel at ~60,000× magnification, corresponding to 2.4 e−/Å2 on the specimen.
4.4. Total imaging area
Table 2 shows the per-pixel sampling, expected maximum reconstruction resolution, and total specimen area (field of view) at various detector magnifications. Since the DE-12 was installed in the film chamber of our microscopes, the detector magnification is approximately equal to the nominal microscope magnification. Note however that CCD cameras are often installed under the film chamber, so there is a post-magnification factor (typically ~1.4×) between the nominal microscope magnification and the CCD magnification.
Table 2.
Comparison of sampling (Å/pixel), best expected reconstruction resolution, and specimen field of view (FOV) for the DE-12 direct detector and a typical CCD camera at various detector magnifications.
| Detector | 3k×4k (6 µm) Direct Detector | 4k×4k (15 µm) CCD | ||||
|---|---|---|---|---|---|---|
| Mag. | Å/pixel | 4/5 Nyq. (Å) | FOV (µm2) | Å/pixel | 2/3 Nyq. (Å) | FOV (µm2) |
| 5,000 | 12.00 | 30.00 | 18.119 | 30.00 | 90.00 | 150.995 |
| 10,000 | 6.00 | 15.00 | 4.530 | 15.00 | 45.00 | 37.749 |
| 20,000 | 3.00 | 7.50 | 1.132 | 7.50 | 22.50 | 9.437 |
| 25,000 | 2.40 | 6.00 | 0.725 | 6.00 | 18.00 | 6.040 |
| 30,000 | 2.00 | 5.00 | 0.503 | 5.00 | 15.00 | 4.194 |
| 40,000 | 1.50 | 3.75 | 0.283 | 3.75 | 11.25 | 2.359 |
| 50,000 | 1.20 | 3.00 | 0.181 | 3.00 | 9.00 | 1.510 |
| 60,000 | 1.00 | 2.50 | 0.126 | 2.50 | 7.50 | 1.049 |
| 80,000 | 0.75 | 1.88 | 0.071 | 1.88 | 5.63 | 0.590 |
| 100,000 | 0.60 | 1.50 | 0.045 | 1.50 | 4.50 | 0.377 |
| 150,000 | 0.40 | 1.00 | 0.020 | 1.00 | 3.00 | 0.168 |
For example, if the target resolution for a project is ~7.5 Å (for visualizing α-helices), one would likely choose a detector magnification of 20,000× using the DE-12, resulting in a total specimen field of view of 1.132 µm2. Alternatively, using a 4k × 4k CCD, one would choose a detector magnification of 60,000×, resulting in a slightly smaller field of view of 1.049 µm2. So although the CCD contains more pixels than this prototype DDD, the better overall resolution of the direct detector results in an effective field of view that is ~8% larger than a 4k × 4k CCD. Recent demonstrations of the benefits of large-format (i.e., 10k × 10k) CCD cameras (Bammes et al., 2011; Lee et al., 2010) may also eventually lead to the development of similar large-format direct electron detectors with even greater benefits.
A larger field of view is advantageous for two reasons: 1) it reduces the total number of frames needed for single-particle imaging by increasing the number of particle images per frame, and 2) it improves the accuracy of CTF parameter determination by including more overall signal per frame. These benefits of a larger field of view are further enhanced by the improved SNR from a direct detector, which allows for reconstructions using fewer total particle images, as illustrated by our ε15 bacteriophage reconstruction from only 50 particle images sufficient to detect -helices (Fig. 5). Furthermore, as these 50 particle images (3,000 asymmetric units) were intentionally selected from 8 different DDD frames to compensate for the CTF. Doing so would not be necessary if the DE-12 were coupled with Zernike phase contrast optics (Murata et. al. 2010), where images can be acquired close to focus to eliminate most CTF oscillations. In such a combination, it may be possible in theory to use the randomly-oriented icosahedral virus particles to generate a subnanometer resolution reconstruction from a single DDD frame (so long as there are a sufficient number of asymmetric units).
