Summary
While significant efforts have been made to understand the mechanisms of caspase activation during apoptosis, many questions remain regarding how and when the executioner caspases get activated. We describe the design and synthesis of an activity-based probe that labels caspases-3/-6 /-7, allowing direct monitoring of all executioner caspases simultaneously. This probe has enhanced in vivo properties and reduced cross-reactivity compared to our previously reported probe AB50. Using this probe we find that caspase-6 undergoes a conformational change and can bind substrates even in the absence of cleavage of the pro-enzyme. We also demonstrate that caspase-6 activation does not require active caspases-3/-7 suggesting that it may auto-activate or be cleaved by other proteases. Together, our results suggest that caspase-6 activation proceeds through a unique mechanism that may be important for its diverse biological functions.
Keywords: caspases, caspase-6, activity-based probes, apoptosis, imaging
Introduction
The caspases are cysteine proteases that play key roles in mediating apoptosis, a highly regulated form of cell death critical for normal development, tissue homeostasis, and the removal of damaged cells. Recently, our group developed fluorescent activity-based probes that can be used to detect caspase activation upon induction of apoptosis both in vitro and in vivo (Edgington et al., 2009). Because these probes form a stable, covalent bond with the active site cysteine, they can be used to monitor caspase activity using a wide variety of detection strategies, including fluorescent SDS-PAGE, flow cytometry, microscopy, and optical imaging of tissues and whole organisms. While our initial peptide acyloxymethyl ketone (AOMK) probe, AB50, is a valuable reagent for the study of caspase activation, it suffers from cross-reactivity with the lysosomal cysteine protease legumain and it also predominantly labels the executioner caspases-3 and -7 (Edgington et al., 2009). Therefore probes with greater selectivity over non-apoptotic proteases and overall broader reactivity within the caspase family would be useful for monitoring multiple caspase activation pathways under different death stimuli.
Caspase-6, like caspase-3 and -7, is dimeric in solution, and cleavage of the prodomain and inter-subunit linker produces the mature enzyme complex composed of a heterotetramer of two large and two small subunits. Caspase-6 has recently been reported to self-activate, at least in vitro (Klaiman et al., 2009; Wang et al., 2010). The roles and mechanism of caspase-6 activation during apoptosis are not well understood and vary depending on the system being analyzed. Caspase-6 activation has been postulated to both precede (Allsopp et al., 2000) and depend on (Inoue et al., 2009; Slee et al., 2001; Slee et al., 1999) caspase-3 activity. It can also become activated in the absence of caspase-3 (Inoue et al., 2009). In primary neurons, caspase-6 has also been reported to act downstream of caspase-1 (Guo et al., 2006). Additionally, once activated by the intrinsic pathway, caspase-6 can cleave the initiator caspase-8 in the cytosol (Cowling and Downward, 2002). Unlike the other executioner caspases, caspase-6 cleaves nuclear Lamin A/C during programmed cell death, which promotes chromatin condensation and the formation of apoptotic bodies (Rao et al., 1996; Ruchaud et al., 2002; Takahashi et al., 1996).
In addition to its roles in apoptosis, caspase-6 is also proposed to be involved in several neurodegenerative disorders. In mouse models of Huntington's disease (HD), resistance to cleavage of the huntingtin protein at a caspase-6 site is sufficient to protect mice from neurological and behavioral abnormalities associated with pathogenesis, as well as NMDA receptor-mediated excitotoxicity, suggesting key roles for caspase-6 in the development of HD (Graham et al., 2010; Graham et al., 2006; Pouladi et al., 2009). In Alzheimer's Disease, caspase-6 has been shown to be active in the early stages of cognitive impairment and mediates cleavage of tau, amyloid-β peptide and other cytoskeletal components leading to plaque formation and neurofibrillary tangles associated with disease progression (Guo et al., 2004; Klaiman et al., 2008). In direct contrast, cleavage of DJ-1 by caspase-6 plays a protective role and mutations of the caspase-6 cleavage site on DJ-1 are associated with pathogenesis of Parkinson's disease (Giaime et al., 2010).
Because of this diversity of biological roles for caspase-6, improved tools to study this protease could lead to a greater understanding of its activity in normal apoptotic conditions and during disease progression. Here we describe the synthesis and testing of a fluorescent activity based probe designed to target caspase-6. The optimal probe that we identified, LE22, efficiently labels caspase-6 but retains activity towards caspases-3 and -7. Therefore it can be used to monitor the activity of all three executioner caspases simultaneously. In addition, LE22 shows enhanced labeling of caspases in vivo as well as overall reduced cross-reactivity towards the off target protease legumain compared to our previous generation probe, AB50 (Edgington et al., 2009). Using this new probe we show that caspase-6 is activated through multiple partially cleaved complexes that productively bind the probe and are in a complex with mature forms of caspase-6. Furthermore, we show that caspase-6 can be activated in the absence of active caspase-3 and -7. These results suggest a unique activation mechanism for caspase-6 compared to the other executioner caspases that may be relevant to its multiple diverse roles in cell biology.
Results and Discussion
Development of activity-based probes for caspase-6
In order to develop a probe to monitor caspase-6 activity, we used AB50 (Edgington et al., 2009) as a starting scaffold and changed the peptide specificity region based on the reported caspase-6 substrate preferences (Figure 1A and Supplemental Figure 1). We chose the sequence Val-Glu-Ile-Asp (VEID) because it is the sequence recognized by caspase-6 on Lamin A/C, a substrate that is not effectively processed by the other executioner caspases (Rao et al., 1996). Furthermore, many commercially available substrates and inhibitors for caspase-6 also make use of this sequence. We also designed a probe containing the sequence Ile-Val-Leu-Asp (IVLD) corresponding to the site on the huntingtin protein that has been shown to be cleaved by caspase-6 to generate neurotoxic fragments (Graham et al., 2010). Finally, we used the optimal substrate sequence for caspase-6, Val-Glu-His-Asp (VEHD), reported from fluorogenic substrate screening assays (Thornberry et al., 1997). We also screened purified, recombinant caspase-6 against our previously reported positional scanning library of AOMK inhibitors (Berger et al., 2006). This screen indicated that caspase-6 strongly preferred threonine at both P2 and P4 positions, while caspase-3 did not (Supplemental Figure 2). Therefore, we also synthesized a probe containing the sequence Thr-Glu-Thr-Asp (TETD). All of the probes were readily synthesized in Cy5-labeled form using a combination of solid phase and solution phase synthesis (Supplemental Scheme 1).
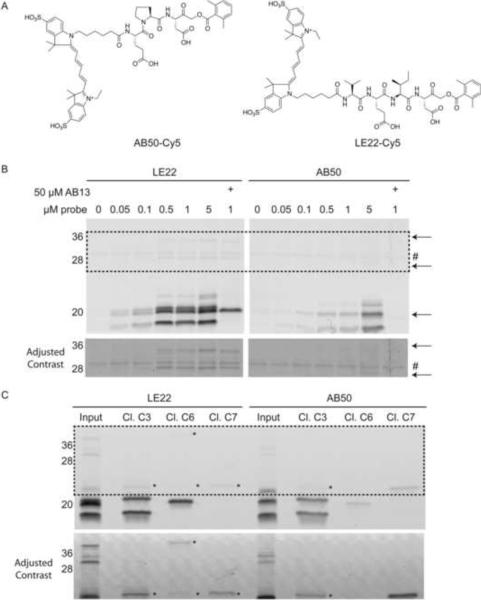
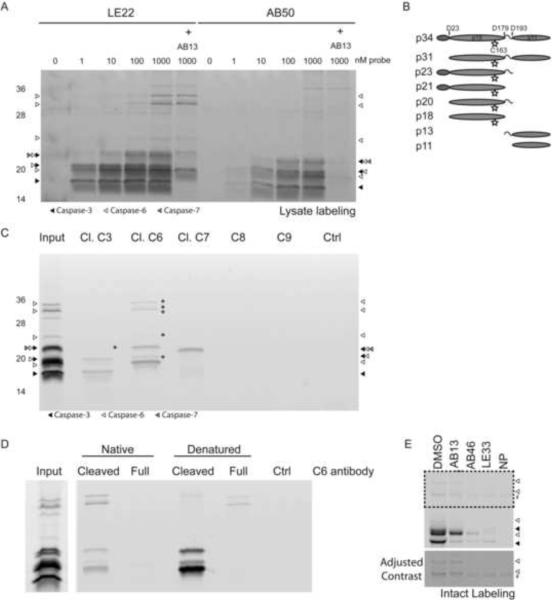
Figure 1. Direct comparison of LE22 and AB50 labeling in intact apoptotic cells.
