Summary
Bone is constantly resorbed and formed throughout life by coordinated actions of osteoclasts and osteoblasts. Here we show that Smurf1, a HECT domain ubiquitin ligase, has a specific physiological role in suppressing the osteogenic activity of osteoblasts. Smurf1-deficient mice are born normal but exhibit an age-dependent increase of bone mass. The cause of this increase can be traced to enhanced activities of osteoblasts, which become sensitized to bone morphogenesis protein (BMP) in the absence of Smurf1. However, loss of Smurf1 does not affect the canonical Smad-mediated intracellular TGFβ or BMP signaling; instead, it leads to accumulation of phosphorylated MEKK2 and activation of the downstream JNK signaling cascade. We demonstrate that Smurf1 physically interacts with MEKK2 and promotes the ubiquitination and turnover of MEKK2. These results indicate that Smurf1 negatively regulates osteoblast activity and response to BMP through controlling MEKK2 degradation.
Introduction
From late stages of embryonic development and continuing during adult life, bone is constantly resorbed and formed by remodeling at microscopic sites throughout the skeleton (Karsenty and Wagner, 2002). Bone resorption is carried out by haematopoietically derived osteoclasts, whereas formation of the mineralized bone is the function of mesenchyme-derived osteoblasts. Concerted action by these two opposing types of cells is required for maintaining proper bone mass and integrity in the adult skeleton.
Bone-forming osteoblasts first arise during embryonic development after patterning of a given skeletal element is complete (Karsenty and Wagner, 2002). Commitment of mesenchymal progenitors to the osteoblastic lineage requires sequential actions of Runx2 and Osterix, two osteoblast-specific transcription factors. Once differentiated, osteoblasts begin to synthesize alkaline phosphatase, type I collagen, osteocalcin, and bone sialoprotein and deposit them to bone extracellular matrix (ECM). Recent genetic studies in mice indicate that maintaining the differentiated osteoblast phenotype and bone-forming function not only requires the input of the commitment factors Runx2 and Osterix, but other transcription factors such as JunB and Fra-1 of the AP-1 family (Kenner et al., 2004; Eferl et al., 2004) and ATF4 of the CREB/ATF family (Yang et al., 2004) also play an indispensable role. These proteins often recognize distinct cis-acting elements in osteoblast-specific promoters and act in conjunction with other transcription factors to activate the expression of bone ECM components. For example, Runx2, ATF4, and AP-1 binding sites are found in the promoter of osteocalcin; all of these contribute in transcriptional activation of osteocalcin (Ducy and Karsenty, 1995; Yang et al., 2004).
In addition to the cell-autonomous controls by transcription factors, osteoblast differentiation and its bone-forming activities are subject to control by many paracrine growth factors, most notably by members of the transforming growth factor β (TGFβ)/bone morphogenetic protein (BMP) superfamily. BMP-2 was first noted in inducing ectopic bone formation when it was implanted in muscles (Wozney et al., 1988). A large body of evidence has now shown that different BMPs, their membrane receptors, and their antagonists all play prominent roles in controlling osteoblast differentiation and deposition of bone ECM (Balemans and Van Hul, 2002; Zhao et al., 2002; Wu et al., 2003). By antagonizing the inductive effect of BMP, TGFβ inhibits expression of Runx2 and osteocalcin (Alliston et al., 2001) and blocks osteoblast differentiation (Maeda et al., 2004). The intracellular signaling of TGFβ and BMP is mediated primarily by Smad proteins (Massagué, 2000; Derynck and Zhang 2003), which normally accumulate in the cytoplasm but traverse into the nucleus when they are phosphorylated by activated receptor serine/threonine kinases. On several occasions, Smads are found to interact with Runx2 to modulate the expression of bone ECM components (Ito and Miyazono, 2003). In addition to the Smad-mediated canonical signaling pathway, TGFβ and BMP can also activate mitogen-activated protein kinases (MAPKs) such as JNK and p38 MAPK (Engel et al., 1999; Yu et al., 2002; Bakin et al., 2002; Itoh et al., 2003). These crosstalks are likely to integrate the input from diverse paracrine signals to allow for cooperative activation of gene expression by Smads and MAPK downstream substrates c-Jun, JunB, and ATFs (Zhang et al., 1998; Liberati et al., 1999; Sano et al., 1999; Wong et al., 1999).
With the identification of Smad ubiquitin regulatory factor (Smurf), a HECT domain ubiquitin ligase, the role of ubiquitin/proteasome system in modulating TGFβ and BMP signaling has received attention. Two highly related Smurf genes exist in vertebrates: Smurf1 and Smurf2 (Zhu et al., 1999; Lin et al., 2000; Kavsak et al., 2000; Zhang et al., 2001). Ectopic expression of Smurf1 in pluripotent mouse C2C12 myoblasts (Ying et al., 2003) as well as in 2T3 osteoblast progenitor cells (Zhao et al., 2004) prevents osteoblast differentiation. Both Smurf1 and Smurf2 have the ability to interact directly with Smad1 and Smad5 of the BMP pathway and mediate their degradation. However, these two proteins can act in another mechanistic mode to mediate ubiquitination of either TGFβ or BMP type I receptors through ligand-activated binding of inhibitory Smad6 or Smad7 as intermediate (Kavsak et al., 2000; Ebisawa et al., 2001; Murakami et al., 2003). Recent in vitro biochemical studies also suggest that Smurf1 can target Runx2 and small GTPase RhoA for ubiquitination and degradation (Zhao et al., 2003; Wang et al. 2003).
Here, we disrupted the mouse Smurf1 allele and found that Smurf1-deficient mice are perinatally normal but exhibit an age-dependent increase of bone mass due to enhanced osteoblast activity. Surprisingly, Smurf1 does not exert its control of osteoblast function through the Smad-dependent canonical TGFβ/BMP signaling; instead, it promotes ubiquitination and destruction of MEKK2, an upstream kinase in the JNK signaling cascade. Our results thus reveal a novel aspect of the molecular underpinning of osteoblast function and identify a physiological substrate of the Smurf1 ubiquitin E3 ligase.
Results
Increased Bone Mass in Smurf1-Deficient Mice
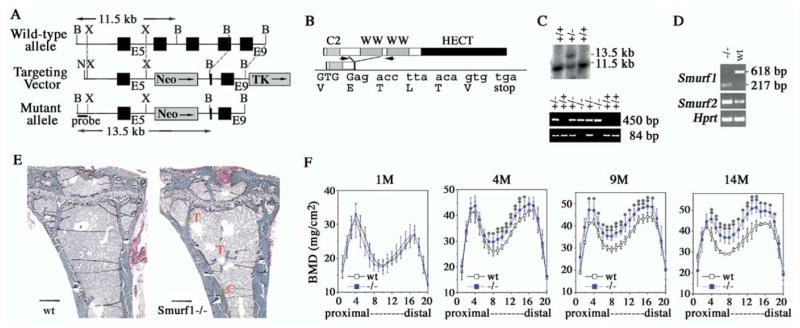
Mouse Smurf1 locus was disrupted through targeted homologous recombination (Figures 1A–1C). This manipulation was expected to generate a genetic null allele, as it would give rise to a truncated protein product that lacks the two highly conserved WW domains and the HECT ubiquitin ligase domain (Figure 1B), which are indispensable for Smurf function (Zhu et al., 1999; Lin et al., 2000; Kavsak et al., 2000; Zhang et al., 2001). Homozygous mutant (Smurf1−/−) mice were born at expected Mendelian ratio (see Table S1 in the Supplemental Data available with this article online), had a normal life span, and were as fertile as their wild-type littermates. At the molecular level, however, the mutated Smurf1 transcript appeared to be less stable than its wild-type counterpart as it accumulated to a much lower level in total cellular RNA in contrast to a marked increase of the Smurf2 transcript (Figure 1D). Given the crucial inductive role by the signaling output of TGFβ/BMP, this compensatory increase of Smurf2 expression was likely to be causative to the seemingly normal embryonic development of Smurf1−/− mice. Despite the lack of gross developmental abnormalities or health problems of the newborns, complete necropsy and histological examination of 20 Smurf1−/− mice between ages 4 and 15 months revealed a thickening of diaphysis in more than 75% of long bones from the cohort (Figure 1E). No other significant alteration in skeletal morphology was observed (data not shown). Measurement of bone mineral density (BMD) showed that this thickening represented a bone mass increase primarily between the topographical sections 4 and 17 of the femurs (Figure 1F). Although newborn Smurf1−/− mice began with a normally mineralized skeleton, the increase of BMD progressed with age, reaching 10%, 17%, and 20% more than that of control littermates by the ages of 4, 9, and 14 months, respectively. Little difference of BMD was seen in metaphysis or epiphysis at the either end of femurs (Figure 1F, sections 1–4 and 17–20), regions primarily consisting of trabecular bones. This bone mass increase is likely due to cell-autonomous causes rather than alteration in the production of bone-metabolizing hormones or a general endocrine disturbance, as no apparent change in the serum levels of parathyroid hormone, calcium, or phosphorus was observed (data not shown).
