Summary
This review focuses on the epigenetic alterations of aberrant promoter hypermethylation of genes, histone modifications or RNA interference in cancer cells. The current knowledge of hypermethylation of allele(s) in classical tumor suppressor genes in inherited and sporadic cancer, candidate tumor suppressor and other cancer genes is summarized gene by gene. Global and array-based studies of tumor cell hypermethylation are discussed. The importance of standardization of scoring of the methylation status of a gene is highlighted. The histone marks associated with hypermethylated genes, and the microRNAs with dysregulated expression, in kidney or bladder tumor cells are also discussed. Kidney cancer has the highest mortality rate of the genitourinary cancers. There are management issues with the high recurrence rate of superficial bladder cancer while muscle invasive bladder cancer has a poor prognosis. These clinical problems are the basis for translational application of gene hypermethylation to the diagnosis and prognosis of kidney and bladder cancer.
Keywords: RCC, bladder cancer, promoter hypermethylation, tumor suppressor gene, methylome, translational application
1. Introduction
There will be an estimated 58,000 new cases and 13,500 deaths from kidney cancer in the United States in 2010 [1]. A quarter of patients with renal cell carcinoma (RCC) present with locally advanced or metastatic disease and a third of patients who undergo resection for local disease will have a recurrence [2]. Over 90% of all kidney cancers are renal cell carcinomas (RCC) originating from the renal parenchyma. The classification of RCC comprises several histological subtypes with different genetic backgrounds and natural histories [3]. Clear cell carcinoma (75%), papillary carcinoma (10-15%) and chromophobe carcinoma (5%) account for the majority of RCC. The remaining <10% of kidney cancers are mainly transitional cell carcinomas (TCC) of the renal pelvis that, in terms of histology, biology and genetics, are similar to TCC of the bladder.
Bladder cancer is the sixth most common cancer in the western world. There will be more than 71,000 new cases of cancer in the bladder in the US this year [1]. Although up to 75-80% of new cases present as non-invasive (pathological stage Ta), stroma invasive (T1) or carcinoma in situ (Tis) disease, the remaining 20-25% of tumors present as muscle invasive or more advanced disease (T2-4) with a poor prognosis. Furthermore, although approximately 20% of Ta and T1 tumors are cured, after initial removal 60-70% recur at least once in 5 years and 10-20% progress to muscle invasive cancer [4]. The established association between tobacco, or certain occupational, exposure and bladder cancer has identified high risk populations. In the Western world, TCC represents 90% of disease. In Africa and the Middle East, squamous cell carcinoma (SCC) is the predominant cell type and is related to bilharzia infection.
Cancer is a disease initiated and driven by clonal selection of cells with either inherited (germline) or acquired (somatic) genetic or epigenetic alteration of key genes that confer a growth advantage. Oncogenes are genes whose increased or altered function can result in neoplastic transformation. To date many oncogenes have been described but only a fraction have been found to be activated in bladder or kidney cancers by somatic mutation [5-7]. Several regions of amplification have been identified by comparative genome hybridization (CGH) studies and further oncogenes likely will be discovered [8, 9]. Tumor suppressor genes are best defined as genes whose loss of function can lead to neoplastic change. Both alleles need to be inactivated by germline or somatic mutation i.e. loss of heterozygosity (LOH), point mutation, homozygous deletion or promoter hypermethylation to initiate tumor formation. Approximately twenty classical tumor suppressor genes have been identified in human cancer. In sporadic clear cell RCC, chromosome 3p deletion, and inactivation of the VHL tumor suppressor gene, is known to be the most common genetic alteration. In bladder cancer, inactivation of the p53, Rb, p16INK4a/p14ARF and PTEN tumor suppressor genes occur at a moderate frequency. Other chromosomal arms have been observed to be frequently lost in RCC and bladder cancer indicating that additional tumor suppressor genes are important in tumorigenesis [3, 5].
2. DNA Methylation
Epigenetic alterations in cancer cells include DNA methylation, histone modification and RNA interference. The most studied epigenetic alteration is DNA methylation that can occur at the cytosine that precedes the guanine in a CpG dinucleotide. While CpG are generally underrepresented in the human genome sequence, the promoter regions of around half of the human genes contain a CpG-rich area termed a CgG island that are generally unmethylated in normal cells [10]. Cancer cells can show both global hypomethylation and localized hypermethylation compared to the normal cell counterpart. To date, there are few reports of hypomethylation of human oncogenes associated with activation by overexpression [11]. In part, this may be due to lack of study. Loss of imprinting (LOI) resulting in aberrant expression of the imprinted allele in cancer cells has been described in Wilms tumor and colorectal cancer [12, 13] but has not yet been clearly demonstrated in renal or bladder cancer [14]. However, it is well-established that aberrant hypermethylation of the promoter region of tumor suppressor genes is associated with transcriptional silencing and that hypermethylation is an alternative mechanism of functional inactivation [10].
2.1 Tumor Suppressor Genes Predisposing to Familial Renal or Bladder Cancer
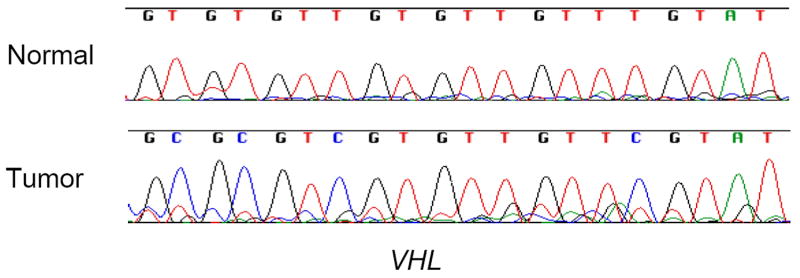
The most frequent form of familial RCC occurs in individuals with inherited von-Hippel Lindau (VHL) syndrome. The identification of the predisposing VHL tumor suppressor gene located on chromosome 3p led to the finding that, in addition to inactivation by point mutation or deletion, the VHL gene also showed allelic inactivation by aberrant hypermethylation of the promoter region (Figure 1) that was associated with transcriptional silencing in 10-15% of familial and sporadic RCC [15]. The VHL tumor suppressor gene was the first gene identified as hypermethylated in RCC. VHL inactivation occurs only in clear cell tumors and similarly, methylation of VHL has been found only in clear cell RCC [15, 16]. Since VHL inactivation is the initiating event in familial clear cell renal tumors, it is likely an early, or even initial, event in sporadic clear cell renal tumorigenesis.