(A) Structure of the previous generation caspase probe, AB50, and the optimal probe from the new series, LE22. (B) Fluorescent SDS-PAGE comparing LE22 and AB50 labeling of apoptotic Human colorectal cancer COLO205 cells. Cells were induced to undergo apoptosis using anti-DR5 and intact cells were labeled with each probe at the indicated concentrations. Where indicated, the caspase-3 and -7 specific inhibitor, AB13 was added prior to labeling with the probes. Total protein lysates were analyzed by SDS-PAGE followed by scanning for Cy5 fluorescence using a flatbed laser scanner. An autofluorescent protein is indicated with a #. Arrows highlight caspase-6 species that are labeled by LE22 but not by AB50. The bottom panel shows enhanced contrast of the boxed region for easier viewing of the high molecular weight bands. (C) Immunoprecipitations using the indicated cleaved caspase antibodies to confirm the identity of labeled proteins in the 5 μM samples in (B). Faint bands in the pulldowns that are difficult to see are noted with an asterisk, and the bottom panel shows enhanced contrast for easier viewing. (See also Figure S1 and S2)
To test these new probes for caspase-6 labeling, we used an in vitro model of apoptosis in which human COLO205 colorectal cancer cells were stimulated with a death receptor 5 agonist antibody (anti-DR5) to induce the extrinsic cell death pathway. Labeling of intact cells with each of the new probes showed similar patterns; however, the probe based on the lamin cleavage sequence, LE22, was by far the most potent (Supplemental Figure 1C). We next compared the labeling of LE22 to our previously reported probe AB50 (Figure 1B). Most strikingly, we found that LE22 was more effective at labeling caspases than AB50. This may be due to a combination of increased cell permeability of the probe as well as increased potency for the executioner caspases. Although the labeling profile of LE22 was similar to AB50, it clearly labeled an 18 kDa protein and several higher molecular weight proteins between 30–36 kDa that were not labeled by AB50 (Figure 1B). Because LE22 was designed to target caspase-6, we hypothesized that these additional labeled proteins may be multiple forms of caspase-6. As an initial test of this hypothesis, we pre-treated cells with the AOMK inhibitor AB13 (Berger et al., 2006) that we previously showed to be selective for mature forms of caspase-3 and -7 prior to labeling with LE22 (Figure 1B). Interestingly, inhibitor pretreatment completely blocked labeling of all of the AB50 labeled proteins but had no effect on the labeling of the LE22-specific 18kDa and 30–36kDa species.
To confirm the identity of the labeled caspases, we performed immunoprecipitation studies using antibodies raised against the cleaved forms of caspase-3, -6 and -7 (Figure 1C). As expected, we were able to precipitate cleaved forms of caspase-3 and -7 in cells labeled with AB50 or LE22. The caspase-6 antibody immunoprecipitated both the 18kDa protein as well as the higher molecular weight (30–36) kDa proteins that were only found in the LE22 labeled cells. These data suggest that LE22 is an overall more potent and sensitive probe than AB50 that labels caspase-3 and -6, and to a lesser extent caspase-7.
Cross-reactivity of caspase probes
In our previous studies, we found that the lysosomal cysteine protease, legumain is the major off-target of the AB50 probe (Edgington et al., 2009). The active site of this enzyme has a similar overall fold to the caspases but is thought to predominantly cleave protein substrates after aspargine residues (Abe et al., 1993). However, legumain is also able to bind aspartic acid-containing probes at the reduced pH of the lysosome (Kato et al., 2005; Sexton et al., 2007). To monitor selectivity, we used both AB50 and LE22 to label RAW cells, an immortalized mouse macrophage line that expresses high levels of active legumain and cathepsins (Figure 2A). As reported previously, we observed significant labeling of legumain by AB50. LE22, on the other hand, showed no labeling of legumain, even when used at high concentrations, indicating a markedly reduced cross-reactivity for this probe.
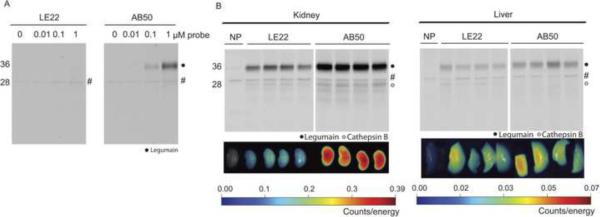
Figure 2. Cross-reactivity of probes towards other cysteine proteases.
Fluorescence SDS-PAGE showing labeling in RAW cells. Intact RAW cell mouse macrophages were labeled with the indicated concentrations of LE22 or AB50. Total protein lysates were harvested and resolved by SDS-PAGE followed by scanning on a flatbed laser scanner for Cy5 fluorescence. An autofluorescent background protein in the no-probe control tumors is marked by an `#' (B) Images of kidney and liver fluorescence after probe injection and corresponding SDS-PAGE analysis. LE22 or AB50 was injected by tail vein into normal BALB/c mice. Kidney and liver were harvested after four hours of circulation and probe fluorescence in the tissues was imaged using a CCD camera. Note differences in scale. Tissues were then homogenized and analyzed by SDS-PAGE followed by fluorescence scanning. Labeled proteases are indicated, and an autofluorescent background protein in the no-probe control tumors is marked by an `#'.
We also evaluated the specificity of both probes in vivo by treating wild type mice and analyzing kidney and liver extracts by fluorescence imaging and SDS-PAGE (Figure 2B). As a result of the high levels of expression of legumain in these organs, LE22 exhibits some cross-reactivity towards this protease, however the cross-reactivity was considerably less than AB50, consistent with the selectivity patterns observed for these probes in RAW cells (Figure 2B).
