Polyketide synthases (PKS) and non-ribosomal peptide synthetases (NRPS) are large multidomain proteins present in microorganisms that produce bioactive compounds. Curacin A is such a bioactive compound with potent anti-proliferative activity. During its biosynthesis the growing substrate is bound covalently to an acyl carrier protein (ACP) that is able to access catalytic sites of neighboring domains for chain elongation and modification. While ACP domains usually occur as monomers, the curacin A cluster codes for a triplet ACP (ACPI-ACPII-ACPIII) within the CurA PKS module. We have determined the structure of the isolated holo ACPI and show that the ACPs are independent of each other within this tri-domain system. In addition, we have determined the structure of the 3-hydroxyl-3-methylglutaryl-loaded holo-ACPI, which is the substrate for the unique halogenase (Hal) domain embedded within the CurA module. We have identified the interaction surface of both proteins using mutagenesis and MALDI-based identification of product formation. Amino acids affecting product formation are located on helices II and III of ACPI and form a contiguous surface. Since the CurA Hal accepts substrate only when presented by one of the ACPs within the ACPI-ACPII-ACPIII tri-domain, our data provide insight into the specificity of the chlorination reaction.
The polyketide synthases (PKS) and the nonribosomal peptide synthetases (NRPS) produce natural products of a huge chemical diversity. PKS and NRPS are multidomain protein assemblies that function by sequentially elongating a growing polyketide or peptide chain by acyl units or amino acids, respectively. The growing product is attached via a thioester linkage to the 4′-phosphopantetheine (4′-Ppant) arm of either a holo acyl carrier protein (ACP) in PKSs or a holo peptidyl carrier protein (PCP) in NRPSs and is passed from one module to another along the chain of reaction centers. The modular arrangement makes PKS and NRPS systems an interesting target for protein engineering (1). More than 200 novel polyketide compounds have already been created; unfortunately, however, engineered PKSs often fail to produce significant amounts of the desired products (2-4). Structural studies may faciliate yield improvement from engineered systems by providing a more complete understanding of the interface between the different domains. One example is the 6-deoxyerythronolide B synthethase (DEBS PKS) from Saccharopolyspora erythraea. Docking experiments of ACP domains with the X-ray structure of the ketosynthase (KS) and acyltransferase (AT) di-domain of module 3 (KS3-AT3) for example suggest that the ACP domain binds in a deep cleft between both domains with interactions mediated by helix II of the ACP (5). Mutations in this helix II affect the specificity of the system (6). Further studies led to the conclusion that helix II is a universal “recognition helix” for interaction with different enzymatic centers (7-11). While some information about domain-domain interactions involving ketosynthase and acyltransferase is starting to emerge (12), little is known about the interaction of ACP domains with other modifying enzymes such as methyltransferases, epimerases or halogenases.
Curacin A is an anticancer natural product derived from the marine cyanobacterium Lyngbya majuscula. It is generated by an unusual mixed PKS/NRPS system with cyclopropane and thiazoline moieties, as well as an internal cis double bond, and a terminal alkene. The biosynthesis of curacin A is mediated by a 2.2 MDa hybrid PKS-NRPS cluster (13, 14) (Figure 1). The first enzyme within the CurA PKS to act on the initial 3-hydroxyl-3-methyl-glutaryl (HMG) linked to ACPI is the curacin halogenase (CurA Hal). This is followed by the discrete dehydratase (ECH1) and decarboxylase (ECH2) that comprise the β-branching steps in formation of the cyclopropane ring (Figure 1) (15). The recently solved crystal structure of CurA Hal in five individual states differing in their substrate and cofactor composition has improved our understanding of its enzymatic mechanism (16). In addition, amino acids essential for the stereo-specific substrate recognition of (S)-HMG-ACP by CurA Hal could be identified by mutagenesis (16). The HMG substrate is presented by an unusual cluster of three tandem ACPs (ACPI,II,III). Little is known about the function of these multiple ACPs but recent studies have demonstrated that they enhance significantly the efficiency of the multienzyme reaction leading to formation of methyl-cyclopropylacetyl-ACP (17).
Figure 1.
The 10-enzyme assembly catalyzing the cyclopropane ring formation. a) The 10 enzymes which are involved in the cyclopropanring are encoded on different proteins. b) Representation of the Biosynthesis of the cyclopropane ring formation. GNATL= loading module, KS= ketosynthethase, AT= acyltransferase, Hal= halogenase, ACP= acetyl-carrier-protein, HCS= HMG-CoA synthase-like enzyme, ECH1= dehydratase, ECH2= decarboxylase, ER= enoyl-reductase.
140×104mm (300 × 300 DPI)
In this study we elucidated the structural basis for the selectivity that drives the interaction between CurA Hal and CurA ACPI,II,III and show that these triplet ACP domains are structurally independent. Furthermore we report the NMR solution structures of holo-ACPI, and HMG-ACPI. NOE experiments indicated that no sequestration of the HMG substrate occurs but it is instead presented on the ACP surface. Using mutational analysis we have mapped the protein-protein interaction surface between CurA HMG-ACPI and CurA Hal that not only includes the recognition helix II but also large parts of helix III.
RESULTS AND DISCUSSION
Impact of ACP repetition on the single domain
The triplet ACP domains (ACPI,II,III) are located at the C-terminus of the CurA module. As shown using negative-stain electron microscopy, ACPI,II,III has a propensity to dimerize through a C-terminal domain (Cd-domain) (17). Our initial objective was to evaluate whether the presence of the triplet ACPI,II,III domains or their dimer leads to domain-domain interactions that adopt a preferred conformation. [15N, 1H]-TROSY spectra of the two proteins CurA-ACPI,II,III with (in black) and without Cd (in red) were recorded and compared (Figure 2). The absence of chemical shift differences indicates that no conformational changes occur. Some peaks decrease in intensity upon adding the 60 aa Cd domain. However, surprisingly we failed to observe new peaks representing the Cd domain. Due to dimerization the overall size increases from 36 kDa to 89 kDa (17). The increased size of the protein, in combination with some conformational exchange of the Cd domain might broaden the resonances of the Cd domain beyond the detection limit. Parts which tumble independently, such as the individual ACP domains are affected less by this increase in size (Figure 2).