Figure 1.
Direct bisulfite sequencing of the promoter region of the VHL gene in normal renal cells and a clear cell renal tumor. The presence of cytosine (C) in the tumor DNA sequence indicates methylation.
The genes that confer predisposition to other inherited forms of RCC were subsequently identified. Activating point mutations of the MET proto-oncogene are a cause of one form of hereditary papillary RCC [17]. No evidence of MET hypomethylation has been reported to date. The Fumarate Hydratase (FH) tumor suppressor gene has been identified as a predisposition gene for a second form of hereditary papillary RCC [18] and the Birt-Hogg-Dube (BHD) tumor suppressor gene for chromophobe RCC [19]. Although both these genes have typical CpG islands in the promoter region, no clear evidence of hypermethylation has been found in familial and sporadic RCC or cancers from other organ sites [16, 20, 21]. Inherited mutation of the succinate dehydrogenase complex, subunit B, iron sulfur (Ip) (SDHB) gene predisposes to early-onset RCC [22] and methylation of SDHB was reported in 1 of 25 sporadic RCC [23].
No common or defined familial form of bladder cancer has been identified. Familial bladder tumors when found are invariably from individuals with Hereditary Nonpolyposis Colorectal Cancer (HNPCC) due to inherited mutation of one of the MSH2, MLH1, MSH6 or PMS1 mismatch repair genes. TCC, predominantly of the upper tract, is the fourth commonest type of cancer in HNPCC and accounts for about 1% of bladder tumors [24]. RCC is rare in HNPCC and when found does not usually show microsatellite instability (MSI) [25]. Methylation of the MLH1 mismatch repair gene is found in the subset of sporadic colorectal, endometrial and gastric tumors with MSI [26]. Neither sporadic bladder nor renal cancers show microsatellite instability (MSI) and MLH1 methylation is absent or rare in renal and bladder cancer (unpublished data and ref [27]). The other mismatch repair genes do not appear to be hypermethylated in human cancer [28, 29].
Cowden syndrome is an autosomal dominant syndrome, which results in a predisposition to certain cancer types, including renal cancer [30, 31]. PTEN, located on chromosome 10q, has been identified as the predisposing gene for this syndrome [32]. Inactivation of PTEN by deletion and point mutation is evident in a minority of primary renal and bladder tumors but studies have shown no evidence of promoter methylation [33]. A PTEN pseudogene located on chromosome 9 can be methylated in human cells. Because of substantial sequence overlap in the promoter region with PTEN it is possible that some reports of PTEN methylation represent methylation of the psuedogene [34].
It is unclear if individuals with the disease tuberous sclerosis have a higher incidence of RCC or not. Methylation of the TSC1 gene has not been well-examined and a solitary report found no promoter methylation of TSC2 in hamartomas from TSC patients [35]. TSC2 contains a typical CpG island in the promoter region but, in a preliminary study, we observed no methylation by bisulfite sequencing of 10 primary RCC and 5 RCC cell lines (unpublished data).
2.2 Classical Tumor Suppressor Genes
Allelic loss of chromosomal region 9p21 is common in most types of solid tumor. The targets of deletion are the p16INK4a and adjacent p14ARF tumor suppressor genes. Homozygous deletion is the most common mechanism of inactivation of these genes. Promoter methylation is found relatively infrequently and point mutation is extremely rare [36]. The frequency of p16INK4a methylation in renal cancer cell lines [37] is higher than in primary RCC [16]. This observation holds true for many genes hypermethylated in cancer. Because cell lines are invariably established from advanced tumors and can undergo clonal selection over a number of passages, the frequency of gene methylation may be unrepresentative of the primary cancer [38]. Promoter hypermethylation of p16INK4a is present in 5-10% of primary RCC and bladder cancer [16, 39]. In renal tumors, p16INK4a mutations are found in both primary and metastatic tumors from the same patient [37] and, in another study, found in all grades and stages of renal tumors [16]. In bladder cancer, p16INK4a hypermethylation appears to be more common in muscle-invasive tumors (unpublished data). The p14ARF gene, which shares a coding region with p16, has a distinct exon 1 and promoter region containing a typical CpG island [40]. The p14ARF promoter and exon 1 CpG island has been reported to be hypermethylated in 5-10% of primary renal and bladder tumors [16, 41].
The Adenomatous Polyposis Coli (APC) tumor suppressor gene is associated with both familial adenomatous polyposis (FAP) and sporadic colorectal tumors and is part of the Wnt signaling pathway. An initial study examined APC promoter hypermethylation in colorectal and other cancers. Hypermethylation was observed in 10% and 8% respectively of a relatively small set of bladder and renal tumors [42]. Two further studies on larger groups of bladder tumors identified APC methylation in 35%-45% of tumors [43, 44]. A profile in renal cancer, using a larger, more representative set of tumors, found the frequency of APC methylation to be 14% [16].
The E-cadherin (CDH1) gene, located on chromosome 16q22.1, has an important role in cell-cell adhesion. Inactivating point mutations of this gene have been identified to predispose to gastric cancer [45, 46] and more rarely to other epithelial tumor types. Methylation of the second allele in tumors arising in individuals with germline mutation has been reported [47]. Loss of E-cadherin function is thought to contribute to tumor progression through increased proliferation, invasion, and metastasis. E-cadherin expression is down-regulated in many human cancer types including bladder and kidney. Hypermethylation of E-cadherin was reported in 64% (9/14) of RCC lines [48]. The same study reported that hypermethylation of the promoter region of E-cadherin and several other tumor suppressor genes was highly, but not always, correlated with loss of expression [48]. Chung et al. found E-cadherin methylation in only A-498, 1 (8%) of 5 kidney and bladder lines examined [49]. In primary renal cancer, Dulaimi et al reported a methylation frequency of 11% [16]. Maruyama et al reported a methylation frequency in 36% of 98 primary bladder tumors. E-cadherin methylation was seen in bladder tumors of all pathological grades and stages [43, 50]. Another cadherin gene, H-cadherin (CDH13) was reported methylated in 29% of the same series of bladder tumors [43]. A detailed study across the promoter region of E-Cadherin revealed significant differences in levels of methylation between individual CpG sites in the same tumor cell line and between different tumor cell lines in the NCI-60 panel [51]. The same study reported that above a threshold of approximately 20% to 30% of promoter CpG sites methylated, E-cadherin mRNA expression was effectively silenced.
