Abstract
We set out to analyze the role of two major non-HLA gene polymorphisms associated with type 1 diabetes (T1D), PTPN22 1858C/T and insulin gene INS−23 A/T in progression to clinical T1D after the appearance of β-cell autoimmunity. The study population comprised 249 children with HLA-associated T1D susceptibility. All subjects were persistently positive for at least one of the T1D-associated biochemically defined autoantibodies (insulin autoantibody, GAD antibody, or IA-2 antibody), and 136 subjects presented with T1D over a median follow-up of 4.3 years (range 0.0–12.5) after the appearance of the first autoantibody. The PTPN22 1858T allele was strongly associated with progression to T1D after the appearance of the first biochemically defined β-cell autoantibody (hazard ratio 1.68 [95% CI 1.09–2.60], P = 0.02 Cox regression analysis, multivariate test), and the effect remained similar when analyzed after the appearance of the second autoantibody (P = 0.013), whereas INS−23 HphI AA genotype was not associated with progression to clinical diabetes after the appearance of the first or second autoantibody (P = 0.38 and P = 0.88, respectively). The effect of the INS risk genotype seems to be limited to the induction and early phases of β-cell autoimmunity, but the PTPN22 1858T allele instead affects the initiation and late progression phase of diabetes-associated autoimmunity.
Type 1 diabetes (T1D) is an autoimmune disease resulting from the destruction of the insulin-producing pancreatic β-cells. The etiology of the disease is not completely understood, but the strong inherited effect of multiple gene loci is a crucial component (1). The HLA gene region on the short arm of chromosome 6 is the most important gene locus affecting susceptibility to T1D. A number of other susceptibility loci have been described outside the HLA region. Of these, the effect of the polymorphisms in the promoter region of the INS gene and the PTPN22 gene that encode the lymphocyte tyrosine phosphatase that regulates T-cell activation on the increased appearance of β-cell autoimmunity and T1D have been confirmed in several populations, and their effect is the strongest one among the non-HLA genes (2).
The diagnosis of T1D is preceded by an autoimmune process that damages the pancreatic β-cells, and circulating autoantibodies can be detected as a marker of the ongoing process. This process is asymptomatic, and the duration of this period varies widely (3). The significance of the various genetic and environmental risk factors in the different stages of the autoimmune disease process is not fully understood. It has been proposed that HLA class II genes determine the initiation of β-cell autoimmunity, whereas other factors (e.g., HLA class I genes) affect the progression of β-cell destruction (4). In support of this hypothesis, we recently reported an enhancing effect of the HLA-B*39 allele and a protecting effect of the *A03 allele on the progression to clinical T1D after the appearance of humoral signs of β-cell autoimmunity (5). Moreover, this effect of the HLA class I genes was affected by the HLA class II genotype, although the high-risk class II genotype did not directly affect the progression to the disease after establishment of advanced β-cell autoimmunity.
Data on the effect of the genetic risk factors outside the HLA region in the different stages of β-cell autoimmunity are limited (6). Thus, in the current study, we analyze the role of the two major non-HLA gene polymorphisms associated with T1D, PTPN22 1858C/T and INS−23 A/T in progression from islet autoimmunity to clinical T1D.
RESEARCH DESIGN AND METHODS
The study subjects were participants in the Finnish Diabetes Prediction and Prevention (DIPP) study and carried HLA-DQB1 genotypes associated with an increased risk for T1D (HLA-DQB1*0302, the HLA-DQB1*0302/DQA1*05-DQB1*02 combination, or DQA1*05-DQB1*02 alone [a subgroup of boys born in Turku] without the presence of the protective alleles [HLA-DQB1*0301 or HLA-DQB1*0602]). According to the study protocol, subjects were prospectively followed up at 3- to 12-month intervals for the appearance of serologic signs of T1D-associated autoantibodies: islet cell antibodies (ICA), and when ICA were detected, for the signs of insulin autoantibodies (IAA), antibodies to the 65 kD isoform of GAD (GADA), and antibodies to the protein tyrosine phosphatase-related IA-2 molecule (IA-2A) (7). All subjects who tested persistently positive for ICA and were positive for at least one of the biochemically defined autoantibodies (IAA, GADA or IA-2A) by May 2004 and whose DNA sample was available for genotyping were included in the current study. In addition, one subject positive for IAA but not ICA and presenting with clinical T1D by May 2004 was added to the study population. The local ethical committees approved the study protocol, and informed consent was obtained from the parents of the study participants.