Figure 2.
Dimerization does not affect the conformation. a) Overlay of [15N, 1H] TROSY spectra of apo ACPI,II,III (red) and apo ACPI,II,III–Cd (black). b) Magnification of the central region. c) Chemical shift differences between the isolated holo ACPI and holo ACPI as part of the triplet ACPI,II,III are plotted for ACPI (black), ACPII (red), ACPIII (green) are plotted against the sequence. The CSPs for Gln2032 (1.95 ppm) and Thr2034 (1.26 ppm), indicated with an asterisk, are truncated at 1.0 ppm.
139×60mm (300 × 300 DPI)
In the absence of clear evidence that the Cd domain can affect the conformation of the entire ACP assembly, we decided to further analyze ACPI,II,III. The [15N, 1H]-TROSY spectrum of ACPI,II,III suffers from severe peak overlap due to the high sequence identity of the individual domains (93-100%). The distinction of the individual domains was achieved by segmental labeling, allowing the assignment of the full length construct (18). Analysis of the excised ACPI, ACPII or ACPIII proteins resulted in [15N, 1H]-TROSY spectra with strong chemical shift perturbations (CSPs). Shift differences Δδ were calculated and plotted against the ACP sequence (Figure 2). For ACPI and ACPII chemical shift perturbations (CSPs) Δδ > 0.4 ppm occurred for amino acids confined to the C-terminus and for ACPIII small CSP occurred at the N-terminus reflecting differences in the environment of the linker region in the isolated domain and the ACPI,II,III tri-domain (Figure 2). These data suggest that no detectable interaction occurs between ACPI, ACPII and ACPIII. To further investigate this question, we performed titration experiments of the 15N-labeled ACPI domain (residues 1946-2043) with the ACPII,III di-domain (2044-2248) and the ACPI,II di-domain with 15N-labeled ACPIII. Both experiments failed to show chemical shift perturbations. Similarly, no inter-domain NOEs were detected in 3D 13C–edited/15N-separated NOESY-HSQC experiments with a [15N]ACPI– [13C]ACPII-ACPIII, labeled version (data not shown). Taken together these data establish that no detectable interactions occur between the ACP domains of the triplet ACP cluster. To investigate the dynamics of the ACP linker regions which were found to be structured in other systems (5, 19, 20) we performed {1H}15N heteronuclear NOE measurements. Nearly all amino acids of the structured domains ACPI, ACPII and ACPIII could be identified, and therefore the additional signals can be associated with the linker region. Heteronuclear NOE values of around 0.8 represent highly structured regions, which is the case for the assigned amino acids within the ACP domains, whereas values around 0.2 were measured for the unassigned peaks indicating that the linkers are unstructured and highly flexible (Supplementary Figure 4).
Taken together we show that the linker region is unstructured and that the ACP domains behave independently from each other in ACPI,II,III as well as in the dimerized form of ACPI,II,III-Cd. This leads to the conclusion that the interaction between Cur Hal and ACP domains can be studied using one representing ACP domain of the triplet, for example ACPI.
The structure of holo ACPI
ACPI was expressed in its apo form and post-translationally modified in vitro. The purity of the apo protein and completeness of conversion to the holo form was investigated using matrix-assisted laser desorption ionization (MALDI) mass spectrometry analysis (Supplementary Figure 3). The structure was solved using standard NMR methods and structural statistics are given in Table 1. Coordinates for holo ACPI have been deposited in the Protein Data Bank under the accession code 2LIU.
Table 1.
Statistics derived from Cyana for the structure of holo and (S)-hydroxyl-methylglutaryl (HMG)-ACPI
| NMR restraints | Holo | HMG | Structural statistics | Holo | HMG |
|---|---|---|---|---|---|
| Number of peaks | Restraint violations | ||||
| 13C NOESY-HSQC | 5260 | 5078 | Max. Dihedral angle restraint violations | / | / |
| 15N NOESY-HSQC | 1557 | 1557 | (Max. distance restraint violations (A) | 0.12 | 0.21 |
| 13CArom NOESY-HSQC | 58 | 57 | Cyana target function value (Å2) | 2.42 | 1.56 |
| Hydroxy-methyl-glutaryl | |||||
| Intramolecular NOEs | - | 86 | |||
| Intermolecular NOEs | - | 37 | |||
|
|
|||||
| Precision for residues 1950-2033 | |||||
|
|
|||||
| Total NOE | 2866 | 2923 | R.m.s.d to mean coordinates (Å) | ||
| Short range ∣i-j∣<=1 | 1307 | 1349 | Heavy atoms | 0.54 ± 0.06 | 1.01 ± 0.29 |
| Medium-range 1<∣i-j∣<5 | 826 | 819 | Backbone | 0.24 ± 0.04 | 0.11 ± 0.02 |
| Long range ∣i-j∣>=5 | 734 | 755 | Ramachandran plot staistics (%) | ||
| Hydrogen bonds | - | most favored | 88.6 | 87.1 | |
| additionally allowed | 11.4 | 12.9 | |||
| Dihedral angle | generously allowed | 0 | 0 | ||
| Talos+ φ/ψ | 162 | 162 | disallowed | 0 | 0 |
Holo ACPI consists of the expected right-hand twisted bundle formed by four major α-helices (I,II,III,VI) connected by three loops (21-25). Figure 3 shows a ribbon diagram of the minimized mean structure. Conformational exchange has been previously reported in the ACP and PCP family (25, 26), however, for ACPI only a single set of cross-peaks was observed with no indication of slow conformational exchange.
Figure 3.
Ribbon diagram of the averaged and minimized NMR structure of holo ACPI from Lyngbya majuscula. Helices I–IV are coloured light blue, yellow, orange and red, respectively. The active site Ser1989 is indicated in red at the N-terminus of helix II.
