Abstract
A recent genome-wide association study identified a common variant (rs798766) on 4p16.3 that confers susceptibility to bladder cancer. The aim of this study was to assess whether rs798766 is associated with risk of bladder cancer in a Chinese population as well. We genotyped this variant using TaqMan technology in a case–control study of 815 histologically confirmed bladder cancer patients and 1141 controls. Normal bladder tissues adjacent to tumors were used to evaluate the functionality of rs798766 using quantitative real-time reverse transcription–polymerase chain reaction. We found that rs798766 CT/TT genotypes were associated with a significantly increased risk of bladder cancer (odds ratio = 1.36, 95% confidence interval = 1.10–1.67), compared with the CC genotype. Furthermore, rs798766 was significantly associated with FGFR3 messenger RNA expression. However, no significant interaction between rs798766 and tobacco smoking on bladder cancer risk was observed (Pmultiplicative = 0.785). Our results suggest that rs798766 on 4p16.3 may contribute to bladder cancer susceptibility in a Chinese population and explains an additional 3.65% of population attributable risk for bladder cancer.
Introduction
Worldwide, bladder cancer is the 7th and 17th most common cancer in men and women, respectively (1). The incidence of bladder cancer varies considerably among countries, with the highest incidence in the USA and Europe compared with Asian countries. Tobacco smoking is a predominant risk factor for bladder cancer, responsible for nearly half of the cases in men and for one-third in women, whereas occupational exposure to carcinogens is the second major risk factor (2). Although many people have been exposed to these risk factors, only a fraction of exposed individuals develop bladder cancer in their lifetime, suggesting that genetic susceptibility may also play a role in bladder carcinogenesis.
During the past few years, genome-wide association studies (GWAS) have identified a large number of robust associations between specific chromosomal loci and complex human diseases (3). For bladder cancer, three recent GWAS identified four common variants, rs9642880 and rs2294008 on chromosome 8q24, rs710521 on 3q28 and rs798766 on 4p16.3, that are associated with risk of bladder cancer in Caucasians (4,5,6). Previously, we performed a validation study of GWAS findings of rs9642880 and rs710521 for their association with bladder cancer risk in our ongoing case–control study, and found that rs9642880, but not rs710521, was significantly associated with an increased risk of bladder cancer (7). Furthermore, rs2294008 on PSCA was also suggested for bladder cancer susceptibility in a Chinese population (8). Recently, Kiemeney et al. (6) discovered an association between rs798766 at the TACC3-FGFR3 locus and increased risk and recurrence of bladder cancer. However, little or nothing is known about whether this significant association between rs798766 and bladder cancer risk exists in non-Caucasian populations.
In the present study, we validated GWAS findings of rs798766 on 4p16.3 associated with bladder cancer and estimated the contribution of rs798766 to bladder cancer risk in a Chinese population.
Materials and methods
Study subjects
This ongoing case–control study of bladder cancer was approved by the institutional review board of Nanjing Medical University and described previously (9). This study included 815 bladder cancer patients and 1141 cancer-free controls. Consecutive bladder cancer patients were recruited from January 2003 at The First Affiliated Hospital of Nanjing Medical University, Nanjing, China. All the bladder cancer cases had a histopathological diagnosis, and ∼95% of eligible patients contacted chose to participate. The cancer-free control subjects were genetically unrelated to the cases and had no individual history of cancer. All controls of appropriate age and sex for frequency matching with the cases were recruited and included if they gave their informed consent. Controls were excluded if they had symptoms suggestive of bladder cancer, such as hematuria. The response rate of control subjects was >85%. All subjects were interviewed, and a 5 ml venous blood sample was obtained from each.
Genotyping
Genomic DNA was isolated from leucocytes of venous blood by proteinase K digestion and phenol/chloroform extraction. Genotyping was performed with the TaqMan single nucleotide polymorphism Genotyping Assay using the 384-well ABI 7900HT Real Time PCR System (Applied Biosystems, Foster City, CA). The sequence of primers and probes for each single nucleotide polymorphism are available upon request. Controls were included in each plate to ensure accuracy of the genotyping. Genotype analysis was performed by two persons independently in a blinded fashion. About 10% of the samples were randomly selected for repeated genotyping for confirmation, and the results were 100% concordant.