Interestingly, although around half of the twenty or so classical tumor suppressor genes identified to date can be hypermethylated in human cancer, several known to be inactivated in renal, bladder and other cancers by point mutation and deletion either lack a typical promoter CpG island, e.g. p53, or have a promoter CpG island that appears to be unmethylated in human cancer e.g. PTEN [33, 34]. The Rb gene, also inactivated in bladder and renal cancer, can have promoter methylation in retinoblastoma [52] but appears unmethylated in urological tumors [16]. Other genes in these tumor suppressor gene pathways are hypermethylated in genitourinary tumors i.e. p16INK4a in the RB/p16 pathway [16, 39] and p14ARF in the p53/p14 pathway [16, 41].
2.3 Candidate Tumor Suppressor and Other Cancer Genes
A number of genes that are not commonly inactivated by genetic alteration, i.e. intragenic point mutation, are transcriptionally inactivated by promoter hypermethylation. Such genes have been considered candidate tumor suppressor genes. Tissue inhibitors of metalloproteinases (TIMPs) regulate extracellular matrix (ECM) degradation by matrix metalloproteinases (MMPs). The finding of loss of expression of the TIMP3 gene in tumor cells led to the investigation of whether the gene was silenced by promoter methylation. TIMP3 was reported to be methylated by Methylation Specific PCR (MSP) analysis in 78% of 36 primary renal cancer tumors across cell type, grade and stage [53]. TIMP3 is therefore one of the most frequently methylated genes known in renal cancer. A more representative study of 100 primary renal tumors, also by conventional gel-based MSP, reported methylation in 58% of RCC [16]. However, as more quantitative technologies for the analysis of gene methylation are available, it is becoming evident that the high frequency of methylation reported for some genes includes cases where only a small proportion of the tumor cells contain methylated alleles. At present, the biological significance of such levels of methylation is unclear. Standardization of scoring a gene as hypermethylated in a tumor specimen is an important issue [54].
Another candidate tumor suppressor gene in renal cancer is the Ras association (RalGDS/AF-6) domain family 1 gene (RASSF1A) [55]. RASSF1A is a microtubule-binding protein, which regulates mitotic progression and functions as a negative regulator of the cell cycle. RASSF1A is methylated in 28-91% of primary renal tumors [16, 56-58]. The differences in the percentage frequency of methylation are likely due to individual studies using primer sequences from different areas of the promoter CpG island, differences in the proportion of cell type, grade and stage of tumor, as well as the methylation analysis technology used in the study. In a large study of RCC broadly representative of cell type, grade and stage at presentation, RASSF1A was methylated in 45% of tumors [16]. While RASSF1A methylation has been identified in both clear cell and papillary RCCs, two studies reported that the frequency of methylation is higher in papillary compared to clear cell [16, 57]. RASSF1A methylation is also found in chromophobe tumors [16] and it is the most frequently methylated gene in early stage (organ-confined) RCC. RASSF1A is frequently methylated (35-60%) and an early event in bladder cancer [43, 59, 60]. RASSF1A expression is either lost or reduced in concordance with promoter methylation [55].
One of the first genes found to be methylated in genitourinary cancer was the carcinogen detoxification enzyme glutathione S-transferase pi-1 (GSTP1) gene. Hypermethylation of the promoter region of the GSTP1 gene is found in the majority (70-90%) of primary prostate carcinomas, but not in normal prostatic tissue or in benign hyperplasia of the prostate [61]. GSTP1 has also been identified as methylated in a minority (<10%) of bladder and renal cancers representative of cell type, grade and stage at presentation [16, 62, 63].
O6-Methylguanine-DNA Methyltransferase (MGMT), a DNA repair gene, has aberrant promoter methylation associated with loss of expression in several cancer types [64]. MGMT promoter hypermethylation is relatively infrequent in both renal (6-8%) and bladder (2-5%) tumors [16, 43, 62, 65].
The death-associated protein kinase (DAPK1) gene is located on chromosome 9q34.1, an area of frequent LOH in bladder cancers [66]. The cellular activities of DAPK1 are critical for antagonizing caspase-dependent apoptosis to promote cell survival under normal cell growth conditions. Kissil et al. found that several cancer cell lines, including bladder and renal, lack DAPK1 mRNA and protein expression. Reactivation of DAPK1 expression following azacytidine treatment was also observed. Methylation was found in 4 of 14 (29%) bladder cancer cell lines and in 2 of 5 (40%) RCC cell lines [66]. Further studies by Katzenellenbogen et al. found a correlation between the loss of DAPK1 expression and promoter hypermethylation [67]. However, most studies report that DAPK1 methylation is uncommon in primary RCC [68, 69] and primary bladder tumors [27, 43]. In contrast, Tada et al. reported that overall, 29% of bladder tumors showed DAPK1 methylation. DAPK1 methylation was identified as a marker of recurrence in stage Ta and T1 bladder cancer. 88% of papillary bladder tumors with DAPK1 methylation recurred within 15 months, while 71% of tumors that are not methylated for DAPK1 had not recurred within 24 months [70]. Another study used qMSP and reported 100% of bladder tumors and 100% of primary RCC to show methylation of DAPK1 albeit of very heterogeneous levels [71]. The apparently conflicting published data on frequency of methylation of DAP-Kinase highlights again the need for standardization of assay and scoring.
Studies have reported 15-19% of bladder tumors to show hypermethylation of the retinoic acid receptor 2 (RARβ2) gene [43, 44, 72]. Methylation of RARβ2 in renal tumors was comparable, with a frequency of 12% [16]. This gene encodes retinoic acid receptor beta, a member of the thyroid-steroid hormone receptor superfamily of nuclear transcriptional regulators. This receptor localizes to the cytoplasm and to subnuclear compartments. It binds retinoic acid, the biologically active form of vitamin A that mediates cellular signaling in embryonic morphogenesis, cell growth and differentiation. The RARβ2 protein is thought to limit growth of many cell types by regulation of transcription.
