Abstract
Whole-genome sequencing of patient DNA can facilitate diagnosis of a disease, but its potential for guiding treatment has been under-realized. We interrogated the complete genome sequences of a 14-year-old fraternal twin pair diagnosed with dopa (3,4-dihydroxyphenylalanine)–responsive dystonia (DRD; Mendelian Inheritance in Man #128230). DRD is a genetically heterogeneous and clinically complex movement disorder that is usually treated with l-dopa, a precursor of the neurotransmitter dopamine. Whole-genome sequencing identified compound heterozygous mutations in the SPR gene encoding sepiapterin reductase. Disruption of SPR causes a decrease in tetrahydrobiopterin, a cofactor required for the hydroxylase enzymes that synthesize the neurotransmitters dopamine and serotonin. Supplementation of l-dopa therapy with 5-hydroxytryptophan, a serotonin precursor, resulted in clinical improvements in both twins.
INTRODUCTION
Subclassification of phenotypically similar but genetically heterogeneous conditions by identifying underlying causative alleles can be pivotal for precise disease diagnosis and treatment. High-throughput sequencing of patient genomes could potentially facilitate the diagnosis of rare diseases. Identified variants can be cross-checked with databases for previous associations with disease, and benign variants with high allele frequency can be eliminated from consideration using population-variation databases (1). The remaining variants can be assessed for their effects on genes and those genes can be assessed for their association with disease. These approaches require integrated and accurate databases as well as best practices guidelines (2).
Dopa (3,4-dihydroxyphenylalanine)–responsive dystonia (DRD) [Mendelian Inheritance in Man (MIM) #128230], formally known as “hereditary dystonia with marked diurnal variation” (Segawa dystonia), is a genetically and clinically heterogeneous movement disorder (3). DRD typically begins in childhood after a period of normal development and frequently manifests variable severity during the day (reduced dystonia upon awakening, increased dystonia by midday). The differential diagnosis for DRD includes early-onset parkinsonism, cerebral palsy, and early-onset primary dystonia (4–6). The clinical diagnosis of DRD is based on neurological presentation, age of onset and progression of the disease, mode of inheritance, concentrations of neurotransmitter metabolites and pterins (cofactors for neurotransmitter-producing enzymes) in the cerebrospinal fluid (CSF), and the degree of responsiveness to l-dopa treatment. Sustained clinical benefit from very low dose l-dopa administration is a clinical hallmark of DRD. However, a range of clinical responses to l-dopa therapy have been documented (7) and l-dopa therapy alone may not be sufficient for complete alleviation of clinical symptoms.
DRD can be inherited as either an autosomal dominant or recessive trait and is associated with mutations in genes encoding guanosine 5′-triphosphate (GTP) cyclohydrolase (GCH1), tyrosine hydroxylase (TH), and sepiapterin reductase (SPR) (Fig. 1). GCH1 and SPR are enzymes of the tetrahydrobiopterin (BH4) biosynthesis pathway. BH4 serves as a cofactor for tyrosine and tryptophan hydroxylases in the initial biosynthesis of the neurotransmitters dopamine, noradrenaline, and serotonin. TH converts tyrosine to l-dopa, a precursor of dopamine and noradrenaline (8) (Fig. 1). In a study of 64 patients diagnosed with DRD, ~83% of cases were caused by autosomal dominant or de novo point mutations and deletions in GTP cyclohydrolase, whereas autosomal recessive cases were caused by mutations in tyrosine hydroxylase (~5%), sepiapterin reductase (~3%), or parkin (encoded by the PARK2 gene, a gene implicated in juvenile-onset Parkinson disease) (~3%). Five percent of DRD cases had unknown genetic causes (9).Molecular genetic testing has proved a valuable tool for diagnosing DRD; however, until recently, clinical molecular genetic assays were limited to the identification of mutations in the TH and GCH1 genes (10). Heterozygous deletion of the entire TH gene, which potentially results in decreased endogenous dopamine production, has also been reported in a patient with adultonset Parkinson disease (11), a common movement disorder caused by loss of dopamine-producing neurons in the brain’s nigrostriatal pathway.
Fig. 1.
Metabolic pathways of neurotransmitter production. DRD has been associated with mutations in the genes encoding GTP cyclohydrolase (GCH1), tyrosine hydroxylase (TH), and sepiapterin reductase (SPR) (boxed), which are enzymes associated with production of the neurotransmitters dopamine and serotonin. The catalytic action of GCH1 is the rate-limiting step in production of tetrahydrobiopterin (BH4), a cofactor for the tyrosine and tryptophan hydroxylases. Disruption of the GCH1 gene can cause autosomal dominant DRD. Autosomal recessive DRD is caused by mutations in TH and SPR. Both 5-hydroxytryptophan (5-HTP) and dopamine production are disrupted by mutations in SPR whereas only dopamine production is disrupted by mutations in TH.
Here, we studied a fraternal twin pair diagnosed with DRD, who had no identified deleterious variants in the TH or GCH1 genes. Sequencing of the SPR gene was not available through a clinical laboratory at the time this study was initiated and was not performed (see Materials and Methods). Because the primary candidate genes for DRD were eliminated, we used high-throughput sequencing (12, 13) to interrogate the whole genomes of the male and female twin to identify potential causative genetic variants.