Real-time analysis of expressions of nearby genes
To further detect the correlation between rs798766 and the expressions of nearby genes (TACC3, FGFR3, TMEM129 and SLBP), RNA from 40 normal samples of urothelial tissue adjacent to the tumors with different genotypes was extracted by using the Trizol Reagent (Invitrogen, Carlsbad, CA). Total RNA was measured by quantitative real-time reverse transcription–polymerase chain reaction. The primers used for amplification are shown in supplementary Table 1, available at Carcinogenesis Online. β-actin gene was used as an internal quantitative control, and each assay was done in triplicate.
Statistical analyses
Differences in the distributions of demographic characteristics, selected variables and frequencies of rs798766 genotypes in cases and controls were evaluated by using the Student's t-test (for continuous variables) or χ2-test (for categorical variables). The associations between rs798766 and risk of bladder cancer were estimated by computing odds ratios (ORs) and their 95% confidence intervals (CIs) from unconditional logistic regression analysis with the adjustment for possible confounders. Hardy–Weinberg equilibrium was tested using a goodness-of-fit χ2-test. Mann–Whitney test was used for analyzing the results of nearby genes messenger RNA expression. Tumor progression analyses were performed by comparing low-risk tumors versus high-risk tumors, as reported previously (6). Briefly, individuals with low-risk tumors were classified as having tumour node metastases stage pTa combined with WHO 1973 differentiation grade 1 or 2 or WHO/ISUP 2004 low grade, and all other tumors were defined as high-risk tumors. Population attributable risk (PAR%) was estimated for rs798766 as follows: PAR% = 100 × p(OR - 1)/[p(OR - 1) + 1], where p is the frequency of the allele among control subjects and the OR is the OR for the allele. A P < 0.05 was considered statistically significant, and all statistical tests were two sided. All the statistical analyses were performed with the software SAS 9.1 (SAS Institute, Cary, NC).
Results
The characteristics of the 815 bladder cancer cases and 1141 controls enrolled in this study are shown in Table I. There were no statistically significant differences between the cases and controls in terms of age and sex (P = 0.299 for age and 0.533 for sex). However, there were more smokers among the cases (53.5%) than among the controls (36.8%, P < 0.001). Of the 815 bladder cancer patients, 534 (65.5%) had low-risk tumors and 281 (34.5%) had high-risk tumors.
Table I.
Distribution of selected variables between the bladder cancer cases and control subjects
| Variables | Cases (n = 815) |
Controls (n = 1141) |
P | ||
| n | % | n | % | ||
| Age (mean ± SD) | 64.5 ± 12.6 | 65.0 ± 8.1 | 0.299 | ||
| Sex | |||||
| Male | 653 | 80.1 | 901 | 79.0 | 0.533 |
| Female | 162 | 19.9 | 240 | 21.0 | |
| Smoking status | |||||
| Never | 379 | 46.5 | 721 | 65.6 | <0.001 |
| Ever | 436 | 53.5 | 420 | 36.8 | |
| Former | 194 | 23.8 | 116 | 10.2 | |
| Current | 242 | 29.7 | 304 | 26.6 | |
| Pack-years of smoking | |||||
| 0 | 379 | 46.5 | 721 | 63.2 | <0.001 |
| ≤20 | 166 | 20.4 | 192 | 16.8 | |
| >20 | 270 | 33.1 | 228 | 20.0 | |
| Progression | |||||
| Low risk | 534 | 65.5 | |||
| High risk | 281 | 34.5 | |||
As shown in Table II, the frequencies of the CC, CT and TT genotypes were 71.8, 26.4 and 1.8%, respectively, among the cases and 77.8, 20.7 and 1.5%, respectively, among the controls (P = 0.009). The rs798766 T allele frequency was 0.150 among the cases and 0.118 among the controls, and the difference was statistically significant (P = 0.004). The observed genotype frequencies among the controls were in agreement with the Hardy–Weinberg equilibrium (P = 0.771). Logistic regression analysis revealed that the CT genotype, but not the TT genotype, was associated with a significantly increased risk of bladder cancer, compared with the CC genotype (OR = 1.36, 95% CI = 1.09–1.68 for CT versus CC and OR = 1.35, 95% CI = 0.66–2.75 for TT versus CC). Furthermore, a significant increased risk of bladder cancer was found in the combined genotypes CT/TT compared with the CC genotype (OR = 1.36, 95% CI = 1.10–1.67). The estimated PAR of rs798766 T allele was 3.65%. In the stratification of tumor progression for bladder cancer cases, no significant association was observed on rs798766 for low-risk tumors versus high-risk tumors (P = 0.509)(supplementary Table 2 is available at Carcinogenesis Online).