A total of 249 subjects fulfilled the inclusion criteria and were accordingly included in the study population; of these, 201 (80.7%) tested positive for IAA, 184 (73.1%) for GADA, and 176 (70.6%) for IA-2A. The median age of the children was 1.8 years (range 0.3–12.0) at the time of seroconversion to positivity for the first biochemically defined autoantibody and 2.0 years (0.8–9.9) for the second biochemically defined autoantibody. Median follow-up from the appearance of the first biochemically defined autoantibody was 6.4 years (0.5–12.5) in the 113 children remaining clinically healthy and 2.8 years (0.0–10.8) among the 136 children who progressed to clinical T1D. The median follow-up times were 6.1 years (1.8–12.0) after the second biochemically defined autoantibody and in the 78 children remaining unaffected and 2.5 years (0.0–10.8) in the 117 children presenting with T1D. All subjects with clinical T1D were diagnosed according to the World Health Organization criteria. At the time of study recruitment, 32 of the 249 study subjects had a first-degree relative with T1D.
Genotyping methods.
HLA genotyping was described in our earlier study (5). The genotyping methods for the PTPN22 1858C/T (rs2476601) and INS−23 A/T (rs689) polymorphisms were based on the use of lanthanide-labeled probes and have been described earlier (8,9). The PTPN22 1858C/T and INS−23 A/T genotypes could be defined in 241 (96.8%) and 240 (96.4%) children, respectively.
Statistical analysis.
Cox regression analysis and Kaplan-Meier analysis with log-rank test were used to compare progression to clinical disease in subjects with various genotypes. The analysis defined time of entry as the time of the appearance of the first or second biochemically defined autoantibody. The effect of the analyzed variants on the progression to clinical T1D was analyzed until the diagnosis of T1D (uncensored) or the end of the follow-up (censored). All statistical analyses were performed with SPSS 19.0 software (IBM SPSS Inc., Chicago, IL).
RESULTS
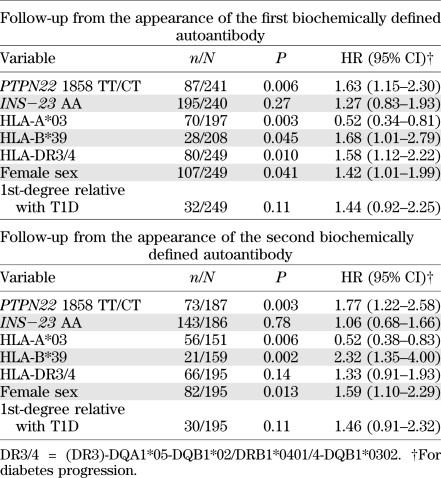
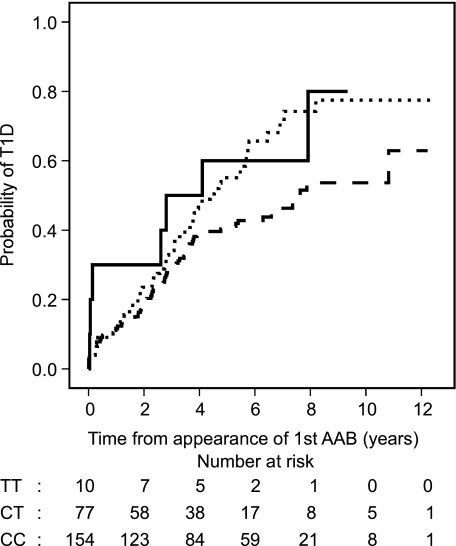
The effect of the PTPN22 1858T allele and INS−23 HphI AA genotype on the progression to clinical T1D after the appearance of humoral β-cell autoimmunity was analyzed. We observed a strong effect of the PTPN22 1858T allele on the progression to clinical T1D after the first signs of humoral β-cell autoimmunity (Table 1, Fig. 1). After the appearance of the first biochemically defined autoantibody, 58 subjects (66.7%) with the TT or CT genotype presented with T1D compared with 71 (46.1%) with the CC genotype (hazard ratio [HR] 1.63 [95% CI 1.15–2.30], P = 0.006, Cox regression analysis, univariate test for the effect of PTPN22 TT/CT vs. CC genotypes). Cumulative risk of T1D at 10 years of follow-up was 78.2% for subjects carrying the T allele and 53.7% for subjects with CC genotype (P = 0.005, Kaplan-Meier analysis, log-rank test).