RESULTS
Clinical presentation
The patients were two affected 14-year-old fraternal twins, who were diagnosed with DRD at age 5 after l-dopa was found to alleviate the clinical symptoms of dystonia in one twin. The subjects were born at 36 weeks of gestation after a pregnancy complicated by a hypercoagulable state in their mother that required heparin treatment. The perinatal history was uneventful. Well-child evaluations in the first year of life revealed generalized hypotonia and global developmental delay. These clinical observations prompted an initial evaluation including imaging studies of the brain (magnetic resonance imaging) that revealed periventricular leukomalacia in the male patient and basic metabolic tests that were normal in both twins. CSF was not obtained before treatment with l-dopa. The female twin was more severely affected and subsequently developed dystonic movements, hypokinesia, rigidity, tremor, oculogyric crises (ocular dystonic movements), and seizures. Her brother had milder disease symptoms and was originally diagnosed with static encephalopathy (cerebral palsy). However, later serial examinations showed the appearance of progressive subtle dystonia at age 5 years. Whereas the female patient had a diurnal fluctuation of neurological symptoms with less severe symptoms in the morning and more severe symptoms in the afternoon, the male patient did not.
At age 5 years, a trial of l-dopa/carbidopa at a ratio of 10:100, one-quarter tablet a day increasing to one-quarter a tablet three times per day over several days, reduced clinical symptoms by day 3 but was accompanied by mild dyskinesia. The dosage, therefore, was reduced initially but then was reinstated. Both patients are in middle school following a regular curriculum and have excellent academic performance despite reportedly a reduced attention span. At age 14 years and on l-dopa/carbidopa 25:100 three times per day, the affected male was found to have mild tremor and dystonic posturing of the hands upon neurological examination. His sister demonstrated slightly unsteady gait; mild choreiform movements in the tongue; mild dysphonia; mild dystonia in the neck, shoulder, and hands; and mild bradykinesia. Her history is also significant for respiratory difficulties thought to be secondary to intermittent laryngospasm. The immediate family history was negative for movement disorders or other neurological diseases apart from fibromyalgia and depression. The diagnosis of recessive dystonia in the probands was complicated by the presence of a first cousin with reported juvenile seizures and a third cousin and her four children diagnosed with an unspecified neurological disorder (Fig. 2).
Fig. 2.
Pedigree of a family segregating recessive DRD, depression, and fibromyalgia. Pedigree of the family with the two DRD-affected probands (shaded), male and female fraternal twins. Their DRD is due to disruption of SPR activity resulting in impaired BH4 cofactor synthesis, leading to disruption in the production of the neurotransmitters dopamine, noradrenaline, and serotonin. In addition to DRD in the probands, the family has a history of depression and fibromyalgia on either side of the pedigree. Segregation of the two SPR mutations is shown for all individuals evaluated.
Genome variation
DNA was extracted from peripheral blood cells obtained from both affected twins, an unaffected sibling, and their parents. DNA from the twins was subjected to whole-genome sequencing on the SOLiD platform. In total, 178.4 giga–base pairs (Gbp) of sequence data was produced and aligned to the human reference genome, resulting in an average sequence coverage of 29.4 and 30.0 for the male and female twin, respectively (59-fold for sites shared by both twins).
A set of putative, high-quality sequence differences between each twin and the reference genome (hg18) was identified, and variants shared by both twins were subsequently analyzed. About 90% of the variants discovered were also identified in the dbSNP database (http://www.ncbi.nlm.nih.gov/projects/SNP/), with one variant discovered per 1100 bp on average across the genome, which is similar to reported values (14). The degree of variant overlap confirmed that the twins were dizygotic, consistent with the model of recessive dystonia in this family. The variant data were next filtered to allow removal of likely benign variants with the dbSNP v.129 (15) and Thousand Genomes (1) databases of common and likely non–disease-causing mutations, as well as the Baylor Human Genome Sequencing Center’s maintained database of common variants from other sequencing projects. Finally, mutations that caused nonsynonymous changes to protein products were classified (Table 1). There were no remaining rare homozygous mutations shared between both twins, and no large genomic regions with stretches of homozygous mutations, which is consistent with the absence of consanguinity.
Table 1.
Single-nucleotide variants in the sequence of the DRD twins. All high-quality variants first observed from primary alignment of sequence reads to the current reference haploid human genome were then filtered for coding regions and annotated if they cause protein coding mutations. These variants were filtered further against databases of known and common variation to enrich for rare variants, which are more likely to be disease-causing. Finally, candidate genes were identified, under a recessive inheritance model, by homozygosity or by identifying genes that harbor two or more variants.
| Nucleotide variants | IV-2 (male subject) |
IV-3 (female subject) |
Shared |
|---|---|---|---|
| All variants | 2,427,038 | 2,504,162 | 1,631,770 |
| % dbSNP129 | 88.7 | 88.1 | |
| Variant density (bp−1) | 1/1112 | 1/1078 | |
| Coding | 13,352 | 14,961 | 9531 |
| Nonsynonymous | 6432 | 7141 | 4605 |
| Rare nonsynonymous | 174 | 175 | 77 |
| Candidate genes | 6 | 9 | 3 |
After overlapping shared mutations, filtering, and genetic annotation, only three genes were identified that contained two or more predicted amino acid–altering heterozygous mutations (table S1). One of these (ZNF544) encodes a computationally predicted zinc finger protein with no known function or targets, another predicts an open reading frame (C2orf16), and the third is the SPR gene encoding sepiapterin reductase. Subsequent automated annotation of these genes by comparison to the MIM disease database (16) indicated a known association of SPR with DRD and no associations of either of the other two genes with any disease.
