Abstract
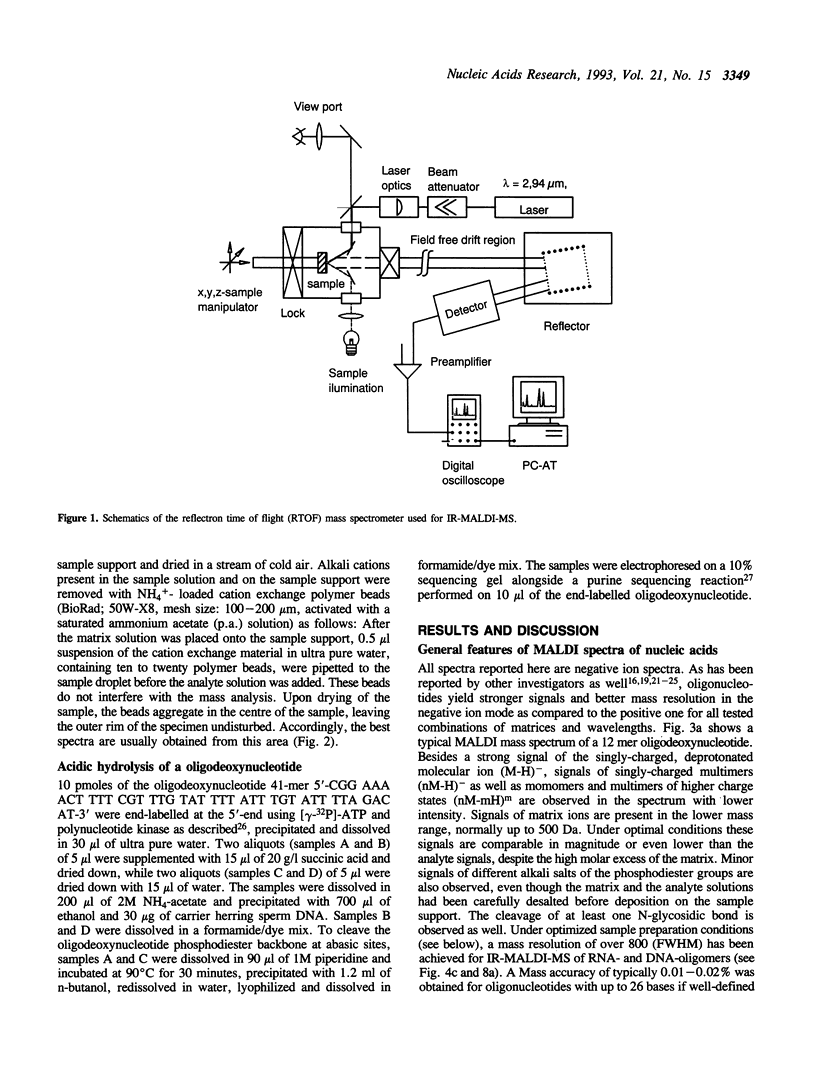
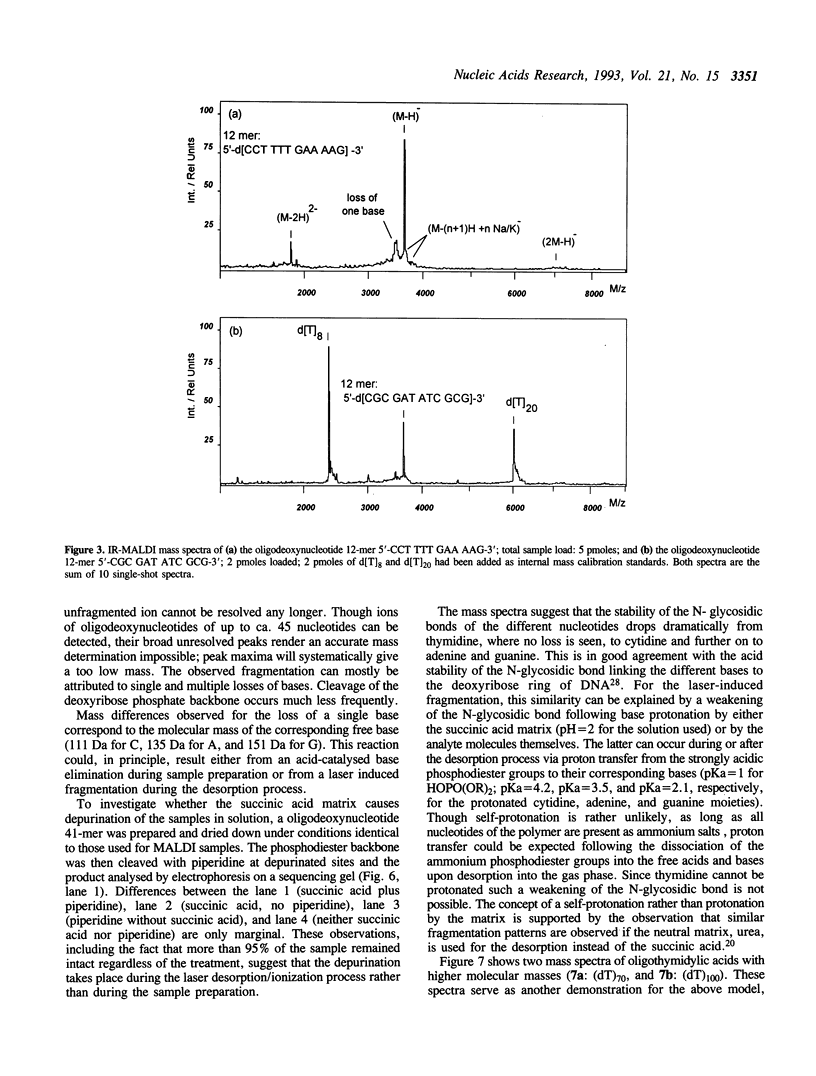
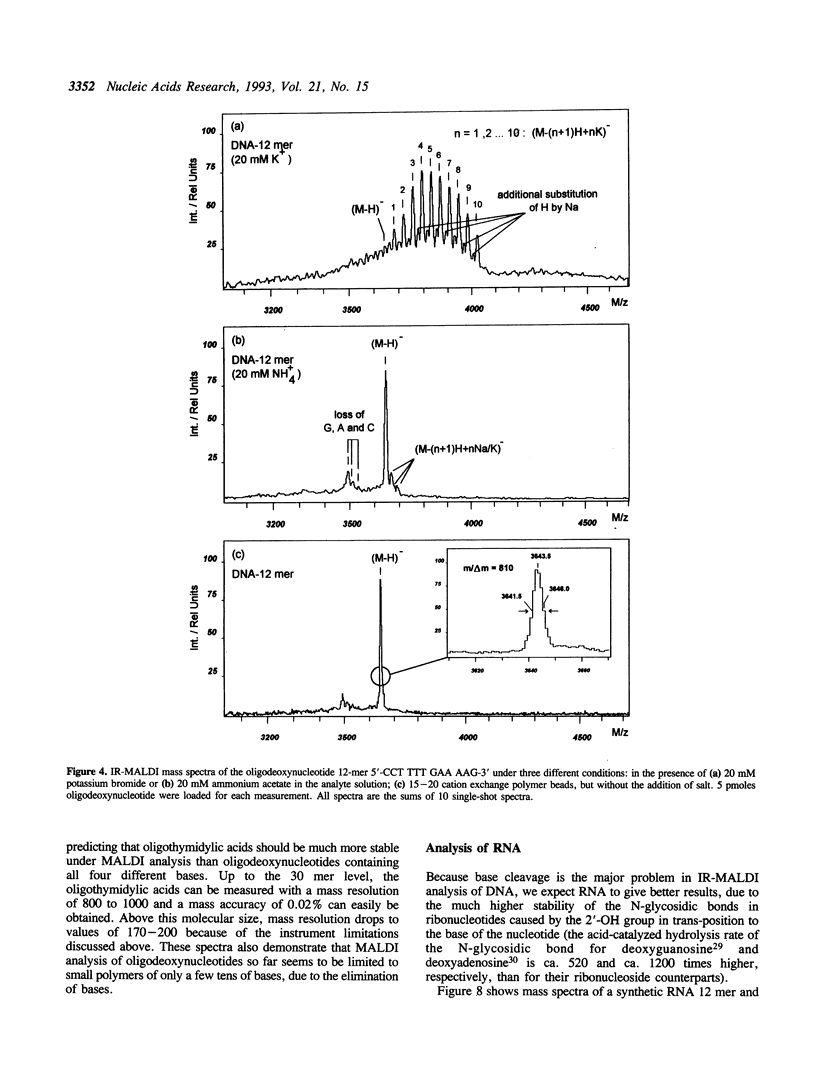
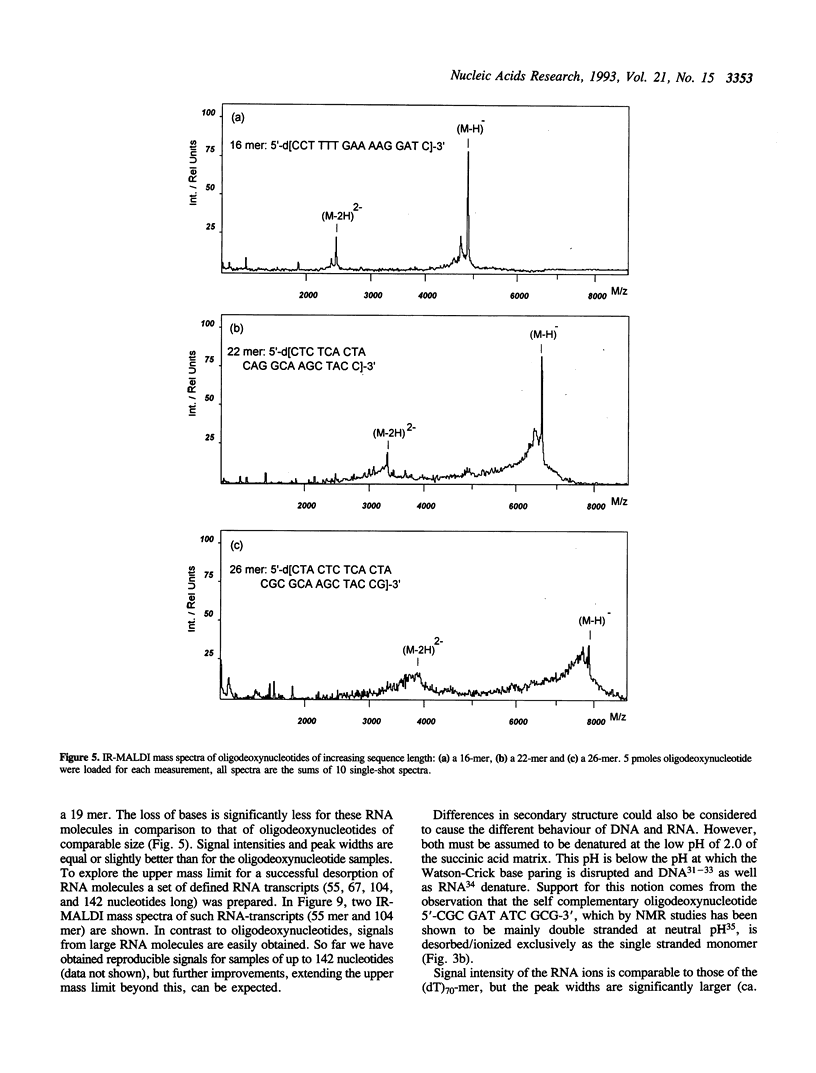
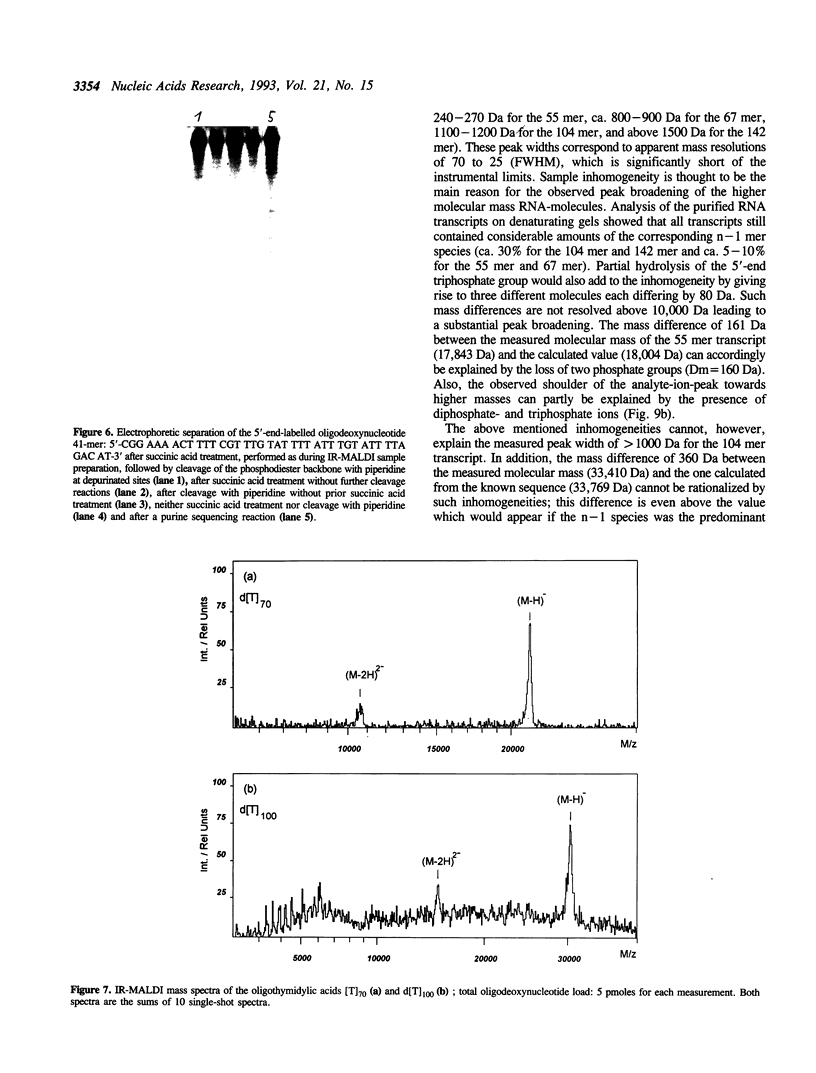
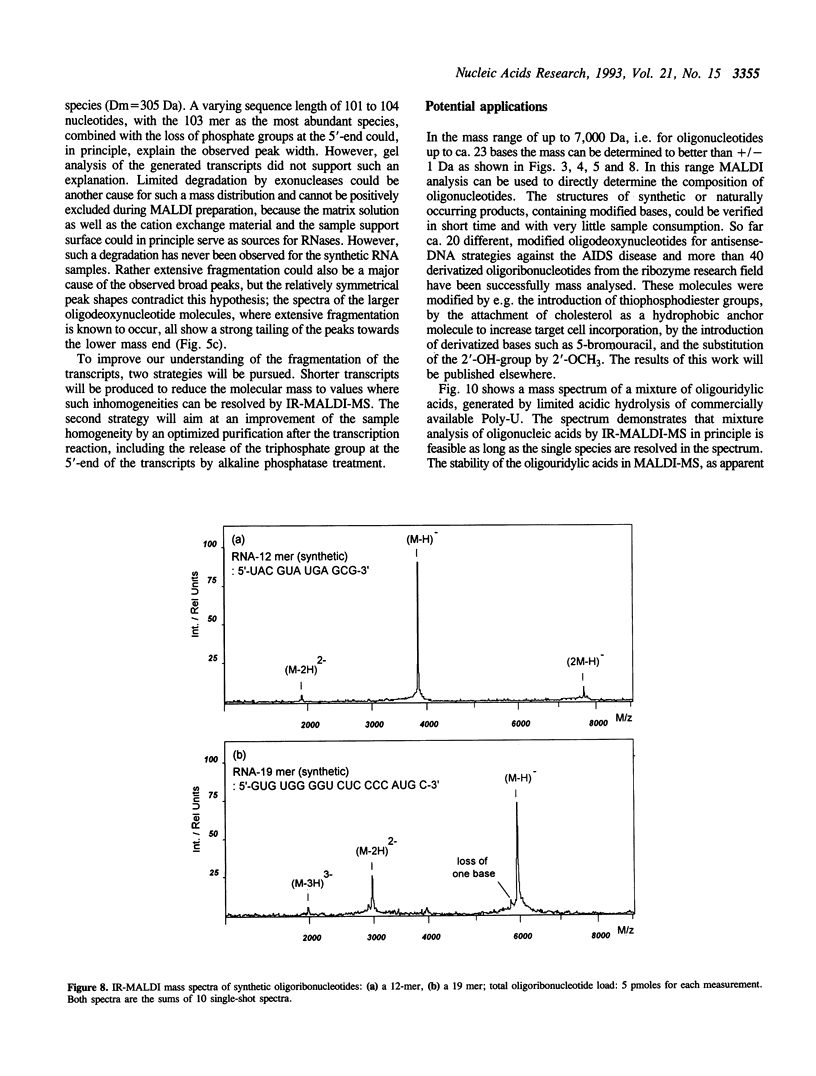
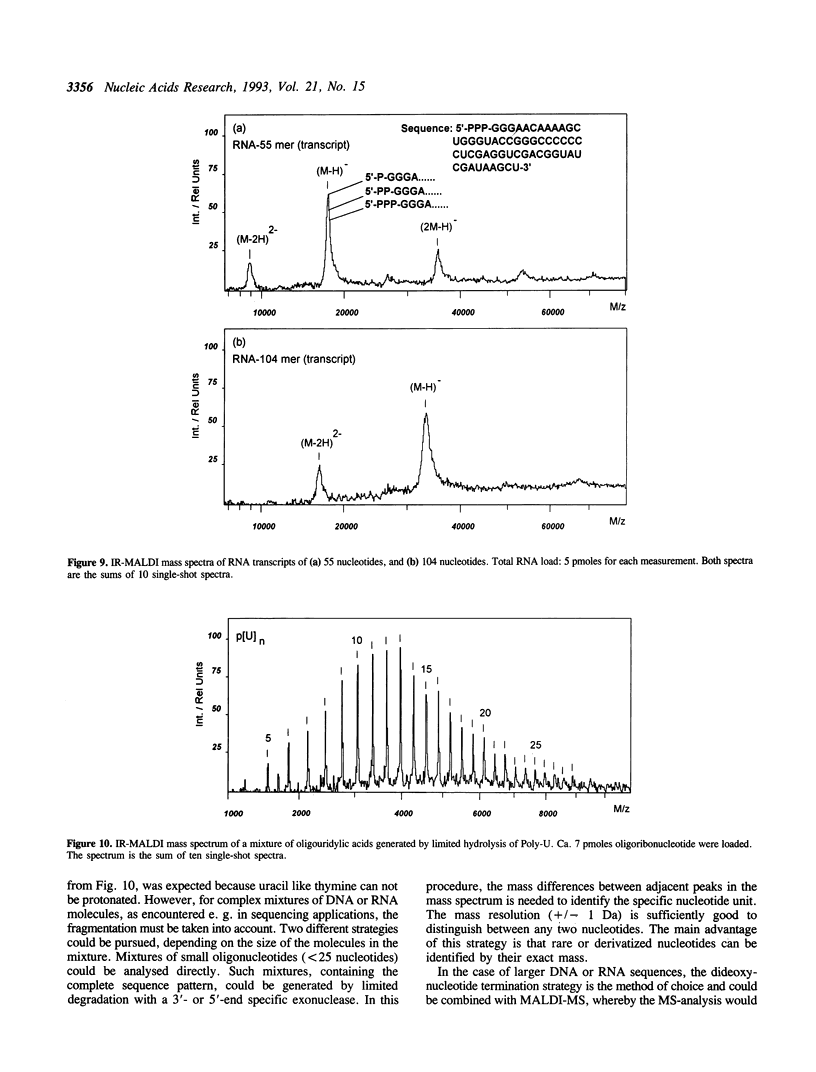
Matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS) with infrared laser light of a wavelength of 2.94 microns has been used for the analysis of nucleic acids. Spectra of oligodeoxynucleotides up to 26 nucleotides, oligothymidylic acids up to 100 nucleotides as well as different synthetic RNA oligomers and RNA transcripts up to 104 nucleotides are presented. A main problem in the analysis of oligodeoxynucleotides was found to be related to the loss of bases. The stability of oligothymidylic acids as opposed to oligodeoxynucleotides containing all four bases indicates that the loss of bases is correlated with A, C and G protonation which decreases the stability of the N-glycosidic bond. Experiments indicate that the breakage of the N-glycosidic bond probably occurs during the desorption process due to proton transfer from the phosphodiester groups to the ionizable bases. RNA displayed a significantly higher stability in MALDI-MS due to the presence of a 2'-OH group. Consequently, signals of RNA transcripts with a length of up to 142 nucleotides could be detected by MALDI-MS. Technical details of the method, including the distribution of positive counterions on the phosphodiester backbone, the upper mass limit and mass accuracy are discussed along with a number of potential analytical applications.