Application of LE22 to in vivo models of apoptosis
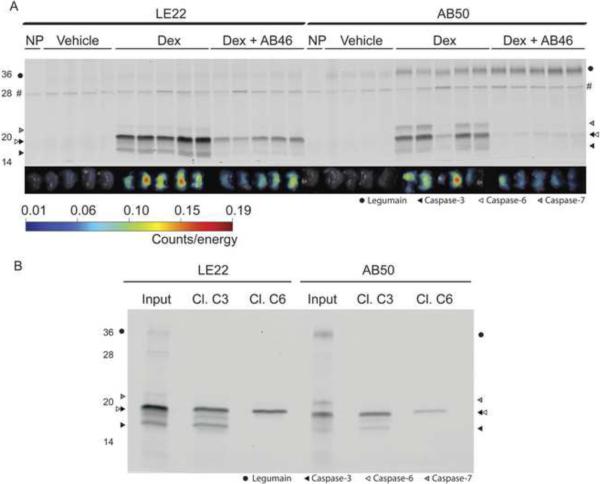
We next determined whether the increased potency of LE22 towards caspases -3, -6 and -7 relative to AB50 was also observed in in vivo models of cell death. We initially tested both probes in a mouse model in which dexamethasone (dex) was used to induce apoptosis in CD4+/CD8+ thymocytes (Figure 3A). For both probes, we observed a dex-dependent increase in both thymus fluorescence and caspase labeling as assessed by SDS-PAGE (Figure 3A). The dex-induced caspase labeling could also be blocked by pre-treating mice with the broad-spectrum caspase inhibitor, AB46. Immunoprecipitation experiments confirmed that AB50 labeled caspase-3 and a small amount of caspase-6, while LE22 labeled caspase-3 and -6 to a similar extent (Figure 3B). Caspase-7 activity was detected by both probes, but to a greater extent by AB50. In addition, in agreement with our results in the mouse RAW cells and normal mice, we only observed legumain labeling by AB50 and not by LE22.
Figure 3. Comparison of LE22 and AB50 in dexamethasone-induced thymocyte apoptosis.
(A) Fluorescent images of apoptotic and control thymi and corresponding SDS-PAGE analysis. BALB/c mice were injected with dexamethasone or vehicle for 12 hours followed by injection of either LE22 or AB50 by tail vein. A subset of the dex-treated mice were pretreated with a caspase inhibitor, AB46, prior to probe injection. After four hours, thymi were harvested and imaged ex vivo for probe accumulation. Thymus proteins were then resolved by SDS-PAGE and scanned for Cy5 fluoresence using a flatbed scanner. An autofluorescent background protein in the no-probe control tumors is marked by an `#' (B) Immunoprecipitations of samples shown in (A). The first sample in each dex-treated lane for LE22 and AB50 was immunoprecipitated with the indicated antibody and analyzed by SDS-PAGE.
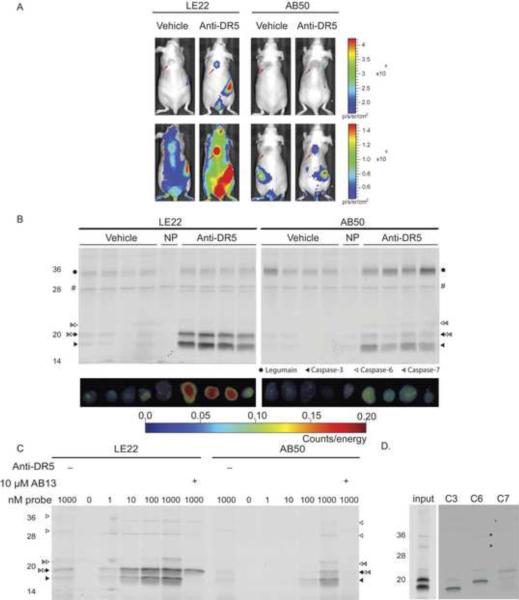
We also used LE22 and AB50 to label caspases in tumors induced to undergo apoptosis by chemotherapy. In this model, human COLO205 colorectal tumor cells were xenografted onto the backs of nude mice and later induced to undergo apoptosis by treatment with the anti-DR5 antibody. While both probes showed an anti-DR5-dependent increase in fluorescence within the tumor, LE22 treated tumors were brighter and showed better contrast over the non-treated controls (Figure 4A). After non-invasive imaging, we removed tumors and performed ex vivo imaging followed by SDS-PAGE analysis of tumor lysates (Figure 4B). Again we found that fluorescence increased in response to anti-DR-5 antibody treatment and signals from LE22-treated tumors were overall much brighter than those from AB50 treated tumors. Biochemical analysis of the tumor tissues verified that fluorescence intensity correlated with levels of caspase labeling. Again, LE22 showed stronger labeling of caspases than AB50 and also labeled less legumain.
Figure 4. Comparison of LE22 and AB50 in COLO205 tumors treated with anti-DR5 anti-body.
(A) Noninvasive imaging of apoptosis in tumor bearing mice. Nude mice bearing COLO205 tumors were treated with anti-DR5 antibody or vehicle for 12 hours followed by intravenous injection of either LE22 or AB50. After one hour of circulation, live mice were imaged using an IVIS 100. Red arrows indicate the location of the tumors. (B) Ex vivo imaging of tumors shown in (A) and corresponding biochemical analysis. Tumors were imaged ex vivo using epifluorescence on a Fluorescence Mediated Tomography (FMT) machine and then homogenized. Total protein lysates were analyzed by SDS-PAGE and scanned for Cy5 fluorescence. An autofluorescent background protein in the no-probe control tumors is marked by an `#' (C) Fluorescent SDS-PAGE analysis of tumors labeled ex vivo. Apoptosis was induced in a COLO205 tumor as in (A) however tumor was removed prior to probe administration and tumor lysates were labeled with LE22 or AB50 at the indicated concentration. Where noted the AB13 inhibitor was added to block caspase-3 and -7 labeling. Vehicle-treated control tumors were also included in the analysis. (D) Immunoprecipitations of tumor lysates labeled with 1 μM LE22 with the indicated antibodies to confirm the identity of labeled caspases. Weak bands in the immunoprecipitations are indicated by an asterisk.
We also wanted to determine if LE22 could be used ex vivo to monitor caspase-6 activation and maturation. We therefore labeled extracts from anti-DR5-treated tumors with AB50 and LE22 and analyzed labeling by SDS-PAGE. As we observed in intact COLO205 cells in vitro, there is a clear increase in labeling intensity of caspases for LE22 compared to AB50 (Figure 4C). We also used the caspase-3 and -7 specific inhibitor AB13, to block the activity of these two proteases. This treatment demonstrated that AB13 blocked labeling of all but three species by LE22 and completely blocked labeling of all species by AB50. Immunoprecipitation using caspase-3, -6 and -7 specific antibodies confirmed that the remaining proteins labeled by LE22 were in fact various forms of caspase-6 (Figure 4D). Overall, the labeling patterns were strikingly similar to what we observed in vivo, except that one of the pro-caspase-6 forms was obscured by legumain in the in vivo samples (legumain was not observed in lysates since labeling was carried out at neutral pH).
Monitoring caspase-6 activation with LE22
To determine whether differences in the sensitivity of AB50 and LE22 were due to permeability or uptake of the probes, we examined probe labeling in apoptotic lysates. As before, COLO205 cells were treated with anti-DR5 for four hours, followed by hypotonic lysis and subsequent probe labeling for 30 minutes (Figure 5A). Overall, we observed approximately 10-fold enhanced potency of LE22 compared to AB50. Interestingly, unlike in intact cells (Figure 1B), AB50 was able to label the mature form of caspase-6 in lysates. This result suggests that part of the reason for the lack of labeling of caspase-6 by AB50 may be due to reduced access of the probe to the intracellular caspase-6 pool. Cleaved caspase-6 has been shown to accumulate in the nucleus of COS cells upon staurosporine treatment (Warby et al., 2008), suggesting that AB50 could have reduced nuclear access.
Figure 5. Direct comparison of LE22 and AB50 labeling in apoptotic lysates.