43×27mm (600 × 600 DPI)
The apo to holo ACPI conversion is accompanied by CSPs. The amino acids surrounding the active site Ser1989 show the strongest perturbations between the apo and holo forms, including amino acids at the N-terminus of helix II and on helix III (all >0.05ppm CSP) (Figure 4). In addition 15N NOESY-HSQCs for apo and holo ACPI were measured. However, comparison of the NOE pattern of amino acids undergoing strong chemical shift perturbations revealed minor differences between both states (Supplementary Figure 5) excluding major conformational changes upon formation of the holo state.
Figure 4.
Comparison of chemical shifts of apo- and holo-ACP . a) An overlay of [15N, 1 IH] TROSY spectra of uniformly 15N-labeled apo ACPI (black) and 15N-labeled holo ACPI (red). The assignments of the amino acids undergoing the strongest chemical shift perturbations are indicated. b) Plot of chemical shift differences between holo ACPI and apo ACPI. The secondary structure elements are indicated below the sequence. No CSP is given for Ile1990, which could not be assigned in the apo form.
140×71mm (300 × 300 DPI)
Acyl-ACP
During curacin A biosynthesis ACPI occurs in its holo, acetyl, HMG and 4-chloro-3-hydroxy-3-methylglutaryl (Cl-HMG) forms. To investigate these different chemical states apo ACPI was loaded by the corresponding acyl-CoAs. For Cl-HMG loading the racemic (R,S)-HMG was used and enzymatically converted to 50% γ-Cl-(S)-HMG-ACPI by Cur Hal and 50% residual R-HMG-ACPI due to the stereo-specificity of the reaction (15). [15N, 1H] TROSY spectra of acetyl, HMG and γ-Cl-HMG were compared to holo ACPI (Figure 5). The CSPs are small compared to the CSPs of the apo to holo conversion and are located near the active site Ser1989 on helices II and III. The CSPs increase from acetyl- to HMG-(~0.05ppm) and γ-Cl-HMG-ACPI (~0.1ppm) (Figure 5).
Figure 5.
Comparison of the different chemical states of ACP . a) An overlay of [15N,1 I H] TROSY spectra of holo-ACPI (red) and acetyl-ACPI (yellow), HMG-ACPI (light blue), Cl-HMG-ACPI (magenta). Assignments of the amino acids undergoing the strongest chemical shift perturbations are indicated and their magnifications are presented on the top panel. The right hand panel presents plots of chemical shift differences between holo-ACPI and acetyl-ACPI (b), HMG-ACPI (c), Cl-HMG-ACPI (d). The secondary structure elements are indicated below.
140×79mm (300 × 300 DPI)
In previous solution NMR investigations of ACPs no NOEs between the protein and the prosthetic group were detected. This led to the assumption that no interactions or only transient interactions are present between the 4′-Ppant arm and the surface of the ACP domain (21-23). If the 4′-Ppant arm is modified with larger substrates, however, association with the surface has been observed with the acyl chain bound in a cleft that is located between helices II and III. Examples are the FAS hexanoyl-ACP of E. coli (27) and octanoyl-ACP from Streptomyces coelicolor (28). For a detailed characterization of the interaction of ACPI with Cur Hal the localization of the substrate on the surface must first be defined. To investigate potential contacts between the 4′-Ppant arm and the protein we expressed 13C/15N-labeled ACPI and modified it in vitro with unlabeled CoA and (R,S)-HMG-CoA. NOEs between 12CH groups of the attached cofactor and the 13CH groups of the protein were detected in a 3D F1-13C/15N filtered, F3-13C-separated NOESY (Figure 6). Surprisingly we could obtain NOEs with similar intensities for both holo and HMG ACPI reflecting that in both cases the 4′-Ppant arm might be located similarly. In a next step we recorded F1/F2-13C/15N double-filtered 2D-NOESY spectra to selectively detect the 12CH-12CH NOEs present within the 4′-Ppant arm loaded with HMG (Figure 6). Using the previous assignments of the 4′-Ppant arm loaded on other PCPs and ACPs (25, 29) we could assign the 4′-Ppant chain attached to ACPI. The 1D spectra of CoA and HMG-CoA then gave rise to the unambiguous assignment of the protons of the HMG methyl group (66, 67,68) (Supplementary Figure 6) and allowed us to assign the NOEs observed between the HMG and the protein. (Supplementary Table 2).
Figure 6.
Isotope filtered NOESY experiments for the structure determination of HMG-ACPI a) F1-F3 strips from a 3D F1-13C/15N filtered, F3-13C-separated NOESY-HSQC of holo and HMG-ACPI filtering the 13CH-15NH NOEs. The protein was loaded in vitro and the substrate is therefore unlabeled giving strong NOEs from 12CH to the 13CH groups of the protein surface. b) The cofactor S-HMG and its numeration; c) Expansion of the two-dimensional F2-filtered NOESY spectrum of 13C,15N-labeled HMG-ACPI. The 4′-Ppant chain and the HMG group are unlabeled and give rise to strong NOE connectivities within the 4′-Ppant arm.
140×100mm (300 × 300 DPI)
To calculate the structure of HMG-ACPI we combined the restraints obtained for holo-ACPI with the NOE-based distance restraints obtained for HMG, assuming that no major conformational changes occur between the different states (Supplementary Figure 5, Table 1 for structural statistics). The structure shows that the HMG-4′-Ppant arm associates with the surface of the ACP, but that this interaction is transient, allowing different orientations of the HMG group to occur (Figure 7). Structures of other ACP-substrate complexes had shown a more tight interaction. X-ray studies of the E. coli FAS II ACP demonstrated that starting from C4 chain length the substrate and the 4′-Ppant arm is sequestered into a hydrophobic pocket (27, 30). These results were similar to the NMR findings for the PKS actinorhodin system of Streptomyces coelicor (28).
Figure 7.
a) Ribbon diagram for the 20 structures of (S)-HMG-ACPI. b) Mean structure of HMG-ACPI. The amino acids which undergo chemical shift perturbations once the 4′-Ppant arm is added are labeled in blue (CSP >0.04ppm), amino acids which undergo slight chemical shift perturbations when HMG is added to the 4′-Ppant arm are indicated in magenta (CSP >0.04ppm). The shifts are located around the attachment site of the substrate on helix II and helix III.