Table II.
Genotype and allele frequencies of rs798766 among the cases and controls and the associations with risk of bladder cancer
| rs798766 | Cases (n = 815) |
Controls (n = 1141) |
P | OR (95% CI)a | PARb (%) | ||
| n | % | n | % | ||||
| CC | 585 | 71.8 | 888 | 77.8 | 1.00 | ||
| CT | 215 | 26.4 | 236 | 20.7 | 0.006 | 1.36 (1.09–1.68) | 6.93 |
| TT | 15 | 1.8 | 17 | 1.5 | 0.411 | 1.35 (0.66–2.75) | 0.52 |
| CT/TT | 230 | 28.2 | 253 | 22.2 | 0.005 | 1.36 (1.10–1.67) | 7.38 |
| C allele | 1385 | 85.0 | 2012 | 88.2 | 1.00 | ||
| T allele | 245 | 15.0 | 270 | 11.8 | 0.004 | 1.32 (1.09–1.59) | 3.65 |
Adjusted for age, sex and pack-years of smoking in logistic regression model.
Population attributable risk.
We evaluated whether an interaction existed between rs798766 and tobacco smoking on bladder cancer risk. As shown in Table III, compared with non-smokers with CC genotype, a significantly increased risk for bladder cancer was observed among smokers with CC and CT/CT genotypes (OR = 2.13, 95% CI = 1.70–2.68 and OR = 2.98, 95% CI = 2.18–4.07, respectively). Nevertheless, no multiplicative interaction between rs798766 and smoking status was detected (P = 0.785).
Table III.
Interaction analyses of rs798766 and smoking status
| Smoking status | Genotypes | Cases |
Controls |
P | OR (95% CI)a | ||
| n | % | n | % | ||||
| Non-smokers | CC | 277 | 34.0 | 564 | 49.4 | 1.00 | |
| Non-smokers | CT/TT | 102 | 12.5 | 157 | 13.8 | 0.058 | 1.32 (0.99–1.76) |
| Smokers | CC | 308 | 37.8 | 324 | 28.4 | <0.001 | 2.13 (1.70–2.68) |
| Smokers | CC/CT | 128 | 15.7 | 96 | 8.4 | <0.001 | 2.98 (2.18–4.07) |
| P for interaction (multiplicative) | 0.785 | ||||||
Adjusted for age and sex in logistic regression model.
We assessed rs798766 in nearby genes expression in 40 bladder tissues using quantitative real-time reverse transcription–polymerase chain reaction. A significant association between rs798766 and FGFR3 expression was observed (P = 0.027) (Figure 1). However, no significant association with messenger RNA expression was identified for other genes examined in this study (P = 0.573 for TACC3, 0.919 for TMEM129 and 0.613 for SLBP).
Fig. 1.
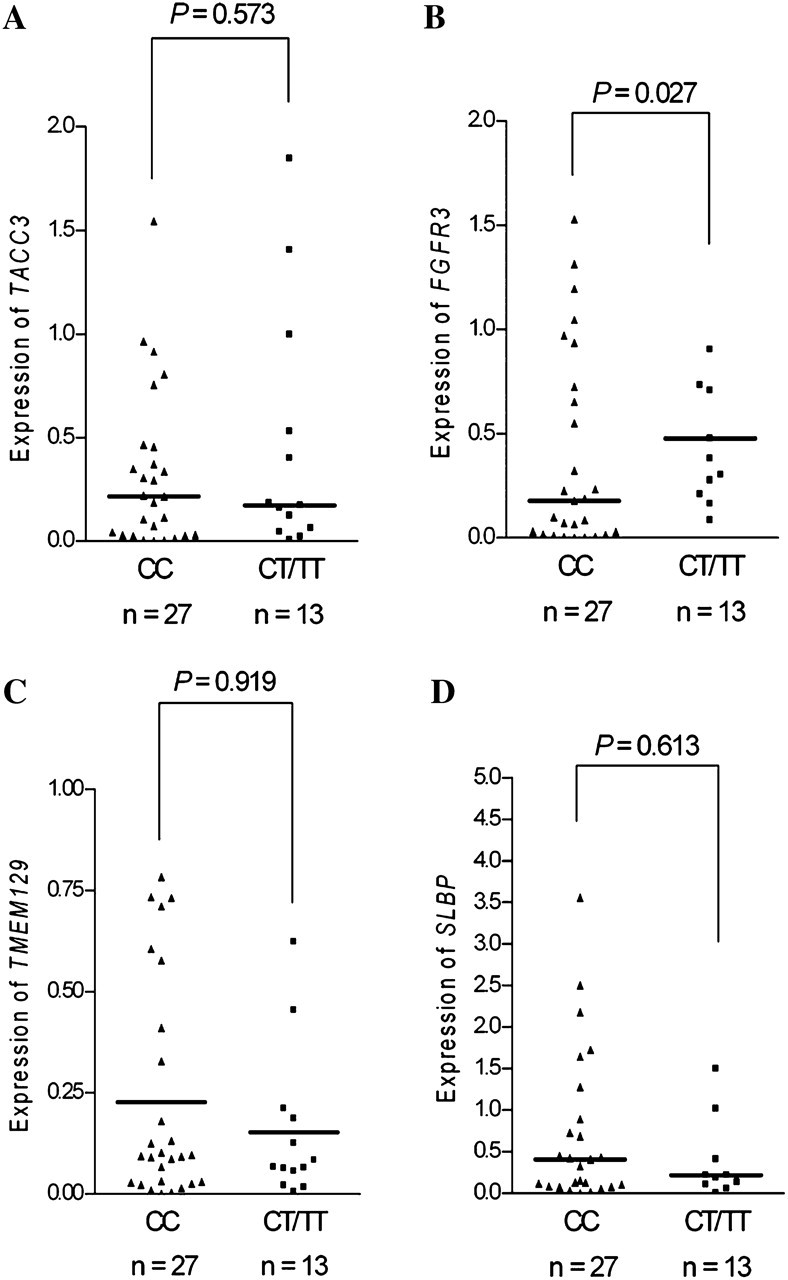
Association between rs798766 and messenger RNA expression of (A) TACC3, (B) FGFR3, (C) TMEM129, (D) SLBP. The transcript in normal urothelium tissues was detected by quantitative real-time reverse transcription–polymerase chain reaction. The frequency distributions of the CC, CT and TT genotypes were 27, 11 and 2, respectively. The fold change was normalized against β-actin.