5. Conclusion
Digital detectors for electron microscopy provide multiple advantages compared to photographic film, however film has continued to maintain better overall SNR performance compared to CCD cameras. The recent development of commercially available CMOS-based direct electron detectors has introduced a new alternative for cryo-EM digital imaging with the potential to provide SNR as good as, or better than, film. We have evaluated the practical performance of a prototype direct electron detector for cryo-EM imaging at both 200 kV and 300 kV. Based on graphitized carbon and thin carbon film images, we have observed that this detector records usable signal to 100% of its Nyquist frequency with SNR values ~5× higher than CCD detectors over nearly all spatial frequencies. The improved resolution and overall SNR performance provide numerous practical benefits, including more accurate CTF parameter estimation, fewer particles needed for reconstructions, and improved alignment accuracy in single-particle imaging and tomography.
Using this detector, we demonstrated for the first time subnanometer resolution reconstructions using images collected at less than 20,000× detector magnification. This also represents the first demonstrated reconstructions with resolution close to 4/5 Nyquist frequency, which extends beyond the current limit of data recorded on either CCD or photographic film. We conclude that CMOS-based direct electron detection is well-suited for low-dose cryo-EM imaging and represents a significant improvement over CCD cameras for digital imaging.
Acknowledgements
This work is supported by the National Institutes of Health through the National Center for Macromolecular Imaging (P41RR002250 and PN2EY016525), the National Library of Medicine (NLM) Training Program in Biomedical Informatics and Computational Biology administered through the Keck Center for Interdisciplinary Bioscience Training of the Gulf Coast Consortia (T15LM007093), and the Robert Welch Foundation (Q1242).
We thank Jonathan King, Cameron Haase-Pettingell, and Peter Weigele at the Massachusetts Institute of Technology (MIT) for the P22 procapsid and ε15 bacteriophage samples. We also thank Direct Electron, LP (San Diego, CA) for providing support and access to their prototype DE-12 camera.
Data Deposition: the 15 and P22 capsid reconstructions have been deposited to the EMDB (EMID-XXXX and EMID-5375 respectively).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baker LA, Smith EA, Bueler SA, Rubinstein JL. The resolution dependence of optimal exposures in liquid nitrogen temperature electron cryomicroscopy of catalase crystals. J. Struct. Biol. 2010;169:431–437. doi: 10.1016/j.jsb.2009.11.014. [DOI] [PubMed] [Google Scholar]
- Bammes BE, Jakana J, Schmid MF, Chiu W. Radiation damage effects at four specimen temperatures from 4 to 100 K. J. Struct. Biol. 2010;169:331–341. doi: 10.1016/j.jsb.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bammes BE, Rochat RH, Jakana J, Chiu W. Practical performance evaluation of a 10k × 10k CCD for electron cryo-microscopy. J. Struct. Biol. 2011;175:384–393. doi: 10.1016/j.jsb.2011.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth CR, Jakana J, Chiu W. Assessing the capabilities of a 4k×4k CCD camera for electron cryo-microscopy. J. Struct. Biol. 2006;156:556–563. doi: 10.1016/j.jsb.2006.08.019. [DOI] [PubMed] [Google Scholar]
- Booth CR, Jiang W, Baker ML, Zhou ZH, Ludtke SJ, et al. A 9 Å single particle reconstruction from CCD captured images on a 200 kV electron cryo-microscope. J. Struct. Biol. 2004;147:116–127. doi: 10.1016/j.jsb.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Böttcher B, Wynne SA, Crowther RA. Determination of the fold of the core protein of hepatitis B virus by electron cryomicroscopy. Nature. 1997;386:88–91. doi: 10.1038/386088a0. [DOI] [PubMed] [Google Scholar]
