Abstract
Exposure to ionizing radiation is a known risk factor for cancer. However, up to now, rigorously defined scientific criteria that could establish case-by-case the radiation-induced (RI) origin of a tumour have been lacking. To identify genes that could constitute a RI signature, we compared the transcriptome of 12 sarcomas arising in the irradiation field of a primary tumour following radiotherapy with the transcriptome of 12 sporadic sarcomas. This learning/training set contained four leiomyosarcomas, four osteosarcomas and four angiosarcomas in each subgroup. We identified a signature of 135 genes discriminating RI from sporadic sarcomas. The robustness of this signature was tested by the blind case-by-case classification of an independent set of 36 sarcomas of various histologies. Thirty-one sarcomas were classified as RI or sporadic; it was not possible to propose an aetiology for the five others. After the code break, it was found that one sporadic sarcoma was misclassified as RI. Thus, the signature is robust with a sensitivity of 96%, a positive and a negative predictive value of 96 and 100%, respectively and a specificity of 62%. The functions of the genes of the signature suggest that RI sarcomas were subject to chronic oxidative stress probably due to mitochondrial dysfunction.
Introduction
An association between the development of malignant neoplasm and exposure to ionizing radiation is now well established by epidemiologic investigations. All types of solid tumours are observed, with a prevalence of sarcomas and thyroid tumours (1). However, up to now, the lack of clearly established differences with tumours that develop in the absence of irradiation has prevented the identification of radiation-induced (RI) tumours using rigorously defined scientific criteria. Nevertheless, in a few situations, it has been possible to establish series of tumours for which a RI nature should be highly probable. Childhood exposure to radioactive fallout from the Chernobyl nuclear power explosion was associated with a strong increase in the incidence of papillary thyroid carcinoma in children and young adults (2). Molecular studies of these tumours did not disclose recurrent genome abnormalities specific to an effect of ionizing radiation (3–5). Transcriptome analysis also failed to define a signature of induction by radiation of post-Chernobyl tumours (6). However, the application of an empirical signature elaborated from previously published oxidation stress-specific signatures was able to roughly discriminate sporadic from post-Chernobyl tumours (7). In addition, the relative abundance of a few proteins made it possible to distinguish post-Chernobyl from sporadic papillary thyroid cancers, although this signature could be more relevant to the aggressiveness of the RI tumours than to their aetiology (8). Another well-defined situation corresponds to second tumours developing within the volume irradiated during previous radiotherapy. We have shown that the high frequency of short deletions observed in the mutation pattern of TP53 in a series of postradiotherapy sarcomas could be related to the introduction of DNA breaks by ionizing radiation (9). However, this mutational signature does not discriminate sporadic from RI sarcomas on a case-by-case basis. Recently, distinct gene expression profiles were observed for radiation-associated breast cancers developing after irradiation for Hodgkin’s lymphoma and sporadic breast cancers (10). However, no blind evaluation of the signature relevance was performed. One major problem encountered in postradiotherapy tumour studies is the shortness of the available series. Global transcriptome or genome studies are particularly affected by this problem since the methods used for data analysis are generally efficient only for large series. In order to solve this problem, we have initiated new strategies to develop methods of classification using transcriptome analysis for a case-by-case tumour diagnosis (11–13). Using these new approaches, the deregulated genes involved in RI tumorigenesis in rat bones were identified (12) and the specificities of adenosquamous lung carcinomas from adenocarcinomas and squamous cell carcinomas characterized (13) and on a series of postradiotherapy thyroid tumours (14). Here, we compared the transcriptome of sporadic sarcomas and postradiotherapy RI sarcomas of various histologies. We show that a signature of 135 genes distinguished the sporadic from the RI sarcomas with high efficiency. The detailed analysis of these genes suggests that chronic oxidative stress could be a hallmark of the RI sarcomas.
Materials and methods
Biological material
Thirty-five secondary sarcomas (RI sarcomas) developing in the field of irradiation of a primary cancer and 25 sarcomas from patients with no irradiation history (sporadic sarcomas: SP-sarcomas) were collected at the Biological Resources Centre of the Institut Curie. Medical and molecular data were previously published for secondary sarcomas up to case 36 (9,15). Data for the other RI sarcomas and the sporadic cases are available in supplementary Table 1, available at Carcinogenesis Online. Radiotherapy was administered by photon or electron beam therapy. Pathological diagnosis was performed according to WHO guidelines. All tumours were of grade II or III. Tumours were diagnosed as RI according to the Cahan criteria (16). He defined three criteria to classify a sarcoma as RI: a formation in the irradiation field of a radiotherapy, a histology clearly different of the primary cancer and a delay of 5 years between the primary and the second cancer. Some patients who developed RI sarcomas were treated by chemotherapy for their primary tumour in addition to the radiotherapy (supplementary Table 2 is available at Carcinogenesis Online). All samples were obtained before treatment of sarcomas. The study was approved by the local ethics committees and informed consent was obtained from all patients.