Laminin-5 (LN5), a glycoprotein that is secreted by epithelial cells, is composed of α3, β3 and γ2 chains encoded by the three LN5 genes: LAMA3, LAMB3, and LAMC2 respectively. The frequency of methylation of the LN5 genes in bladder tumors is reported to range from 21-45%. LAMA3, the most frequently methylated of the three genes, had a frequency of 45% in bladder tumors. LAMB3 was methylated in 25% and LAMC2 in 23% of the same subset of tumors. It was reported that patients with LAMC2 methylation had a shorter survival than patients that did not have methylation. [73]
Reprimo, a gene involved in the p53-mediated cell cycle arrest at the G2/M checkpoint, was reported to be frequently hypermethylated with associated loss of gene expression in a number of varying cancers (>30%). The methylation frequency in bladder cancer was lower (19%) [74]. The methylation status of Reprimo in renal cancer has not been studied.
Promoter hypermethylation of the fragile histidine triad gene (FHIT) was identified in 11-16% of bladder tumors and was correlated with a poor survival rate in patients [43, 75]. FHIT hypermethylation has also been reported in 54% of clear cell RCC [76]. This gene, a member of the histidine triad gene family, encodes a protein involved in purine metabolism. The gene encompasses the common fragile site FRA3B on chromosome 3p, where carcinogen-induced damage can lead to translocations and aberrant transcripts of this gene.
HAI-2/SPINT2 encodes Kunitz-type protease inhibitor, which functions as a regulator of hepatocyte growth factor (HGF) activity. Tumor suppressor activity as well as inactivation by hypermethylation of SPINT2 has been identified in both the clear cell (30%) and papillary (40%) subtypes of RCC. [77]
Another candidate TSG, BLU also known as ZMYND10 for zinc finger, MYND-type containing 10, has been identified on the short arm of chromosome 3, located upstream of the RASSF1A gene. BLU was reported to have a methylation frequency of 50% in kidney cell lines. It was noted that, although RASSF1A and BLU are in close proximity to one another, no correlation was found between promoter methylation of these two genes. [78]
The secreted frizzled receptor protein (SFRP) family, involved in the Wnt signaling pathway, has been studied in both kidney and bladder cancer. Promoter hypermethylation of SFRP1 has been reported to be 29% in papillary bladder cancer [79]. In a large set of bladder tumors, the frequency of methylation was SFRP1 (18%), SFRP2 (52%), SFRP4 (9%) and SFRP5 (37%) [80]. In RCC, hypermethylation of SFRP1 was reported in 68% of 38 RCC and correlated with loss of expression by immunohistochemical analysis [81] and hypermethylation of SFRP2 correlated with loss of expression in RCC cell lines [82]. A study of SFRP1, 2, 4 and 5 and other Wnt antagonist genes DKK3 and Wif1 found each gene to be methylated in around 50% and Wif-1 in 73% of RCC [83]. Standardized technology and scoring will be necessary to determine the true frequency of biologically relevant levels of methylation of SFRP and other genes.
The transforming growth factor β (TGFβ) family of genes regulates a variety of cellular functions. Regulation of the TGFβ genes has been identified in cancer cells during different stages of pathogenesis [84-86]. Suzuki et al. examined the methylation status of three TGFβ-related genes; DRM/Gremlin a member of the bone morphogenic protein antagonist family implicated in cellular hypertrophy, the transcription factor RUNX3, and HPP1/TMEFF2 transmembrane protein with EGF-like and two follistatin domains in human cancers with various clinicopathologic features. [87] In bladder cancer, DRM/Gremlin was reported as methylated in 51% of tumors examined, while HPP1 has a methylation frequency of 35% and RUNX3 showed methylation in 42% of tumors. In all three genes, methylation was tumor-specific [87].
The Human homeo-box B13 (HOXB13) gene has been identified as methylated in 30% of RCC. Loss of expression of HOXB13 gene in primary RCC and cell lines correlated with methylation status. [88]
DAL-1/4.1B, an actin binding protein, was found to be methylated in 47% of 19 renal cancer cell lines and 45% of 55 clear cell renal tumors [89].
The promoter region of ABCG2, an ATP-binding cassette transmembrane protein implicated in clinical drug resistance, has been reported to be densely methylated in RCC cell lines and to show reactivation of expression after azacytidine treatment [90].
The location of an imprinted gene DLK1 in the chromosomal region 14q, commonly deleted in RCC, prompted a report that DLK1 and GTL2 are reciprocally imprinted genes in the manner of IGF2/H19 since methylation status of GTL2 correlated with expression of DLK1 in RCC lines [91].
The apoptosis-associated interferon response gene XAF1 is methylated and reactivated by azacytidine treatment in the ACHN renal tumor cell line [92]. Methylation associated with down-regulation of expression has been reported in 6 of 7 (86%) kidney and 16 of 18 (89%) bladder primary tumors by conventional MSP [93]. However, qMSP analysis of primary RCC detected methylation in 10% of 91 cases, suggesting other mechanisms for transcriptional downregulation of XAF1 [94].
2.4 Global Gene Hypermethylation Studies
To date, the majority of genes known to undergo aberrant methylation in cancer cells have been identified by a candidate approach. By definition, this has resulted in the examination of a limited number of genes. Recently, a global profile of genes silenced by hypermethylation in RCC was generated by an expression microarray-based analysis of genes reactivated in 4 RCC lines after treatment with the demethylating drug 5Aza-2 deoxycytidine (5Aza-dC) and histone deacetylation inhibiting drug trichostatin A (TSA) [95]. Between 111 to 170 genes were found to have at least 3-fold upregulation of expression after treatment in each cell line. To establish the specificity of the screen for identification of genes epigenetically silenced in cancer cells, a subset of 12 upregulated genes was validated. The promoter methylation status and transcription status of the 12 genes were validated by semi-quantitative RT-PCR of untreated and treated cell line cDNA and by bisulfite sequencing and methylation specific PCR (MSP) of tumor and normal cell DNA. Three of the 12 genes (IGFBP1, IGFBP3 and COL1A1) showed promoter methylation in tumor DNA but were unmethylated in normal cell DNA, 1 gene (GDF15) was methylated in normal cells but more densely methylated in tumor cells, and 1 gene (PLAU) showed cancer cell specific methylation that did not correlate well with expression status. The remaining seven genes had unmethylated promoters. However, there is evidence for at least one of these genes (TGM2) to be regulated by another gene, RASSF1A [96], which was methylated in the RCC lines. It is likely that the epigenetic reactivation of particular genes leads to a cascade of upregulation in diverse pathways and networks. Other genes may be upregulated as a direct response to the stress of 5Aza-dC treatment.