(A) Fluorescent SDS-PAGE of labeled COLO205 lysates. Apoptosis was induced in COLO205 cells with an anti-DR5 antibody and cells were harvested. Lysates were then labeled with LE22 or AB50 at the indicated concentrations. AB13 was added to samples as indicated to selectively block caspase-3 and -7 labeling. Protein was resolved by SDS-PAGE and scanned for Cy5 fluorescence. (B) Schematic of the proposed forms of caspase-6 based on known cleavage sites. The pro-domain is indicated in red, the large subunit is green, and the small subunit is blue. The intersubunit linker is depicted by a curved line, and the active site cysteine (the site at which the probe binds) is marked by a yellow star. Predicted sizes are listed at the left. (C) Immunoprecipitations of lysates labeled with 1 μM LE22. Five times as much protein was used as in (A) to ensure adequate pulldown of less abundantly labeled proteins. Faintly labeled species that are difficult to see in the pulldowns are marked by an asterisk. Ctrl indicates an immunoprecipitation that was performed in the absence of antibody to account for nonspecific sticking of proteins to the Protein A/G beads. (D) Immunoprecipations of LE22 labeled lysates with antibodies for the cleaved and full-length caspase-6 species in their native, folded state (left lanes) or denatured by boiling in SDS (right lanes). (E) Full-length capase-6 labeling by LE22 in intact apoptotic cells was blocked by pretreatment with AB46 and biotinylated LE22 (LE33), but not by pretreatment with AB13. An autofluorescent band is denoted with a #.
In lysates, we saw more pronounced labeling of the larger forms of caspase-6, which were appropriately sized to be proforms that had not been cleaved between the large and small subunits (Figure 5B). We were initially surprised to see labeling of these forms, as activation of caspase-6 is thought to depend on removal of the prodomain and cleavage of the intersubunit linker. We also did not expect these forms to immunoprecipitate with an antibody that recognizes cleaved forms of caspase-6 (Figure 5C). This polyclonal antibody was raised against the c-terminus of the large subunit of caspase-6, and therefore should not detect full-length forms. The most plausible explanation for our results was that we were precipitating caspase-6 dimers in which a labeled, full-length uncleaved monomer was in a complex with a cleaved monomer. This half-cleaved complex would be similar to what has previously been reported for caspase-7 (Berger et al., 2006; Denault et al., 2006).
To test this hypothesis, we performed immunoprecipitation using probe-labeled apoptotic lysates that were denatured by boiling in SDS using an antibody that recognizes only cleaved caspase-6 (Figure 5D). Under denaturing conditions, the cleaved caspase-6 antibody precipitated negligible amounts of the full-length forms, suggesting that proforms of caspase-6 can be labeled by active site probes and that at least some fraction of these proforms can be isolated in complex with cleaved forms of caspase-6. To further confirm that the single chain (uncleaved) forms of caspase-6 possess catalytic activity, we pretreated lysates with two different inhibitors, AB46 and LE33, a version of LE22 in which the Cy-5 tag was replaced with biotin (Figure 5E). These results confirmed that active site directed inhibitors could block the labeling of the higher molecular weight proforms of caspase-6 by LE22. AB13, the caspase-3/-7 selective inhibitor, however, was unable to block labeling of those same pro-forms.
A conformational change is permissive for activation in the absence of cleavage
To gain more insight into the activation mechanism of caspase-6, we generated several cleavage site mutants in which the endogenous aspartic acid (`D') residues at three cleavage sites in the linker region were converted to alanine (`A') (Supplemental Figure 3). Compared to wildtype, the reduction in activity of the D23A or D179A single mutants was minimal, and even the D23A/D179A double mutant showed only a two-fold decrease in Kcat/KM (Supplemental Table 1). The most dramatic reduction in activity, however, was observed for the D193A (~20-fold reduction in Kcat/KM). D23A or D179A mutations further reduced the activity of D193A. This confirms the earlier finding that caspase-6 can auto-process at D193, whereas D179 requires an additional enzyme for cleavage (Wang et al., 2010). Cleaving the D193A mutants with 1% caspase-3 overnight increased the activity by about 40 fold. However, activity failed to reach levels expected for fully processed caspase-6, indicating that processing of the linker at D193 is absolutely essential to generate a fully active caspase-6 species. Interestingly, the amount of active enzyme in the preparations of non-cleaved caspase-6 as determined by titration with Z-VAD-FMK was only about 10–15% of what was expected based on absorption at 280 nm (A280), while incubation with caspase-3 overnight led to activation of ~100% of the caspase-6 species in the preparations (not shown). Thus, in the total pool of non-cleaved caspase-6, only a fraction has activity, while the entire pool has the capacity to generate activity.
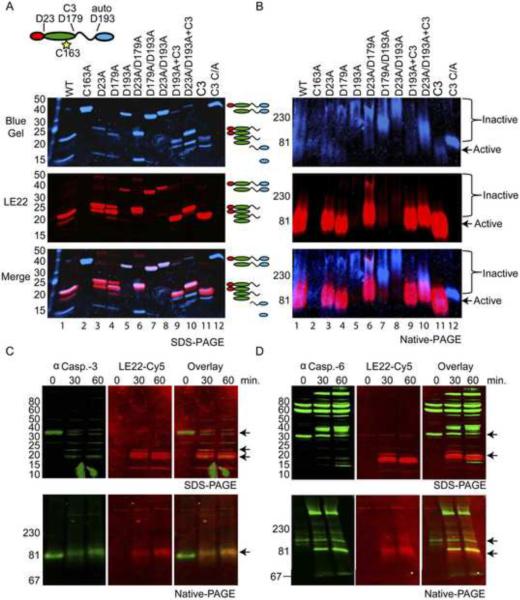
Since the non-cleavable caspase-6 mutants retained activity, albeit very little, we decided to utilize LE22 to visualize the active species of recombinant caspase-6 (Figure 6A). The full-length caspase-6 species containing D193A readily bound the probe (lanes 5, 7, and 8), albeit to a lesser degree than those species processed at D193 (lanes 1, 3, 4, and 6). Addition of 1% active caspase-3 to the D193A or D23A/D193A mutants dramatically increased probe labeling (lanes 9 and 10). The active site mutants of both caspase-6 and caspase-3 (lanes 2 and 12, respectively) were not labeled by the probe at all, indicating that probe binding is specific and depends on activity. In addition, several higher molecular weight species were also labeled by the probe. These are not likely to be contaminants carried over from E. coli, since they are not detected in the Cys mutant species and could represent aggregates of active caspase-6.
Figure 6. Caspase-6 undergoes a conformational change upon activation.
(A and B) Recombinant caspase-6 as well as wild type and the active-site mutant (C/A) of caspase-3 were incubated with an excess of LE22 (4 μM) for 3 hours and analyzed by SDS-PAGE (A) or on pore limit native PAGE (B). D193A mutants of caspase-3 were cleaved with 1% caspase-3 overnight where indicated (+C3). The gel was first scanned on a fluorescent scanner to detect the probe signal and then stained with coomassie and re-scanned to detect total protein. The two figures were then overlaid to visualize the species of caspase-6 that were active (i.e. bound the probe; red) relative to species that were not catalytically active (i.e. only stained blue; blue). (C and D) Hypotonic lysates from 293T cells were activated by the addition of cytochrome c and dATP. Samples were taken before, after 30 minutes and after 60 minutes of cytochrome c addition, labeled with LE22 for 30 minutes at 37°C and analyzed by Western blot after separating the proteins either by SDS-PAGE (upper panels) or by pore limit native PAGE (lower panels). Blots were probed for total and active caspase-3 (C) or caspase-6 (D). The signal obtained for the caspase blots is displayed in green, signal from LE22 in red. Full-length and cleaved caspase species are indicated by arrows. Notice that on the pore limit gel, full length and cleaved caspase-3 run at the same molecular weight. (See also Figure S3, S4 and ST1)
Executioner caspases have been suggested to fluctuate between an `active' and an `inactive' conformation, with processing of the inter subunit linker stabilizing the `active' conformation (Fuentes-Prior and Salvesen, 2004; Gray et al., 2010). In theory, an inhibitor or active site probe such as LE22 could stabilize this active conformation. If this were the case, we would expect to see a steady increase in labeling over time when the uncleavable caspase-6 is labeled with the probe. However, we found that although the uncleaved species labels more slowly than the wild type (as expected from its decreased kcat), maximum labeling of all species was achieved after ~90 minutes of incubation with an excess of probe (Supplemental Figure 4A). This labeling was significantly lower (~7 fold) than the maximum labeling of WT caspase-6, even though equal amounts of total protein were used in the experiment (Supplemental Figure 4B). This suggests that only a fraction of the uncleaved caspase-6 species is in an active conformation and, although this fraction labels more slowly than processed caspase-6, it does not increase over time.