RNA extraction, labelling and hybridization
RNA samples were prepared and hybridized as described (9). All samples were hybridized on human 25K 50-52mer oligo-microarrays [Resogen Program, RNG/MCR, Evry, (17)]. Each tumour sample was co-hybridized with a common pool of tumour cell lines (T47D, A549, MCF7 and Boleth) used as reference, and all hybridizations were duplicated in dye swap.
Microarray analysis
After hybridization, each spot was defined automatically using image analysis spot-tracking software (US patent 10/173,672; CA 2,389,901). Fluorescence intensity values for both dyes (Alexa Fluor 555 and Alexa Fluor 647), and local background-subtracted values for individual spots, were obtained using an expectation-maximization algorithm (US patent 10/173 672; CA 2,389,901). After microarray hybridization, the hybridization signals were acquired and normalized, and the expression ratios were calculated as described previously (11). Microarray data were entered in the ArrayExpress database (accession: E-MEXP-2687).
Data analysis
The 60 sarcomas analysed in this study were divided into two sets: a learning/training set of known aetiology, comprising 12 RI- and 12 SP-sarcomas, with four leiomyosarcomas, four osteosarcomas and four angiosarcomas in each subgroup of aetiology (supplementary Table 2 is available at Carcinogenesis Online), and a testing set that comprised the remaining 36 sarcomas of various histologies, for which the aetiology was not known by experimenters before the end of the data analysis (supplementary Table 3 is available at Carcinogenesis Online).
Most methods used to analyse microarray data want to identify groups of genes that have coherent patterns of expression with large variance across groups of samples. Unfortunately, using these methods, we did not find any signature of aetiology. Since gene shaving is a useful alternative method, but not adapted to small series of samples, we have developed an approach based on a similar strategy but adapted to limited number of cases. It finds, if they exist, criteria discriminating two groups of tumours. The method that is detailed in supplementary Data 1, available at Carcinogenesis Online comprised the following four steps:
A learning step to select sets of candidate genes whose expression discriminates between the two aetiology subgroups.
A training step to select from the sets of candidate genes those with the highest potential to classify training tumours correctly.
The compilation of a unique discriminating set of genes and standardization of their expressions according to gene expression variability in the subgroups.
The blind classification, case-by-case, of testing tumours.
In addition, we used a decision-making tool based on calculation of the root mean square (RMS) to attribute each tumour to either the SP or RI group.
Learning step
The whole process for identifying the signature of aetiology was focused on the search for differentially expressed genes permitting to distinguish SP versus RI tumours, independently of tumour histology. For that, we searched for genes with a common level of expression within each group and with a difference in expression when comparing the two groups. Matrices of all combinations of 5 of the 12 sporadic cases and of 5 of the 12 RI cases were built from the learning/training set of tumours (combinatorial 10 tumour matrices), the 14 remaining tumours of each combinatorial matrix being used as training tumours. To avoid bias due to histology, each half matrix (5 sporadic or RI tumours) should include at least one tumour of each histology. Finally, we obtained ninety-eight 10 tumour-training matrices, which are composed on average of 386 genes with a standard deviation of 514 genes.
Training step
Each of the 98 training matrices was used to classify the 14 remaining tumours. Rules to select a matrix were applied so that at least one of the training tumours was correctly classified and none was misclassified, otherwise the matrix was discarded. The process continued if at least 90% of the training tumour classifications were validated by at least one of the training matrices. In these conditions, 10% of the tumours may not be validated, but none must be rejected by the selected training matrices. The training step selected 26 matrices containing 1074 genes able to classify at least one training tumour, without false classification.
Compilation of a unique discriminating set of genes and standardization of their expressions according to gene expression variability in the subgroups
The search for a final unique set of genes (signature) needed a standardization step to attenuate gene expression heterogeneity. The 26 selected training matrices are compiled in one matrix. Among the 1074 genes belonging to this matrix, we finally selected for the final signature 135 genes that were retained after applying a cut-off of 70% to the frequency of relevance, which is defined as the frequency at which a given gene and a given tumour are found together in a selected training matrix, weighted by the number of training tumours correctly classified by this training matrix, plus twice the standard deviation. Beside the signature, we also applied a less stringent criterion (cut-off of 33% for the frequency of relevance) for gene selection, given an additional list of 282 genes to have a better overview of potentially dysregulated pathways in RI tumours.
Blind classification, case-by-case, of testing tumours.
The relevance of the final signature was tested by blind classification of the 36 testing tumours.
Decision-making tool based on calculation of the RMS
In the classic approach by multidimensional scaling, the two or three most informative dimensions are used to visualize and define the relative positions of the tumours in a two- or three-dimensional space. However, in our case, the positioning of the tumours necessitated more than three dimensions. Since it is no longer possible to visualize the positioning of the tumour using multidimensional scaling, we used a decision-making tool based on calculation of the RMS (the RMS of a set of values is the square root of the arithmetic mean of the squares of the values, see supplementary Data 1, available at Carcinogenesis Online). During the learning/training step, the learning tumours were projected in a two-dimensional space (classification space), thus defining the aetiology subgroups RI and SP spaces. The RMS of each validation tumour for the barycentres of RI (RMSRI) and SP (RMSSP) subgroups was then calculated. The efficient discrimination between the two subgroups was validated if no tumour of a subgroup had an RMS greater than the RMS of one tumour of the other subgroup. The training tumours were then projected in the classification space. In order to assign a tumour to the aetiological subgroup RI or SP during the blind classification step, the RMS of the tumour must be lower that at least one RMS of the considered subgroup SP or RI, taking into account the RMS variance. In practise, the aetiology of a tumour could be predicted if the difference between the RMSRI and the RMSSP was >10%.