51×40mm (600 × 600 DPI)
Interaction studies of ACPI with Cur Hal
The curacin A biosynthetic pathway differs from other β-branching pathways in the introduction of a γ-chlorination step on (S)-HMG-ACPI mediated by Cur Hal, a non-heme Fe(II), α-ketoglutarate-dependent enzyme (15). Recent studies showed that the chlorination step takes place before dehydration catalyzed by ECH1 and ECH2–mediated decarboxylation followed. Previous work has shown that ECH1 and ECH2 in the CurA β-branching cassette are more promiscuous enzymes. For example ECH1 can accommodate either S-HMG- or γ-Cl-(S)-HMG bound to ACPI,II,III (15), CurB (31) or in the acyl-CoA form, albeit with lower efficiency (32). By contrast, the halogenase CurA Hal was shown to be stereo-specific as it distinguishes between R- and S-HMG (15, 16) and accepts S-HMG only in an ACP-bound form. In order to assess the specificity determinants for this interaction Cur Hal was expressed, purified and reconstituted anaerobically as described (15, 16) and its activity was confirmed by MALDI mass spectrometry (Supplementary Figure 7). To identify the specific interaction surface on ACPI we titrated (R,S)-HMG-[15N]ACPI with active, unlabeled CurA Hal, but no chemical shift perturbations could be observed. The experiment was repeated under non-reactive conditions: without Cl−, α-ketoglutarate or O2 or missing all components. Subsequently the inactive mutant R241A (16) or Fe3+-reconstituted, inactive halogenase were tested. Finally, ACPI in apo, holo or loaded with the substrates acetyl or malonyl were titrated with functional halogenase. In all cases, despite using a high excess of the enzyme, no CSPs or line broadening effects were observed which would indicate formation of an HMG-ACPI/Cur Hal complex. To investigate if the reaction occurs even in the absence of CSPs and to probe the specificity of the reaction, we performed activity assays using CurA ACPI, CurB–ACP and TycB1 PCP as a substrate donor. Only in the case of ACPI all S-HMG-ACPI was chlorinated within 10 min (Supplementary Figure 7). Even after 1 hr HMG-CurB and HMG-PCP did not convert to the chlorinated product (data not shown). This result demonstrates that Cur Hal distinguishes between these ACPs, leading to the conclusion that an interface must be present, which cannot be detected by typical NMR titrations.
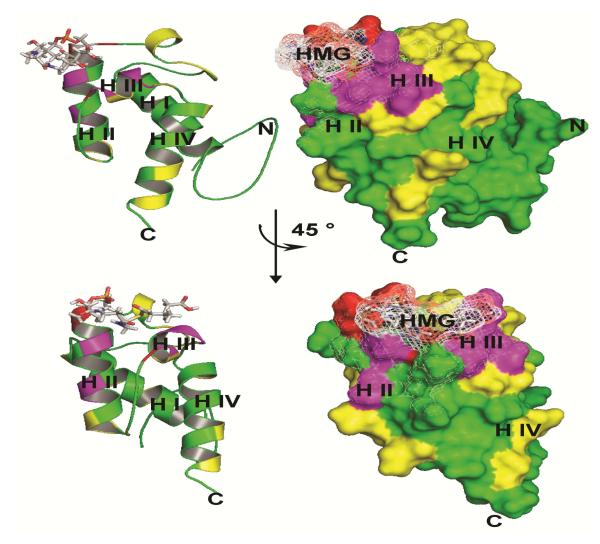
To further investigate the CurA Hal/ACPI interaction surface, we mutated solvent exposed, not conserved amino acid (Supplementary Figure 8). Since we used (R,S)-HMG-ACPI as a substrate, activity was determined by assuming that chlorination of 100% S-HMG results in two peaks, one representing the unreacted R-HMG-ACP and the other one γ-Cl-(S)-HMG-ACP, separated by 34 Da and of equal intensities (see MALDI MS in Supplementary Figure 9 and the derived activities are listed in Supplementary Table 1). The mutations D1988A and I1990A neighboring the active site Ser1989 and A2009R on helix III had the strongest impact on the chlorination efficiency (activity: 29 ± 10%; 18 ± 7%; 15 ± 12%, respectively). In these cases no product peaks were observed. However, the first and second sodium adduct peaks (~+22 and ~+44 Da) might overlap with a small product peak. To avoid underestimation of the product yield we assumed the peak intensity at a distance of 34 Da to be product (Supplementary Figure 9). This leads to an overestimation rather than an underestimation of the activity, making clear that these mutations reduce the activity significantly. The single point mutations T2010A, Y2013A and D2014A on helix III decrease the activity to 37 ± 7%, 35% ± 2% and 59 ± 2%, respectively. Circular dichroism (CD) curves of the less active ACPI mutants showed that the mutations did not have an impact on the global fold. Melting temperatures (TM) derived from CD melting curves showed that the mutations D1988A and D2014A destabilize the protein (Supplementary Table 1, Supplementary Figure 10). To ensure that a strong reduction by a mutation is not simply caused by structural perturbations we compared [15N, 1H] TROSY spectra of mutants with the WT (Supplementary Figure 11) revealing for D1988A and I1990A CSPs for only 5-6 amino acids located near the mutation site, suggesting that these mutations do not affect the structure. The mutation A2009R leads to stronger CSPs which might be due to structural rearrangements. We were interested whether these mutations might lead to a displacement of the cofactor which in turn affects the Cur Hal/ACP interaction. We loaded the mutants D1988A, I1990A and A2009R with the 4′-Ppant arm, acetyl and HMG, mapped the CSPs against the sequence and compared these results with the WT (Supplementary Figure 12). Interestingly in all cases the same amino acids showed CSPs, suggesting that the localization of the cofactor is not affected (Supplementary Figure 12). Mapping the mutations with the strongest effects onto the structure of HMG-ACPI shows that these amino acids surround the substrate and form a contiguous surface suggesting its importance for Cur Hal recognition and selectivity (Figure 8).