Discussion
Recent GWAS have identified many genetic variants associated with bladder cancer risk (4,5,6). However, most studies were conducted in Caucasian populations. Because there are significant differences in the prevalence of bladder cancer and the frequencies of genetic variants among different ethnic populations (1), it is of importance to explore the effects of these variants in other diverse ethnic groups. Previously, we confirmed that GWAS findings of rs9642880 on 8q24, rs710521 on 3q28 and rs2294008 on PSCA are associated with risk of bladder cancer in a Chinese population. In the present study, we examined the association of rs798766 on 4q16.3 with risk of bladder cancer and found that this variant was significantly associated with bladder cancer risk in our study population. We have also provided in vivo evidence that rs798766 was associated with FGFR3 messenger RNA expression. These findings supports that the variant rs798766 on 4q16.3 may contribute to the etiology of bladder cancer carcinogenesis.
The locus rs798766 on 4q16.3 resides in an intronic region of TACC3, the transforming acidic coiled-coil-containing protein 3, which recently emerged as an important factor in mitotic spindles organization (10,11,12). Linkage disequilibrium analysis using HapMap showed that TACC3 shares a linkage disequilibrium block with two other genes, TMEM129 and SLBP. However, little is known about the involvement of these three genes in bladder cancer. In our study, we did not find any association of bladder cancer risk between rs798766 and the expression of these three genes. In contrast, a significant association between rs798766 and FGFR3 expression was observed for bladder cancer risk. FGFR3 encodes fibroblast growth factor receptor 3, which is adjacent to TACC3 and 70 kb away from rs798766. About 70% of low-grade non-muscle-invasive bladder cancer patients have a mutation in FGFR3 (13,14), and it was shown that patients with an FGFR3 mutation have a good prognosis (15,16,17). Therefore, it is biologically plausible that rs798766 could alter distant regulatory elements of FGFR3 expression. Kiemeney et al. (6) did not detect any significant association between FGFR3 expression and rs798766 in 27 low passage normal human urothelial cell strains, whereas a significant correlation between rs798766 and increased expression of FGFR3 in adipose tissues was observed, which was consistent with our findings in normal bladder tissues, suggesting that potential mechanism in different tissues for rs798766 and its association with the FGFR3 expression.