Gene ontology and functional analysis
The 135 genes of the signature, the additional list of 282 genes and all probes present on the microarray were categorized by using Gene Ontology (http://www.geneontology.org/). Significant deregulated biological processes in RI compared with SP tumours were checked by means of the chi-square test.
Results
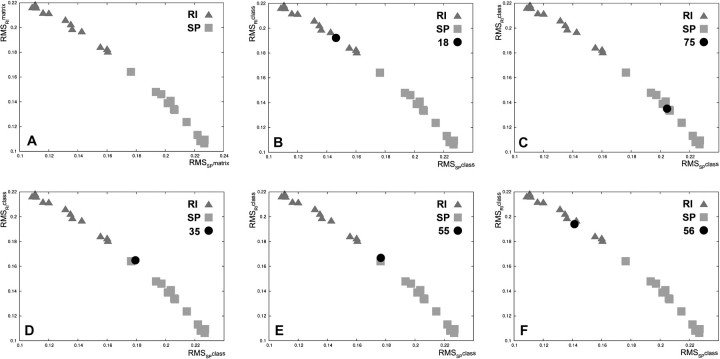
We analysed the different histological subtypes of sarcomas, either sporadic or RI. A learning/training set of sarcomas of known aetiology (12 RI and 12 SP) comprising four leiomyosarcomas, four osteosarcomas and four angiosarcomas in each subgroup was used (supplementary Table 2 is available at Carcinogenesis Online). After standardization steps, necessary to attenuate gene expression heterogeneity of the tumours, a set of 135 genes (63 downregulated and 72 upregulated) was selected, with the greatest frequencies of relevance, to define a single stable signature used to further blindly classify the remaining sarcomas (genes with log ratio, P-values and frequency of relevance are listed in the supplementary Table 4, available at Carcinogenesis Online). As shown in Figure 1A, this signature efficiently discriminates between the two subgroups of RI- and SP-sarcomas in the validation space.
Fig. 1.
(A) Discrimination of the two subgroups of RI- and sporadic sarcomas of the learning tumours. Scatter plot of the RMSRImatrix as a function of the RMSSPmatrix [RMSmatrix is the RMS between each learning tumour and the barycentre of all RI-learning tumours (RMSRmatrix) or the barycentre of all SP-learning tumours (RMSSmatrix)]. (B–F) Examples of testing sarcomas classification. Each graph illustrates the positioning of a given testing tumour (RMSRIclass and RMSSPclass) [RMSRIclass is the RMS between a given testing tumour and the barycentre of all RI-learning tumours (RMSRclass) or the barycenter barycentre of all SP-learning tumours (RMSSclass)] in the scatter plot of the learning sarcomas. Triangle: RI sarcoma; square: sporadic sarcoma (SP); circle: testing sarcoma. (B) one example of correctly classified RI-sarcoma (case 18); (C) one example of correctly classified SP-sarcoma (case 75); (D–E) outlier classification of the two sarcomas (cases 35 and 55), the aetiology of the tumours could not be predicted; (F) mis-classification of SP-sarcoma 56 in the RI subgroup. SP, sporadic sarcoma.
To blindly validate this molecular signature, each of the 36 testing sarcomas was projected into the validation space, allowing us to propose an aetiology depending on the sarcoma’s position relative to the tumours of the learning group. Thirty-one of the 36 sarcomas were classified in the RI or SP subgroup (Figure 1B and C; supplementary Table 5 and Figure 1 are available at Carcinogenesis Online). The five remaining tumours were positioned between the two subgroups (Figure 1D and E; supplementary Table 5 and Figure 1 are available at Carcinogenesis Online), and thus, it was not possible to propose any aetiology for these tumours. After the codebreak, of the 31 classified sarcomas, one SP was found to be misclassified as RI (Figure 1F, supplementary Table 5 and Figure 1 are available at Carcinogenesis Online). Overall, the classification presents a sensitivity of 96%, a specificity of 62% with a positive predictive value of 96% and a negative predictive value of 100% calculated from the summary tumour classification (Table I).
Table I.
Classifier performances according to the blind classification of the testing tumours
| Clinical aetiology |
Predicted aetiology |
|||
| RI | SP | RI | SP | Not classified |
| 23 | 22 | 0 | 1 | |
| 13 | 1 | 8 | 4 | |
Clinical aetiology: aetiology determined on the basis of the medical records, sarcoma developed in patients with (radiation-induced, RI) or without (sporadic, SP) irradiation history. Predicted aetiology: aetiology predicted using the transcriptome signature. Sensitivity 96%: proportion of RI sarcomas correctly classified among RI sarcomas. Specificity 62%: proportion of SP sarcomas correctly classified among SP sarcomas. Positive predictive value 96%: proportion of sarcomas with a positive test (RI) and correctly classified. Negative predictive value 100%: proportion of sarcomas with negative test (SP) and correctly classified.