The executioner caspases-3 and -7 have been described to form obligate dimers (Boatright et al., 2003) and dimerization is an absolute requirement for caspase activation. We wondered whether the uncleavable caspase-6 mutants were primarily expressed as inactive monomers, while only the fraction that was labeled by LE22 formed active dimers. To investigate this hypothesis, we analyzed all caspase-6 species on a native pore limit gel after labeling with LE22 (Figure 6B). On such a gel, a native protein sample is resolved on a 4–20% polyacrylamide gradient. Migration of a protein or protein complex is limited by the pore size of the polyacrylamide, thus a protein complex will reach equilibrium higher up in the gel (at a larger pore-size) than the individual proteins would (Barrett et al., 1979; Boatright et al., 2003). To our surprise, the inactive, unlabeled species ran higher in the gel than the labeled species, which ran at approximately the same molecular weight as the caspase-3 dimer irrespective of linker or pro-domain processing. Probe labeled species resolved lower on the gel, suggesting that either native caspase-6 forms a higher oligomer or that inactive, single chain, caspase-6 is partially unfolded and therefore migrates slower through the gel. In support of the latter hypothesis, the crystal structure of ligand-free caspase-6 reveals a misalignment of active site residues, which is not observed for caspases-3 or -7 (Baumgartner et al., 2009). When we compared the different species of single chain caspase-6 with WT on native gel either with or without LE22 labeling, we observed a significant shift upon probe binding (Supplemental Figure 4D), suggesting that caspase-6 folds around its ligand, as has been suggested (Vaidya et al., 2011).
To study the behavior of natural (endogenous) caspase-6 upon activation, we activated caspase-9 in cytosolic extracts from HEK293T cells by adding cytochrome c and dATP, a commonly used model for caspase activation (Figure 6C, D; Stennicke et al., 1999). As indicated by probe-labeling, both caspase-3 and caspase-6 are activated within 30 minutes and are fully processed within 60 minutes. At 30 minutes, a partially processed species of caspase-3 can also be observed that has activity, as indicated by probe binding, and is detected by the active caspase-3 antibody. We then probed the blots successively with antibodies against full length and cleaved caspase-3 and -6 respectively to distinguish between specific and non-specific antibody recognition. Although both caspase-6 and -3 are cleaved during activation, only caspase-6 undergoes a dramatic change in conformation as observed on the native gel (lower panels). Caspase-3 migrates at the same MW, whether or not it is cleaved/active, whereas active caspase-6 shifts to a lower MW relative to inactive caspase-6 upon activation. Furthermore, labeling of uncleaved caspase-6 can also be observed, both on the denatured and the native gel.
Altogether, our data suggest that a conformational change is permissive for initial caspase-6 activation, while cleavage, in particular auto-processing of D193, further enhances activity and stabilizes the active conformation.
Kinetic studies of caspase activation using activity based probes
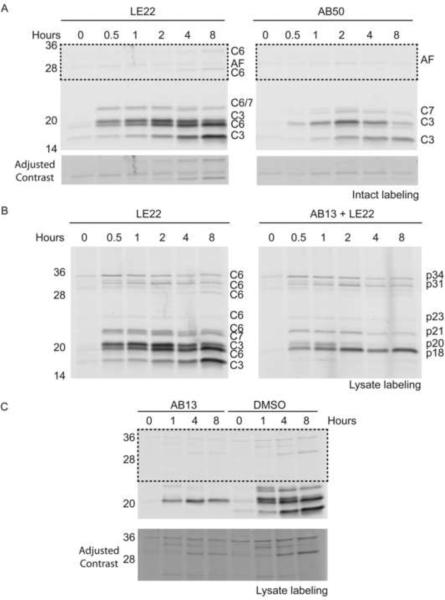
We next used LE22 to examine the dynamics of executioner caspase activation in intact cells after induction of a death stimulus. For these studies, we treated COLO205 cells with anti-DR5 antibody over an eight hour time period and labeled with LE22 or AB50 in the final 30 minutes of the experiment (Figure 7A). We found that the p19 form of caspase-3 is active at early time points and matures over time, leading to increased activity of the p17 form. Interestingly, Caspase-6 activates more slowly than caspase-3, and shows the most activity at the late time points. In intact cells, the proforms of caspase-6 gradually show increased activity over time. In parallel, we also conducted a similar time-course in which lysates were used in place of intact cells (Figure 7B). In these samples we observed a sharp increase in activity of the proforms of caspase-6 immediately upon anti-DR5 stimulation. This activity remained relatively constant over time (Figure 7B). To obtain a clearer picture of caspase-6 activity we also used AB13 to block labeling of caspase-3 and -7 in the lysates before labeling with LE22 (Figure 7B). Using this method we were able to clearly identify all of the 6 major predicted forms of caspase-6 (see figure 5B). Additionally, we performed analogous studies in Jurkat cells using a variety of intrinsic and extrinsic death stimuli and observed similar trends in caspase-6 activation (Supplemental Figure 5). The kinetics of activation were slower with anti-DR5, anti-Fas, or Staurosporine than with Etoposide. However, the pro-forms of caspase-6 increase in activity under all stimuli.
Figure 7. Monitoring maturation of executioner caspases during apoptosis.
(A) COLO205 cells were stimulated with anti-DR5 to initiate apoptosis for the indicated period of time. During the last 30 minutes of treatment, intact cells were labeled with 1 μM LE22 or AB50. Cells were then lysed and subject to SDS-PAGE analysis. The bottom panel shows enhanced contrast of the boxed region for easier viewing of the high molecular weight proteins. (B) Cells were treated in parallel to those in (A) however, labeling with 1μM LE22 was performed post-lysis. In the right panel, 10 μM AB13 was added prior to LE22 to allow for specific labeling of caspase-6. (C) Cells were treated as above, except AB13 was added at the time of anti-DR5 stimulation to block caspase-3/-7 activity throughout the course of the experiment. The bottom panel shows enhanced contrast of the boxed region for easier viewing of the high molecular weight bands. (See also Figure S5)
Finally, we performed a kinetic study in which AB13 was added at the time of anti-DR5 treatment to block caspase-3/-7 activity throughout the experiment (Figure 7C). If these executioner caspases were essential for caspase-6 activation in this model, we would expect to see dramatic changes in the kinetics of its maturation. However, the caspase-6 labeling pattern remained relatively unchanged. While auto-processing occurs at D193, full maturation to the p18 form of caspase-6 would not be expected to occur without cleavage at D179. This suggests that either caspase-6 is capable of autoactivation, or that other caspases are able to cleave at D179.