TABLE 1.
The effect of the PTPN22 1858C/T, INS−23 HphI A/T, HLA DR3/4, HLA-A*03, and *B39 genotypes, sex, and presence of first-degree relative with T1D on the progression to clinical T1D after the appearance of the first and second biochemically defined autoantibody (Cox regression analysis, univariate test)
FIG. 1.
Kaplan-Meier survival analysis shows the effects of the PTPN22 1858TT (solid line), 1858CT (dotted line), and 1858CC (dashed line) genotypes on the progression to clinical T1D after the appearance of the first biochemically defined autoantibody (AAB).
The effect of the T allele marking disease risk remained strong also after the appearance of the second biochemically defined autoantibody. Here, 53 of the subjects (72.6%) carrying the T allele presented with T1D, whereas 57 (50.0%) with the CC genotype progressed to clinical T1D (P = 0.003, Cox regression analysis; Table 1). In contrast, no effect of the T1D-risk associated INS−23 HphI AA genotype on progression to clinical T1D after the appearance of β-cell autoimmunity was observed in Cox regression analysis (Table 1) or in Kaplan-Meier survival analysis (data not shown). A total of 102 of the subjects (55.1%) with the AA genotype presented with T1D after the appearance of the first biochemically defined autoantibody compared with 28 of the subjects (50.9%) with the AT or TT genotype (P = 0.27), and the result remained similar also after the appearance of the second biochemically defined autoantibody (85 [59.4%] vs. 26 [60.5%], respectively; P = 0.78).
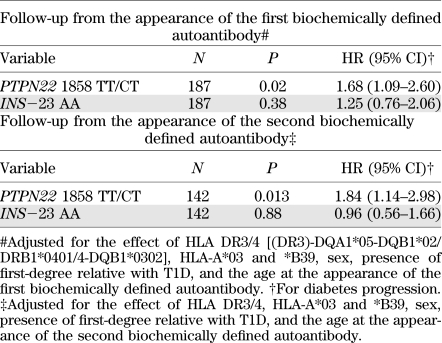
The effect of the major HLA class II risk genotypes, the two HLA class I alleles, which in the same series affected progression of β-cell autoimmunity to clinical disease, HLA-A*03 and *B39 (5), and sex, presence of first-degree relative with T1D, and the age at autoantibody seroconversion were then included in the analysis. After these adjustments, the strong effect of the PTPN22 T allele and the absent effect of the INS genotype on the disease progression remained. After the appearance of the first and second biochemically defined autoantibody, the PTPN22 T disease-risk allele was associated with enhanced progression to clinical disease (P = 0.02 and P = 0.013, respectively), whereas no effect of the INS genotype on the appearance of clinical T1D was observed (P = 0.38 and P = 0.88, follow-up after the appearance of the first and second autoantibody, respectively; Table 2).