Conventional MSP analysis of 32 primary, mainly organ-confined (stage I or II), renal tumors of the most common histological cell types (20 clear cell, 10 papillary and 2 chromophobe) was performed. IGFBP1 was methylated in 31%, IGFBP3 in 37%, and COL1A1 in 56% of the primary RCC. Because conventional MSP is not quantitative, we cannot be certain the methylation of the genes in the primary renal tumors is clonal, as we observed in the RCC cell lines, without further studies. The frequent methylation of these 3 genes in early stage tumors of the most common histological subtypes of RCC implicates these genes in renal tumorigenesis
In regard to the putative role of these genes in cancer, the insulin like growth factor binding proteins 1 (IGFBP1) and 3 (IGFBP3) are major forms of the IGF-binding protein family that can inhibit the growth promoting activity of both IGF I and IGF II. IGFBP3 is known to inhibit cell growth by sequestering IGF I, however, the mechanism by which IGFBP1 exerts its activity is less well understood. Clearly, methylation-based silencing of IGFBP1 and IGFBP3 could provide growth advantages to the neoplastic cell. Activation of this pathway may be of therapeutic advantage in limiting tumor growth.
COL1A1 is the human gene coding for the α1 chain of type I collagen, the major extracellular matrix component of skin and bone. Changes in the synthesis of type I collagen are associated with normal growth and tissue repair processes. Alterations in extracellular matrix composition have been implicated in tumor progression and metastasis. The COL1A2 gene has been reported as hypermethylated in bladder cancer [97]. Both the IGFBP and the COL1A gene families appear prone to hypermethylation and it is interesting that other global epigenetic screens have shown reactivation of gene families e.g. IFN in bladder [98] and SFRP family members in colorectal cancer [99]. Another epigenetic reactivation study reported hypermethylation of IGFBP3 and PLAU as well as newly identified hypermethylation of KRT19 and CXCL16 in RCC [100]. A more extensive study of 11 RCC cell lines by the same group identified eight genes (BNC1, PDLIM4, RPRM, CST6, SFRP1, GREM1, COL14A1 and COL15A1) that were frequently (>30%) methylated in primary RCC [101]. A global reactivation study of the ACHN renal tumor cell line identified the Ubiquitin carboxyl-terminal esterase L1 (UCHL1) gene, also called PGP9.5, as hypermethylated in RCC. The same study also reported upregulation of COL1A1 in ACHN cells after demethylating drug treatment [82]. Another, independent, study reported UCHL1 as hypermethylated in around a third of primary RCC [102]. A mass spectrometry analysis of a set of genes down-regulated in RCC compared to normal renal tissue in RCC identified SCNN1B, SYT6, DACH1 and TFAP2A as hypermethylated in RCC [103]. A first generation methylation array analysis of RCC identified many genes not previously reported to be methylated in RCC including TWIST and SFRP3 [104].
Global epigenetic screens of several bladder cancer cell lines identified the lysyl oxidase-like 1 and 4 (LOXL1 and LOXL4) genes as frequently hypermethylated in primary bladder cancer [105] and FGF18 and MMP11 as hypermethylated in the untreated cell lines [106].
2.5 Differing Frequencies of Methylation of a Gene in the Literature
There are several likely explanations for the discrepancies in the reported frequency of methylation of a gene between different studies. These include differences in the characteristics of the tumor set, i.e. number studied, cell type, grade, stage, and percentage tumor cell content of biopsy [107]. The frequency of methylation of a gene in tumor cell lines does not always correspond with the frequency in a representative set of primary tumors [38]. The efficiency of the bisulfite modification and the technology used for analysis is relevant since cases where only a small proportion of the tumor cells (5-10%) contain methylated alleles could be missed by direct bisulfite sequencing or pyrosequencing for example. Conventional gel-based MSP cannot readily distinguish between a tumor with clonal (in 100% of cells) methylation and a tumor with methylation in only 1% of cells. As previously mentioned, the biological significance of low levels of methylation is unclear. Primer design and location can differ between studies and heterogeneity in the methylation of individual CpG sites between different tumors can lead to MSP, which interrogates only a few CpG sites, scoring the same tumor specimen as methylated or unmethylated depending upon which particular CpG sites the primers are directed to. The stringency of a MSP reaction depends on both the salt concentration in the PCR buffer and the annealing temperature which can vary between laboratories [54].
These points again raise the question of how methylation should be scored. The increasing use of quantitative real time PCR for methylation analysis provides more information than conventional MSP but there is still no standardization for reporting of results. A direct sequence readout that is quantitative remains the gold standard [54]. Direct bisulfite sequencing which is only semi-quantitative, provides a long sequence read, and is not subject to cloning bias or direct pyrosequencing, which is more quantitative but gives a short read, are at present the closest to that goal. Current methylation arrays also have issues including that only a small minority of CpG sites of a gene promoter are represented. Additional issues are that some array-based studies do not independently validate the methylation by a different technology and that some gene methylation publications do not show any primary data.
There is also the question of whether only methylation of functional significance i.e. at or above the threshold (by extent and/or position within a given gene promoter) for loss of expression be scored? This will likely vary from one gene to another. Reexpression of a methylated gene after demethylating drug treatment of cultured cells is only an indication that the methylation of that particular gene is of functional significance as it is possible that demethylation of a transcription factor or upstream regulatory gene has restored expression. Even if methylation of a gene can be shown to have functional significance that does not imply a causal role in tumor initiation or progression. If a CpG island methylator phenotype (CIMP) [108] is clearly demonstrated in bladder cancer or RCC it will complicate the identification between driver and passenger gene hypermethylation in the same way that has occurred with small insertions or deletions inactivating genes in mismatch repair deficient tumors [109].