Discussion
The process of activating apoptosis is complex and involves multiple caspase proteases. While a great deal is known about the multiple steps in the signaling cascade, still relatively little is known about the details of how and when specific caspases are activated. In particular, the executioner caspase-6 has been proposed to play roles in classical death signaling but it also may play important functions in regulation of protein turnover and neuronal function. In addition, it is not clear how and when this protease gets activated. In this study we present a newly developed activity based probe that is capable of labeling all of the executioner caspases, including caspase-6, both in intact cells and also in vivo. Using this new probe we show that caspase-6 is activated independently of the other downstream caspases-3 and -7. Furthermore, our results confirm that caspase-6 activation proceeds thru a series of partially processed intermediates that have an ordered active site and a unique structural conformation relative to the other executioner caspases. This suggests that caspase-6 may exist in multiple forms in the cell and these forms may have relevance to its diverse functional roles in the cell.
One of our most surprising findings was that LE22 was able to label uncleaved, immature forms of caspase-6. These proforms have not previously been shown to be active in living cells or lysates. Recent studies using recombinant caspase-6 suggested that cleavage of at least one site within the intersubunit linker, either Asp 179 or 193, is required to activate the enzyme (Klaiman et al., 2009). A non-cleavable form of caspase-6 (D179A/D193A) showed weak activity against a fluorogenic VEID substrate in vitro; however, this activity was not detected in lysates or cells (Klaiman et al., 2009).
Recent crystal structures of caspase-6 indicate that both the zymogen and the unbound processed form adopt a conformation unique among all caspases in which the active site residues are misaligned (Baumgartner et al., 2009; Wang et al., 2010). It is only upon substrate binding that a conformational change occurs, allowing the formation of a canonical caspase active site (Vaidya et al., 2011). Caspase-6 has also been suggested to be capable of intramolecular auto-activation, a phenomenon not observed by other executioner caspases (Wang et al., 2010). This auto-activation occurs through intramolecular cleavage at Asp193, a site that is located near the catalytic Cys163 of the active site. If the shift in conformation is able to initiate intramolecular cleavage, it is feasible that this shift could also promote substrate processing, even without D193 cleavage. A half-cleaved intermediate would result if intramolecular cleavage at D193 did not occur in both molecules of the dimer simultaneously. Most of the active pro-caspase-6 detected in apoptotic cells appears to be in complex with a cleaved molecule, as indicated by immunoprecipation experiments, suggesting that cleavage occurs through a stepwise process. Whether or not these intermediates are active against physiological substrates remains unclear and further studies are warranted.
While caspase-6 is able to auto-activate, it is clear from our studies that the activity of caspase-6 increases sharply upon introduction of a death stimulus. It is currently unclear how caspase-6 is prevented from auto-activating in the cell under normal conditions. In addition to a conformational shift and subsequent autocleavage, other factors likely contribute to its full activation in vivo. Previous studies suggest that caspase-3 and -7 are responsible for this activation (Inoue et al., 2009; Slee et al., 2001; Slee et al., 1999). However, our data suggest that full maturation of caspase-6 can occur independently of these proteases. Full activation may require cleavage by an initiator protease or the release of an inhibitor, although endogenous inhibitors of caspase-6 have yet to be described.
Significance <300 words
In this study, we have improved upon the previous generation of fluorescent activity-based probes to detect caspase activity in lysates, intact cells, tissues, and whole organisms. Compared to our previous probe AB50, LE22 shows increased potency towards caspases with reduced off-target labeling. Furthermore, LE22 is a fluorescent activity-based probe capable of simultaneously monitoring the dynamics of all executioner caspase activity during apoptosis in living cells and tissues. Given these properties, LE22 is potentially valuable for both preclinical and clinical applicaitons where detection of apoptosis may be beneficial (e.g. assessing efficacy of chemotherapy response during cancer treatment and identifying off-target effects, etc.) Caspase-6 has also been demonstrated to have non-apoptotic roles in a number of neurological disorders such as Huntington's, Parkinson's, and Alzheimer's disease. Therefore, LE22 could also be an important tool to assist in further dissection of the contribution of caspase-6 activity during progression of these diseases.
In addition to its potential clinical value, LE22 is also an effective tool to aid in understanding basic mechanisms of executioner caspase activation during apoptosis. In this study, we used LE22 to demonstrate that caspase-6 is activated in a manner distinct from caspase-3. Unlike caspase-3, caspase-6 undergoes a conformational change upon activation and substrate binding and can cleave substrates in its full-length form. Additionally, we showed that caspase-6 can reach full maturation and activation in the absence of caspase-3 or caspase-7 activity, suggesting highly regulated autoactivation or cleavage by other proteases. Future studies will utilize LE22 to better define the activation mechanisms and function of caspase-6 during apoptosis.
Experimental Procedures
Compound synthesis
All caspase probes and inhibitors were synthesized using solid-phase peptide synthesis methods previously reported for P1-Asp-AOMK compounds. (Berger et al., 2006; Edgington et al., 2009) The Cy5 fluorophore was synthesized in house according to (Mujumdar et al., 1993) and coupled to probes using established methods (Berger et al., 2006; Edgington et al., 2009). Purity and identity of all compounds were assessed by LC-MS using an Agilent HPLC in tandem with an API 150 mass spectrometer (Applied Biosystems/SCIEX) equipped with an electrospray interface.
Labeling of intact cells
Human colorectal cancer COLO205 cells (ATCC) were seeded in six-well plates (1.5×106/well) 24 hours prior to stimulation. Media was refreshed and anti-DR5 (20μg/mL) was added for the indicated time. For intact cell labeling, probe was added during the last 30 minutes from 1000× concentrated DMSO stock solutions, yielding a final DMSO concentration of 0.1%. AB13 inhibitor, where indicated, was added 30 minutes prior to probe addition. At t = 0, cells were scraped, washed once with PBS and resuspended in hypotonic lysis buffer containing 50 mM PIPES pH 7.4, 10 mM KCl, 5 mM KCl2, 2 mM EDTA, 4 mM DTT, and 1% NP-40. Samples were snap frozen in liquid N2, centrifuged at 4°C and supernatants were solubilized with 4× sample buffer. 50 μg total protein was resolved by 15% SDS-PAGE and scanned using a Typhoon flatbed laser scanner (633 nm excitation/670 nm emission). RAW cells were labeled by a similar procedure except anti-DR5 was not added. 293T cell extracts were prepared in a similar fashion and caspase activation in these extracts was initiated by the addition of purified cytochrome c from horse heart (Sigma-Aldritch) at a concentration of 5 μM and dATP (Sigma-Aldritch; 60 μM).
Lysate labeling
For labeling of lysates, experiments were carried out as above except inhibitor and probe were added after hypotonic lysis from 100× DMSO stock solutions, where the final DMSO concentration is 1%. Inhibitor was added just prior to addition of probe, and 4× sample buffer was added after 30 minutes of labeling. 10 μg total protein was analyzed by SDS-PAGE as above.
Immunoprecipitations
Samples from intact labeling experiments were immunoprecipitated from the same samples, which had been boiled in SDS sample buffer. Samples from lysate labeling were not boiled unless indicated. 50–100μg total protein was diluted in 500 μL RIPA buffer (PBS, 1 mM EDTA, 0.5% NP-40, pH 7.4). 5 μL of the indicated antibody was added to diluted lysates for 10 minutes on ice before addition of 40 μL slurry of prewashed Protein A/G agarose beads (Santa Cruz). Samples were shaken overnight at 4°C followed by washing four times with RIPA buffer and once with 0.9% NaCl. 2× sample buffer was added and beads were boiled followed by SDS-PAGE analysis. All antibodies were purchased from Cell Signaling Technologies. Cleaved Caspase-6 (9761), Caspase-6 (9162), Cleaved Caspase-3 (9661), Cleaved Caspase-7 (9491), Caspase 8 (9746), Caspase 9 (9502).