Figure 1. Increased Bone Mass in Smurf1−/− Mice.
(A) Generation of Smurf1-deficient mice. Closed box, exon; shaded box, selective marker, pGKneo or pGKtk; N, NotI; X, XhoI; B, BamHI.
(B) Structural organization of Smurf1 protein. Sequence surrounding the acquired splicing junction between Exons 5 (upper case) and Exon 9 (lower case) was confirmed by sequencing the RT-PCR products in (D).
(C) (Upper panel) Southern blot analysis of the homologous recombination using the 5′ external probe (shown in [A]). Wt, 11.5 kb; mutant, 13.5 kb. (Lower panel) PCR genotyping. Wt and mutated alleles yield 84 bp and 450 bp products, respectively.
(D) RT-PCR analyses of Smurf1 and Smurf2 expression using total RNA isolated from newborn wild-type and Smurf1−/− mice. For Smurf1 expression, a primer pair from E5 and E9 (arrows in [B]) was used. Amplification of Hprt was used as an internal control.
(E) Villanueva Goldner staining of plastic sections of tibiae from mice at age of 9 months. Scale bar, 0.4 mm. C, cortical bone; T, trabecular bone.
(F) BMD measurement in 20 divisions from the proximal to distal femora of male mice at ages of 1, 4, 9, and 14 months. Wt: n = 4; Smurf1−/−: n = 8 (1 month old), or n = 6 (4, 9, 14 month old). Statistical difference of Smurf1−/− mice from Wt mice was assessed by Student’s t test. *p < 0.05, **p < 0.01.
Enhanced Osteoblast Activity in Smurf1−/− Mice
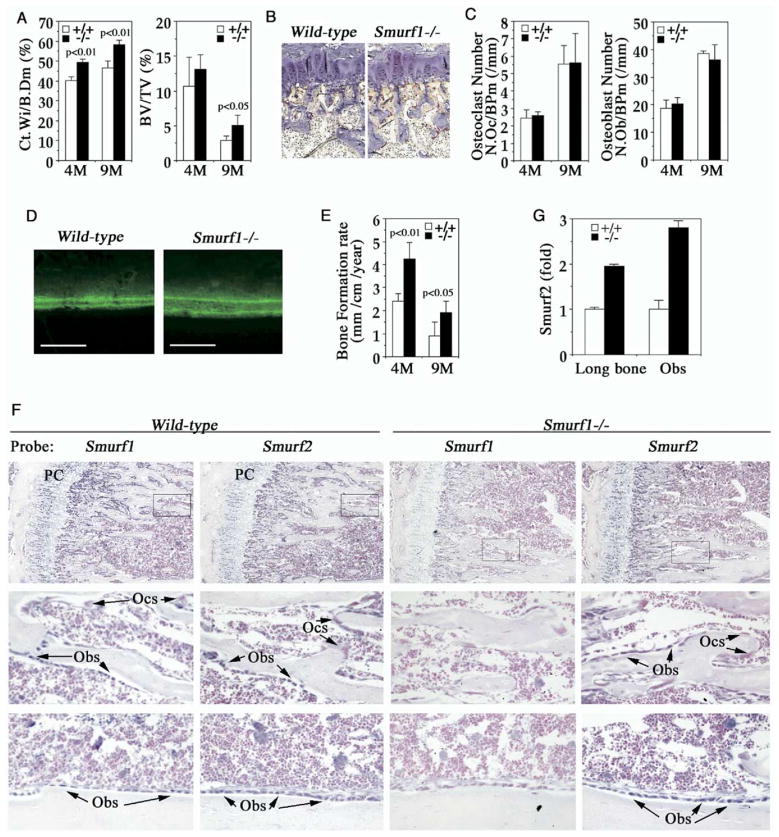
To investigate the cellular basis of bone mass increase associated with loss of Smurf1 function, we performed detailed histomorphometric measurements in sections of undecalcified tibiae that were collected at the ages of 4 and 9 months. Similar to femurs, Smurf1−/− tibiae also exhibited a marked thickening of cortical bone. The ratio of cortex width to total bone diameter at the midpoint of tibiae, a two-dimensional approximation of cortical bone volume, was about 20% higher (Figure 2A, left panel). Likewise, the ratio of trabecular bone volume to tissue volume, a surrogate measure of trabecular bone mass, increased slightly at 4 months of age (13.13% versus 11.73%) but more dramatically at 9 months (4.54% versus 2.99%) (Figure 2A, right panel). The hitherto-described bone mass increase could not be attributed to the cartilage-dependent endochondral bone formation, as the length and the growth plate anatomic structure of Smurf1−/− long bones were indistinguishable from those of wild-type mice (Figures 1E and 2B and data not shown). In contrast, change was observed neither in the number of multinucleated osteoclasts that were stained positive for tartrate-resistant acid phosphatase (TRAP) (Figures 2B and 2C, left panel) nor in the number of osteoblasts in 1% Toluidine blue stained bone sections (Figure 2C, right panel). Thus, this ruled out an imbalance in osteoblast/osteoclast differentiation, which would have altered the ratio of the respective cell numbers as a likely cause of the observed bone mass increase. However, calcein double labeling analysis, a histomorphometrical measurement of osteoblast activity in vivo, revealed a significant increase in bone formation rate associated with loss of Smurf1 (Figures 2D and 2E) while urinary elimination of deoxypyridinoline, a biochemical marker of osteoclast activity, was normal (data not shown). Taken together, these results indicate that the bone mass increase in Smurf1−/− mice likely emanates from an enhanced bone-forming activity of osteoblasts.
Figure 2. Enhanced Osteoblast Activity in Smurf1−/− Mice.
(A) Bone morphometric analyses of 4- and 9-month-old male mice (n = 4). Ct.Wi/B. Dm, cortex bone width per bone diameter; BV/TV, trabecular bone volume per tissue volume. Statistical difference between two mice groups was assessed by Student’s t test.
(B) TRAP staining of osteoclasts in proximal epiphysis of tibia from 4-month-old mice.
(C) Quantification of osteoclast (left) and osteoblast (right) cell numbers.
(D) Calcein-labeled mineralization fronts in tibiae cortex bone from 4-month-old mice by fluorescent micrography. Scale bar = 0.2 mm.
(E) Quantification of bone formation rate measured with calcein double-labeling.
(F) In situ hybridization analyses of Smurf1 and Smurf2 expression in tibia at the metaphysis (top panels) and near cortical bone (bottom panels). (Middle panels) Insets of top panels at higher magnification. Sections were counterstained with nuclear fast red. PC, proliferative chondrocytes; Obs, osteoblasts; Ocs, osteoclasts.
(G) Real-time RT-PCR measurement of Smurf2 transcript from lone bones or osteoblasts.
To ascertain that the expression pattern of Smurf genes is compatible with a role in osteoblast, we conducted in situ hybridization analyses in tibiae sections. Signals of Smurf1 transcript were concentrated in osteoblasts and proliferative chondrocytes but not in osteoclasts (Figure 2F). Smurf2 transcript exhibited a similar cellular distribution, but its level in Smurf1−/− tibiae was markedly increased (Figure 2F). Real-time RT-PCR quantification of mRNA isolated from bone extracts or purified osteoblasts confirmed the increase of Smurf2 expression (Figure 2G). Apparently, the elevated Smurf2 was not sufficient to completely compensate for the loss of Smurf1 function that enhanced osteoblast activity.
Smurf1 Negatively Regulates Osteoblast Function and Response to BMP
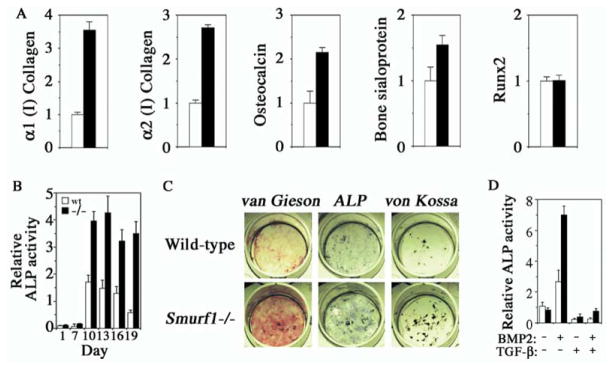
A consequence of enhanced osteoblast activity would be acceleration of bone ECM production. Real-time RT-PCR analyses revealed that this indeed was the case. The mRNA levels of α1 collagen type 1, α2 collagen type 1, osteocalcin, and bone sialoprotein were all increased in bone extracts of Smurf1−/− mice (Figure 3A). In contrast, the mRNA level of Runx2, the early differentiation marker of commitment to osteoblastic lineage, was unchanged (Figure 3A). This observation was consistent with the normal number of osteoblasts described in above histomorphometric measurements (Figure 2C).
Figure 3. Enhanced ECM Production in Smurf1−/− Mice and BMP Sensitivity of Smurf1−/− Osteoblasts.