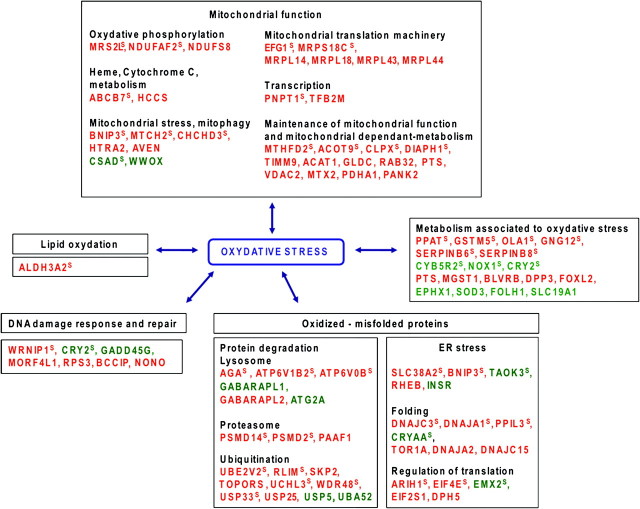
To define if biological functions or pathways are specifically deregulated in RI compared with SP sarcomas, we grouped the 135 genes of the signature as a function of the literature data and Gene Ontology pathways. Moreover, to get a better overview of RI-specific deregulation, we also perform this clustering on an additional series of 282 genes selected with less stringent criteria (supplementary Table 4 is available at Carcinogenesis Online). Remarkably, the only function found to be significantly deregulated in both lists, as compared with the clustering of all genes spotted on the microarray, was mitochondria (supplementary Table 6 is available at Carcinogenesis Online).
Discussion
The constitution of series of RI and sporadic tumours supposes that it is possible to define criteria able to discriminate the tumours as function of their aetiology. Here, we compare sarcomas developed in the irradiation field after radiotherapy with sarcomas that appeared in patients without any irradiation history. Sarcomas are rare tumours which accounts for roughly 1% of all cancers and the average annual incidence is estimated to be 6/100 000 inhabitants (18). Sarcomas are overrepresented in the RI cancers and, in the irradiation field of a radiotherapy, the prevalence of secondary sarcomas is 0.14–0.20% (19). The frequencies of the different histological subtypes are similar in RI and sporadic tumours, except for a lower percentage of RI liposarcomas. Moreover, no specific pathological features are observed between RI and sporadic sarcomas (20,21). In 1948, Cahan defined three criteria to classify a sarcoma as RI: formation in the irradiation field, histology clearly different from the primary cancer and an interval of at least 5 years between the primary and the secondary cancer (16). Secondary sarcomas developed in the irradiation field included in this study strictly met these criteria, and thus have a high probability of being RI.
Here, we compare the transcriptome of RI sarcomas selected according to these criteria with sarcomas that developed in patients without a radiation history. Using a learning/training set of 12 RI- and 12 SP sarcomas, we defined a signature of 135 genes, which efficiently discriminates these tumours as a function of their aetiology (Figure 1A). This signature was blindly validated using a set of 36 other sarcomas. The predicted aetiology was correct for 30 of the 36 tumours (Figure 1B–E; supplementary Table 5 and Figure 1 are available at Carcinogenesis Online). One of 23 RI-sarcomas (4.3%) and 4 of 13 SP-sarcomas (30%) were not classified by the signature (they were positioned between the two subgroups) and one case was misclassified since the tumour was deemed as RI but developed in a patient with no history of irradiation (Figure 1F; supplementary Table 5 and Figure 1 are available at Carcinogenesis Online). Thus, using the present method of tumour classification, the prediction of the aetiology of the sarcomas as either RI or SP was very robust (sensitivity of 96%, positive predictive value of 96% and a negative predictive value of 100%). However, a few sarcomas could not be classified which explains the rather low specificity (62%) (Table I). Indeed, to find a signature of tumour aetiology independent of tumour histology, we pay special attention in the distribution of tumour histology in the learning/training matrix (see Materials and Methods and supplementary Figure 2, available at Carcinogenesis Online). The whole process for identifying the signature of aetiology was focused on the search for differentially expressed genes that distinguish SP versus RI tumours, independently of tumour histology and that have a common level of expression within each aetiology group. For this purpose, at the learning/training step, the combinatorial 10 tumour matrices were built such that each half matrix (five sporadic or five RI tumours) included at least one tumour of each histology to avoid any impact of the histology on the gene selection. Thus, after the normalization step, whatever the tumour histology within each subgroup, the principal information included in each gene profile is relative to the difference of expression of RI versus SP. Moreover, for the final 135-gene signature, we kept only the genes with the greatest frequencies of relevance again strongly minimizing the likehood that the RI signature is affected by the histology. Finally, even if some genuine radiation-related genes may be associated with histology, the success of this exploration is proved by the fact that the 135 gene signature is able blindly to classify 30 of 36 sarcomas correctly according to the RI and SP criteria and independently of tumour histology. It should be noted that signatures of aetiology were also independently obtained within each histological subgroup of sarcomas, but the low number of available tumours prevented us from testing their robustness by blind validation (data not shown). Our series of tumours are also heterogeneous in terms of TP53 status and adjuvant treatment of patients (see supplementary Table 2 and Figure 2 are available at Carcinogenesis Online). So, as for histology, it was of major importance to check the potential influence of these parameters on the present signature of tumour aetiology. Four RI sarcomas with a biallelic inactivation of TP53 were included in the learning/training set together with 20 sarcomas with at least one active allele (supplementary Table 2, available at Carcinogenesis Online). The use of the combinatorial 10 tumour matrix avoided any impact of TP53 on the signature. Half of the RI-sarcomas developed in patients who received chemotherapy for the treatment of their primary tumours (supplementary Table 2 is available at Carcinogenesis Online) and because of the combinatorial 10 tumour matrix strategy, we can exclude a major role of the chemotherapy in the selection of the genes belonging to the signature.