Dexamethasone-induced thymocyte apoptosis
All animal experiments were performed according to specific guidelines approved by the Stanford Administrative Panel on Laboratory Animal Care. Six-week old female BALB/c mice were obtained from Jackson Laboratories. Each treatment group contained five mice. Mice were injected intraperitoneally with water-soluble dexamethasone (Sigma) (50mg/kg dissolved in 100 μL sterile PBS) or PBS vehicle. Twelve hours later, mice were injected intravenously by tail vein with 40 nmol probe in 10% DMSO/PBS (~1mg/kg). For inhibitor-treated mice, AB46 (125 nmol in 20% DMSO/PBS, ~3.3 mg/kg) was administered by tail vein 20 minutes prior to probe injection. Four hours later, mice were anesthetized with isofluorane and killed by cervical dislocation. Thymi were removed from all mice and kidney and liver from the vehicle control mice for assessment of off-target labeling. Organs were imaged immediately using Perkin Elmer's FMT machine with a Cy5.5 filter and then homogenized in muscle lysis buffer (1% Triton X-100, 0.1% SDS, 0.5% sodium deoxycholate in PBS, pH 7.4). Total protein (50 μg) was analyzed by SDS-PAGE as described above.
Anti-DR5 antibody-induced tumor apoptosis
Six-week old, female nude mice were obtained from Charles Rivers. COLO205 xenografted tumors were established on their backs as described previously (Edgington et al., 2009). Anti-DR5 antibody (10 mg/kg, Genentech) or vehicle (10 mM histidine, 0.8% sucrose, and 0.02% Tween-20, pH 6) was administered via tail vein in 100 μL volume. Twelve hours later, 40 nmol probe was administered as above. After one hour, mice were imaged noninvasively using an IVIS 100 system. Mice were then killed as above. Tumors were removed, imaged ex vivo using an FMT and processed as above. For tumor lysate labeling, tumors were harvested without administering probe and labeled according to the protocol above. Four mice were used for each treatment group and representative images are shown.
Recombinant caspase-6 and enzyme kinetics
Full length human caspase-6 was cloned in a pET23b expression vector with a C-terminal 6×His-tag for expression in E. coli and purified as described (Stennicke et al., 1999). Mutants were generated by quickchange site-directed mutagenesis. Active site concentration in the purified protein preparations were determined by titration with the irreversible caspase inhibitor Z-VAD-FMK, and Vmax and KM values were determined with the substrate Ac-VEID-AFC, as described (Stennicke et al., 1999). Recombinant caspases were labeled at a total protein concentration of 4 μM, as determined by A280, with 10 μM LE22 for up to 3 hours at 37°C. Gels were scanned on the Li-Cor Odyssey infrared scanner at 700 nM to detect the Cy5 signal of the probe, stained with GelCode Blue reagent (Thermo Scientific) and re-scanned. The two images were colored and overlaid in Adobe Photoshop 7.0.
Pore limit PAGE and Western blot
For pore limit PAGE of native proteins, 4%–20% polyacrylamide gels were prepared and run at constant voltage (80V) for 16 h with cooling and buffer recirculation between the reservoirs in 89 mM Tris, 2 mM, EDTA, 89 mM boric acid (pH 9) (Barrett et al., 1979). Size standards were catalase (232 kDa), transferrin (81 kDa), BSA (67 kDa), CrmA Thr345Arg mutant [Tewari et al., 1995)] (39 kDa).
For Western blotting, the proteins were transferred to nitrocellulose membrane and probed for total and cleaved caspase-3 (Cell Signaling Technology; #9662 and #9661, respectively) or full length and cleaved caspase-6 (Upstate Biotechnology; #06-691 and Millipore; AB10512, respectively). Secondary antibody was Donkey anti-Rabbit IRDye 800CW from Li-Cor. Blots were scanned on an Odyssey infrared scanner (Li-Cor).
Supplementary Material
Acknowledgements
We thank D. Ehrnhoefer, N. Skotte, and M. Hayden of the University of British Columbia for critical discussions of caspase-6 biology, C. Pop, formerly of the Sanford-Burnham Medical Research Institute for helpful advice on the enzyme kinetics experiments and S. Snipas for his help in cloning the caspase-6 mutants. We also thank T. Doyle at the Stanford Small Animal Facility at Stanford for assistance with optical imaging studies. This work was supported by grants from the NIH (R01 EB005011). BJvR is supported by a Rubicon fellowship from the Netherlands Organization for Scientific Research (NWO) and a fellowship from the Barth Syndrome Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no conflicts of interest.
References
- Abe Y, Shirane K, Yokosawa H, Matsushita H, Mitta M, Kato I, Ishii S. Asparaginyl endopeptidase of jack bean seeds. Purification, characterization, and high utility in protein sequence analysis. J Biol Chem. 1993;268:3525–3529. [PubMed] [Google Scholar]
- Allsopp TE, McLuckie J, Kerr LE, Macleod M, Sharkey J, Kelly JS. Caspase 6 activity initiates caspase 3 activation in cerebellar granule cell apoptosis. Cell Death Differ. 2000;7:984–993. doi: 10.1038/sj.cdd.4400733. [DOI] [PubMed] [Google Scholar]
- Barrett AJ, Brown MA, Sayers CA. The electrophoretically `slow' and `fast' forms of the alpha 2-macroglobulin molecule. Biochem J. 1979;181:401–418. doi: 10.1042/bj1810401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgartner R, Meder G, Briand C, Decock A, D'Arcy A, Hassiepen U, Morse R, Renatus M. The crystal structure of caspase-6, a selective effector of axonal degeneration. Biochem J. 2009;423:429–439. doi: 10.1042/BJ20090540. [DOI] [PubMed] [Google Scholar]
- Berger AB, Witte MD, Denault JB, Sadaghiani AM, Sexton KM, Salvesen GS, Bogyo M. Identification of early intermediates of caspase activation using selective inhibitors and activity-based probes. Mol Cell. 2006;23:509–521. doi: 10.1016/j.molcel.2006.06.021. [DOI] [PubMed] [Google Scholar]
- Boatright KM, Renatus M, Scott FL, Sperandio S, Shin H, Pedersen IM, Ricci JE, Edris WA, Sutherlin DP, Green DR, et al. A unified model for apical caspase activation. Mol Cell. 2003;11:529–541. doi: 10.1016/s1097-2765(03)00051-0. [DOI] [PubMed] [Google Scholar]
- Cowling V, Downward J. Caspase-6 is the direct activator of caspase-8 in the cytochrome c-induced apoptosis pathway: absolute requirement for removal of caspase-6 prodomain. Cell Death Differ. 2002;9:1046–1056. doi: 10.1038/sj.cdd.4401065. [DOI] [PubMed] [Google Scholar]
- Denault JB, Bekes M, Scott FL, Sexton KM, Bogyo M, Salvesen GS. Engineered hybrid dimers: tracking the activation pathway of caspase-7. Mol Cell. 2006;23:523–533. doi: 10.1016/j.molcel.2006.06.020. [DOI] [PubMed] [Google Scholar]