Figure 8.
Effect of ACPI mutation on its activity. According to the strength of activity decrease we applied a colour code. Mutations which decreased the activity to 0-30% are marked in red (D1988A and I1990A amino acids neighbouring the active site Ser1989 and A2009R located on helix III), mutations which decreased the activity to 30-70% are marked in magenta (e.g. the multiple mutant V1993N/T1998M/T1999M located on helix II, the single mutants T2010A, Y2013A and D2014A all located on Helix III) and mutations with minor or no effect with an activity of over 70% are marked in yellow
84×107mm (600 × 600 DPI)
Overall our data demonstrate that Cur Hal interacts with ACPI specifically, which is likely applicable to Cur Hal interaction also with ACPII and ACPIII of the tri-domain. Cur Hal recognizes an interface consisting mainly of the N-terminus of helix II, the 4′-Ppant arm with its substrate, and helix III. Substrate attached to a different carrier domain (either ACP or PCP) is not recognized by the halogenase. The importance of presenting the substrate as part of an entire substrate-ACP complex is also demonstrated by the fact that the isolated HMG-CoA does not serve as a substrate for CurA Hal. In contrast to high specificity of the Hal, it is evident that ECH1 and ECH2 β-branching cassette enzymes are less specific and can accommodate acyl-CoAs, albeit with significantly lower efficiency. The radical mechanism involved in the chlorination step (inherently highly reactive and less specific) is possibly the driving force for the Hal acyl-ACP specificity. The requirement of presenting the correct substrate within a specific environment might be necessary to prevent non-specific chlorination or a competing hydroxylation reaction of substrates presented by other ACPs. For example, CurB is a discrete ACP that can freely diffuse, and should therefore be able to interact with Cur Hal.
Surprisingly, it was not possible to directly detect the interactions between ACPI and Cur Hal by NMR spectroscopy. Weak or transient interactions are difficult to detect using the standard NMR experiments due to the low population and transiency of the complex. One technique for investigating these transient interactions is paramagnetic relaxation enhancement (PRE) measurements. Spin labels attached to one partner result in relaxation enhancement in the other partner even when the contact time is short and the complex unstable (33-36). We therefore attempted to apply PRE experiments to our system, in order to visualize low populated states. Unfortunately no spin label could be attached to Cur Hal because the removal of the WT cysteine (Cys19Ala) as well as the insertion of new cysteines into Cur Hal (C19S, I61C, H109C, G111C, Y274C, H168C and S178C) resulted in protein precipitation.
Our results demonstrate that the individual CurA ACPs of the ACPI,II,III tri-domain do not interact with each other. Previous studies have shown that the advantage of a triplet assembly over a single ACP is the increase of the efficiency of the entire enzyme reaction (17). It has been proposed that this clustering of ACP-linked substrates might result in binding of adjacent enzymes to corresponding substrates through non-specific enzyme– substrate interactions (17). Further kinetic and enzymatic investigation of Cur ACPI,II,III tri-domain involving intact CurA module might reveal the basis for tandem ACP function and whether increasing the local concentration of potential substrate rather than distinct conformational effects affect higher efficiency during polyketide chain elongation in the curacin A pathway.
METHODS
Protein Expression and Purification
apo ACP (amino acids 1946-2034), the mutants, the triplet ACPI,II,III (1946–2248) and ACPI,II,III – Cd (1946–2311) were expressed in the E. coli strain BL21(DE3) (Novagen) using the pET28a(+)plasmid (17, 18) at 15°C for 7-10 hrs. Media were supplemented with 0.8 ml 0.05 M Fe-II-solution in order to prevent modifications by an endogenous PPTase (37). All proteins were purified by Ni-NTA affinity chromatography. The constructs of Cur Hal and CurB were described previously (15, 31). Hal was expressed, purified and reconstituted as described previously (31) (Supplemantary materials and Methods). CurB was expressed in BL21 star-pRARE (Novagen) and expressed and purified like ACPI. TycB1 PCP was cloned into pBH4 using and expressed and purified as ACPI.
Loading of ACP
The recombinant ACPs (200 μM) were artificially loaded with either coenzyme A (CoA) or analogs (Acetyl-, Malonyl-, HMG-CoA) by using the promiscuous Bacillus subtilis phosphopantetheinyl (Ppant) transferase Sfp (3 μM). The loading reaction was incubated at room temperature for 1 hr in buffered aqueous solution (pH 7.5, 20 mM HEPES, 50 mM NaCl, 20 mM MgCl2). To ensure that complete loading was achieved the reaction mixture was analyzed directly by MALDI-MS (Supplemantary Figure 3).
Activity Assay
For the reactivity test Hal was purified and reconstituted as described. 100 μl of 100 μM HMG-ACP in 20 mM Hepes, 50 mM NaCl, pH 7.2 were supplemented with 0.5 mM fresh α-KG and 75 μM Fe-II-SO4 and 5 μM Hal. The reaction was incubated at 25 °C under shaking (800 rpm), after 10 min the reaction was completed. The reaction products were analysed using MALDI-MS. The experiments were performed three times.
Mass Spectrometry
Mass spectra were recorded on a MALDI-TOF mass analyzer (Voyager STR, Applied Biosystems) in linear positive ion mode. Samples were diluted with water to a concentration of 5–10 μM and 1 μl was mixed with 1 μl MALDI matrix (sDHB, Bruker) directly on the sample target. Spectra were accumulated over 50-100 laser shots.