The identified variants in SPR were NM_003124:c.448A>G (chromosome 2: 72,969,094, p.Arg150Gly) and NM_003124:c.751A>T (chromosome 2:72,972,139, p.Lys251X). The former mutation occurs in a β strand secondary structure element in close proximity to a substrate binding region, and the latter results in a truncation of the last 10 amino acid residues of SPR destroying one entire β strand (17). Both mutations have been previously identified in two Caucasian families (18, 19), but in both cases, the mutation was homozygous rather than a compound heterozygote. Functional studies for each of the putative pathogenic variants were reported and found to be deleterious to SPR activity (18, 19). In these studies, SPR activity was measured with a biochemical assay either using skin fibroblasts taken from the patient or by cloning the mutated gene in a bacterial vector and subsequent purification. Disruption of SPR prevents the regeneration of BH4, which is an important cofactor for the production of both dopamine and serotonin (Fig. 1). Thus, the recommended treatment of DRD caused by SPR mutations is with both the dopamine precursor l-dopa, which the twins were already prescribed, and the serotonin precursor 5-hydroxytryptophan (5-HTP), which the twins were not receiving. Both compounds can readily cross the blood-brain barrier. The serotonin pathway may be further enhanced by the addition of selective serotonin reuptake inhibitors (SSRIs) used to treat depression.
Validation and segregation
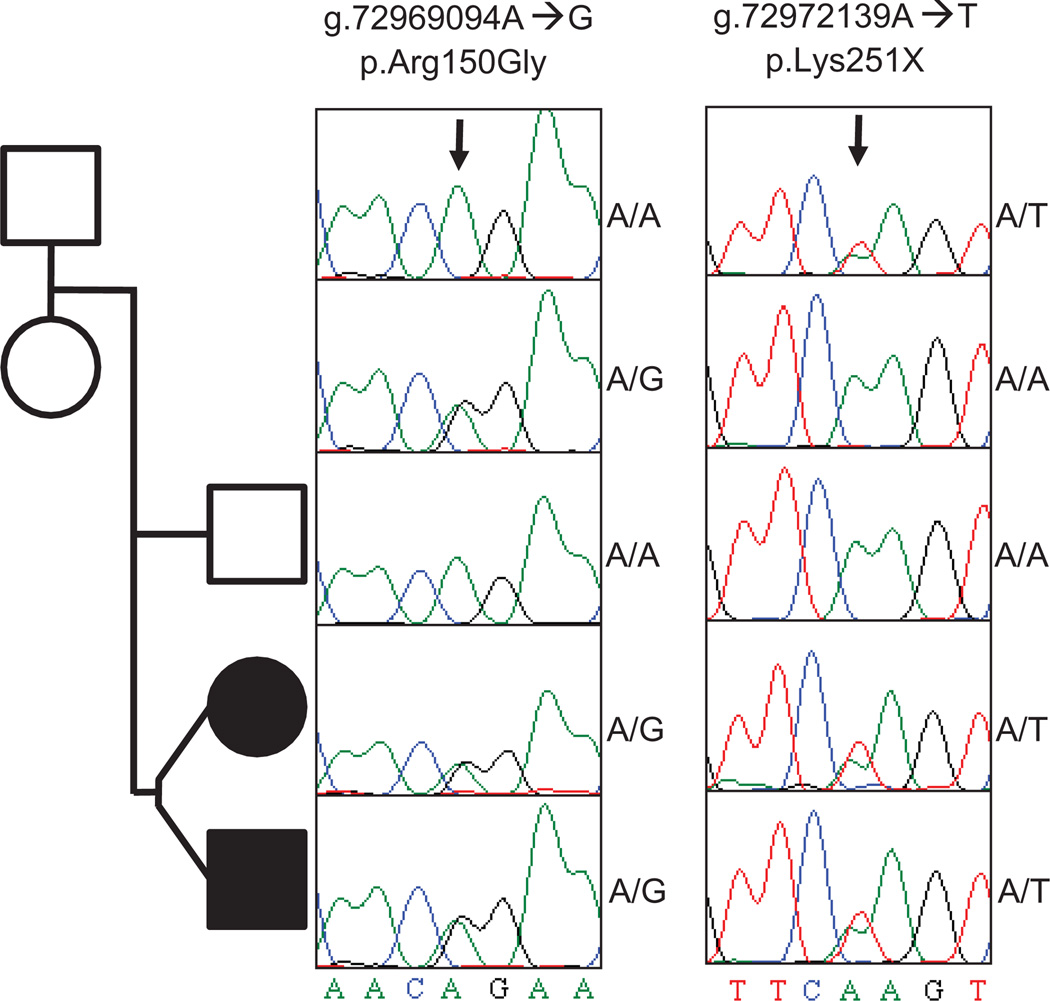
To test for a pattern of segregation of these alleles that is consistent with their causative role in DRD, we designed oligonucleotide primers to correspond to sequences that flank both mutations, and we used them to PCR (polymerase chain reaction) amplify and capillary sequence all members of the immediate family. Both mutations were confirmed as compound heterozygous mutations in the affected twins, and the c.751A>T (p.Lys251X) nonsense mutation and the c.448A>G (p.Arg150Gly) missense mutation were found in the heterozygous state in the mother and father, respectively. Neither mutation was found in the unaffected sibling, although the individual alleles were identified in members of previous generations (Fig. 3).