3. Histone Modifications
The crosstalk between DNA methylation, histone deacetylation and the chromatin state reinforces the expression status of a gene promoter. Particular patterns of histone marks are found at hypermethylated gene promoters in cancer cells [110]. The nucleosome is a subunit of chromatin that comprises a short length of 146bp of DNA wrapped around a core of histone proteins consisting of two subunits each of H2A, H2B, H3 and H4 forming an octamer. Histone core subunits share a common structure including an extended tail that is the site of post-translation modifications. The most common modifications to histone tails include acetylation, methylation, phosphorylation and ubiquitylation. These modifications have the ability to enhance or block transcription factor binding and thereby initiation of transcription. Profiles of the many potential histone modifications in cancer cells are only beginning. Histone H3-lysine 9 methylation has been associated with aberrant gene silencing in the T24 bladder tumor cell line [111]. Modifications of histone H4 have been reported in several tumor types although bladder or renal tumors were not examined [112]. A study of characteristic patterns of expression of selected histone modifier genes reported that EZH2 gene expression distinguished renal tumor from paired normal renal tissue. The pattern of expression over all 12 genes studied could discriminate bladder tumor from normal bladder. Tissue-specific patterns of expression across the 12 genes was also evident [113]. A comparison of array CGH and transcriptome analysis in bladder carcinomas identified chromosomal regions with down-regulation of expression but no loss of copy number and so yielded an overview of regional epigenetic alteration. One such copy number-independent region was validated as a region of epigenetic alteration in that loss of expression was due to tumor-specific aberrant histone methylation in the absence of DNA methylation [114]. Lower global levels of histone H3 lysine 4 dimethylation (H3K4me2) and H3K18 acetylation, and to a lesser extent H3K9me2, are reported to predict poorer prognosis in kidney cancer patients [115]. A study of histone H3 lysine 4 mono-methyl (H3K4me1), -di-methyl (H3K4me2) and -trimethyl (H3K4me3) levels on a tissue microarray of 193 RCC reported an inverse correlation with tumor grade and stage and patient survival [116].
4. MicroRNAs
MicroRNAs (miRNAs) are short (22 nucleotide) noncoding RNAs that base pair 2-8 nucleotides of their sequence to the 3’-UTR of complementary mRNA transcripts and facilitate target mRNA degradation. A single miRNA can pair to and post-transcriptionally regulate the expression of many mRNAs. Several hundred miRNAs have been identified in the human cell. The availability of array technology has led to profiles of differences in miRNA expression levels between; normal and cancer cells, grade and stage of a cancer, histological cell types, and prognostic subgroups. MiRNAs have been shown to growth-promoting or growth-inhibitory. An early study examined the relationship between the chromosomal location of miRNAs and alterations in copy number and reported that more miRNAs were located in areas deleted, rather than amplified, in human bladder cancer cells [117]. An early study profiled miRNA expression in the T24 bladder cancer cell line that showed >3-fold upregulation of 17 of 313 human miRNAs after treatment with 5Aza-2 deoxycytidine (5Aza-dC). One of the upregulated miRNAs, miR-127, is expressed in normal cells but not in tumor cells, is embedded in a CpG island and highly induced by its own promoter. This suggests it is epigenetically silenced in cancer cells and may have a tumor suppressor function [118]. Several profiles of miRNA expression in normal cells compared to renal or bladder cancer and also by tumor stage and patient outcome have been reported. Friedman et al examined pooled 9 TCCs and a pool of the matched normal transitional cells to identify a signature of miRNA expression in TCC. miR-101 tumor suppressor by down-regulation of EZH2 and consequent genome-wide effects on chromatin state [119]. Two independent studies have reported up-regulation of miR-21 or down-regulation of miR-145 in bladder cancer compared to normal urothelium [119-121]. A recent study describes miR-200c expression as a marker of progression in bladder cancer [122]. In RCC, more than one independent study has reported downregulation of miR-141 and miR-200c [123, 124] and upregulation of miR-210 [123, 125] in clear cell RCC compared to normal renal parenchymal tissue. MiR-141 and miR-200c can inhibit Epithelial to Mesenchymal Transition (EMT) by directly targeting ZEB1 and SIP1, which are repressors of E-cadherin [126]. As the field emerges, the impact of differences in source and preparation of normal cells or tumor cells, array and next generation sequencing platforms, statistical analysis, and extent of validation on the published findings to date will become clearer.
5. Translational Applications of Gene Methylation
5.1 Diagnosis and Prognosis
Aberrant methylation of cancer genes has been found in different histological cell types and all pathologic grades and stages of genitourinary cancer across patients of both sexes and of all ages and ethnicities [107]. The natural history of sporadic renal cancer is unclear but the finding of hypermethylation in kidney tumors of the lowest pathological stage (T1a) and grade (I), including tumors as small as 2cm in size, indicates that methylation can be a relatively early event in renal tumorigenesis [16]. Similarly, gene methylation is present in grade I, stage Ta tumors and carcinoma in situ (CIS) of the bladder [39, 62]. In general, classical tumor suppressor genes and some candidate tumor suppressor genes have been found to be unmethylated in normal transitional cells and normal renal cells although age-related gene methylation will be an increasingly important focus of study [107].