NMR spectroscopy of holo ACP
0.6-1 mM solutions of ACPI were prepared in 50 mM arginine/50 mM glutamate buffer at pH 6.8 containing 5% D2O and 0,15 mM DSS (4, 4-dimethyl-4-silapentane-1-sulfonate). NMR spectra were collected at a temperature of 291 K on 950, 900, 800 and 600 MHz on Bruker Avance spectrometers equipped with cryogenic 5mm z-axis gradient triple resonance probes and a 500 MHz Bruker Avance spectrometer equipped with 5 mm x,y,z-axis gradient triple resonance room temperature probe. For backbone and aliphatic side chain assignments [15N, 1H] TROSY (38) version of HSQC, HNCACO, HNCO, HNCACB, (H)C(CCO)NH-TOCSY, and H(CCCO)NH-TOCSY were recorded. Aromatic ring resonances were assigned with (H)CB(CGCC-TOCSY)Har experiments (39). 15N and 13C separated 3D NOESY experiments were recorded with 70 ms mixing time. All spectra were processed with TopSpin (Bruker Biospin) and analysed with Sparky (40). For the acyl chain binding studies, standard sensitivity-enhanced [15N, 1H]-TROSY spectra were acquired. Steady state {1H}15N hetNOE measurements were performed on a 600 MHz instrument using a TROSY-type pulse sequence (41) .
NMR of HMG-ACP and holo ACP
To observe NOEs between the 4′ Ppant arm and HMG loaded 4′ Ppant arm and the ACP domain highly 13C, 15N enriched ACPI was loaded with unlabeled 12C CoA or HMG-CoA. The protein was concentrated to 1 mM in 50 mM arginine/ 50 mM glutamate buffer at a pH of 6.8 and a 3D F1-13C/15N-filtered, F3-13C-seperated NOESY-HSQC was acquired at 800 MHz using a mixing time of 80 ms and filter delays of 93 Hz for 1JNH and to 125 Hz and 147 Hz for 1JCH. For the assignment of the unlabeled cofactor a 2D F1/F2-13C/15N double-filtered NOESY experiment (42) with 150 ms mixing time was acquired at 950 MHz. Fully 13C/15N-labeled ACPI was loaded with unlabeled 12C HMG-CoA and the protein was concentrated to 1mM in 25 mM NaPi and 50 mM NaCl at a pH of 6.8 and measured at 291K.
Structure Calculation
Backbone torsion angle restraints were obtained from chemical shift data using the TALOS+ algorithm (43). NOE-based distance restraints from 3D 15N-NOESY-HSQC and 13C-NOESY HSQC spectra were assigned automatically by CYANA (44, 45), which was also used for the structure calculations by torsion angle dynamics (46). The final structure calculations included 2852 distance restraints and 162 backbone torsion angle restraints (Table 1). 100 conformers were computed using 10000 torsion angle dynamics steps. The 20 conformers with the lowest target function values were subjected to restrained energy refinement with the program OPALP (47) using the AMBER force field (48). The quality of the structures was checked by PROCHECK (49) and WHATCHECK (50).
Structure calculations with HMG
A model of HMG was created using the online version of the program CORINA for 3D structure generation (http://www.molecular-networks.com/online_demos/corina_demo). HMG was attached covalently to the Ser1989 residue. For the structure calculation with CYANA (51), a residue consisting of a serine backbone connected to the HMG cofactor was added to the standard residue library and the dataset was complemented with additional NOE information for the cofactor. The resulting structures were subjected to restrained energy minimization using OPALp (47). The necessary partial charges for HMG-serine were calculated using the PRODRG server (52).
Supplementary Material
Acknowledgement
Special thanks go to U. Bahr for mass spectrometry analysis. We thank the Müller group for allowing us to use their equipment for the work in anaerobe conditions. This work was supported by the Deutsche Forschungsgemeinschaft (DO545/6-1), the Center for Biomolecular Magnetic Resonance (BMRZ) and the Cluster of Excellence Frankfurt (Macromolecular Complexes), and by the Lichtenberg program of the Volkswagen Foundation. We gratefully acknowledge NIH grant R01 CA108874 (D.H.S.) to support studies of cyanobacterial natural product pathways.
Footnotes
Accession code- Coordinates for holo ACPI have been deposited in the protein databank under the code 2LIU. The coordinates for (S)-hydroxy-methylglutaryl ACPI have been deposited in the protein databank under the code 2LIW.
Supporting information available: This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Fischbach MA, Walsh CT. Assembly-line enzymology for polyketide and nonribosomal Peptide antibiotics: logic, machinery, and mechanisms. Chem. Rev. 2006;106:3468–3496. doi: 10.1021/cr0503097. [DOI] [PubMed] [Google Scholar]
- 2.Menzella HG, Carney JR, Santi DV. Rational design and assembly of synthetic trimodular polyketide synthases. Chem. Biol. 2007;14:143–151. doi: 10.1016/j.chembiol.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 3.Menzella HG, Reeves CD. Combinatorial biosynthesis for drug development. Curr. Opin. Microbiol. 2007;10:238–245. doi: 10.1016/j.mib.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 4.Kittendorf JD, Sherman DH. Developing tools for engineering hybrid polyketide synthetic pathways. Curr. Opin. Biotechnol. 2006;17:597–605. doi: 10.1016/j.copbio.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 5.Tang Y, Chen AY, Kim CY, Cane DE, Khosla C. Structural and mechanistic analysis of protein interactions in module 3 of the 6-deoxyerythronolide B synthase. Chem. Biol. 2007;14:931–943. doi: 10.1016/j.chembiol.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alekseyev VY, Liu CW, Cane DE, Puglisi JD, Khosla C. Solution structure and proposed domain domain recognition interface of an acyl carrier protein domain from a modular polyketide synthase. Protein Sci. 2007;16:2093–2107. doi: 10.1110/ps.073011407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang YM, Wu B, Zheng J, Rock CO. Key residues responsible for acyl carrier protein and beta-ketoacyl-acyl carrier protein reductase (FabG) interaction. J. Biol. Chem. 2003;278:52935–52943. doi: 10.1074/jbc.M309874200. [DOI] [PubMed] [Google Scholar]