TABLE 2.
The effect of the PTPN22 1858C/T and INS−23 HphI A/T genotypes on the progression to clinical T1D after the appearance of the first and second biochemically defined autoantibody (Cox regression analysis, multivariate test)
DISCUSSION
This study describes the role of the two major non-HLA gene polymorphisms associated with T1D, PTPN22 1858C/T and INS−23 A/T, in progression to clinical T1D after the appearance of β-cell autoimmunity. The effect of these polymorphisms on the T1D risk has been repeatedly confirmed (2). However, data describing their effect on the disease risk in the different stages of the disease process is scarce. Butty et al. (6) reported results on the effect of the various genetic factors, including PTPN22 C1858T (R620W) and INS−23 HphI A/T polymorphisms, in the progression to clinical T1D among at-risk relatives of T1D patients with autoimmune manifestations in the Diabetes Prevention Trial–Type 1 (DPT-1) trial. They observed no differences in the PTPN22 and INS allele frequencies between the nonprogressors who remained clinically healthy and progressors who presented with clinical T1D in their study cohort. Neither did the recent report on the Diabetes Autoimmunity Study in the Young (DAISY) follow-up cohort see any significant effect in progression from persistent autoimmunity to clinical diabetes in the smaller cohort of 112 subjects with persistent autoimmunity and in 47 diagnosed with T1D, although they observed a significant effect of the PTPN22 risk genotypes on the progression to persistent islet autoimmunity (10).
There are obvious differences between the study cohort in the DPT-1 trial and in our study. The DPT-1 cohort comprised ICA- and IAA-positive relatives of T1D patients, and the presence of other autoantibodies was not taken into account. The subjects were older at screening (mean age, 9.5 years), and the exact autoantibody seroconversion age could not be defined. HLA-associated risk was also lower in the DPT-1 study because only the presence of the protective DQB1*0602 allele was excluded. It is thus conceivable that a subgroup with high disease risk and rapid progression to clinical disease in early childhood was less well represented in the DPT-1 than in our DIPP follow-up cohort. The setting of the DAISY study, with follow-up from birth on, was closer to our study. An apparent difference is the higher power of our study owing to higher numbers in the follow-up cohort and probably a more homogeneous population. In addition, we point out that the frequency of the risk-associated PTPN22 minor T allele is clearly more common among both the Finnish T1D patients and the background population than in the respective U.S. populations, thus further increasing the power of the DIPP study in this context (11).
The results indicate differences in the rate of progression between the various susceptibility genes in the process leading to the development of β-cell autoimmunity and eventually to loss of insulin-secreting β-cells. The INS disease-risk genotype is associated with lower levels of insulin expression in the thymus, leading to ineffective deletion of insulin-specific T cells (12–14). In addition, the INS disease-risk genotype has been associated with IAA (15,16), which is usually the first autoantibody reactivity to be detected, especially among young children (17,18). Thus, in accordance with our findings, the INS T1D-risk genotype is likely to affect the initial breakdown of insulin-specific immunologic tolerance rather than to affect the immunoregulation leading to β-cell destruction. The PTPN22 disease allele is shown to alter T cell and to some extent also B cell function, but exactly how it affects the process leading to β-cell destruction is not known (19–21). A major hypothesis suggests that poor T-cell receptor–mediated signaling in the thymus is associated with defective negative selection and maintenance of an autoreactive T-cell repertoire. Alternative mechanisms might be related to the function of high levels of Lyp-expressing dendritic cells, which are important in antigen presentation and activation of T helper cells or further, that the decreased function of naturally occurring self-reactive regulatory T cells allows the generation of autoreactive T-effector cells (22). The last alternative might best fit our finding of more rapid course of β-cell destruction after establishment of islet-specific autoimmunity. In line with this, the 1858T allele has been associated with a diminished residual β-cell function and metabolic control or with increased residual β-cell stress as measured by elevated proinsulin/C-peptide level at the time of T1D diagnosis and during the early period of insulin treatment (23,24). These findings, together with the results observed in this study, suggest that the PTPN22+1858T allele has a role in the β-cell destruction leading to clinical T1D in addition to the initiation of β-cell autoimmunity.