Because tumor suppressor and other cancer gene hypermethylation is a common, early and cancer specific alteration as well as amenable to detection by the sensitive MSP technique capable of detecting one methylated allele from a neoplastic cell in a background of several thousand unmethylated alleles from normal cells, a number of feasibility studies of methylation-based detection of cancer in body fluids were performed [107]. Using conventional MSP, an identical pattern of gene hypermethylation to that in the RCC was detected in 44 of 50 (88%) matched urine DNA. Gene methylation was positively detected in pre-operative urine from patients with organ-confined (stage I and II) RCC, including tumors as small as 2.2 cm in size. In contrast, methylation was absent from normal renal tissue and urine obtained from normal and non-neoplastic disease controls [127]. Other investigators confirmed these findings in a subsequent quantitative real time MSP study of gene methylation in RCC patient urine DNA [65]. Several studies have used panels of genes methylated in bladder cancer, commonly including the RASSF1A, p16, p14ARF, DAPK1, APC genes as well as laminin-5, apoptosis and Wnt-antagonist family genes, to demonstrate sensitive and specific detection of gene methylation in the paired pre-operative urine [39, 62, 73, 83, 128-130]. A recent study found the TWIST1 and NID2 genes to be frequently hypermethylated in bladder tumors and by qMSP of urine sediment DNA from several hundred patients with bladder cancer detected methylation of these genes with 90% sensitivity and 93% specificity [131].
One barrier to translational application of gene methylation for early detection is that the vast majority of genes identified to date can be methylated in all the genitourinary cancer types as well as cancers in other organ sites. Relatively few genes have been identified to have organ specific or cell type specific methylation that would facilitate differential diagnosis. VHL methylation is restricted to clear cell RCC. Methylation of Timp-3 is more common in RCC than in some other tumor types although this has not been well studied [53]. The RASSF1A and SPINT2 genes are more frequently methylated in papillary than clear cell RCC [16, 57, 77, 104, 132] while IGFBP1 and COL1A1 gene methylation is more common in clear cell RCC [95]. CDH1 methylation was reported significantly higher in clear cell RCC compared to chromophobe RCC or oncocytoma [132]. The <10% of bladder cancer that is non-transitional cell has not been extensively profiled for gene methylation. One study reported that RASSF1A, APC, p16, DAPK1 and RARβ2 are also hypermethylated in SCC of the bladder [133]. More genes methylated at different frequencies in different cell types will likely be identified [95].
Gene methylation may disrupt critical pathways, and thus likely plays an important role in the progression of renal and bladder tumorigenesis. The potential association of methylation status of specific genes with the biology of the tumor may facilitate prognostic classification in terms of response to targeted therapy and disease outcome and therefore merits study. For example, the γ-catenin gene has been associated with poor prognosis in RCC patients [134] and methylation of particular genes associated with progression risk in bladder cancer [135].
5.2 Epigenetic Therapy
Because demethylating drugs such as azacytidine and histone deacetylase (HDAC) inhibitors such as suberoylanilide hydroxamic acid (SAHA) can reactivate expression of epigenetically silenced genes there is much interest in the therapeutic potential of these agents and some early studies. A Phase I trial of low dose 5-Aza-dC and high dose interleukin-2 (IL-2) in 5 RCC patients reported a temporal overlap between decitabine-induced DNA hypomethylation and re-expression of methylated genes, and immune activation by high-dose IL-2. The study demonstrated this combination could be safely administered although no antitumor activity was noted in the small number of RCC patients [136]. A phase II trial of the DNMT inhibitor MG98 in metastatic RCC patients is underway [137]. Another phase II study is of the VEGF-inhibitor bevacizumab and the HDAC inhibitor vorinostat in metastatic RCC patients [138]. Combination therapy of an mTOR inhibitor and an HDAC inhibitor reduced HIF1a expression and showed growth inhibitory effects on a VHL-deficient RCC cell line greater than with single agents [139]. Treatment with azacytidine and cisplatin showed synergistic growth suppression in five bladder cancer cell lines [140]. Another bladder cancer cell line-based study reported upregulation of 17 of 313 human miRNAs after treatment with 5Aza-dC [118]
6. Future Perspective
The study of cancer epigenetics is still in a formative period. DNA methylation is the best studied of the types of epigenetic alteration present in the cancer cell. The number of genes with aberrant methylation in the human cancer cell is not known but a reasonable estimate might be that 1%, or 250 genes, of the human genome can be aberrantly methylated in a tumor cell [141, 142]. Approximately 50 genes are discussed in this chapter. Subsequent examination of further individual cancer genes, as well as array-based discovery [143] and high-throughput-based global profiles [104] of gene methylation, in larger numbers of specific genitourinary tumor types will almost certainly reveal more classical tumor suppressor genes and other important cancer genes to be methylated in kidney and bladder cancer. Studies of histone modifications, chromatin remodeling and microRNAs in cancer are emerging. Such studies combined with the methylome and transcriptome should ultimately lead to an integrated epigenome of kidney and bladder cancer. Changes in the epigenome of the normal progenitor cells of kidney and bladder cancer through ageing [144] and environmental influences [145] and a better understanding of the earliest steps in the development of kidney and bladder cancer lesions will be other major areas of study.
Executive Summary
Introduction
Epigenetic alterations are evident in bladder and renal tumor cells compared with the normal progenitor cell.
DNA hypermethylation of CpG islands in the promoter regions of genes is well-studied. Initial studies of histone modification or miRNA expression report differences between tumor and normal cells.
Tumor Suppressor Genes Predisposing to Familial Renal or Bladder Cancer
The promoter region of the VHL gene is hypermethylated in a subset of clear cell RCC
Classical Tumor Suppressor Genes
Why some genes with a promoter CpG island e.g. Pten are unmethylated in human tumor cells and whether other epigenetic alterations silence transcription of these genes remains unclear.
Candidate Tumor Suppressor and Other Cancer Genes
Clonal selection suggests that hypermethylation of cancer genes confers a growth advantage but the driver versus passenger question is pertinent.
Global Gene Hypermethylation Studies
Azacytidine reactivation coupled with expression array analysis as well as first generation gene methylation array studies have produced large sets of genes, however only a small subset have been validated. Consortium initiatives such as TCGA will be a source of potential biomarkers.
Differing Frequencies of Methylation of a Gene in the Literature
For standardization, a consensus is needed on the definition of whether an individual gene in a particular tumor is ‘methylated” or not based on normal and tumor, extent and position of methylation of CpG sites, effect on transcription and whether a minor or major clone within the tumor.
Translational Applications of Gene Methylation
Currently, there is evidence for a limited number of methylated genes to have clinical utility for early detection, prognostic and predictive classification of response in cancer. Better designed validation studies are required.
Combinations of epigenetic drugs with a standard therapy are beginning to be investigated.
Conclusions
Elucidation of the DNA methylome in cancer appears possible.