Detailed analysis of the genes of the signature and of the additional series of genes selected with less stringent criteria indicated that several mitochondrial functions are differentially deregulated, generally by upregulation of the genes in the RI-sarcomas as compared with SP-sarcomas (Figure 2 and Table I). The function of these genes may suggest that mitochondria were subject to oxidative stress related to an overproduction of reactive oxygen species (Figure 2 and Table I; analysis of dysregulated gene functions is presented in supplementary Data 2, available at Carcinogenesis Online). In parallel, the analysis of the other deregulated genes may indicate a more general oxidative stress since several other genes involved in detoxification or antioxidant functions are also deregulated (Figure 2 and Table I). Defence mechanisms including protein degradation, specifically ubiquitination–deubiquitination and proteasome pathways (22,23), lipid turnover and DNA repair sustain cellular functions by repairing or removing the oxidized macromolecules. Several genes of these pathways, including response to endoplasmic reticulum stress, are deregulated suggesting an excess of abnormal proteins and biomolecules in RI-sarcomas. Overall, it seems that the fingerprint of the ionizing radiation in the RI-sarcomas could be a mitochondrial dysfunction together with a chronic oxidative stress hallmark. Enzymes important in reactive oxygen species detoxification, such as catalase, glutathione peroxidase, thioredoxin reductase and glutathione reductase, were not differentially expressed in RI-sarcomas as compared with SP-sarcomas. These genes are known to be involved in the acute oxidative stress response, for example in cells treated with H2O2. In the RI-sarcomas, cells were selected and adapted to survive a probably low chronic oxidative stress that probably involved genes distinct from those known to respond to acute oxidative stress. The origin of the mitochondrial alterations remains a question open to speculation. They could be distant consequences of the RI carcinogenesis, although a known direct biological effect of ionizing radiation is associated with mitochondria alterations. Radiation increase in reactive oxygen species generation or oxidative stress or both were observed in vivo within several days after exposure in rodent models (24,25). Moreover, RI oxidative stress has been confirmed clinically in lung cancer irradiated patients (26,27). Interestingly, oxidative stress has been recurrently associated with the persistent transgenerational mutagenic effects called non-targeted or bystander effects of ionizing radiation (28). The number of surviving cells exhibiting increased delayed mutation frequencies, following irradiation, is greater than would be predicted considering the cells directly exposed to radiation. The progeny of the bystander cells is characterized by genomic instability linked to oxidative stress due to dysfunction of the mitochondrial electron transport chain complexes (29,30). It has been proposed that this oxidative stress-induced genomic instability participates in the development of tumours, a hypothesis fully compatible with our results.
Fig. 2.
Deregulated genes related to oxidative stress in RI-sarcomas as compared with SP-sarcomas. Red: up-regulated in RI-sarcomas, green: downregulated in RI-sarcomas. The genes of the signature, corresponding to the genes with the greatest frequencies of relevance for the classification of the training tumours, are noted with a superscript S. The other genes belong to the additional list obtained using less stringent criteria. The names of the genes are listed in Table II.
Table II.