Full text
PDF



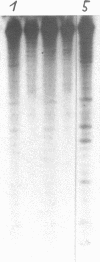
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ambrose B. J., Pless R. C. DNA sequencing: chemical methods. Methods Enzymol. 1987;152:522–538. doi: 10.1016/0076-6879(87)52059-6. [DOI] [PubMed] [Google Scholar]
- Beavis R. C., Chait B. T. Cinnamic acid derivatives as matrices for ultraviolet laser desorption mass spectrometry of proteins. Rapid Commun Mass Spectrom. 1989 Dec;3(12):432–435. doi: 10.1002/rcm.1290031207. [DOI] [PubMed] [Google Scholar]
- Beavis R. C., Chait B. T. Factors affecting the ultraviolet laser desorption of proteins. Rapid Commun Mass Spectrom. 1989 Jul;3(7):233–237. doi: 10.1002/rcm.1290030708. [DOI] [PubMed] [Google Scholar]
- Beavis R. C., Chait B. T. Matrix-assisted laser-desorption mass spectrometry using 355 nm radiation. Rapid Commun Mass Spectrom. 1989 Dec;3(12):436–439. doi: 10.1002/rcm.1290031208. [DOI] [PubMed] [Google Scholar]
- Funk W. D., Pak D. T., Karas R. H., Wright W. E., Shay J. W. A transcriptionally active DNA-binding site for human p53 protein complexes. Mol Cell Biol. 1992 Jun;12(6):2866–2871. doi: 10.1128/mcb.12.6.2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillenkamp F., Karas M. Mass spectrometry of peptides and proteins by matrix-assisted ultraviolet laser desorption/ionization. Methods Enzymol. 1990;193:280–295. doi: 10.1016/0076-6879(90)93420-p. [DOI] [PubMed] [Google Scholar]
- Karas M., Hillenkamp F. Laser desorption ionization of proteins with molecular masses exceeding 10,000 daltons. Anal Chem. 1988 Oct 15;60(20):2299–2301. doi: 10.1021/ac00171a028. [DOI] [PubMed] [Google Scholar]
- Keough T., Baker T. R., Dobson R. L., Lacey M. P., Riley T. A., Hasselfield J. A., Hesselberth P. E. Antisense DNA oligonucleotides. II: The use of matrix-assisted laser desorption/ionization mass spectrometry for the sequence verification of methylphosphonate oligodeoxyribonucleotides. Rapid Commun Mass Spectrom. 1993 Mar;7(3):195–200. doi: 10.1002/rcm.1290070306. [DOI] [PubMed] [Google Scholar]
- Nordhoff E., Ingendoh A., Cramer R., Overberg A., Stahl B., Karas M., Hillenkamp F., Crain P. F. Matrix-assisted laser desorption/ionization mass spectrometry of nucleic acids with wavelengths in the ultraviolet and infrared. Rapid Commun Mass Spectrom. 1992 Dec;6(12):771–776. doi: 10.1002/rcm.1290061212. [DOI] [PubMed] [Google Scholar]
- Record M. T., Jr Electrostatic effects on polynucleotide transitions. II. Behavior of titrated systems. Biopolymers. 1967;5(10):993–1008. doi: 10.1002/bip.1967.360051011. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schieltz D. M., Chou C. W., Luo C. W., Thomas R. M., Williams P. Mass spectrometry of DNA mixtures by laser ablation from frozen aqueous solution. Rapid Commun Mass Spectrom. 1992 Oct;6(10):631–636. doi: 10.1002/rcm.1290061009. [DOI] [PubMed] [Google Scholar]
- Shapiro R., Danzig M. Acidic hydrolysis of deoxycytidine and deoxyuridine derivatives. The general mechanism of deoxyribonucleoside hydrolysis. Biochemistry. 1972 Jan 4;11(1):23–29. doi: 10.1021/bi00751a005. [DOI] [PubMed] [Google Scholar]
- Spengler B., Pan Y., Cotter R. J., Kan L. S. Molecular weight determination of underivatized oligodeoxyribonucleotides by positive-ion matrix-assisted ultraviolet laser-desorption mass spectrometry. Rapid Commun Mass Spectrom. 1990 Apr;4(4):99–102. doi: 10.1002/rcm.1290040402. [DOI] [PubMed] [Google Scholar]
- Wu K. J., Steding A., Becker C. H. Matrix-assisted laser desorption time-of-flight mass spectrometry of oligonucleotides using 3-hydroxypicolinic acid as an ultraviolet-sensitive matrix. Rapid Commun Mass Spectrom. 1993 Feb;7(2):142–146. doi: 10.1002/rcm.1290070206. [DOI] [PubMed] [Google Scholar]
- Zoltewicz J. A., Clark D. F., Sharpless T. W., Grahe G. Kinetics and mechanism of the acid-catalyzed hydrolysis of some purine nucleosides. J Am Chem Soc. 1970 Mar 25;92(6):1741–1749. doi: 10.1021/ja00709a055. [DOI] [PubMed] [Google Scholar]