(A) Real-time PCR of osteoblast marker genes from long bone mRNA of 4-month-old wild-type (open column) or Smurf1−/− mice (closed column). Values are presented as relative expression.
(B) Time course of ALP activity in differentiating calvaria cell culture.
(C) Calvaria-derived osteoblasts were differentiated for 12 days in vitro and stained for collagen matrix formation (van Gieson) and ALP activity. Von Kossa staining for mineralized nodules was done after culturing for 21 days.
(D) ALP activity of calvaria-derived cells after 7 days of differentiation in the presence of exogenous BMP-2 and/or TGFβ.
To specifically address the impact of loss of Smurf1 on bone-forming activity of osteoblasts, we established an osteoblast culture using cells isolated from calvaria bones. In vitro differentiation of mesenchymal progenitor cells or immature osteoblasts that are enriched in this culture faithfully recapitulates the osteoblast maturation process in expressing alkaline phosphatases (ALP), depositing type I collagen to ECM, and forming mineralized bone nodules (Bhargava et al., 1988). In Smurf1−/− osteoblasts, although ALP activity was comparable to that of wild-type osteoblasts initially after culturing ex vivo for 7 days, it became significantly higher after 10 days (Figures 3B and 3C). Similarly, production of collagen matrix (van Gieson staining) was dramatically increased after 12 days (Figure 3C), and more and bigger mineralized bone nodules (von Kossa staining) appeared after 21 days (Figure 3C). Thus, disruption of Smurf1 clearly has an augmentative effect on osteoblast activity, indicating that Smurf1 normally is a negative regulator of osteoblast function.
Both TGFβ and BMP play important roles in osteoblast differentiation and function, and, as implicated by previous in vitro studies, Smurf1 has the potential to degrade the BMP pathway-specific Smads and BMP or TGFβ type I receptors (Zhu et al., 1999; Ebisawa et al., 2001; Murakami et al., 2003; Ying et al., 2003). We thus examined the effect of TGFβ or BMP on osteoblastic function in calvaria cell culture. At the seventh day of ex vivo culturing, little ALP activity was detected without ligand treatment (Figures 3B and 3D). In the presence of BMP-2, however, ALP activity was induced in wild-type cells at this early time point, but a much robust induction was observed in Smurf1−/− cells (Figure 3D). In the presence of TGFβ, which inhibits osteoblast differentiation and function, the basal level of ALP activity was suppressed, and, when added together with BMP-2, TGFβ also suppressed the BMP-induced ALP activity by more than 80% in both types of cells (Figure 3D). These results indicate that Smurf1-deficient osteoblasts were sensitized to BMP signaling while probably having a normal response to TGFβ.
Normal Smad-Dependent Responses in Smurf1-Deficient Osteoblasts
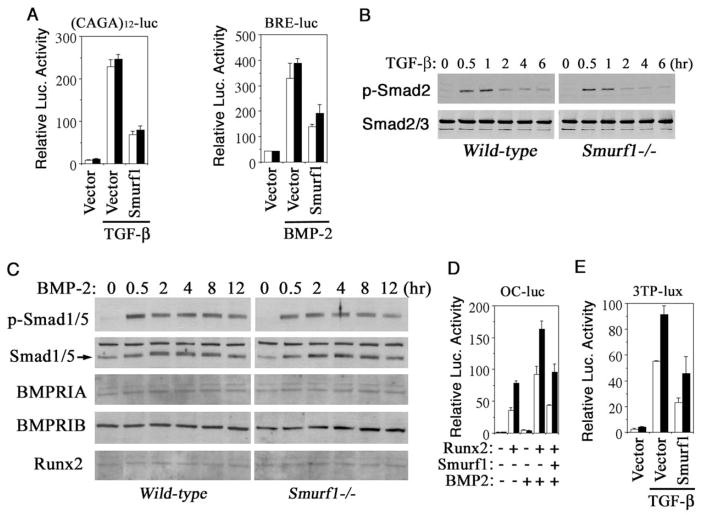
To further delineate the effect of loss of Smurf1 on TGFβ or BMP signaling in osteoblasts, we analyzed the signaling output of a number of pathway-specific transcriptional reporters that were transfected into calvaria cells. We began with (CAGA)12-luc, which can be activated only by TGFβ through its type I receptor (Dennler et al., 1998), and BRE-luc, a BMP-specific transcription reporter driven by BMP-responsive elements of the Id1 gene (Korchynskyi and ten Dijke, 2002). While addition of either TGFβ or BMP-2 stimulated their respective reporters, the extent to which these reporters were activated was independent of the genetic background of the cells in which they were tested (Figure 4A). Western blot analyses showed little difference in the levels of total Smad2 and Smad3 (Figure 4B), Smad1 and Smad5, and the BMP type IA and IB receptors (Figure 4C). Although the BMP-2-mediated Smad1 and Smad5 phosphorylation lasted longer than that of the TGFβ-mediated Smad2 phosphorylation, no significant difference was observed between wild-type and Smurf1−/− osteoblasts (Figures 4B and 4C). Thus, even though Smurf1-deficient osteoblasts are sensitized to BMP for controlling bone-forming activity, the Smad-dependent TGFβ and BMP signaling per se is not affected. Notwithstanding, Smurf1 did display the ability to down-regulate the activities of these pathway-specific reporters when it was overexpressed (Figure 4A), in agreement with previous reports (Zhu et al., 1999; Ebisawa et al., 2001; Murakami et al., 2003).
Figure 4. Normal Smad-Dependent TGFβ and BMP Response in Smurf1−/− Osteoblasts.
(A) Smad-dependent TGFβ or BMP signaling in osteoblasts measured by (CAGA)12-Luc or BRE-luc transcription reporter assay.
(B) Western analyses of TGFβ-induced Smad2 phosphorylation and the steady-state levels of the endogenous Smad2 and Smad3.
(C) Western analyses of BMP-2-induced Smad1/5 phosphorylation and the steady-state levels of the endogenous total Smad1/5, BMP receptors (BMPRIA and BMPRIB), and Runx2.
(D) Effect of loss of Smurf1 on transcription from the Runx2-dependent osteocalcin promoter (OC-Luc).
(E) Effect of loss of Smurf1 on transcription from 3TP-Lux. Open bar, wild-type; closed bar, Smurf1−/−.
Surprisingly, when the signaling response was measured with a set of more complex transcriptional reporters—OC-luc for monitoring BMP response and 3TP-lux for TGFβ response—a much potent transcriptional activation was now observed in Smurf1−/− cells (Figures 4D and 4E). The control elements of OC-luc reporter were derived from the osteocalcin promoter (Ducy and Karsenty, 1995), whose activation requires osteoblast-specific transcription factor Runx2 and is influenced by interaction between Runx2 and other coactivators, including Smads and AP-1 (Franceschi and Xiao, 2003; Ito and Miyazono, 2003). Without culturing in differentiation medium or introduction of exogenous Runx2, OC-luc remained silent (Figure 4D). In the presence of exogenous Runx2, however, OC-luc was activated by BMP in both wild-type and Smurf1−/− cells, but the extent of activation was much higher in Smurf1−/− cells (Figure 4D). Although it was reported that Smurf1 could cause degradation of Runx2 (Zhao et al., 2003), and overexpression of Smurf1 did decrease the overall transcription activity from this reporter (Figure 4D), the augmented activation of OC-luc in Smurf1−/− cells could not be accounted solely by such a mechanism because the protein level of Runx2 was unchanged (Figure 4C). Similarly, an enhancement of TGFβ-induced transcription activation was observed in Smurf1−/− cells when assayed by 3TP-lux (Figure 4E), whose promoter contains three tandem repeats of the AP-1 binding element and a segment of the TGFβ-inducible PAI-1 promoter (Wrana et al., 1992). Because the AP-1 family of transcription factors can cooperate with Runx2 or Smad to increase Runx2- or Smad-dependent transcription (Zhang et al., 1998; Wong et al., 1999; Hess et al., 2001; D’Alonzo et al., 2002), the above results suggest that Smurf1 may regulate osteoblast function and response to BMP by modulating AP-1 activity.