- Brink J, Chiu W. Applications of a slow-scan CCD camera in protein electron crystallography. J. Struct. Biol. 1994;113:23–34. doi: 10.1006/jsbi.1994.1029. [DOI] [PubMed] [Google Scholar]
- Carragher B, Kisseberth N, Kriegman D, Milligan RA, Potter CS, et al. Leginon: an automated system for acquisition of images from vitreous ice specimens. J. Struct. Biol. 2000;132:33–45. doi: 10.1006/jsbi.2000.4314. [DOI] [PubMed] [Google Scholar]
- Chen DH, Baker ML, Hryc CF, DiMaio F, Jakana J, et al. Structural basis for scaffolding-mediated assembly and maturation of a dsDNA virus. Proc. Natl. Acad. Sci. USA. 2011;108:1355–1360. doi: 10.1073/pnas.1015739108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen DH, Jakana J, Liu X, Schmid MF, Chiu W. Achievable resolution from images of biological specimens acquired from a 4k×4k CCD camera in a 300-kV electron cryomicroscope. J. Struct. Biol. 2008;163:45–52. doi: 10.1016/j.jsb.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway J, Cheng N, Wingfield PT, Stahl SJ, Steven AC. Visualisation of a 4-helix bundle in the hepatitis B virus capsid by cryo-electron microscopy. Nature. 1997;385:91–94. doi: 10.1038/386091a0. [DOI] [PubMed] [Google Scholar]
- Daberkow I, Herrmann KH, Liu L, Rau WD. Performance of electron image converters with YAG single-crystal screen and CCD sensor. Ultramicroscopy. 1991;38:215–223. [Google Scholar]
- Deptuch G, Besson A, Rehak P, Szelezniak M, Wall J, et al. Direct electron imaging in electron microscopy with monolithic active pixel sensors. Ultramicroscopy. 2007;107:674–684. doi: 10.1016/j.ultramic.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Downing KH, Hendrickson FM. Performance of a 2k CCD camera designed for electron crystallography at 400 kV. Ultramicroscopy. 1999;75:215–233. doi: 10.1016/s0304-3991(98)00065-5. [DOI] [PubMed] [Google Scholar]
- Edelstein A, Amodaj N, Hoover K, Vale R, Stuurman N. Computer control of microscopes using μ Manager. Curr. Protoc. Mol. Biol. 2010;14:20.1–20.17. doi: 10.1002/0471142727.mb1420s92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan GY, Datte P, Beuville E, Beche JF, Millaud J, et al. ASIC-based event-driven 2D digital electron counter for TEM imaging. Ultramicroscopy. 1998;70:107–113. doi: 10.1016/s0304-3991(97)00109-5. [DOI] [PubMed] [Google Scholar]
- Faruqi AR, Cattermole DM, Raeburn C. Direct electron detection methods in electron microscopy. Nucl. Instr. and Meth. A. 2003;513:317–321. [Google Scholar]
- Faruqi AR, Subramaniam S. CCD detectors in high-resolution biological electron microscopy. Q. Rev. Biophys. 2000;33:1–27. doi: 10.1017/s0033583500003577. [DOI] [PubMed] [Google Scholar]
- Frank J. Three-dimensional electron microscopy of macromolecular assemblies: visualization of biological molecules in their native state. New York: Oxford University Press; 2006. [Google Scholar]
- Gilliland RL. Details of noise sources and reduction processes. Astronomical CCD observing and reduction techniques. 1992;23:68–89. [Google Scholar]
- Gipson B, Zeng X, Zhang ZY, Stahlberg H. 2dx - user-friendly image processing for 2D crystals. J. Struct. Biol. 2007;157:64–72. doi: 10.1016/j.jsb.2006.07.020. [DOI] [PubMed] [Google Scholar]
- Glaeser RM, Downing K, DeRosier D, Chiu W, Frank J. Electron Crystallography of Biological Macromolecules. New York: Oxford University Press; 2007. [Google Scholar]