Fig. 3.
Validation and segregation of two deleterious SPR alleles. Shown is the pedigree of the two DRD-affected probands (shaded), their unaffected sibling, and their parents. Squares indicate male subjects and circles female subjects. The Sanger sequencing traces showing the SPR genotype of each member of the pedigree are shown. p.Arg150Gly refers to the A→G mutation on chromosome 2 at nucleotide 72,969,094, leading to the amino acid missense mutation; p.Lys251X indicates the A→T mutation on chromosome 2 at nucleotide 72,972,139, causing an amino acid nonsense mutation. The variants are deleterious to SPR activity but have only been observed, separately, as homozygous mutations from presumed consanguineous kindreds. The unaffected mother is homozygous (A/A) for the wild-type allele at the first locus but heterozygous (A/T) for the pathogenic p.Lys251X allele at the second locus. Similarly, the unaffected father is heterozygous (A/G) for the pathogenic p.Arg150Gly allele at the first locus and homozygous (A/A) for the wild-type allele at the second locus. Each DRD-affected proband is heterozygous (A/G and A/T) for the pathogenic mutations at both alleles, indicating a new compound heterozygous combination of mutant alleles that is causative for DRD. The mutations result in reduction of SPR-mediated synthesis of BH4, a coenzyme needed for neurotransmitter production. Absence or reduction of the neurotransmitter dopamine causes distinct clinical symptoms including dystonia.
Efficacy of 5-HTP treatment
As a consequence of the molecular diagnosis, the treatments for the male and female twin were modified to include 5-HTP (0.8 and 1.2 mg/kg, respectively). They have been on this therapy for ~4 months at the time of writing. Both patients underwent periodic follow-up visits at the same time of day with one physician (J.F.) who assessed the impact of the medications. According to the physician report, both patients showed the first signs of improvement after 1 to 2 weeks, and their condition reached a plateau after 2 months of therapy. The male DRD patient reported improved focus in school, as well as improved coordination in athletics. Further, the male showed reduced drooling and hand tremor, and objective evidence for the latter was provided by serial handwriting samples (fig. S1). The female twin reported reduced frequency of laryngeal spasms, improved sleep and focus, and improved tolerance for exercise and was able to resume participation in sports after a 14-month absence. In the female DRD patient, there were also reduced choreiform movements of the tongue by objective physical examination (J.F.). Neither twin reported significant side effects from the therapy.
DISCUSSION
This work demonstrates the application of whole-genome sequencing to the discovery of the cause of autosomal recessive DRD in an affected fraternal twin pair. Whole-genome sequencing of both twins implicated three genes with variants potentially causative of autosomal recessive DRD. Only one of these genes, SPR was a high-priority candidate by functional criteria. This gene encodes the enzyme sepiapterin reductase, which catalyzes the reaction of 6-pyruvoyl-tetrahydropterin to BH4, and has been implicated previously in DRD. BH4 is an important coenzyme for the production of the neurotransmitters dopamine, noradrenalin, and serotonin. The involvement of SPR was unexpected in the twin pair given their clinical symptoms including the lack of intellectual disability, their sustained response to monotherapy with l-dopa (18, 20), and a family history of neurological disorders that complicated the interpretation of the potential inheritance pattern (Fig. 2). Our observations highlight both the challenges of clinical phenotyping, especially for rare diseases by nonspecialists, and the ability of whole-genome sequencing to identify susceptibility variants, especially when initial candidate genes have been eliminated by more traditional locus-specific gene testing. Whole-genome sequencing facilitated both the diagnostic workup and the medical management of the fraternal twins with DRD.
The nonsense (p.Lys251X) and missense (p.Arg150Gly) mutations in the SPR gene are predicted to be deleterious by disrupting the secondary structure and binding to NADP (nicotinamide adenine dinucleotide phosphate) of the SPR protein, respectively. Critically, both variants have been previously reported and deemed to be deleterious for SPR activity through functional biochemical assays (18, 19, 21). Both SPR mutations were originally reported as homozygous alleles but, in this study, were found to be compound heterozygous alleles.