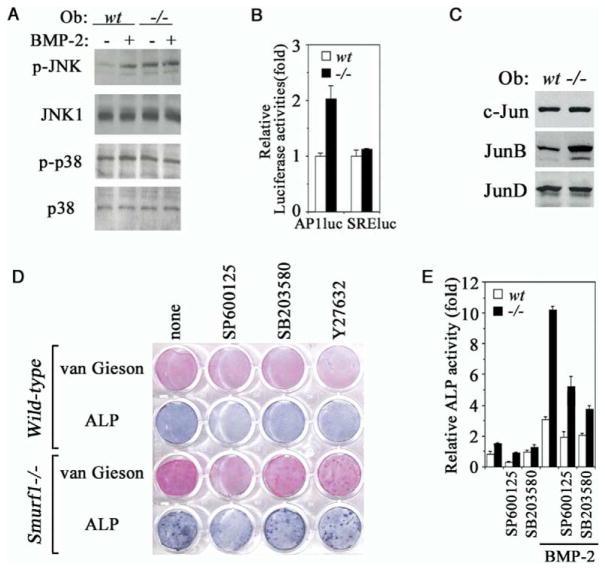
Activation of JNK Kinase Cascade in Smurf1-Deficient Osteoblasts
The overt increase of osteoblast activity in Smurf1−/− mice and the seeming lack of change in the Smad-dependent signaling prompted us to look beyond the orthodox TGFβ/BMP intracellular signaling pathway for physiological target of Smurf1 function. The crosstalks between TGFβ/BMP signaling and various MAP kinases are well substantiated (Massagué, 2000; Derynck and Zhang 2003). Two MAP kinases, JNK and p38 MAPK, which act upstream of the AP-1 family transcription factors, have recently been shown to play a role in controlling bone ECM production, expression of osteoblast specific markers, and other aspects of osteoblast function (Guicheux et al., 2003). Indeed, Western blot analyses showed that, while JNK could become phosphorylated in response to BMP in wild-type osteoblasts, it was already phosphorylated without the ligand in Smurf1−/− cells (Figure 5A). The level of total JNK, however, remained constant (Figure 5A), suggesting that the appearance of phosphorylated JNK was due to post-translational activation. In contrast, little change was seen in the levels of either total or phosphorylated p38 MAPK (Figure 5A). Consequential to JNK activation, downstream JNK-dependent transcription was enhanced in Smurf1−/− cells, as indicated by AP1-luc reporter assay (Figure 5B). In addition, the protein level of one of JNK targets, JunB, but not c-Jun or JunD was increased in Smurf1−/− osteoblasts (Figure 5C), although neither c-Jun nor JunB itself constitutes as direct target for Smurf1 (Figure S1). Functional cooperation between Smads and other families of transcription activators, including members of the AP-1 or ATF family, is a recurring theme in activation of TGFβ/BMP target genes (Massagué, 2000; Derynck and Zhang 2003). Thus, the activation of JNK and its downstream AP-1 or ATF effectors may account for the enhanced bone-forming activity as well as the increased sensitivity to BMP signaling in Smurf1−/− osteoblasts. To test this hypothesis, we applied SP600125, a JNK-specific inhibitor, to the calvaria cell culture. Blocking JNK by SP600125 suppressed the accelerated bone ECM production and reduced the augmented ALP activity in Smurf1−/− osteoblasts to a level that was comparable to that in wild-type cells (Figure 5D). This was specific to the inhibition of JNK because treating calvaria cells with SB203580, a p38 MAPK inhibitor, or Y27632, a Rho-dependent kinase inhibitor, had little effect (Figure 5D). Blocking JNK also specifically desensitized the osteoblasts to BMP signaling, as the amplitude of BMP-induced ALP activity was curtailed by the addition of SP600125 (Figure 5E). Based on the results above, we conclude that activation of the JNK kinase cascade is an essential molecular change that leads to enhanced bone-forming activity and sensitizes BMP response associated with the loss of Smurf1 function. Interestingly, although blocking p38 MAPK by SB203580 had little effect on basal level of bone ECM production and ALP activity (Figures 5D and 5E), it nevertheless dampened the BMP-induced ALP activity (Figure 5E), suggesting that p38 MAPK may play a role in the BMP-stimulated osteoblast activity.
Figure 5. Elevated JNK Activity in Smurf1−/−Osteoblasts.
(A) Western analyses of JNK and p38 MAPK phosphorylation in isolated osteoblasts. Where indicated, BMP-2 was added at 100 ng/ml for 30 min prior to sample preparation.
(B) JNK-dependent AP-1 transcription as measured by AP-1-luc reporter. SRE-luc, a control reporter containing the serum response element.
(C) Western analyses of endogenous c-Jun, JunB, and JunD expression.
(D) Blocking the augmented collagen matrix production and ALP activity by JNK inhibitor in Smurf1−/− osteoblasts. Experiments were carried out as in Figure 3C except for the addition of SP600125, SB203580, or Y27632.
(E) ALP quantification in osteoblasts cultured for 12 days. The relative activity was expressed as fold to the ALP activity of wild-type osteoblasts in the absence of inhibitors and BMP-2.
Smurf1 Physically Interacts with and Targets MEKK2 for Ubiquitination
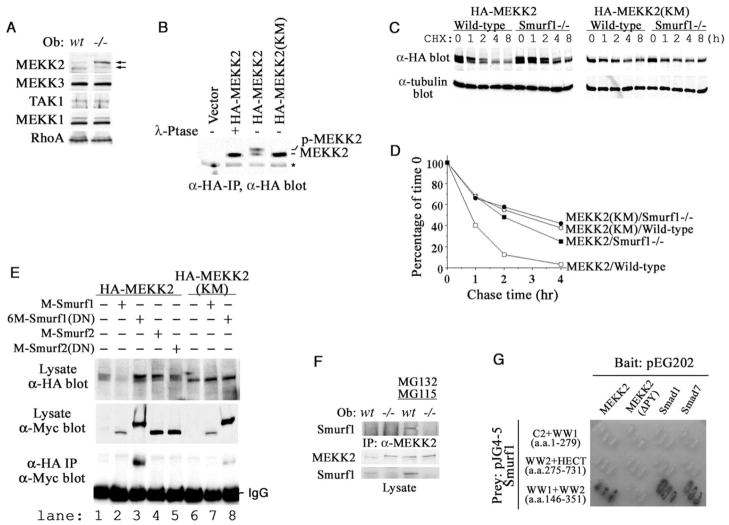
Having established a requirement for the JNK kinase cascade, we set out to identify a direct target of Smurf1 that controls osteoblast activity and BMP response. Smurfs are known to recognize a “PY” motif that has the ability to interact with WW domains of the HECT family E3 ligases (Sudol and Hunter, 2000). No such motif was present in JNK, nor did JNK interact physically with Smurf1 (data not shown). These preclude JNK itself as a direct Smurf1 target. We then scanned the sequence of several kinases that are known to act upstream of JNK, including MKK3, -4, -6, and -7; MEKK1, -2, -3, and -4; and TAK1 (Davis, 2000). Among these, only MEKK2 and MEKK3 contain a PY motif. Western blot showed that a slow-migrating band of MEKK2 accumulated in Smurf1−/− but not wild-type osteoblasts (Figure 6A). This slow-migrating band is likely to be the phosphorylated MEKK2, because introduction of the wild-type but not a kinase-deficient MEKK2 cDNA (Su et al., 2001) into mouse embryonic fibroblasts (MEFs) gave rise to a similar slow-migrating band, which could be collapsed to the normal migrating band by λ-phosphatase treatment (Figure 6B). In contrast to the accumulation of phosphorylated MEKK2, no difference in the level of the related MEKK3 was observed (Figure 6A). No discernible change was seen in two other kinases, MEKK1 and TAK1, which were implicated previously in TGFβ/BMP signaling (Brown et al., 1999; Yamaguchi et al., 1999) (Figure 6A). Nor was the level of RhoA (Figure 6A), a small GTPase that was recently reported as a target of Smurf1 (Wang et al., 2003) and could function upstream of JNK.
Figure 6. Accumulation of Phosphorylated MEKK2 in Smurf1−/− Osteoblasts and Physical Interaction between Smurf1 and MEKK2.
(A) Western analyses of MEKK2, MEKK3, MEKK1, TAK1, and RhoA in osteoblasts.
(B) Western analyses of HA-tagged MEKK2 or kinase-deficient MEKK2 (KM) after immunoprecipitation from transfected Smurf1−/− MEFs.
(C) Western analyses of HA-MEKK2 and HA-MEKK2(KM) in transfected MEFs at the indicated intervals following cycloheximide treatment.
(D) [35S]-methionine labeling and chase studies of MEKK2 and MEKK2 (KM) in wild-type Smurf1−/− MEFs. The amount of each labeled protein was expressed as the percentage of that at the beginning of chase (time 0).
(E) Interaction between exogenous Smurfs and MEKK2 in Smurf1−/− MEFs. Expression of the HA-tagged MEKK2s and the Myc-tagged Smurfs was shown in top and middle panel, respectively. Only Smurf1(DN) coimmunoprecipitated with the wild-type MEKK2 and weakly with MEKK2(KM) (bottom panel, lanes 3 and 8).
(F) Interaction between endogenous Smurf1 and MEKK2. MEKK2 was precipitated from wild-type and Smurf1−/− osteoblasts that were pre-treated with proteasome inhibitors and subjected to Western analysis with anti-Smurf1 antibody (top panel). The amount of total MEKK2 and Smurf1 present in cell lysates was shown at two bottom panels.
(G) Requirements for direct interaction between Smurf1 and MEKK2 as revealed by yeast two-hybrid assays. Binding of the WW domains of Smurf1 to Smad1 or Smad7 was included as positive controls.