Names of the genes related to oxidative stress presented in Figure 2
| Symbol | Name | Symbol | Name |
| ABCB7 | ATP-binding cassette, subfamily B (MDR/TAP), member 7 | MRPL43 | Mitochondrial ribosomal protein L43 |
| ACAT1 | Acetyl-Coenzyme A acetyltransferase 1 | MRPL44 | Mitochondrial ribosomal protein L44 |
| ACOT9 | Acyl-CoA thioesterase | MRPS18C | Mitochondrial ribosomal proteinS18C |
| AGA | Aspartylglucosaminidase | MRS2L | MRS2 magnesium homeostasis factor homolog |
| ALDH3A2 | Aldehyde dehydrogenase 3 family, member A2 | MTCH2 | Mitochondrial carrier homolog 2 |
| ARIH1 | Ariadne ubiquitin-conjugating enzyme E2 binding protein, 1 | MTHFD2 | Mitochondrial carrier homolog 2 |
| ATG2A | Autophagy related 2 homolog A | MTX2 | Metaxin |
| ATP6V0B | ATPase, H+ transporting, lysosomal 21kDa, V0 subunit b | NDUFAF2 | NADH dehydrogenase 1 alpha, assembly factor 2 |
| ATP6V1B2 | ATPase, H+ transporting, lysosomal 56/58kDa, V1 subunit B2 | NDUFS8 | NADH dehydrogenase (ubiquinone) Fe-S protein 8 |
| AVEN | Apoptosis, caspase activation inhibitor | NONO | non-POU domain containing, octamer-binding |
| BCCIP | BRCA2 and CDKN1A interacting protein | NOX1 | NADPH oxidase 1 |
| BLVRB | Biliverdin reductase B (flavin reductase (NADPH)) | OLA1 | Obg-like ATPase 1 |
| BNIP3 | BCL2/adenovirus E1B 19kDa interacting protein 3 | PAAF1 | Proteasomal ATPase-associated factor 1 |
| CHCHD3 | Coiled-coil-helix-coiled-coil-helix domain containing 3 | PANK2 | Pantothenate kinase |
| CLPX | ClpX caseinolytic peptidase X homolog (Escherichia coli) | PDHA1 | Pyruvate dehydrogenase alpha 1, variant 1 |
| CRY2 | Cryptochrome 2 (photolyase-like) | PNPT1 | Polyribonucleotide nucleotidyltransferase 1 |
| CRYAA | Crystallin, alpha A | PPAT | Phosphoribosyl pyrophosphate amidotransferase |
| CSDA | Cold shock domain protein A | PPIL3 | Peptidylprolyl isomerase (cyclophilin)-like 3 |
| CYB5R2 | Cytochrome b5 reductase 2 | PSMD14 | Proteasome 26S subunit, non-ATPase, 14 |
| DIAPH1 | Diaphanous homolog 1 (Drosophila) | PSMD2 | Proteasome 26S subunit, non-ATPase, 2 |
| DNAJA1 | DnaJ (Hsp40) homolog, subfamily A, member 1 | PTS | 6-pyruvoyltetrahydropterin synthase |
| DNAJA2 | DnaJ (Hsp40) homolog, subfamily A, member 2 | RAB32 | RAB32, member RAS oncogene family |
| DNAJC15 | DnaJ (Hsp40) homolog, subfamily C, member 15 | RHEB | Ras homolog enriched in brain |
| DNAJC3 | DnaJ (Hsp40) homolog, subfamily C, member 3 | RLIM | Ring finger protein, LIM domain interacting |
| DPH5 | DPH5 homolog (Saccharomyces. cerevisiae) | RPS3 | Ribosomal protein S3 |
| DPP3 | Dipeptidyl-peptidase 3 | SERPINB6 | Serpin peptidase inhibitor, clade B (ovalbumin), member 6 |
| EFG1 | EFG1 Elongation Factor G, Mitochondrial 1 | SERPINB8 | Serpin peptidase inhibitor, clade B (ovalbumin), member 8 |
| EIF2S1 | Eukaryotic translation initiation factor 2, subunit 1 alpha | SKP2 | S-phase kinase-associated protein 2 (p45) |
| EIF4E | Eukaryotic translation initiation factor 4E | SLC19A1 | Solute carrier family 19 (folate transporter), member 1 |
| EMX2 | Empty spiracles homeobox 2 | SLC38A2 | Solute carrier family 38, member 2 |
| EPHX1 | Epoxide hydrolase 1, microsomal (xenobiotic) | SOD3 | Superoxide dismutase 3, extracellular |
| FOLH1 | Folate hydrolase 1 | TAOK3 | TAO kinase 3 |
| FOXL2 | Forkhead box L2 | TFB2M | Transcription factor B2, mitochondrial |
| GABARAPL1 | GABA(A) receptor-associated protein like 1 | TIMM9 | Translocase of inner mitochondrial membrane 9 homolog |
| GABARAPL2 | GABA(A) receptor-associated protein-like 2 | TOPORS | Topoisomerase I binding, arginine/serine-rich |
| GADD45G | Growth arrest and DNA-damage-inducible, gamma | TOR1A | Torsin family 1, member A (torsin A) |
| GLDC | Glycine dehydrogenase (decarboxylating) | UBA52 | Ubiquitin A-52 residue ribosomal protein fusion product 1 |
| GNG12 | Guanine nucleotide binding protein (G protein), gamma 12 | UBE2V2 | Ubiquitin-conjugating enzyme E2 variant 2 |
| GSTM5 | Glutathione S-transferase mu 5 | UCHL3 | Ubiquitin carboxyl-terminal esterase L3 |
| HCCS | Holocytochrome c synthase (cytochrome c heme-lyase) | USP25 | Ubiquitin specific peptidase 25 |
| HTRA2 | HtrA serine peptidase 2 | USP33 | Ubiquitin specific peptidase 33 |
| INSR | Insulin receptor | USP5 | Ubiquitin specific peptidase 5 (isopeptidase T) |
| MGST1 | Microsomal glutathione S-transferase 1 | VDAC2 | Voltage-dependent anion channel 2 |
| MORF4L1 | Mortality factor 4 like 1 | WDR48 | WD repeat domain 48 |
| MRPL14 | Mitochondrial ribosomal protein L14 | WRNIP1 | Werner helicase interacting protein1 |
| MRPL18 | Mitochondrial ribosomal protein L18 | WWOX | WW domain containing oxidoreductase |
The symbols of the genes of the signature are underlined; the other genes come from the additional list (see supplementary Table 4, available at Carcinogenesis Online).