The identification of the involvement of the SPR gene in this family manifesting DRD has directly influenced clinical management of the individuals with DRD. Although DRD caused by mutations in SPR can be treated with dopamine therapy alone, typically the response is limited when compared to treatment of DRD caused by autosomal dominant mutations in GCH1. The administration of 5-HTP, a serotonin precursor, in addition to l-dopa has been shown to greatly improve cognitive abilities and gait and to minimize pyramidal signs in DRD caused by mutations in SPR (18). Moreover, supplementation with 5-HTP potentially allows tapering of the dosage of l-dopa in these patients. Treatment with BH4 has also been shown to reduce clinical symptoms in DRD (22). The identification of these medically actionable alleles in the fraternal twin pair in this study prompted administration of 5-HTP as a prelude to other possible adjuvant therapies that directly stimulate the serotonin pathways. 5-HTP therapy led to improvements in the male twin’s fine motor skills, including handwriting and athletic ability. Furthermore, in the more severely affected female twin, 5-HTP therapy appears to have led to abatement of a severe recurrent laryngospasm, which would often end in vomiting, preventing physical activity and greatly reducing quality of life. Although identification of mutations that cause recessive disorders has previously been demonstrated (12, 23) and in some cases has led to the alteration of treatment (24, 25), the application of whole-genome sequencing to a direct alteration in clinical management has not yet been fully realized. This study demonstrates the utility of whole-genome sequencing and databases of deleterious variants to facilitate both diagnostic and therapeutic medical management decisions.
Because genetic diagnosis using whole-genome sequencing in individual families is becoming more accessible, the debate concerning the underlying value of the extensive information that is generated has intensified (26). Here, we demonstrate the application of the technique to a recessive disease of unknown cause. Further, the new knowledge afforded by the molecular testing resulted in the implementation of additional treatment options and further optimized patient care.
The identification of mutations in SPR accounts for the previous history and diagnosis of DRD in this family. With hindsight, SPR deficiency is a logical potential differential diagnosis. However, before whole-genome sequencing, implication of SPR mutations in this disease was hampered by an atypical DRD presentation and the presence of other neurological disorders in the family that were initially thought to be related. Further, it is tempting to suggest the use of candidate gene sequencing rather than whole-genome sequencing for this family now that the identified causative gene is a known good candidate. Candidate gene sequencing, however, suffers from several disadvantages that whole-genome sequencing does not. Candidate gene sequencing may fail to analyze the correct genes, typically does not interrogate all of the biologically important regions of a gene (for example, untranslated regions and promoters), does not eliminate other genes from consideration, and rapidly becomes more expensive and time-consuming compared with whole-genome sequencing as the number of loci deemed clinically relevant increases. Although candidate gene sequencing would have been successful in this case, clinical testing for SPR mutations was unavailable at the time of the initial evaluation and initiation of therapy, and attention to the potential gene involved was not directed by clinically observed phenotypic information.
In addition to providing comprehensive analysis of the potential underlying genetic contributions to specific diseases, whole-genome sequencing may provide insights into other phenotype/genotype relationships and potentially illuminate the genetic susceptibility of other family members. Initial pedigree analysis of the family of the DRD twins identified a family history of depression in three generations on the paternal side of the family and a diagnosis of fibromyalgia in two generations on the maternal side. The individual pathogenic alleles in the SPR gene cosegregate with the fibromyalgia phenotype (Fig. 2), although not with the depression phenotype. Because fibromyalgia can respond to serotonin reuptake inhibitor drugs, suggesting that pathogenesis of this disease may be related to reduced serotonin, we hypothesize that heterozygous loss-of-function SPR mutations may contribute to a genetic susceptibility to fibromyalgia.
Challenges in clinical application of whole-genome sequencing include informed consent of subjects, unknown medical significance of most variants, uncovering both susceptibility variants and recessive risk alleles, development of databases of medically actionable variants, and counseling based on variant interpretation (26). The current case study of a fraternal twin pair with DRD, however, argues for the clinical utility of information gained from whole-genome sequencing in place of molecular genetic testing of individual genes, where there is even minimal suspicion of genetic heterogeneity for the clinical phenotype.
MATERIALS AND METHODS
Study participants
The study family consisted of affected twins, an unaffected older brother, unaffected parents, and cousins (Fig. 2). The affected twins have been followed by a single physician (J.F.), who initiated therapy with 5-HTP, in addition to the l-dopa/carbidopa that the children were already receiving, and evaluated the patients’ response to medications. The patients’ mother (III-3) and maternal grandmother (II-6) were diagnosed with fibromyalgia and responded well to conventional therapies. The paternal aunt (III-1) and paternal grandmother (II-2) had a history of major depression that was ameliorated by treatment with SSRIs. All adult members of this investigation provided written informed consent for participation with additional assent provided by the twins. The study was approved by the institutional review board at Baylor College of Medicine.
DNA sequence analysis
Whole-genome sequencing was conducted with the SOLiD 4 sequencing platform (27) on genomic DNA isolated from the blood of both twins. Sequencing consisted solely of 50-bp reads generated from 1-kbp mate-pair libraries, which were subsequently aligned to the human reference genome with the BFast application (28). High-resolution oligonucleotide aCGH (array comparative genomic hybridization) was conducted with Agilent 1M oligonucleotide arrays and sex-matched controls for both patients with DRD. We did not observe any potentially pathogenic copy number variants in candidate genes for dystonia. Sequence data are available in dbGAP pending reconsent by the twins at age 18.
Bioinformatic discovery and analysis of variants
Single-nucleotide variants were discovered with the Pileup (29) application and were subsequently filtered for variants shared between the twins. Variants that were uncommon in the population (minor allele frequency <0.5%) and were nonsynonymous or otherwise affected protein translation, microRNA, or transcription factor binding were preferentially investigated (30). Assuming recessive inheritance, we identified genes that contained either homozygous or two or more mutations presumed to represent compound heterozygous alleles. These genes were then annotated with known functional data and known association with disease and subsequently cross-referenced to a database of known variants affecting genes involved in BH4 production and utilization (20).