To investigate if MEKK2 accumulates in the absence of Smurf1 due to impairment of protein turnover, we measured the turnover rate of exogenous MEKK2 in MEFs. Western analyses showed that the rate of MEKK2 turnover was indeed impeded in Smurf1−/−MEFs (Figure 6C). This was especially true to the phosphorylated MEKK2; however, the turnover rate of kinase-deficient MEKK2(KM) was unaffected (Figure 6C). Similar results were obtained with pulse-chase labeling experiments. In wild-type MEFs, MEKK2 displayed a half-life of less than 1 hr, which was prolonged to about 2 hr in Smurf1−/− MEFs, similar to that of MEKK2(KM) (Figure 6D). Taken together, the above results suggest that MEKK2 is a target of Smurf1.
To determine if Smurf1 specifically interacts with MEKK2, we coexpressed MEKK2 with Smurf1, Smurf2, or dominant-negative mutants carrying an inactivating point mutation in their respective HECT E3 ligase domain in Smurf1−/− MEFs. In agreement with the protein turnover studies, MEKK2 and its phosphorylated form ceased to accumulate when it was coexpressed with the wild-type Smurf1 (Figure 6E, lane 2) but accumulated to high level with Smurf1(DN) (lane 3). Once again, accumulation of the kinase-deficient MEKK2(KM) was unaffected by Smurf1 or Smurf1(DN) (Figure 6E, lanes 6–8). Coexpression of Smurf2 or Smurf2(DN) had little effect on MEKK2 (Figure 6E, lanes 4 and 5), indicating that turnover of MEKK2 was specifically controlled by Smurf1. Immunoprecipitation experiments showed that Smurf1(DN) interacts strongly with the HA-tagged MEKK2 (Figure 6E, lane 3) but weakly with MEKK2(KM) (lane 7). Consistent with the resistance to Smurf2, MEKK2 did not coprecipitate with Smurf2 (Figure 6E, lanes 4 and 5). We did not detect wild-type Smurf1 in the anti-HA-MEKK2 pellet (Figure 6E, lane 2), possibly because MEKK2 was rapidly degraded once it was recruited to Smurf1. To demonstrate the in vivo interaction between these two proteins, we pretreated calvaria cells with the proteasome inhibitors MG132 and MG115. A slow-migrating phosphorylated MEKK2 accumulated under this condition in the otherwise wild-type osteoblasts, and the endogenous Smurf1 was co-precipitated with the accumulated MEKK2 (Figure 6F). The hitherto described interaction appears to be of direct nature, as it is corroborated in yeast two-hybrid assay (Figure 6G) in which no homolog of TGFβ/BMP receptors or Smads exists. Mapping studies showed that MEKK2 specifically interacts with the two WW domains of Smurf1 between residues 146 and 351 and that this binding requires the PY motif of MEKK2 (Figure 6G).
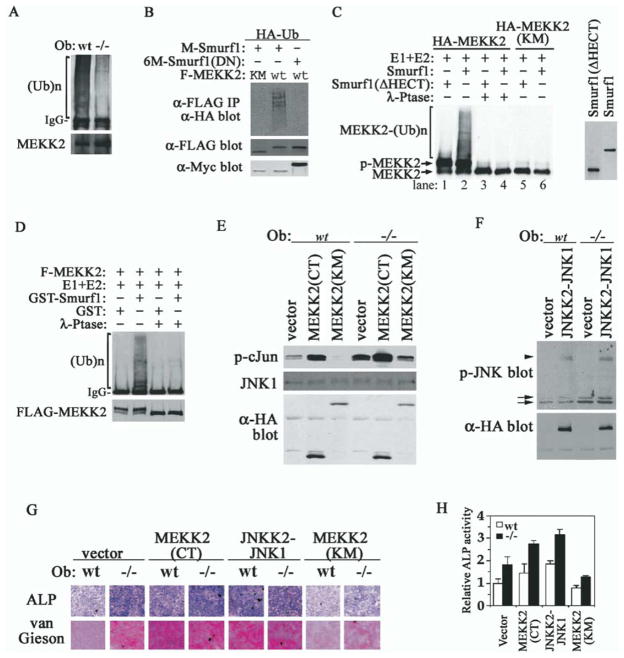
To formally demonstrate that MEKK2 is a substrate of Smurf1, we performed a series of ubiquitination assays. First, in wild-type calvaria cells, polyubiquitinated MEKK2 accumulated to a higher level than in Smurf1−/− cells in the presence of proteasome inhibitors (Figure 7A). Second, in transfected Smurf1−/− MEFs, we detected polyubiquitinated MEKK2 only when it was coexpressed with Smurf1 (Figure 7B). Accumulation of polyubiquitinated MEKK2 was dependent on its intrinsic kinase activity and the functionality of Smurf1 HECT E3 ligase (Figure 7B). Third, we reconstituted the ubiquitination of MEKK2 in vitro using MEKK2 isolated from transfected Hep3B cells, purified E1 and E2, and in vitro-translated Smurf1. In this reconstituted system, only the wild-type MEKK2 became polyubiquitinated when it was mixed with Smurf1 but not a truncated Smurf1 lacking the HECT domain (Figure 7C, lanes 2 and 1). The kinase-deficient MEKK2(KM) was refractory to Smurf1-mediated ubiquitination (Figure 7C, lanes 5 and 6). Pretreating the isolated MEKK2 with λ-phosphatase abolished its ubiquitination (Figure 7C, lanes 3 and 4), suggesting that Smurf1-mediated ubiquitination of MEKK2 requires phosphorylation. Similar results were obtained using purified recombinant GST-Smurf1 fusion protein from E. coli and FLAG-MEKK2 from Drosophila S2 cells (Figure 7D).
Figure 7. MEKK2 Is a Substrate of Smurf1-Mediated Ubiquitination and Activation of MEKK2-JNK Pathway Is Sufficient to Enhance Osteoblast Activity.
(A) Ubiquitination of endogenous MEKK2 in osteoblasts. Polyubiquitinated MEKK2 was detected by anti-ubiquitin Western analysis after precipitation of MEKK2 from cells that were pretreated with proteasome inhibitors.
(B) Smurf1 promotes MEKK2 ubiquitination. Ubiquitinated F-MEKK2 in Smurf1−/− MEFs was visualized by anti-HA-ubiquitin Western analysis after anti-FLAG immunoprecipitation. Expression of Myc-tagged Smurfs is shown in bottom panel.
(C) In vitro polyubiquitination of MEKK2 by Smurf1 detected by anti-HA blot. HA-MEKK2 and HA-MEKK2(KM) were isolated by anti-HA antibody from transfected Hep3B cells. Myc-tagged Smurf1 and Smurf1(ΔHECT) were in vitro translated and are shown in the right panel.
(D) In vitro ubiquitination of MEKK2 by purified GST-Smurf1 detected by anti-ubiquitin blot. FLAG-MEKK2 purified from transfected Drosophila S2 cells is shown at bottom panel.
(E) In vitro kinase assay of JNK1 after immunoprecipitation from osteoblasts expressing MEKK2 mutants. GST-c-Jun was used as the kinase substrate and was analyzed by anti-p-c-Jun blot. Total JNK1, HA-MEKK2(CT), and MEKK2(KM) are shown below.
(F) Western analyses of phosphorylated JNK in osteoblasts expressing HA-JNKK2-JNK1, which does not affect endogenous JNK activity. Arrows, endogenous p-JNK1/2; arrowhead, p-JNKK2-JNK1 fusion.
(G) Ectopic expression of activated MEKK2 and JNK1 is sufficient to enhance osteoblast activity. Staining for ALP activity and collagen matrix was carried out after incubating in differentiation medium for 16 days.
(H) ALP quantification of cells in (G) after differentiation for 12 days.
Finally, we expressed constitutively active MEKK2 or JNK, or kinase-deficient MEKK2 in osteoblasts by retrovirus-mediated transduction (Figures 7E and 7F), and examined the effect of the ectopic expression on osteoblast activity. MEKK2(CT), the constitutively active form containing only the kinase domain (Cheng et al., 2000), strongly activated JNK, as revealed by the ability of JNK1 from the infected calvaria cells to phosphorylate GST-c-Jun in an in vitro kinase assay (Figure 7E). MEKK2(CT) also induced high levels of ALP and collagen matrix production in both wild-type and Smurf1−/− osteoblasts after they were put in differentiation medium (Figures 7G and 7H). Similarly, high levels of ALP activity and collagen matrix production were induced by JNKK2-JNK1, the constitutively active fusion protein (Zheng et al., 1999) (Figures 7G and 7H). However, the kinase-deficient MEKK2(KM) reduced the production of ALP and collagen matrix in Smurf1−/− cells while having little effect in wild-type osteoblasts (Figures 7G and 7H). We therefore conclude that MEKK2 is a bona fide substrate of Smurf1 and activation of the MEKK2-JNK signaling cascade in Smurf1−/− mice is sufficient to enhance osteoblast activity.