ACKNOWLEDGMENTS
The study was supported by the Juvenile Diabetes Research Foundation, the Academy of Finland, the Sigrid Jusélius Foundation, and the Foundation for Paediatric Research.
No potential conflicts of interest relevant to this article were reported.
J.L. acquired and reviewed research data used in the analysis, undertook statistical analysis and interpretation of the results, and drafted the manuscript. R.H. and R.V. were involved in the interpretation of the results and critically reviewed the manuscript. O.S. and M.K. were two of the principal investigators, designed the DIPP study, were involved in the interpretation of the results, and critically reviewed the manuscript. J.I. was one of the principal investigators, was involved in the interpretation of the results, designed the DIPP study, provided the genotyping results of the research subjects, and critically reviewed the manuscript. J.L. is the guarantor of this work and, as such, had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Preliminary data from this study were presented in poster form at the 46th Annual Meeting of the European Association for the Study of Diabetes, Stockholm, Sweden, 21–24 September 2010.
REFERENCES
- 1.Eisenbarth GS. Update in type 1 diabetes. J Clin Endocrinol Metab 2007;92:2403–2407 [DOI] [PubMed] [Google Scholar]
- 2.Concannon P, Rich SS, Nepom GT. Genetics of type 1A diabetes. N Engl J Med 2009;360:1646–1654 [DOI] [PubMed] [Google Scholar]
- 3.Knip M. Natural course of preclinical type 1 diabetes. Horm Res 2002;57 (Suppl. 1):6–11 [DOI] [PubMed] [Google Scholar]
- 4.Tait BD, Colman PG, Morahan G, et al. HLA genes associated with autoimmunity and progression to disease in type 1 diabetes. Tissue Antigens 2003;61:146–153 [DOI] [PubMed] [Google Scholar]
- 5.Lipponen K, Gombos Z, Kiviniemi M, et al. Effect of HLA class I and class II alleles on progression from autoantibody positivity to overt type 1 diabetes in children with risk-associated class II genotypes. Diabetes 2010;59:3253–3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butty V, Campbell C, Mathis D, Benoist C; DPT-1 Study Group Impact of diabetes susceptibility loci on progression from pre-diabetes to diabetes in at-risk individuals of the diabetes prevention trial-type 1 (DPT-1). Diabetes 2008;57:2348–2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kupila A, Muona P, Simell T, et al. ; Juvenile Diabetes Research Foundation Centre for the Prevention of Type I Diabetes in Finland Feasibility of genetic and immunological prediction of type I diabetes in a population-based birth cohort. Diabetologia 2001;44:290–297 [DOI] [PubMed] [Google Scholar]
- 8.Hermann R, Lipponen K, Kiviniemi M, et al. Lymphoid tyrosine phosphatase (LYP/PTPN22) Arg620Trp variant regulates insulin autoimmunity and progression to type 1 diabetes. Diabetologia 2006;49:1198–1208 [DOI] [PubMed] [Google Scholar]
- 9.Laine AP, Holmberg H, Nilsson A, et al. ; Finnish Paediatric Diabetes Registry Two insulin gene single nucleotide polymorphisms associated with type 1 diabetes risk in the Finnish and Swedish populations. Dis Markers 2007;23:139–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Steck AK, Zhang W, Bugawan TL, et al. Do non-HLA genes influence development of persistent islet autoimmunity and type 1 diabetes in children with high-risk HLA-DR,DQ genotypes? Diabetes 2009;58:1028–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gregersen PK, Lee HS, Batliwalla F, Begovich AB. PTPN22: setting thresholds for autoimmunity. Semin Immunol 2006;18:214–223 [DOI] [PubMed] [Google Scholar]