Variant validation and segregation analysis
The genomic regions flanking chromosome 2:72,969,094 (SPR:R150) and chromosome 2:72,972,139 (SPR:K251) were amplified by PCR and were capillary sequenced directly in all members of the study. Analysis of the Sanger sequence was conducted by the SNP-detector application (31) and by manual visual inspection of the trace sequence with the examiner masked as to the subject’s affected status.
Supplementary Material
Acknowledgments
We thank the patients and their family for their participation.
Funding: Support for the sequencing and analysis was shared by Life Technologies and National Human Genome Research Institute (NHGRI). Supported in part by grants from the NHGRI (5 U54 HG003273, to R.A.G.), the National Institute of Neurological Disorders and Stroke (R01 NS058529, to J.R.L.), the Natural Sciences and Engineering Research Council of Canada (Postgraduate Doctoral Scholarship, to M.N.B.), the Department of Veterans Affairs (Merit Review Award), and NIH R01 NS069700 (to J.K.F.).
Footnotes
SUPPLEMENTARY MATERIAL
www.sciencetranslationalmedicine.org/cgi/content/full/3/87/87re3/DC1
Fig. S1. Handwriting samples.
Table S1. The six identified compound heterozygous mutations shared by both twins.
Author contributions: M.N.B., W.W., D.R.M., J.R.L., and R.A.G. authored the manuscript. M.N.B., D.R.M., C.G.-J., and J.G.R. conducted the data analysis. L.D.H., S.Y., M.N.B., and R.A.G. conceived the study. J.F., J.R.L., W.W., and J.K.F. provided medical interpretation. I.N., M.B.M.,M.-C.G., C.G.-J., and D.M.M. conducted data generation.
Competing interests: R.A.G. has co-investments with Life Technologies, which developed the SOLiD sequencer. J.K.F. is a paid consultant for Athena Diagnostics. The other authors declare that they have no competing interests.
REFERENCES AND NOTES
- 1.1000 Genomes Project Consortium. Durbin RM, Abecasis GR, Altshuler DL, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME, McVean GA. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Richards CS, Bale S, Bellissimo DB, Das S, Grody WW, Hegde MR, Lyon E, Ward BE Molecular Subcommittee of the ACMG Laboratory Quality Assurance Committee. ACMG recommendations for standards for interpretation and reporting of sequence variations: Revisions 2007. Genet. Med. 2008;10:294–300. doi: 10.1097/GIM.0b013e31816b5cae. [DOI] [PubMed] [Google Scholar]
- 3.Fink JK, Ravin PD, Filling-Katz M, Argoff CE, Hallett M. Clinical and genetic analysis of progressive dystonia with diurnal variation. Arch. Neurol. 1991;48:908–911. doi: 10.1001/archneur.1991.00530210034019. [DOI] [PubMed] [Google Scholar]
- 4.Nygaard TG, Marsden CD, Fahn S. Dopa-responsive dystonia: Long-term treatment response and prognosis. Neurology. 1991;41:174–181. doi: 10.1212/wnl.41.2_part_1.174. [DOI] [PubMed] [Google Scholar]
- 5.Geyer HL, Bressman SB. The diagnosis of dystonia. Lancet Neurol. 2006;5:780–790. doi: 10.1016/S1474-4422(06)70547-6. [DOI] [PubMed] [Google Scholar]
- 6.Tarsy D, Simon DK. Dystonia. N. Engl. J. Med. 2006;355:818–829. doi: 10.1056/NEJMra055549. [DOI] [PubMed] [Google Scholar]
- 7.de Carvalho Aguiar PM, Ozelius LJ. Classification and genetics of dystonia. Lancet Neurol. 2002;1:316–325. doi: 10.1016/s1474-4422(02)00137-0. [DOI] [PubMed] [Google Scholar]
- 8.Ichinose H, Suzuki T, Inagaki H, Ohye T, Nagatsu T. Molecular genetics of dopa-responsive dystonia. Biol. Chem. 1999;380:1355–1364. doi: 10.1515/BC.1999.175. [DOI] [PubMed] [Google Scholar]
- 9.Clot F, Grabli D, Cazeneuve C, Roze E, Castelnau P, Chabrol B, Landrieu P, Nguyen K, Ponsot G, Abada M, Doummar D, Damier P, Gil R, Thobois S, Ward AJ, Hutchinson M, Toutain A, Picard F, Camuzat A, Fedirko E, Sân C, Bouteiller D, LeGuern E, Durr A, Vidailhet M, Brice A French Dystonia Network. Exhaustive analysis of BH4 and dopamine biosynthesis genes in patients with Dopa-responsive dystonia. Brain. 2009;132:1753–1763. doi: 10.1093/brain/awp084. [DOI] [PubMed] [Google Scholar]
- 10.Athena Diagnostics. [accessed July 2010]; http://www.athenadiagnostics.com.