- 8.Zhang YM, Rao MS, Heath RJ, Price AC, Olson AJ, Rock CO, White SW. Identification and analysis of the acyl carrier protein (ACP) docking site on beta-ketoacyl-ACP synthase III. J. Biol. Chem. 2001;276:8231–8238. doi: 10.1074/jbc.M008042200. [DOI] [PubMed] [Google Scholar]
- 9.Rafi S, Novichenok P, Kolappan S, Zhang X, Stratton CF, Rawat R, Kisker C, Simmerling C, Tonge PJ. Structure of acyl carrier protein bound to FabI, the FASII enoyl reductase from Escherichia coli. J. Biol. Chem. 2006;281:39285–39293. doi: 10.1074/jbc.M608758200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gong H, Murphy A, McMaster CR, Byers DM. Neutralization of acidic residues in helix II stabilizes the folded conformation of acyl carrier protein and variably alters its function with different enzymes. J. Biol. Chem. 2007;282:4494–4503. doi: 10.1074/jbc.M608234200. [DOI] [PubMed] [Google Scholar]
- 11.Arthur CJ, Williams C, Pottage K, Ploskon E, Findlow SC, Burston SG, Simpson TJ, Crump MP, Crosby J. Structure and malonyl CoA-ACP transacylase binding of streptomyces coelicolor fatty acid synthase acyl carrier protein. ACS Chem. Biol. 2009;4:625–636. doi: 10.1021/cb900099e. [DOI] [PubMed] [Google Scholar]
- 12.Buchholz TJ, Geders TW, Bartley FE, 3rd, Reynolds KA, Smith JL, Sherman DH. Structural basis for binding specificity between subclasses of modular polyketide synthase docking domains. ACS Chem. Biol. 2009;4:41–52. doi: 10.1021/cb8002607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang Z, Sitachitta N, Rossi JV, Roberts MA, Flatt PM, Jia J, Sherman DH, Gerwick WH. Biosynthetic pathway and gene cluster analysis of curacin A, an antitubulin natural product from the tropical marine cyanobacterium Lyngbya majuscula. J. Nat. Prod. 2004;67:1356–1367. doi: 10.1021/np0499261. [DOI] [PubMed] [Google Scholar]
- 14.Verdier-Pinard P, Lai JY, Yoo HD, Yu J, Marquez B, Nagle DG, Nambu M, White JD, Falck JR, Gerwick WH, Day BW, Hamel E. Structure-activity analysis of the interaction of curacin A, the potent colchicine site antimitotic agent, with tubulin and effects of analogs on the growth of MCF-7 breast cancer cells. Mol. Pharmacol. 1998;53:62–76. doi: 10.1124/mol.53.1.62. [DOI] [PubMed] [Google Scholar]
- 15.Gu L, Wang B, Kulkarni A, Geders TW, Grindberg RV, Gerwick L, Hakansson K, Wipf P, Smith JL, Gerwick WH, Sherman DH. Metamorphic enzyme assembly in polyketide diversification. Nature. 2009;459:731–735. doi: 10.1038/nature07870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khare D, Wang B, Gu L, Razelun J, Sherman DH, Gerwick WH, Hakansson K, Smith JL. Conformational switch triggered by alpha-ketoglutarate in a halogenase of curacin A biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 2010;107:14099–14104. doi: 10.1073/pnas.1006738107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gu L, Eisman EB, Dutta S, Franzmann TM, Walter S, Gerwick WH, Skiniotis G, Sherman DH. Tandem acyl carrier proteins in the curacin biosynthetic pathway promote consecutive multienzyme reactions with a synergistic effect. Angew. Chem. Int. Ed. Engl. 2011;50:2795–2798. doi: 10.1002/anie.201005280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Busche AE, Aranko AS, Talebzadeh-Farooji M, Bernhard F, Dötsch V, Iwai H. Segmental isotopic labeling of a central domain in a multidomain protein by protein trans-splicing using only one robust DnaE intein. Angew. Chem. Int. Ed. Engl. 2009;48:6128–6131. doi: 10.1002/anie.200901488. [DOI] [PubMed] [Google Scholar]
- 19.Maier T, Leibundgut M, Boehringer D, Ban N. Structure and function of eukaryotic fatty acid synthases. Q. Rev. Biophys. 2010;43:373–422. doi: 10.1017/S0033583510000156. [DOI] [PubMed] [Google Scholar]
- 20.Tang Y, Kim CY, Mathews II, Cane DE, Khosla C. The 2.7-Angstrom crystal structure of a 194-kDa homodimeric fragment of the 6-deoxyerythronolide B synthase. Proc. Natl. Acad. Sci. U.S.A. 2006;103:11124–11129. doi: 10.1073/pnas.0601924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim Y, Kovrigin EL, Eletr Z. NMR studies of Escherichia coli acyl carrier protein: dynamic and structural differences of the apo- and holo-forms. Biochem. Biophys. Res. Commun. 2006;341:776–783. doi: 10.1016/j.bbrc.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 22.Xu GY, Tam A, Lin L, Hixon J, Fritz CC, Powers R. Solution structure of B. subtilis acyl carrier protein. Structure. 2001;9:277–287. doi: 10.1016/s0969-2126(01)00586-x. [DOI] [PubMed] [Google Scholar]
- 23.Wong HC, Liu G, Zhang YM, Rock CO, Zheng J. The solution structure of acyl carrier protein from Mycobacterium tuberculosis. J. Biol. Chem. 2002;277:15874–15880. doi: 10.1074/jbc.M112300200. [DOI] [PubMed] [Google Scholar]
- 24.Li Q, Khosla C, Puglisi JD, Liu CW. Solution structure and backbone dynamics of the holo form of the frenolicin acyl carrier protein. Biochemistry. 2003;42:4648–4657. doi: 10.1021/bi0274120. [DOI] [PubMed] [Google Scholar]
- 25.Koglin A, Mofid MR, Löhr F, Schäfer B, Rogov VV, Blum MM, Mittag T, Marahiel MA, Bernhard F, Dötsch V. Conformational switches modulate protein interactions in peptide antibiotic synthetases. Science (New York, N.Y. 2006;312:273–276. doi: 10.1126/science.1122928. [DOI] [PubMed] [Google Scholar]