- Goddard TD, Huang CC, Ferrin TE. Software extensions to UCSF chimera for interactive visualization of large molecular assemblies. Structure. 2005;13:473–482. doi: 10.1016/j.str.2005.01.006. [DOI] [PubMed] [Google Scholar]
- Grigorieff N, Harrison SC. Near-atomic resolution reconstructions of icosahedral viruses from electron cryo-microscopy. Curr. Opin. Struct. Biol. 2011;21:265–273. doi: 10.1016/j.sbi.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrini N, Turchetta R, Van Hoften G, Henderson R, McMullan G, et al. A high frame rate, 16 million pixels, radiation hard CMOS sensor. J. Inst. 2011;6:C03003. [Google Scholar]
- Harauz G, van Heel M. Exact filters for general geometry three dimensional reconstruction. Optik. 1986;73:146–156. [Google Scholar]
- Houska CR, Warren BE. X-ray study of the graphitization of carbon black. J. Appl. Phys. 1954;25:1503–1509. [Google Scholar]
- Howell SB. Handbook of CCD astronomy. New York: Cambridge University Press; 2000. [Google Scholar]
- Hryc CF, Chen D, Chiu W. Near-atomic resolution cryo-EM for molecular virology. Opin. Virol. 2011;1:110–117. doi: 10.1016/j.coviro.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizuka K. Coma-free alignment of a high-resolution electron microscope with three-fold astigmatism. Ultramicroscopy. 1994;55:407–418. doi: 10.1016/0304-3991(95)00155-7. [DOI] [PubMed] [Google Scholar]
- Jiang W, Baker ML, Jakana J, Weigele PR, King J, et al. Backbone structure of the infectious epsilon 15 virus capsid revealed by electron cryomicroscopy. Nature. 2008;451:1130–1134. doi: 10.1038/nature06665. [DOI] [PubMed] [Google Scholar]
- Jiang W, Chang J, Jakana J, Weigele P, King J, et al. Structure of epsilon 15 bacteriophage reveals genome organization and DNA packaging/injection apparatus. Nature. 2006;439:612–616. doi: 10.1038/nature04487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Bilhorn R. Performance of the DDD as a direct electron detector for low dose electron microscopy. MicroscMicroanal. 2010;16(S2):854–855. [Google Scholar]
- Jin L, Milazzo AC, Kleinfelder S, Li S, Leblanc P, et al. Applications of direct detection device in transmission electron microscopy. J. Struct. Biol. 2008;161:352–358. doi: 10.1016/j.jsb.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kivioja T, Ravantti J, Verkhovsky A, Ukkonen E, Bamford D. Local average intensity-based method for identifying spherical particles in electron micrographs. J. Struct. Biol. 2000;131:126–134. doi: 10.1006/jsbi.2000.4279. [DOI] [PubMed] [Google Scholar]
- Lawson CL, Baker ML, Best C, Bi C, Dougherty M, et al. EMDataBank.org: unified data resource for cryoEM. Nucleic Acids Res. 2011;39:D456–D464. doi: 10.1093/nar/gkq880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B, Mollon B, Mooney P. 300 kV performance of a 10k × 10k scintillator- and fiber-coupled CCD camera for transmission electron microscopes. Microsc. Microanal. 2010;16(S2):64–65. [Google Scholar]
- Liu X, Jiang W, Jakana J, Chiu W. Averaging tens to hundreds of icosahedral particle images to resolve protein secondary structure elements using a Multi-path Simulated Annealing optimization algorithm. J. Struct. Biol. 2007;160:11–27. doi: 10.1016/j.jsb.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W. EMAN: semi-automated software for high resolution single particle reconstructions. J. Struct. Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- Milazzo AC, Cheng A, Moeller A, Lyumkis D, Jacovetty E, et al. Initial evaluation of a direct detection device detector for single particle cryo-electron microscopy. J. Struct. Biol. 2011;176:404–408. doi: 10.1016/j.jsb.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milazzo AC, Leblanc P, Duttweiler F, Jin L, Bouwer JC, et al. Active pixel sensor array as a detector for electron microscopy. Ultramicroscopy. 2005;104:152–159. doi: 10.1016/j.ultramic.2005.03.006. [DOI] [PubMed] [Google Scholar]