Discussion
In this study, we have uncovered a novel aspect of the molecular underpinning of the bone homeostasis through target inactivation of the mouse Smurf1 locus and showed that loss of Smurf1 results in accumulation of phosphorylated MEKK2 and activation of the downstream JNK kinase cascade, which has the tenacity to mobilize the AP-1/ATF family of transcription factors to regulate osteoblast activity.
Smurf1 as a Negative Regulator of BMP Signaling
Biochemical interaction and functional studies have uncovered a long list of Smurf targets including Smad1/5, Smad2, Smad6/7, and type I receptors of both TGFβ and BMP pathways. However, despite intense scrutiny through the use of transcription reporters and direct protein analyses, no indication of Smurf1 affecting the Smad-dependent BMP/TGFβ signaling was found. It is possible that Smurf1 normally serves a very restricted role in clamping down BMP signaling regardless of its other potentials. Alternatively, Smurf1 could have overlapping functions with Smurf2, which also has the ability to degrade R-Smads or type I receptors. The latter notion was corroborated by the 2-fold increase of Smurf2 transcripts in Smurf1−/− mice. Indeed, our preliminary analysis showed that the compounded Smurf1 and Smurf2 doubly deficient embryos die prior to E9.5 (M.Y. and Y.E.Z., unpublished data), underscoring the critical requirement of a common Smurf function during embryogenesis. This embryonic lethality could be brought about by deregulated activation of Smads or TGFβ/BMP type I receptors, as previous in vitro studies would suggest. The final depiction of Smurf1 physiological function thus will be multifaceted, with specific function in a given tissue mediated by a specialized biochemical mechanism.
Smurf1 as a Negative Regulator of Osteoblast Function
The role of Smurf1 in osteoblasts was implicated previously in studies employing in vitro differentiation of C2C12 myoblasts or 2T3 osteoblasts and transgenic mice carrying a Smurf1 transgene driven by a type I collagen promoter (Ying et al., 2003, Zhao et al., 2004). The outcomes of these gain-of-function perturbations, however, were quite different from the current loss-of-function study. When ectopically expressed, Smurf1 interacts with and directs ubiquitin/proteasome-mediated degradation of BMP pathway-specific Smads and/or Runx2 in osteoblast progenitor cells and reduces both the number and proliferative potential of mature osteoblasts in the transgenic mice. As such, it affects the commitment of osteoblast precursors during early stage of osteoblast differentiation rather than bone-forming activities of mature osteoblasts. These mechanisms could operate in cells upon overexpression of Smurf1 but may not do so obligatorily in its physiological function.
It is unlikely that Smurf1 plays an indispensable role in early commitment of osteoblast cell lineage because the number of mature osteoblasts remains the same in Smurf1−/− mice as in wild-type littermates (Figure 2C). Studies of molecular markers seem to corroborate this notion, as no change was detected in the level of Runx2 expression, whereas expression of α1 collagen type 1, α2 collagen type 1, osteocalcin, and bone sialoprotein, all of which are markers of late-stage osteoblast maturation and function, was increased (Figure 3A). Thus, our results indicate Smurf1 is a physiological negative regulator of osteoblast function and place it in the same group of genes as ATF4 and Fra-1, which are required for maintaining normal osteoblasts physiology and function (Eferl et al., 2004; Yang et al., 2004).
Activation of MEKK2-JNK Kinase Cascade Sensitizes Osteoblasts to BMP Signaling
It is increasingly appreciated that TGFβ/BMP signals through Smads and Smad-independent JNK or p38 MAPK kinases that play an important role in osteoblast differentiation and function. The JNK downstream effector AP-1 regulates osteoblast biology by binding to AP-1 elements in osteoblast-specific gene promoters and by interaction with Runx2 (D’Alonzo et al., 2002; Hess et al., 2001). This paradigm is exemplified in the transcription of osteocalcin, the best-characterized marker of osteoblast maturation, which is controlled by at least three cis-acting elements: OSE1, OSE2, and AP-1 sites that are recognized by Runx2, ATF4, and Jun/Fos, respectively (Ducy and Karsenty, 1995; Franceschi and Xiao, 2003; Yang et al., 2004). Mice engineered to lack JunB or Fra-1 selectively in bone cells are osteopenic due to defects in osteoblast differentiation and proliferation (Kenner et al., 2004) or reduced bone ECM production (Eferl et al., 2004). The current finding extends the above observations by revealing a causal relationship between heightened osteoblast activity and elevated JNK in Smurf1−/− osteoblasts. As AP-1/ATF factors are capable of interacting synergistically with Smads in controlling TGFβ and BMP-induced transcription (Zhang et al., 1998; Sano et al., 1999; Wong et al., 1999), one could speculate that Smurf1 normally functions to keep the MEKK2-JNK pathway of the Smad-independent arm from overactivating bone ECM production as the consequence of BMP signaling.
MEKK2 Is a Substrate of Smurf1 E3 Ubiquitin Ligase
Four lines of evidence from the present study directly support MEKK2 as a physiological substrate of Smurf1: (1) Smurf1 forms direct physical contact with MEKK2, (2) the interaction depends on the integrity of the “WW” domains of Smurf1 and the PY motif of MEKK2, (3) Smurf1 promotes MEKK2 ubiquitination both in vivo and in a reconstituted in vitro system, and (4) loss of Smurf1 impedes MEKK2 turnover. The interaction between Smurf1 and MEKK2 requires a functional kinase domain (Figure 6E), which often autophosphorylates. Moreover, phosphorylated MEKK2 accumulates in Smurf1−/− cells (Figure 6A) and is preferentially ubiquitinated by Smurf1 (Figures 7A–7D). These data suggest that the interaction between MEKK2 and Smurf1 is phosphorylation dependent. Previously, WW domains of Pin1, a peptidyl-prolyl isomerase, were shown to specifically bind to phosphoSer-Pro or phosphoThr-Pro motifs (Lu et al., 1999). However, phosphorylation-dependent binding is not known for WW domains of the HECT family E3 ligases. It is not clear at present if phosphorylated residues surrounding the PY motif constitute part of the binding requirement or if phosphorylation per se causes a conformational change in MEKK2 that exposes the PY motif to the WW domains of Smurf1. Regardless of which mechanism, our data indicate that phosphorylation is an obligatory step in substrate recognition by Smurf1, suggesting a possible way of controlling Smurf1-mediated ubiquitination. Moreover, these data also provide the first example of a MAPKKK that is targeted by a HECT family E3 ligase for ubiquitin-mediated degradation.
The mechanism by which MEKK2 is activated in Smurf1−/− osteoblasts is not clear, and, for that matter, nor is it clear how MEKKs are activated in general. Nevertheless, the fact that Smurf1−/− mice mature to adulthood with normal stature indicates that Smurf1 is not essential for maintaining skeletal integrity; this may have implication in targeting Smurf1 as a therapeutic strategy for treating age-related bone loss such as osteoporosis.
Experimental Procedures
Bone Morphological and Histological Analysis
BMD was determined using a dual energy X-ray absorptiometry apparatus (DCS-600R, Aloka, Japan). For in vivo fluorescent labeling, intraperitoneal injections of calcein (20 mg/kg body weight) were twice administrated to mice with a 7 day interval. Tibiae dissected at day 10 were fixed in 70% ethanol and embedded in glycolmethacrylate without decalcification, and longitudinal serial sections were prepared. Unstained sections were visualized by fluorescence microscopy, and the ratio of mineralized surface to bone surface and mineral appositional rate were measured to calculate bone formation rate. Bone sections stained with Villanueva Goldner method or 1% Toluidine blue were used to discriminate between mineralized and unmineralized bone and to identify cellular components. Similar sections were used for staining TRAP (specific for osteoclasts) with leukocyte acid phosphatase kit (Sigma). The numbers of osteoblast and osteoclast were quantified 0.2–4 mm from growth plates in bone marrow cavity and endosteum. Trabecular bone and cortex bone parameters were measured in bone marrow cavity 0.2–2 mm from growth plates or at the midpoint of tibiae, respectively. Nomenclature, symbols, and units used hereof are those according to field standard (Parfitt et al., 1987).
Cell Culture, Protein Analysis, and Pulse Labeling
Primary calvaria cells isolated from 2- to 4-daya-old mice (Ducy et al., 1999; Zhao et al., 2002) were cultured in 24-well plates in α-MEM plus 10% FBS. Two days after confluence, differentiation was induced with addition of 100 μg/ml ascorbic acid and 5 mM β-glycerophosphate in the absence or presence of 100 ng/ml BMP-2 or 5 ng/ml TGFβ. Histochemical staining was performed using van Gieson and von Kossa method (Bhargava et al., 1988). ALP staining and quantification were described (Ying et al., 2003). SP600125, SB203580, and Y27632 (Calbiochem) were used at 10 μM, 5 μM, and 10 μM, respectively. MEFs were derived from E14.5 embryos (Hogan et al., 1994).
MSCV-derived retroviral vector (Clontech) was used for expressing HA-tagged MEKK2(CT), MEKK2(KM), and JNKK2-JNK1. Primary calvaria cells were infected within two passages. Two days after infection, puromycin (2.5 μg/ml) was added for 3 days, and the selected cells were then pooled and plated in 24-well plates for differentiation analysis.