The paucity of published data on the transcriptome specificities of RI tumours limits comparisons. For radiation-associated breast cancers (10), no pathway was specifically deregulated as a function of aetiology and we did not find any overlap with the present signature of the RI-sarcomas. The involvement of chronic oxidative stress was also suspected in a series of post-Chernobyl and postradiotherapy thyroid tumours (7,14). However, the pathways were poorly characterized. Upregulation of oxidoreductases was observed in another series of post-Chernobyl thyroid cancers, but the authors associated this characteristic with the aggressiveness of the tumours rather than directly with their RI origin (31). In the present study, all sarcomas were of high grade and the oxidative stress could not be associated with a higher aggressiveness of the RI-sarcomas. Thus, even if deregulation of the redox process also seems to be involved in post-Chernobyl thyroid cancers, similarity with RI-sarcomas is not yet established.
In conclusion, we have blindly validated a transcriptome signature able to diagnose with high efficiency the RI origin of postradiotherapy sarcomas of various histologies. This signature suggested that the RI sarcomas are characterized by chronic endogenous oxidative stress probably generated by mitochondrial dysfunction. More experimental data, analysed with similar approaches, will be necessary to understand whether part of the present RI signature may constitute a general signature of ionizing RI carcinogenesis of if it is specific to RI sarcomas.
Supplementary material
Supplementary Tables 1–6, Figures 1 and 2 and Data 1 and 2 can be found at http://carcin.oxfordjournals.org/
Funding
Institut Curie ‘Programme Incitatif et Coopératif Retinoblastome: transcriptome et cibles thérapeutiques’; Electricité de France: ‘Signature fonctionnelle de la radio-induction dans les cancers humains’ (RB 2009-02 and CEA DSV RB 2010-10) and the European Union 6th Framework GenRiskT.
Supplementary Material
Acknowledgments
N.-S.H.-H. and N.B.-L. were fellows of the Ministère de l’éducation Nationale et de la Recherche and of the Association pour la Recherche contre le Cancer (ARC).
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- RMS
root mean square
- RI
radiation-induced
References
- 1.Suit H, et al. Secondary carcinogenesis in patients treated with radiation: a review of data on radiation-induced cancers in human, non-human primate, canine and rodent subjects. Radiat. Res. 2007;167:12–42. doi: 10.1667/RR0527.1. [DOI] [PubMed] [Google Scholar]
- 2.Williams ED. Chernobyl and thyroid cancer. J. Surg. Oncol. 2006;94:670–677. doi: 10.1002/jso.20699. [DOI] [PubMed] [Google Scholar]
- 3.Williams ED, et al. Thyroid carcinoma after Chernobyl latent period, morphology and aggressiveness. Br. J. Cancer. 2004;90:2219–2224. doi: 10.1038/sj.bjc.6601860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Powell N, et al. Frequency of BRAF T1796A mutation in papillary thyroid carcinoma relates to age of patient at diagnosis and not to radiation exposure. J. Pathol. 2005;205:558–564. doi: 10.1002/path.1736. [DOI] [PubMed] [Google Scholar]
- 5.Kimmel RR, et al. Microarray comparative genomic hybridization reveals genome-wide patterns of DNA gains and losses in post-Chernobyl thyroid cancer. Radiat. Res. 2006;166:519–531. doi: 10.1667/RR0547.1. [DOI] [PubMed] [Google Scholar]
- 6.Detours V, et al. Absence of a specific radiation signature in post-Chernobyl thyroid cancers. Br. J. Cancer. 2005;92:1545–1552. doi: 10.1038/sj.bjc.6602521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Detours V, et al. Genome-wide gene expression profiling suggests distinct radiation susceptibilities in sporadic and post-Chernobyl papillary thyroid cancers. Br. J. Cancer. 2007;97:818–825. doi: 10.1038/sj.bjc.6603938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boltze C, et al. Sporadic and radiation-associated papillary thyroid cancers can be distinguished using routine immunohistochemistry. Oncol. Rep. 2009;22:459–467. doi: 10.3892/or_00000457. [DOI] [PubMed] [Google Scholar]
- 9.Gonin-Laurent N, et al. Specific TP53 mutation pattern in radiation-induced sarcomas. Carcinogenesis. 2006;27:1266–1272. doi: 10.1093/carcin/bgi356. [DOI] [PubMed] [Google Scholar]