- 26.Zornetzer GA, Fox BG, Markley JL. Solution structures of spinach acyl carrier protein with decanoate and stearate. Biochemistry. 2006;45:5217–5227. doi: 10.1021/bi052062d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roujeinikova A, Simon WJ, Gilroy J, Rice DW, Rafferty JB, Slabas AR. Structural studies of fatty acyl-(acyl carrier protein) thioesters reveal a hydrophobic binding cavity that can expand to fit longer substrates. J. Mol. Biol. 2007;365:135–145. doi: 10.1016/j.jmb.2006.09.049. [DOI] [PubMed] [Google Scholar]
- 28.Evans SE, Williams C, Arthur CJ, Ploskon E, Wattanaamorn P, Cox RJ, Crosby J, Willis CL, Simpson TJ, Crump MP. Probing the Interactions of early polyketide intermediates with the Actinorhodin ACP from S. coelicolor A3(2) J. Mol. Biol. 2009;389:511–528. doi: 10.1016/j.jmb.2009.03.072. [DOI] [PubMed] [Google Scholar]
- 29.Ploskon E, Arthur CJ, Evans SE, Williams C, Crosby J, Simpson TJ, Crump MP. A mammalian type I fatty acid synthase acyl carrier protein domain does not sequester acyl chains. J. Biol. Chem. 2008;283:518–528. doi: 10.1074/jbc.M703454200. [DOI] [PubMed] [Google Scholar]
- 30.Roujeinikova A, Baldock C, Simon WJ, Gilroy J, Baker PJ, Stuitje AR, Rice DW, Slabas AR, Rafferty JB. X-ray crystallographic studies on butyryl-ACP reveal flexibility of the structure around a putative acyl chain binding site. Structure. 2002;10:825–835. doi: 10.1016/s0969-2126(02)00775-x. [DOI] [PubMed] [Google Scholar]
- 31.Geders TW, Gu L, Mowers JC, Liu H, Gerwick WH, Hakansson K, Sherman DH, Smith JL. Crystal structure of the ECH2 catalytic domain of CurF from Lyngbya majuscula. Insights into a decarboxylase involved in polyketide chain beta-branching. J. Biol. Chem. 2007;282:35954–35963. doi: 10.1074/jbc.M703921200. [DOI] [PubMed] [Google Scholar]
- 32.Gu L, Jia J, Liu H, Hakansson K, Gerwick WH, Sherman DH. Metabolic coupling of dehydration and decarboxylation in the curacin A pathway: functional identification of a mechanistically diverse enzyme pair. J. Am. Chem. Soc. 2006;128:9014–9015. doi: 10.1021/ja0626382. [DOI] [PubMed] [Google Scholar]
- 33.Iwahara J, Tang C, Clore GM. Practical aspects of (1)H transverse paramagnetic relaxation enhancement measurements on macromolecules. J. Magn. Reson. 2007;184:185–195. doi: 10.1016/j.jmr.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iwahara J, Clore GM. Detecting transient intermediates in macromolecular binding by paramagnetic NMR. Nature. 2006;440:1227–1230. doi: 10.1038/nature04673. [DOI] [PubMed] [Google Scholar]
- 35.Clore GM, Iwahara J. Theory, practice, and applications of paramagnetic relaxation enhancement for the characterization of transient low-population states of biological macromolecules and their complexes. Chem. Rev. 2009;109:4108–4139. doi: 10.1021/cr900033p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang C, Iwahara J, Clore GM. Visualization of transient encounter complexes in protein-protein association. Nature. 2006;444:383–386. doi: 10.1038/nature05201. [DOI] [PubMed] [Google Scholar]
- 37.Lambalot RH, Gehring AM, Flugel RS, Zuber P, LaCelle M, Marahiel MA, Reid R, Khosla C, Walsh CT. A new enzyme superfamily - the phosphopantetheinyl transferases. Chem. Biol. 1996;3:923–936. doi: 10.1016/s1074-5521(96)90181-7. [DOI] [PubMed] [Google Scholar]
- 38.Pervushin K, Riek R, Wider G, Wüthrich K. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc. Natl. Acad. Sci. U.S.A. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Löhr F, Hänsel R, Rogov VV, Dötsch V. Improved pulse sequences for sequence specific assignment of aromatic proton resonances in proteins. J. Biomol. NMR. 2007;37:205–224. doi: 10.1007/s10858-006-9128-4. [DOI] [PubMed] [Google Scholar]
- 40.Goddard TD, Kneller DG. SPARKY 3. University Of California; San Francisco: [Google Scholar]
- 41.Ferrage F, Piserchio A, Cowburn D, Ghose R. On the measurement of 15N-{1H} nuclear Overhauser effects. J. Magn. Reson. 2008;192:302–313. doi: 10.1016/j.jmr.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ogura K, Terasawa H, Inagaki F. An improved double-tuned and isotope-filtered pulse scheme based on a pulsed field gradient and a wide-band inversion shaped pulse. J. Biomol. NMR. 1996;8:492–498. doi: 10.1007/BF00228150. [DOI] [PubMed] [Google Scholar]
- 43.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Güntert P. Automated structure determination from NMR spectra. Eur. Biophys. J. 2009;38:129–143. doi: 10.1007/s00249-008-0367-z. [DOI] [PubMed] [Google Scholar]
- 45.Herrmann T, Güntert P, Wüthrich K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 2002;319:209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- 46.Güntert P, Mumenthaler C, Wüthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 47.Koradi R, Billeter M, Güntert P. Point-centered domain decomposition for parallel molecular dynamics simulation. Comput. Phys. Commun. 2000;124:139–147. [Google Scholar]
- 48.Ponder JW, Case DA. Force fields for protein simulations. Adv. Protein Chem. 2003;66:27–85. doi: 10.1016/s0065-3233(03)66002-x. [DOI] [PubMed] [Google Scholar]
- 49.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 50.Hooft RW, Vriend G, Sander C, Abola EE. Errors in protein structures. Nature. 1996;381:272. doi: 10.1038/381272a0. [DOI] [PubMed] [Google Scholar]
- 51.Güntert P. Calculating protein structures from NMR data. Methods Mol. Biol. 1997;60:157–194. doi: 10.1385/0-89603-309-0:157. [DOI] [PubMed] [Google Scholar]
- 52.Schüttelkopf AW, van Aalten DM. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta crystallogr. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.