- Edgington LE, Berger AB, Blum G, Albrow VE, Paulick MG, Lineberry N, Bogyo M. Noninvasive optical imaging of apoptosis by caspase-targeted activity-based probes. Nat Med. 2009;15:967–973. doi: 10.1038/nm.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes-Prior P, Salvesen GS. The protein structures that shape caspase activity, specificity, activation and inhibition. Biochem J. 2004;384:201–232. doi: 10.1042/BJ20041142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaime E, Sunyach C, Druon C, Scarzello S, Robert G, Grosso S, Auberger P, Goldberg MS, Shen J, Heutink P, et al. Loss of function of DJ-1 triggered by Parkinson's disease-associated mutation is due to proteolytic resistance to caspase-6. Cell Death Differ. 2010;17:158–169. doi: 10.1038/cdd.2009.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham RK, Deng Y, Carroll J, Vaid K, Cowan C, Pouladi MA, Metzler M, Bissada N, Wang L, Faull RL, et al. Cleavage at the 586 amino acid caspase-6 site in mutant huntingtin influences caspase-6 activation in vivo. J Neurosci. 2010;30:15019–15029. doi: 10.1523/JNEUROSCI.2071-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham RK, Deng Y, Slow EJ, Haigh B, Bissada N, Lu G, Pearson J, Shehadeh J, Bertram L, Murphy Z, et al. Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cell. 2006;125:1179–1191. doi: 10.1016/j.cell.2006.04.026. [DOI] [PubMed] [Google Scholar]
- Gray DC, Mahrus S, Wells JA. Activation of specific apoptotic caspases with an engineered small-molecule-activated protease. Cell. 2010;142:637–646. doi: 10.1016/j.cell.2010.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Albrecht S, Bourdeau M, Petzke T, Bergeron C, LeBlanc AC. Active caspase-6 and caspase-6-cleaved tau in neuropil threads, neuritic plaques, and neurofibrillary tangles of Alzheimer's disease. Am J Pathol. 2004;165:523–531. doi: 10.1016/S0002-9440(10)63317-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Petrin D, Zhang Y, Bergeron C, Goodyer CG, LeBlanc AC. Caspase-1 activation of caspase-6 in human apoptotic neurons. Cell Death Differ. 2006;13:285–292. doi: 10.1038/sj.cdd.4401753. [DOI] [PubMed] [Google Scholar]
- Inoue S, Browne G, Melino G, Cohen GM. Ordering of caspases in cells undergoing apoptosis by the intrinsic pathway. Cell Death Differ. 2009;16:1053–1061. doi: 10.1038/cdd.2009.29. [DOI] [PubMed] [Google Scholar]
- Kato D, Boatright KM, Berger AB, Nazif T, Blum G, Ryan C, Chehade KA, Salvesen GS, Bogyo M. Activity-based probes that target diverse cysteine protease families. Nat Chem Biol. 2005;1:33–38. doi: 10.1038/nchembio707. [DOI] [PubMed] [Google Scholar]
- Klaiman G, Champagne N, LeBlanc AC. Self-activation of Caspase-6 in vitro and in vivo: Caspase-6 activation does not induce cell death in HEK293T cells. Biochim Biophys Acta. 2009;1793:592–601. doi: 10.1016/j.bbamcr.2008.12.004. [DOI] [PubMed] [Google Scholar]
- Klaiman G, Petzke TL, Hammond J, Leblanc AC. Targets of caspase-6 activity in human neurons and Alzheimer disease. Mol Cell Proteomics. 2008;7:1541–1555. doi: 10.1074/mcp.M800007-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mujumdar R, Ernst L, Mujumdar S, Lewis C, Waggoner A. Cyanine dye labeling reagents: Sulfoindocyanine succinimidyl esters. Bioconj Chem. 1993;4 doi: 10.1021/bc00020a001. [DOI] [PubMed] [Google Scholar]
- Pouladi MA, Graham RK, Karasinska JM, Xie Y, Santos RD, Petersen A, Hayden MR. Prevention of depressive behaviour in the YAC128 mouse model of Huntington disease by mutation at residue 586 of huntingtin. Brain. 2009;132:919–932. doi: 10.1093/brain/awp006. [DOI] [PubMed] [Google Scholar]
- Rao L, Perez D, White E. Lamin proteolysis facilitates nuclear events during apoptosis. J Cell Biol. 1996;135:1441–1455. doi: 10.1083/jcb.135.6.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruchaud S, Korfali N, Villa P, Kottke TJ, Dingwall C, Kaufmann SH, Earnshaw WC. Caspase-6 gene disruption reveals a requirement for lamin A cleavage in apoptotic chromatin condensation. Embo J. 2002;21:1967–1977. doi: 10.1093/emboj/21.8.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton KB, Kato D, Berger AB, Fonovic M, Verhelst SH, Bogyo M. Specificity of aza-peptide electrophile activity-based probes of caspases. Cell Death Differ. 2007;14:727–732. doi: 10.1038/sj.cdd.4402074. [DOI] [PubMed] [Google Scholar]
- Slee EA, Adrain C, Martin SJ. Executioner caspase-3, -6, and -7 perform distinct, non-redundant roles during the demolition phase of apoptosis. J Biol Chem. 2001;276:7320–7326. doi: 10.1074/jbc.M008363200. [DOI] [PubMed] [Google Scholar]
- Slee EA, Harte MT, Kluck RM, Wolf BB, Casiano CA, Newmeyer DD, Wang HG, Reed JC, Nicholson DW, Alnemri ES, et al. Ordering the cytochrome c-initiated caspase cascade: hierarchical activation of caspases-2, -3, -6, -7, -8, and -10 in a caspase-9-dependent manner. J Cell Biol. 1999;144:281–292. doi: 10.1083/jcb.144.2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stennicke HR, Deveraux QL, Humke EW, Reed JC, Dixit VM, Salvesen GS. Caspase-9 can be activated without proteolytic processing. J Biol Chem. 1999;274:8359–8362. doi: 10.1074/jbc.274.13.8359. [DOI] [PubMed] [Google Scholar]
- Takahashi A, Alnemri ES, Lazebnik YA, Fernandes-Alnemri T, Litwack G, Moir RD, Goldman RD, Poirier GG, Kaufmann SH, Earnshaw WC. Cleavage of lamin A by Mch2 alpha but not CPP32: multiple interleukin 1 beta-converting enzyme-related proteases with distinct substrate recognition properties are active in apoptosis. Proc Natl Acad Sci U S A. 1996;93:8395–8400. doi: 10.1073/pnas.93.16.8395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornberry NA, Rano TA, Peterson EP, Rasper DM, Timkey T, Garcia-Calvo M, Houtzager VM, Nordstrom PA, Roy S, Vaillancourt JP, et al. A combinatorial approach defines specificities of members of the caspase family and granzyme B. Functional relationships established for key mediators of apoptosis. J Biol Chem. 1997;272:17907–17911. doi: 10.1074/jbc.272.29.17907. [DOI] [PubMed] [Google Scholar]
- Vaidya S, Velazquez-Delgado EM, Abbruzzese G, Hardy JA. Substrate-induced conformational changes occur in all cleaved forms of caspase-6. J Mol Biol. 2011;406:75–91. doi: 10.1016/j.jmb.2010.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XJ, Cao Q, Liu X, Wang KT, Mi W, Zhang Y, Li LF, LeBlanc AC, Su XD. Crystal structures of human caspase 6 reveal a new mechanism for intramolecular cleavage self-activation. EMBO Rep. 2010;11:841–847. doi: 10.1038/embor.2010.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warby SC, Doty CN, Graham RK, Carroll JB, Yang YZ, Singaraja RR, Overall CM, Hayden MR. Activated caspase-6 and caspase-6-cleaved fragments of huntingtin specifically colocalize in the nucleus. Hum Mol Genet. 2008;17:2390–2404. doi: 10.1093/hmg/ddn139. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.