- Milazzo AC, Moldovan G, Lanman J, Jin L, Bouwer JC, et al. Characterization of a direct detection device imaging camera for transmission electron microscopy. Ultramicroscopy. 2010;110:741–744. doi: 10.1016/j.ultramic.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullan G, Chen S, Henderson R, Faruqi AR. Detective quantum efficiency of electron area detectors in electron microscopy. Ultramicroscopy. 2009;109:1126–1143. doi: 10.1016/j.ultramic.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney PE. Optimization of image collection for cellular electron microscopy. Methods Cell Biol. 2007;79:661–719. doi: 10.1016/S0091-679X(06)79027-6. [DOI] [PubMed] [Google Scholar]
- Mooney PE, Contarato D, Denes P, Gubbens A, Lee B, et al. A high-speed electron-counting direct detection camera for TEM. Microsc. Microanal. 2011;17:1004–1005. [Google Scholar]
- Murata K, Liu X, Danev R, Jakana J, Schmid MF, et al. Zernike phase contrast cryo-electron microscopy and tomography for structure determination at nanometer and subnanometer resolutions. Structure. 2010;18:903–912. doi: 10.1016/j.str.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N, Shimizu Y, Shinkawa T, Nakata M, Bammes B, et al. Automated specimen search in cryo-TEM observation with DIFF-defocus imaging. J. Electron Microsc. (Tokyo) 2010;59:299–310. doi: 10.1093/jmicro/dfq009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlova EV, Dube P, Harris JR, Beckmann E, Zemlin F, et al. Structure of Keyhole Limpet Hemocyanin Type 1 (KLH1) at 15 Å resolution by electron cryomicroscopy and angular reconstitution. J. Mol. Biol. 1997;271:417–437. doi: 10.1006/jmbi.1997.1182. [DOI] [PubMed] [Google Scholar]
- Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- Sander B, Golas MM, Stark H. Advantages of CCD detectors for de novo three-dimensional structure determination in single-particle electron microscopy. J. Struct. Biol. 2005;151:92–105. doi: 10.1016/j.jsb.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, et al. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Xuong NH, Milazzo AC, LeBlanc P, Duttweiler F, Bouwer J, et al. First use of a high-sensitivity active pixel sensor array as a detector for electron microscopy. Proc. SPIE. 2004;5301:242–249. [Google Scholar]
- Yang C, Jiang W, Chen DH, Adiga U, Ng EG, et al. Estimating contrast transfer function and associated parameters by constrained non-linear optimization. J. Microsc. 2009;233:391–403. doi: 10.1111/j.1365-2818.2009.03137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Baker ML, Schröder GF, Douglas NR, Reissmann S, et al. Mechanism of folding chamber closure in a group II chaperonin. Nature. 2010;21:379–383. doi: 10.1038/nature08701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Nakamura N, Shimizu Y, Liang N, Liu X, et al. JADAS: a customizable automated data acquisition system and its application to ice-embedded single particles. J. Struct. Biol. 2009;165:1–9. doi: 10.1016/j.jsb.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Borgnia MJ, Mooney P, Shi D, Pan M, et al. Automated image acquisition and processing using a new generation of 4K × 4K CCD cameras for cryo electron microscopic studies of macromolecular assemblies. J. Struct. Biol. 2003;143:135–144. doi: 10.1016/s1047-8477(03)00124-2. [DOI] [PubMed] [Google Scholar]
- Zhou ZH, Chiu W. Prospects for using an IVEM with a FEG for imaging macromolecules towards atomic resolution. Ultramicroscopy. 1993;49:407–416. doi: 10.1016/0304-3991(93)90246-t. [DOI] [PubMed] [Google Scholar]