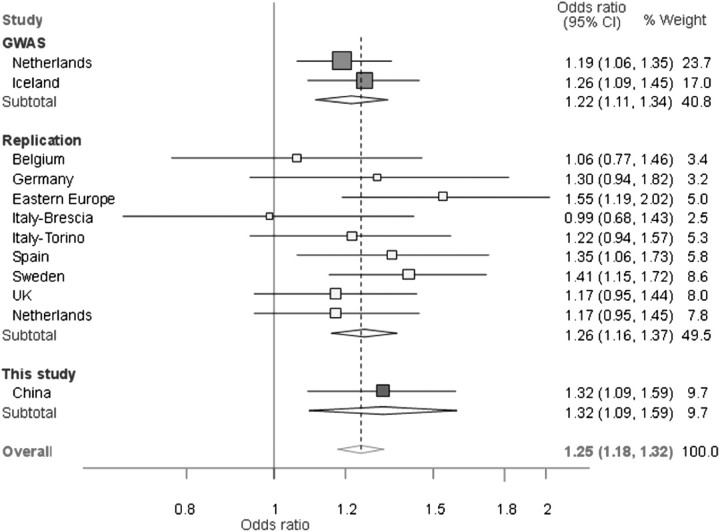
It has been postulated that tumors with different progression phenotype undergo different mechanisms of carcinogenesis (18). Kiemeney et al. (6) found that the frequency of rs798766 T allele was significantly higher in bladder cancer cases with low risk of progression than in those with high risk of progression. However, our stratified analysis of low-risk and high-risk tumors demonstrated that there was no significant difference between rs798766 and tumor progression. This inconsistent result may be due to our relatively small sample size; therefore, this finding should be interpreted with caution and validated in larger studies in Asian populations. Tobacco smoking was found to be a risk factor for bladder cancer (19). However, our results indicated that there was no multiplicative interaction between rs798766 and bladder cancer risk among smokers, which again may be due to the small sample size. In our present study, we replicated the results from the GWAS identified by Kiemeney et al. in a Chinese population and found the same results of rs798766 T allele acting as a risk allele for bladder cancer. Furthermore, when we performed a meta-analysis on the GWAS in Caucasians and our data in Asians, we found the estimated overall risk of bladder cancer with rs798766 T allele was 1.25 times that of non-carriers (OR = 1.25, 95% CI = 1.18–1.32) (Figure 2). In addition, we found that the minor allele frequency of rs798766 was varied across different ethnicities. The frequency of rs798766 T allele was 19.0% among Caucasian controls, which was higher than that in Asian controls (11.8%).
Fig. 2.
Forest plot of cancers risk associated with rs798766 (T allele versus C allele). The squares and horizontal lines correspond to the study-specific OR and 95% CI. The area of the squares reflects the study-specific weight (inverse of the variance). The diamond represents the pooled OR and 95% CI.
In conclusion, we validated that the chromosome 4p16.3 variant rs798766 was associated with bladder cancer risk in a Chinese population, which further supports that rs798766 may be a biomarker for genetic susceptibility in bladder cancer. Further investigation in larger populations and functional characterizations are needed to validate our findings.
Supplementary material
Supplementary Tables 1 and 2 can be found at http://carcin.oxfordjournals.org/
Funding
This study was partly supported by National Natural Science Foundation of China (30872084 and 30972444); National Natural Science Foundation of Jiangsu Province (2010080); the Key Program for Basic Research of Jiangsu Provincial Department of Education (08KJA330001); the Postdoctoral Science Foundation of China (20100481164) and ‘Qinglan Project’ Foundation for the Young Academic Leader of Jiangsu Province (Z.Z.).