For protein analysis, cells were lyzed in 10 mM HEPES (pH 7.9) containing 300 mM NaCl, 0.1 mM EGTA, 20% glycerol, 0.2% NP-40, and protease inhibitors or in RIPA buffer. In cases of proteasome inhibitor treatment, cells at 95% confluency were incubated with MG132 and MG115 at 25 μM each for 12–14 hr. For pulse-chase labeling experiment, MEFs were transfected with 2 μg of indicated cDNA plasmids in two 60 mm plates, replated into five wells of a 6-well plate, and labeled with 35S-pro-mix (NEN) (Zhang et al., 2001).
Transcription Reporter Assays
Transcription reporter assays in osteoblast cells were performed in 12-well plates by transfecting 0.25 μg of testing vectors and 0.2 μg of pRL-TK (Promega) control vector per well. Luciferase activity was determined after treating with BMP-2 (200 ng/ml) or TGFβ (5 ng/ml) for 18–20 hr. Results were obtained from at least two independent experiments using cells isolated from different mice with duplicate samples for each data point.
In Vitro Ubiquitination Assay
HA-tagged MEKK2 or MEKK2 (KM) or FLAG-tagged MEKK2 was purified by immunoprecipitation from lysates of transfected Hep3B cells (ATCC) or Drosophila S2 cells (Invitrogen), respectively. Isolated MEKK2 were divided into two equal aliquots, one of which was subjected to λ-phosphatase treatment (New England Biolabs). In vitro ubiquitination was carried out in 30 μl 50 mM HEPES-HCl (pH 7.6), 0.7 μg of E1, 1 μg of recombinant Ubc-H5c, 15 μg ubiquitin, ATP regeneration system, and MG132 (5 μM) (all from Boston Biochem), and either in vitro-translated Smurf1 or Smurf1 (ΔHECT) or 2 μg GST or GST-Smurf1 fusion protein purified from E. coli was added. The reaction mixture was incubated at 25°C for 60 min before termination with SDS-sample buffer.
Creation of Smurf1−/− mice, RT-PCR, In Situ Analysis, Antibodies, Plasmids, and Yeast Two-Hybrid Assay
See Supplemental Data.
Supplementary Material
Acknowledgments
We are in debt to Drs. B. Su, A. Lin, P. ten Dijke, C-H. Heldin, G. Karsenty, T. Alliston, and K. Miyazono for various plasmid and antibody reagents; V. Barr for assistance with microscope and computer software; and D. Smith for pathology service. We also thank Dr. L. Samelson for critically reading the manuscript. M.Y. is a fellow of Japan Society for the Promotion of Science.
Footnotes
Supplemental Data include three figures, two tables, Supplemental Experimental Procedures, and Supplemental References and can be found with this article online at http://www.cell.com/cgi/content/full/121/1/101/DC1/.
References
- Alliston T, Choy L, Ducy P, Karsenty G, Derynck R. TGF-β-induced repression of CBFA1 by Smad3 decreases cbfa1 and osteocalcin expression and inhibits osteoblast differentiation. EMBO J. 2001;20:2254–2272. doi: 10.1093/emboj/20.9.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakin AV, Rinehart C, Tomlinson AK, Arteaga CL. p38 mitogen-activated protein kinase is required for TGFβ-mediated fibroblastic transdifferentiation and cell migration. J Cell Sci. 2002;115:3193–3206. doi: 10.1242/jcs.115.15.3193. [DOI] [PubMed] [Google Scholar]
- Balemans W, Van Hul W. Extracellular regulation of BMP signaling in vertebrates: a cocktail of modulators. Dev Biol. 2002;250:231–250. [PubMed] [Google Scholar]
- Bhargava U, Bar-Lev M, Bellows CG, Aubin JE. Ultrastructural analysis of bone nodules formed in vitro by isolated fetal rat calvaria cells. Bone. 1988;9:155–163. doi: 10.1016/8756-3282(88)90005-1. [DOI] [PubMed] [Google Scholar]
- Brown JD, DiChiara MR, Anderson KR, Gimbrone MA, Jr, Topper JN. MEKK-1, a component of the stress (stress-activated protein kinase/c-Jun N-terminal kinase) pathway, can selectively activate Smad2-mediated transcriptional activation in endothelial cells. J Biol Chem. 1999;274:8797–8805. doi: 10.1074/jbc.274.13.8797. [DOI] [PubMed] [Google Scholar]
- Cheng J, Yang J, Xia Y, Karin M, Su B. Synergistic interaction of MEK kinase 2, c-Jun N-terminal kinase (JNK) kinase 2, and JNK1 results in efficient and specific JNK1 activation. Mol Cell Biol. 2000;20:2334–2342. doi: 10.1128/mcb.20.7.2334-2342.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Alonzo RC, Selvamurugan N, Karsenty G, Partridge NC. Physical interaction of the activator protein-1 factors c-Fos and c-Jun with Cbfa1 for collagenase-3 promoter activation. J Biol Chem. 2002;277:816–822. doi: 10.1074/jbc.M107082200. [DOI] [PubMed] [Google Scholar]
- Davis RJ. Signal transduction by the JNK group of MAP kinases. Cell. 2000;103:239–252. doi: 10.1016/s0092-8674(00)00116-1. [DOI] [PubMed] [Google Scholar]
- Dennler S, Itoh S, Vivien D, ten Dijke P, Huet S, Gauthier JM. Direct binding of Smad3 and Smad4 to critical TGFβ-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J. 1998;17:3091–3100. doi: 10.1093/emboj/17.11.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-β family signalling. Nature. 2003;425:577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- Ducy P, Karsenty G. Two distinct osteoblast-specific cis-acting elements control expression of a mouse osteocalcin gene. Mol Cell Biol. 1995;15:1858–1869. doi: 10.1128/mcb.15.4.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducy P, Starbuck M, Priemel M, Shen J, Pinero G, Geoffroy V, Amling M, Karsenty G. A Cbfa1-dependent genetic pathway controls bone formation beyond embryonic development. Genes Dev. 1999;13:1025–1036. doi: 10.1101/gad.13.8.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebisawa T, Fukuchi M, Murakami G, Chiba T, Tanaka K, Imamura T, Miyazono K. Smurf1 interacts with transforming growth factor-β type I receptor through Smad7 and induces receptor degradation. J Biol Chem. 2001;276:12477–12480. doi: 10.1074/jbc.C100008200. [DOI] [PubMed] [Google Scholar]
- Eferl R, Hoebertz A, Schilling AF, Rath M, Karreth F, Kenner L, Amling M, Wagner EF. The Fos-related antigen Fra-1 is an activator of bone matrix formation. EMBO J. 2004;23:2789–2799. doi: 10.1038/sj.emboj.7600282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel ME, McDonnell MA, Law BK, Moses HL. Interdependent SMAD and JNK signaling in transforming growth factor-β-mediated transcription. J Biol Chem. 1999;274:37413–37420. doi: 10.1074/jbc.274.52.37413. [DOI] [PubMed] [Google Scholar]
- Franceschi RT, Xiao G. Regulation of the osteoblast-specific transcription factor, Runx2: responsiveness to multiple signal transduction pathways. J Cell Biochem. 2003;88:446–454. doi: 10.1002/jcb.10369. [DOI] [PubMed] [Google Scholar]
- Guicheux J, Lemonnier J, Ghayor C, Suzuki A, Palmer G, Caverzasio J. Activation of p38 mitogen-activated protein kinase and c-Jun-NH2-terminal kinase by BMP-2 and their implication in the stimulation of osteoblastic cell differentiation. J Bone Miner Res. 2003;18:2060–2068. doi: 10.1359/jbmr.2003.18.11.2060. [DOI] [PubMed] [Google Scholar]
- Hess J, Porte D, Munz C, Angel P. AP-1 and Cbfa/runt physically interact and regulate parathyroid hormone-dependent MMP13 expression in osteoblasts through a new osteoblast-specific element 2/AP-1 composite element. J Biol Chem. 2001;276:20029–20038. doi: 10.1074/jbc.M010601200. [DOI] [PubMed] [Google Scholar]
- Hogan B, Beddington R, Costantini F, Lacy E. Manipulating the Mouse Embryos: A Laboratory Manual. 2. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- Ito Y, Miyazono K. RUNX transcription factors as key targets of TGF-β superfamily signaling. Curr Opin Genet Dev. 2003;13:43–47. doi: 10.1016/s0959-437x(03)00007-8. [DOI] [PubMed] [Google Scholar]
- Itoh S, Thorikay M, Kowanetz M, Moustakas A, Itoh F, Heldin C-H, ten Dijke P. Elucidation of Smad requirement in transforming growth factor-β type I receptor-induced responses. J Biol Chem. 2003;278:3751–3761. doi: 10.1074/jbc.M208258200. [DOI] [PubMed] [Google Scholar]