- 11.Bademci G, Edwards TL, Torres AL, Scott WK, Züchner S, Martin ER, Vance JM, Wang L. A rare novel deletion of the tyrosine hydroxylase gene in Parkinson disease. Hum. Mutat. 2010;31:E1767–E1771. doi: 10.1002/humu.21351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lupski JR, Reid JG, Gonzaga-Jauregui C, Rio Deiros D, Chen DC, Nazareth L, Bainbridge M, Dinh H, Jing C, Wheeler DA, McGuire AL, Zhang F, Stankiewicz P, Halperin JJ, Yang C, Gehman C, Guo D, Irikat RK, Tom W, Fantin NJ, Muzny DM, Gibbs RA. Whole-genome sequencing in a patient with Charcot–Marie–Tooth neuropathy. N. Engl. J. Med. 2010;362:1181–1191. doi: 10.1056/NEJMoa0908094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, McGuire A, He W, Chen YJ, Makhijani V, Roth GT, Gomes X, Tartaro K, Niazi F, Turcotte CL, Irzyk GP, Lupski JR, Chinault C, Song XZ, Liu Y, Yuan Y, Nazareth L, Qin X, Muzny DM, Margulies M, Weinstock GM, Gibbs RA, Rothberg JM. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 14.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, Skupski M, Subramanian G, Thomas PD, Zhang J, Gabor Miklos GL, Nelson C, Broder S, Clark AG, Nadeau J, McKusick VA, Zinder N, Levine AJ, Roberts RJ, Simon M, Slayman C, Hunkapiller M, Bolanos R, Delcher A, Dew I, Fasulo D, Flanigan M, Florea L, Halpern A, Hannenhalli S, Kravitz S, Levy S, Mobarry C, Reinert K, Remington K, Abu-Threideh J, Beasley E, Biddick K, Bonazzi V, Brandon R, Cargill M, Chandramouliswaran I, Charlab R, Chaturvedi K, Deng Z, Di Francesco V, Dunn P, Eilbeck K, Evangelista C, Gabrielian AE, Gan W, Ge W, Gong F, Gu Z, Guan P, Heiman TJ, Higgins ME, Ji RR, Ke Z, Ketchum KA, Lai Z, Lei Y, Li Z, Li J, Liang Y, Lin X, Lu F, Merkulov GV, Milshina N, Moore HM, Naik AK, Narayan VA, Neelam B, Nusskern D, Rusch DB, Salzberg S, Shao W, Shue B, Sun J, Wang Z, Wang A, Wang X, Wang J, Wei M, Wides R, Xiao C, Yan C, Yao A, Ye J, Zhan M, Zhang W, Zhang H, Zhao Q, Zheng L, Zhong F, Zhong W, Zhu S, Zhao S, Gilbert D, Baumhueter S, Spier G, Carter C, Cravchik A, Woodage T, Ali F, An H, Awe A, Baldwin D, Baden H, Barnstead M, Barrow I, Beeson K, Busam D, Carver A, Center A, Cheng ML, Curry L, Danaher S, Davenport L, Desilets R, Dietz S, Dodson K, Doup L, Ferriera S, Garg N, Gluecksmann A, Hart B, Haynes J, Haynes C, Heiner C, Hladun S, Hostin D, Houck J, Howland T, Ibegwam C, Johnson J, Kalush F, Kline L, Koduru S, Love A, Mann F, May D, McCawley S, McIntosh T, McMullen I, Moy M, Moy L, Murphy B, Nelson K, Pfannkoch C, Pratts E, Puri V, Qureshi H, Reardon M, Rodriguez R, Rogers YH, Romblad D, Ruhfel B, Scott R, Sitter C, Smallwood M, Stewart E, Strong R, Suh E, Thomas R, Tint NN, Tse S, Vech C, Wang G, Wetter J, Williams S, Williams M, Windsor S, Winn-Deen E, Wolfe K, Zaveri J, Zaveri K, Abril JF, Guigó R, Campbell MJ, Sjolander KV, Karlak B, Kejariwal A, Mi H, Lazareva B, Hatton T, Narechania A, Diemer K, Muruganujan A, Guo N, Sato S, Bafna V, Istrail S, Lippert R, Schwartz R, Walenz B, Yooseph S, Allen D, Basu A, Baxendale J, Blick L, Caminha M, Carnes-Stine J, Caulk P, Chiang YH, Coyne M, Dahlke C, Mays A, Dombroski M, Donnelly M, Ely D, Esparham S, Fosler C, Gire H, Glanowski S, Glasser K, Glodek A, Gorokhov M, Graham K, Gropman B, Harris M, Heil J, Henderson S, Hoover J, Jennings D, Jordan C, Jordan J, Kasha J, Kagan L, Kraft C, Levitsky A, Lewis M, Liu X, Lopez J, Ma D, Majoros W, McDaniel J, Murphy S, Newman M, Nguyen T, Nguyen N, Nodell M, Pan S, Peck J, Peterson M, Rowe W, Sanders R, Scott J, Simpson M, Smith T, Sprague A, Stockwell T, Turner R, Venter E, Wang M, Wen M, Wu D, Wu M, Xia A, Zandieh A, Zhu X. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 15.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McKusick VA. Mendelian Inheritance in Man and its online version, OMIM. Am. J. Hum. Genet. 2007;80:588–604. doi: 10.1086/514346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boeckmann B, Bairoch A, Apweiler R, Blatter MC, Estreicher A, Gasteiger E, Martin MJ, Michoud K, O’Donovan C, Phan I, Pilbout S, Schneider M. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Echenne B, Roubertie A, Assmann B, Lutz T, Penzien JM, Thöny B, Blau N, Hoffmann GF. Sepiapterin reductase deficiency: Clinical presentation and evaluation of long-term therapy. Pediatr. Neurol. 