- 10.Broeks A, et al. Radiation-associated breast tumors display a distinct gene expression profile. Int. J. Radiat. Oncol. Biol. Phys. 2010;76:540–547. doi: 10.1016/j.ijrobp.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 11.Chevillard S, et al. Gene expression profiling of differentiated thyroid neoplasms: diagnostic and clinical implications. Clin. Cancer Res. 2004;10:6586–6597. doi: 10.1158/1078-0432.CCR-04-0053. [DOI] [PubMed] [Google Scholar]
- 12.Daino K, et al. Silencing of Cited2 and Akap12 genes in radiation-induced rat osteosarcomas. Biochem. Biophys. Res. Commun. 2009;390:654–658. doi: 10.1016/j.bbrc.2009.10.022. [DOI] [PubMed] [Google Scholar]
- 13.Bastide K, et al. Are adenosquamous lung carcinomas a simple mix of adenocarcinomas and squamous cell carcinomas, or more complex at the molecular level? Lung Cancer. 2009;63:348–353. doi: 10.1016/j.lungcan.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 14.Ory C, et al. Gene expression signature discriminates sporadic from post-radiotherapy-induced thyroid tumors. Endocr. Relat. Cancer. 2011;18:193–206. doi: 10.1677/ERC-10-0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gonin-Laurent N, et al. RB1 and TP53 pathways in radiation-induced sarcomas. Oncogene. 2007;26:6106–6112. doi: 10.1038/sj.onc.1210404. [DOI] [PubMed] [Google Scholar]
- 16.Cahan WG, et al. Sarcoma arising in irradiated bone. Cancer. 1948;1:3–29. doi: 10.1002/1097-0142(194805)1:1<3::aid-cncr2820010103>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 17.Le Brigand K, et al. An open-access long oligonucleotide microarray resource for analysis of the human and mouse transcriptomes. Nucleic Acids Res. 2006;34:e87. doi: 10.1093/nar/gkl485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clark MA, et al. Soft-tissue sarcomas in adults. N. Engl. J. Med. 2005;353:701–711. doi: 10.1056/NEJMra041866. [DOI] [PubMed] [Google Scholar]
- 19.Huvos AG, et al. Postradiation osteogenic sarcoma of bone and soft tissues. A clinicopathologic study of 66 patients. Cancer. 1985;55:1244–1255. doi: 10.1002/1097-0142(19850315)55:6<1244::aid-cncr2820550617>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 20.Lagrange JL, et al. Sarcoma after radiation therapy: retrospective multiinstitutional study of 80 histologically confirmed cases. Radiation Therapist and Pathologist Groups of the Federation Nationale des Centres de Lutte Contre le Cancer. Radiology. 2000;216:197–205. doi: 10.1148/radiology.216.1.r00jl02197. [DOI] [PubMed] [Google Scholar]
- 21.Penel N, et al. Frequency of certain established risk factors in soft tissue sarcomas in adults: a prospective descriptive study of 658 cases. Sarcoma. 2008;2008:459386. doi: 10.1155/2008/459386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jung T, et al. The proteasomal system. Mol. Aspects Med. 2009;30:191–296. doi: 10.1016/j.mam.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 23.Reyes-Turcu FE, et al. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu. Rev. Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robbins ME, et al. Radiation-induced kidney injury: a role for chronic oxidative stress? Micron. 2002;33:133–141. doi: 10.1016/s0968-4328(01)00006-3. [DOI] [PubMed] [Google Scholar]
- 25.Fleckenstein K, et al. Temporal onset of hypoxia and oxidative stress after pulmonary irradiation. Int. J. Radiat. Oncol. Biol. Phys. 2007;68:196–204. doi: 10.1016/j.ijrobp.2006.12.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jack CI, et al. Indicators of free radical activity in patients developing radiation pneumonitis. Int. J. Radiat. Oncol. Biol. Phys. 1996;34:149–154. doi: 10.1016/0360-3016(95)00209-x. [DOI] [PubMed] [Google Scholar]
- 27.Beinert T, et al. Oxidant-induced lung injury in anticancer therapy. Eur. J. Med. Res. 1999;4:43–53. [PubMed] [Google Scholar]
- 28.Rzeszowska-Wolny J, et al. Ionizing radiation-induced bystander effects, potential targets for modulation of radiotherapy. Eur. J. Pharmacol. 2009;625:156–164. doi: 10.1016/j.ejphar.2009.07.028. [DOI] [PubMed] [Google Scholar]
- 29.Prise KM, et al. Radiation-induced bystander signalling in cancer therapy. Nat. Rev. Cancer. 2009;9:351–360. doi: 10.1038/nrc2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim GJ, et al. A role for mitochondrial dysfunction in perpetuating radiation-induced genomic instability. Cancer Res. 2006;66:10377–10383. doi: 10.1158/0008-5472.CAN-05-3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Port M, et al. A radiation-induced gene signature distinguishes post-Chernobyl from sporadic papillary thyroid cancers. Radiat. Res. 2007;168:639–649. doi: 10.1667/RR0968.1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.