- Karsenty G, Wagner EF. Reaching a genetic and molecular understanding of skeletal development. Dev Cell. 2002;2:389–406. doi: 10.1016/s1534-5807(02)00157-0. [DOI] [PubMed] [Google Scholar]
- Kavsak P, Rasmussen RK, Causing CG, Bonni S, Zhu H, Thomsen GH, Wrana JL. Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGFβ receptor for degradation. Mol Cell. 2000;6:1365–1375. doi: 10.1016/s1097-2765(00)00134-9. [DOI] [PubMed] [Google Scholar]
- Kenner L, Hoebertz A, Beil T, Keon N, Karreth F, Eferl R, Scheuch H, Szremska A, Amling M, Schorpp-Kistner M, et al. Mice lacking JunB are osteopenic due to cell-autonomous osteoblast and osteoclast defects. J Cell Biol. 2004;164:613–623. doi: 10.1083/jcb.200308155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korchynskyi O, ten Dijke P. Identification and functional characterization of distinct critically important bone morphogenetic protein-specific response elements in the Id1 promoter. J Biol Chem. 2002;277:4883–4891. doi: 10.1074/jbc.M111023200. [DOI] [PubMed] [Google Scholar]
- Liberati NT, Datto MB, Frederick JP, Shen X, Wong C, Rougier-Chapman EM, Wang XF. Smads bind directly to the Jun family of AP-1 transcription factors. Proc Natl Acad Sci USA. 1999;96:4844–4849. doi: 10.1073/pnas.96.9.4844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Liang M, Feng XH. Smurf2 is a ubiquitin E3 ligase mediating proteasome-dependent degradation of Smad2 in transforming growth factor-β signaling. J Biol Chem. 2000;275:36818–36822. doi: 10.1074/jbc.C000580200. [DOI] [PubMed] [Google Scholar]
- Lu PJ, Zhou XZ, Shen M, Lu KP. Function of WW domains as phosphoserine- or phosphothreonine-binding modules. Science. 1999;283:1325–1328. doi: 10.1126/science.283.5406.1325. [DOI] [PubMed] [Google Scholar]
- Maeda S, Hayashi M, Komiya S, Imamura T, Miyazono K. Endogenous TGF-β signaling suppresses maturation of osteoblastic mesenchymal cells. EMBO J. 2004;23:552–563. doi: 10.1038/sj.emboj.7600067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massagué J. How cells read TGF-β signals. Nat Rev Mol Cell Biol. 2000;1:169–178. doi: 10.1038/35043051. [DOI] [PubMed] [Google Scholar]
- Murakami G, Watabe T, Takaoka K, Miyazono K, Imamura T. Cooperative inhibition of bone morphogenetic protein signaling by Smurf1 and inhibitory Smads. Mol Biol Cell. 2003;14:2809–2817. doi: 10.1091/mbc.E02-07-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parfitt AM, Drezner MK, Glorieux FH, Kanis JA, Malluche H, Meunier PJ, Ott SM, Recker R. Bone histomorphometry: standardization of nomenclature, symbols, and units. J Bone Miner Res. 1987;2:595–610. doi: 10.1002/jbmr.5650020617. [DOI] [PubMed] [Google Scholar]
- Sano Y, Harada J, Tashiro S, Gotoh-Mandeville R, Maekawa T, Ishii S. ATF-2 is a common nuclear target of Smad and TAK1 pathways in transforming growth factor-β signaling. J Biol Chem. 1999;274:27161–27167. doi: 10.1074/jbc.274.13.8949. [DOI] [PubMed] [Google Scholar]
- Su B, Cheng J, Yang J, Guo Z. MEKK2 is required for T-cell receptor signals in JNK activation and interleukin-2 gene expression. J Biol Chem. 2001;276:14784–14790. doi: 10.1074/jbc.M010134200. [DOI] [PubMed] [Google Scholar]
- Sudol M, Hunter T. NeW wrinkles for an old domain. Cell. 2000;103:1001–1004. doi: 10.1016/s0092-8674(00)00203-8. [DOI] [PubMed] [Google Scholar]
- Wang HR, Zhang Y, Ozdamar B, Ogunjimi AA, Alexandrova E, Thomsen GH, Wrana JL. Regulation of cell polarity and protrusion formation by targeting RhoA for degradation. Science. 2003;302:1775–1779. doi: 10.1126/science.1090772. [DOI] [PubMed] [Google Scholar]
- Wong C, Rougier-Chapman EM, Frederick JP, Datto MB, Liberati NT, Li JM, Wang X-F. Smad3-Smad4 and AP-1 complexes synergize in transcriptional activation of the c-Jun promoter by transforming growth factor beta. Mol Cell Biol. 1999;19:1821–1830. doi: 10.1128/mcb.19.3.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozney JM, Rosen V, Celeste AJ, Mitsock LM, Whitters MJ, Kriz R, Hewick R, Wang EA. Novel regulators of bone formation: molecular clones and activities. Science. 1988;242:1528–1534. doi: 10.1126/science.3201241. [DOI] [PubMed] [Google Scholar]
- Wrana JL, Attisano L, Carcamo J, Zentella A, Doody J, Laiho M, Wang XF, Massagué J. TGFβ signals through a heteromeric protein kinase receptor complex. Cell. 1992;71:1003–1014. doi: 10.1016/0092-8674(92)90395-s. [DOI] [PubMed] [Google Scholar]
- Wu XB, Li Y, Schneider A, Yu W, Rajendren G, Iqbal J, Yamamoto M, Alam M, Brunet LJ, Blair HC, et al. Impaired osteoblastic differentiation, reduced bone formation, and severe osteoporosis in noggin-overexpressing mice. J Clin Invest. 2003;112:924–934. doi: 10.1172/JCI15543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi K, Nagai S, Ninomiya-Tsuji J, Nishita M, Tamai K, Irie K, Ueno N, Nishida E, Shibuya H, Matsumoto K. XIAP, a cellular member of the inhibitor of apoptosis protein family, links the receptors to TAB1–TAK1 in the BMP signaling pathway. EMBO J. 1999;18:179–187. doi: 10.1093/emboj/18.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Matsuda K, Bialek P, Jacquot S, Masuoka HC, Schinke T, Li L, Brancorsini S, Sassone-Corsi P, Townes TM, et al. ATF4 is a substrate of RSK2 and an essential regulator of osteoblast biology: implication for Coffin-Lowry Syndrome. Cell. 2004;117:387–398. doi: 10.1016/s0092-8674(04)00344-7. [DOI] [PubMed] [Google Scholar]
- Ying SX, Hussain ZJ, Zhang YE. Smurf1 facilitates myogenic differentiation and antagonizes the bone morphogenetic protein-2-induced osteoblast conversion by targeting Smad5 for degradation. J Biol Chem. 2003;278:39029–39036. doi: 10.1074/jbc.M301193200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Hebert MC, Zhang YE. TGF-β receptor-activated p38 MAP kinase mediates Smad-independent TGF-β responses. EMBO J. 2002;21:3749–3759. doi: 10.1093/emboj/cdf366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Feng XH, Derynck R. Smad3 and Smad4 cooperate with c-Jun/c-Fos to mediate TGF-β-induced transcription. Nature. 1998;394:909–913. doi: 10.1038/29814. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Chang C, Gehling DJ, Hemmati-Brivanlou A, Derynck R. Regulation of Smad degradation and activity by Smurf2, an E3 ubiquitin ligase. Proc Natl Acad Sci USA. 2001;98:974–979. doi: 10.1073/pnas.98.3.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Harris SE, Horn D, Geng Z, Nishimura R, Mundy GR, Chen D. Bone morphogenetic protein receptor signaling is necessary for normal murine postnatal bone formation. J Cell Biol. 2002;157:1049–1060. doi: 10.1083/jcb.200109012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Qiao M, Oyajobi BO, Mundy GR, Chen D. E3 ubiquitin ligase Smurf1 mediates core-binding factor alpha1/Runx2 degradation and plays a specific role in osteoblast differentiation. J Biol Chem. 2003;278:27939–27944. doi: 10.1074/jbc.M304132200. [DOI] [PubMed] [Google Scholar]
- Zhao M, Qiao M, Harris SE, Oyajobi BO, Mundy GR, Chen D. Smurf1 inhibits osteoblast differentiation and bone formation in vitro and in vivo. J Biol Chem. 2004;279:12854–12859. doi: 10.1074/jbc.M313294200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng C, Xiang J, Hunter T, Lin A. The JNKK2–JNK1 fusion protein acts as a constitutively active c-Jun kinase that stimulates c-Jun transcription activity. J Biol Chem. 1999;274:28966–28971. doi: 10.1074/jbc.274.41.28966. [DOI] [PubMed] [Google Scholar]
- Zhu H, Kavsak P, Abdollah S, Wrana JL, Thomsen GH. A SMAD ubiquitin ligase targets the BMP pathway and affects embryonic pattern formation. Nature. 1999;400:687–693. doi: 10.1038/23293. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.