- 12.Pugliese A. Central and peripheral autoantigen presentation in immune tolerance. Immunology 2004;111:138–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pugliese A, Zeller M, Fernandez A, Jr, et al. The insulin gene is transcribed in the human thymus and transcription levels correlated with allelic variation at the INS VNTR-IDDM2 susceptibility locus for type 1 diabetes. Nat Genet 1997;15:293–297 [DOI] [PubMed] [Google Scholar]
- 14.Vafiadis P, Bennett ST, Todd JA, et al. Insulin expression in human thymus is modulated by INS VNTR alleles at the IDDM2 locus. Nat Genet 1997;15:289–292 [DOI] [PubMed] [Google Scholar]
- 15.Graham J, Hagopian WA, Kockum I, et al. ; Diabetes Incidence in Sweden Study Group; Swedish Childhood Diabetes Study Group Genetic effects on age-dependent onset and islet cell autoantibody markers in type 1 diabetes. Diabetes 2002;51:1346–1355 [DOI] [PubMed] [Google Scholar]
- 16.Hermann R, Laine AP, Veijola R, et al. The effect of HLA class II, insulin and CTLA4 gene regions on the development of humoral beta cell autoimmunity. Diabetologia 2005;48:1766–1775 [DOI] [PubMed] [Google Scholar]
- 17.Ziegler AG, Hummel M, Schenker M, Bonifacio E. Autoantibody appearance and risk for development of childhood diabetes in offspring of parents with type 1 diabetes: the 2-year analysis of the German BABYDIAB Study. Diabetes 1999;48:460–468 [DOI] [PubMed] [Google Scholar]
- 18.Kukko M, Kimpimäki T, Korhonen S, et al. Dynamics of diabetes-associated autoantibodies in young children with human leukocyte antigen-conferred risk of type 1 diabetes recruited from the general population. J Clin Endocrinol Metab 2005;90:2712–2717 [DOI] [PubMed] [Google Scholar]
- 19.Vang T, Congia M, Macis MD, et al. Autoimmune-associated lymphoid tyrosine phosphatase is a gain-of-function variant. Nat Genet 2005;37:1317–1319 [DOI] [PubMed] [Google Scholar]
- 20.Rieck M, Arechiga A, Onengut-Gumuscu S, Greenbaum C, Concannon P, Buckner JH. Genetic variation in PTPN22 corresponds to altered function of T and B lymphocytes. J Immunol 2007;179:4704–4710 [DOI] [PubMed] [Google Scholar]
- 21.Aarnisalo J, Treszl A, Svec P, et al. Reduced CD4+T cell activation in children with type 1 diabetes carrying the PTPN22/Lyp 620Trp variant. J Autoimmun 2008;31:13–21 [DOI] [PubMed] [Google Scholar]
- 22.Burn GL, Svensson L, Sanchez-Blanco C, Saini M, Cope AP. Why is PTPN22 a good candidate susceptibility gene for autoimmune disease? FEBS Lett 2011;585:3689–3698 [DOI] [PubMed] [Google Scholar]
- 23.Petrone A, Spoletini M, Zampetti S, et al. ; Immunotherapy Diabetes (IMDIAB) Group The PTPN22 1858T gene variant in type 1 diabetes is associated with reduced residual beta-cell function and worse metabolic control. Diabetes Care 2008;31:1214–1218 [DOI] [PubMed] [Google Scholar]
- 24.Nielsen LB, Pörksen S, Andersen ML, et al. ; Hvidoere Study Group on Childhood Diabetes The PTPN22 C1858T gene variant is associated with proinsulin in new-onset type 1 diabetes. BMC Med Genet 2011;12:41. [DOI] [PMC free article] [PubMed] [Google Scholar]