Supplementary Material
Acknowledgments
We would like to thank Oluf D. Røe (Department of Cancer Research and Molecular Medicine, Norwegian University of Science and Technology, Trondheim, Norway) for scientific editing.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- CI
confidence interval
- GWAS
genome-wide association study
- OR
odds ratio
References
- 1.Kakehi Y, et al. Bladder Cancer Working Group report. Jpn. J. Clin. Oncol. 2010;40(suppl. 1):i57–i64. doi: 10.1093/jjco/hyq128. [DOI] [PubMed] [Google Scholar]
- 2.Murta-Nascimento C, et al. Epidemiology of urinary bladder cancer: from tumor development to patient's death. World J. Urol. 2007;25:285–295. doi: 10.1007/s00345-007-0168-5. [DOI] [PubMed] [Google Scholar]
- 3.Hardy J, et al. Genomewide association studies and human disease. N. Engl. J. Med. 2009;360:1759–1768. doi: 10.1056/NEJMra0808700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kiemeney LA, et al. Sequence variant on 8q24 confers susceptibility to urinary bladder cancer. Nat. Genet. 2008;40:1307–1312. doi: 10.1038/ng.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu X, et al. Genetic variation in the prostate stem cell antigen gene PSCA confers susceptibility to urinary bladder cancer. Nat. Genet. 2009;41:991–995. doi: 10.1038/ng.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kiemeney LA, et al. A sequence variant at 4p16.3 confers susceptibility to urinary bladder cancer. Nat. Genet. 2010;42:415–419. doi: 10.1038/ng.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang M, et al. Common genetic variants on 8q24 contribute to susceptibility to bladder cancer in a Chinese population. Carcinogenesis. 2009;30:991–996. doi: 10.1093/carcin/bgp091. [DOI] [PubMed] [Google Scholar]
- 8.Wang S, et al. Genetic variation in PSCA and bladder cancer susceptibility in a Chinese population. Carcinogenesis. 2010;31:621–624. doi: 10.1093/carcin/bgp323. [DOI] [PubMed] [Google Scholar]
- 9.Wang M, et al. A novel XPF -357A>C polymorphism predicts risk and recurrence of bladder cancer. Oncogene. 2010;29:1920–1928. doi: 10.1038/onc.2009.484. [DOI] [PubMed] [Google Scholar]
- 10.Kinoshita K, et al. Aurora A phosphorylation of TACC3/maskin is required for centrosome-dependent microtubule assembly in mitosis. J. Cell Biol. 2005;170:1047–1055. doi: 10.1083/jcb.200503023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pascreau G, et al. Phosphorylation of maskin by Aurora-A participates in the control of sequential protein synthesis during Xenopus laevis oocyte maturation. J. Biol. Chem. 2005;280:13415–13423. doi: 10.1074/jbc.M410584200. [DOI] [PubMed] [Google Scholar]
- 12.Peset I, et al. Function and regulation of Maskin, a TACC family protein, in microtubule growth during mitosis. J. Cell Biol. 2005;170:1057–1066. doi: 10.1083/jcb.200504037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cappellen D, et al. Frequent activating mutations of FGFR3 in human bladder and cervix carcinomas. Nat. Genet. 1999;23:18–20. doi: 10.1038/12615. [DOI] [PubMed] [Google Scholar]
- 14.Zuiverloon TC, et al. Fibroblast growth factor receptor 3 mutation analysis on voided urine for surveillance of patients with low-grade non-muscle-invasive bladder cancer. Clin. Cancer Res. 2010;16:3011–3018. doi: 10.1158/1078-0432.CCR-09-3013. [DOI] [PubMed] [Google Scholar]
- 15.Burger M, et al. Prediction of progression of non-muscle-invasive bladder cancer by WHO 1973 and 2004 grading and by FGFR3 mutation status: a prospective study. Eur. Urol. 2008;54:835–843. doi: 10.1016/j.eururo.2007.12.026. [DOI] [PubMed] [Google Scholar]
- 16.Hernandez S, et al. Prospective study of FGFR3 mutations as a prognostic factor in nonmuscle invasive urothelial bladder carcinomas. J. Clin. Oncol. 2006;24:3664–3671. doi: 10.1200/JCO.2005.05.1771. [DOI] [PubMed] [Google Scholar]
- 17.van Rhijn BW, et al. Molecular grading of urothelial cell carcinoma with fibroblast growth factor receptor 3 and MIB-1 is superior to pathologic grade for the prediction of clinical outcome. J. Clin. Oncol. 2003;21:1912–1921. doi: 10.1200/JCO.2003.05.073. [DOI] [PubMed] [Google Scholar]
- 18.Wu XR. Urothelial tumorigenesis: a tale of divergent pathways. Nat. Rev. Cancer. 2005;5:713–725. doi: 10.1038/nrc1697. [DOI] [PubMed] [Google Scholar]
- 19.Cohen SM, et al. Epidemiology and etiology of premalignant and malignant urothelial changes. Scand. J. Urol. Nephrol. Suppl. 2000;205:105–115. doi: 10.1080/00365590050509869. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.