2006;35:308–313. doi: 10.1016/j.pediatrneurol.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 19.Bonafé L, Thöny B, Penzien JM, Czarnecki B, Blau N. Mutations in the sepiapterin reductase gene cause a novel tetrahydrobiopterin-dependent monoamine-neurotransmitter deficiency without hyperphenylalaninemia. Am. J. Hum. Genet. 2001;69:269–277. doi: 10.1086/321970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blau N, Barnes I, Dhondt JL. International database of tetrahydrobiopterin deficiencies. J. Inherit. Metab. Dis. 1996;19:8–14. doi: 10.1007/BF01799342. [DOI] [PubMed] [Google Scholar]
- 21.Friedman J, Hyland K, Blau N, MacCollin M. Dopa-responsive hypersomnia and mixed movement disorder due to sepiapterin reductase deficiency. Neurology. 2006;67:2032–2035. doi: 10.1212/01.wnl.0000247274.21261.b4. [DOI] [PubMed] [Google Scholar]
- 22.Fink JK, Ravin P, Argoff CE, Levine RA, Brady RO, Hallett M, Barton NW. Tetrahydrobiopterin administration in biopterin-deficient progressive dystonia with diurnal variation. Neurology. 1989;39:1393–1395. doi: 10.1212/wnl.39.10.1393. [DOI] [PubMed] [Google Scholar]
- 23.Choi M, Scholl UI, Ji W, Liu T, Tikhonova IR, Zumbo P, Nayir A, Bakkaloğlu A, Ozen S, Sanjad S, Nelson-Williams C, Farhi A, Mane S, Lifton RP. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc. Natl. Acad. Sci. U.S.A. 2009;106:19096–19101. doi: 10.1073/pnas.0910672106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones SJ, Laskin J, Li YY, Griffith OL, An J, Bilenky M, Butterfield YS, Cezard T, Chuah E, Corbett R, Fejes AP, Griffith M, Yee J, Martin M, Mayo M, Melnyk N, Morin RD, Pugh TJ, Severson T, Shah SP, Sutcliffe M, Tam A, Terry J, Thiessen N, Thomson T, Varhol R, Zeng T, Zhao Y, Moore RA, Huntsman DG, Birol I, Hirst M, Holt RA, Marra MA. Evolution of an adenocarcinoma in response to selection by targeted kinase inhibitors. Genome Biol. 2010;11:R82. doi: 10.1186/gb-2010-11-8-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Worthey EA, Mayer AN, Syverson GD, Helbling D, Bonacci BB, Decker B, Serpe JM, Dasu T, Tschannen MR, Veith RL, Basehore MJ, Broeckel U, Tomita-Mitchell A, Arca MJ, Casper JT, Margolis DA, Bick DP, Hessner MJ, Routes JM, Verbsky JW, Jacob HJ, Dimmock DP. Making a definitive diagnosis: Successful clinical application of whole exome sequencing in a child with intractable inflammatory bowel disease. Genet. Med. 2011;13:255–262. doi: 10.1097/GIM.0b013e3182088158. [DOI] [PubMed] [Google Scholar]
- 26.Ormond KE, Wheeler MT, Hudgins L, Klein TE, Butte AJ, Altman RB, Ashley EA, Greely HT. Challenges in the clinical application of whole-genome sequencing. Lancet. 2010;375:1749–1751. doi: 10.1016/S0140-6736(10)60599-5. [DOI] [PubMed] [Google Scholar]
- 27.McKernan KJ, Peckham HE, Costa GL, McLaughlin SF, Fu Y, Tsung EF, Clouser CR, Duncan C, Ichikawa JK, Lee CC, Zhang Z, Ranade SS, Dimalanta ET, Hyland FC, Sokolsky TD, Zhang L, Sheridan A, Fu H, Hendrickson CL, Li B, Kotler L, Stuart JR, Malek JA, Manning JM, Antipova AA, Perez DS, Moore MP, Hayashibara KC, Lyons MR, Beaudoin RE, Coleman BE, Laptewicz MW, Sannicandro AE, Rhodes MD, Gottimukkala RK, Yang S, Bafna V, Bashir A, MacBride A, Alkan C, Kidd JM, Eichler EE, Reese MG, De La Vega FM, Blanchard AP. Sequence and structural variation in a human genome uncovered by short-read, massively parallel ligation sequencing using two-base encoding. Genome Res. 2009;19:1527–1541. doi: 10.1101/gr.091868.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Homer N, Merriman B, Nelson SF. BFAST: An alignment tool for large scale genome resequencing. PloS One. 2009;4:e7767. doi: 10.1371/journal.pone.0007767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang K, Li M, Hakonarson H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang J, Wheeler DA, Yakub I, Wei S, Sood R, Rowe W, Liu PP, Gibbs RA, Buetow KH. SNPdetector: A software tool for sensitive and accurate SNP detection. PLoS Comput. Biol. 2005;1:e53. doi: 10.1371/journal.pcbi.0010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.