An integrated methylome, mRNA and miRNA transcriptome of kidney and bladder cancer will be produced by the TCGA.
The role of epigenetic alterations arising from environmental interaction and ageing in the risk of developing cancer will be an important area of study.
Table 1.
Selected classical tumor suppressor genes identified as hypermethylated in cancer. Function is from Gene Ontology. TSG = classical tumor suppressor gene. Methylation frequency refers to primary tumors unless stated. ND = Not done. Invariably, bisulfite sequencing was used as an initial assay followed by MSP of a larger number of tumors. COBRA is also used by some researchers.
| Gene Name | Chromosomal Location | Function | Methylation Frequency | References | |
|---|---|---|---|---|---|
| Kidney | Bladder | ||||
| VHL | 3p25.3 | TSG, transcription factor binding | 10-15% | ND | [15, 16] |
| BHD | 17p11.2 | TSG, cell cycle | None | ND | [20, 21] |
| FH | 1q43 | TSG, cell cycle | None | ND | [16] |
| SDHB | 1p36.1 | Mitochondrion respiratory chain | 4% | ND | [23] |
| MLH1 | 3p21.3 | TSG, DNA mismatch repair | None | None | [27, 68] |
| PTEN | 10q2.31 | TSG, Regulation of AKT1 signaling pathway | None | None | [33] |
| p16INK4a | 9p21.3 | TSG, Cell cycle regulation | 5-10% | 5-10% | [16, 27, 39, 43] |
| p14ARF | 9p21.3 | TSG, Cell cycle regulation | 5-10% | 5-10% | [16, 27, 41] |
| APC | 5q22.2 | TSG, Wnt receptor signaling pathway | 8-14% | 10-45% | [16, 42-44] |
Table 2.
Selected candidate tumor suppressor and cancer genes identified as hypermethylated in cancer. Function is from Gene Ontology. Methylation frequency refers to primary tumors unless stated. ND = Not done. Invariably, bisulfite sequencing was used as an initial assay followed by MSP of a larger number of tumors. COBRA is also used by some researchers.
| Gene Name | Chromosomal Location | Function | Methylation Frequency | References | |
|---|---|---|---|---|---|
| Kidney | Bladder | ||||
| E-cadherin | 16q22.1 | TSG, Cell-cell adhesion | 11% | 36% | [16, 43] |
| H-cadherin | 16q23.3 | Cell adhesion protein; negative regulator of neural cell growth | ND | 29% | [43] |
| Timp-3 | 22q12.3 | Inhibitor of proteolytic matrix metalloproteinase activity | 58-78% | ND | [16, 53] |
| RASSF1A | 3p21.31 | Negative cell cycle regulation | 28-91% | 35-60% | [16, 39, 43, 56-58, 60] |
| GSTP1 | 11q13.2 | Detoxification | <10% | <10% | [16, 27, 63, 68] |
| MGMT | 10q26.3 | DNA repair | 6-8% | 2-5% | [16, 43, 64] |
| DAP-Kinase | 9q34.1 | Positive regulator of apoptosis | None | 4-9% | [27, 41, 43, 68] |
| RARβ2 | 3p24.2 | Transcriptional regulation | 12% | 15-19% | [16, 43, 44, 72] |
| LAMA3 | 18q11.2 | Cell adhesion, Signal transduction | ND | 45% | [73] |
| LAMB3 | 1q32.2 | Mediator of cellular attachment, migration and organization | ND | 25% | |
| LAMC2 | 1q25.3 | Mediator of cellular attachment, migration and organization | ND | 23% | |
| Reprimo | 2q23.3 | Cell cycle control | ND | 19% | [74] |
| FHIT | 3p14.2 | Cell cycle | ND | 11-16% | [43, 75] |
| SPINT2 | 19q13.2 | Inhibitor of HGF activator | 33% | ND | [77] |
| BLU | 3p21.3 | Unknown | 50% cell lines | ND | [78] |
| SFRP1 | 8p11.1 | Modulator of Wnt signaling. | 47-68% | 18-29% | [79-81, 83] |
| SFRP2 | 4q31.3 | Modulator of Wnt signaling. | 53% | 52% | [80, 83] |
| SFRP3 | 2q32.1 | Modulator of Wnt signaling. | >30% | ND | [104] |
| SFRP4 | 7p14.1 | Modulator of Wnt signaling. | 53% | 9% | [80, 83] |
| SFRP5 | 10q24.1 | Modulator of Wnt signaling. | 56% | 37% | [80, 83] |
| DRM/Gremlin | 15q13.3 | Inbibits TGFb signaling | >30% | 51% | [87, 101] |
| RUNX3 | 1p36.11 | Facilitates TGFb signaling | ND | 35% | [87] |
| HPP1 | 2q32.3 | Inbibits TGFb Signaling | ND | 42% | [87] |
| HOXB13 | 17q21.32 | Vertebrate Development | 30% | ND | [88] |
| DAL-1/4.1B | 18p11.31 | Actin Binding Protein | 45% | ND | [89] |
| ABCG2 | 4q22.1 | Xenobiotic transporter | 67% cell lines | ND | [90] |
| XAF1 | 17p13.1 | Zinc ion binding | 10-86% | 89% | [93, 94] |
| IGFBP1 | 7p13 | IGF binding protein | 31% | ND | [95] |
| IGFBP3 | 7p13 | IGF binding protein, inhibitor of cell growth | 37% | ND | [95] |
| COL1A1 | 17q21.33 | Extracellular matrix component; growth and tissue repair | 56% | ND | [95] |
| COLlA2 | 7q21.3 | Extracellular matrix component; growth and tissue repair | ND | 63% | [97] |
| KRT19 | 17q21.2 | Cytoskeleton structure | 38% | ND | [100] |
| CXCL16 | 17p13.2 | Transmembrane chemokine | 42% | ND | [100] |
| UCHL1/PGP9.5 | 4p13 | Processing of ubiquitin precursors and of ubiquitinated proteins | 37-41% | ND | [82, 102] |
| LOXL1 | 15q24.1 | Extracellular matrix | ND | 70% | [105] |
| LOXL4 | 10q24.2 | Extracellular matrix | ND | 40% | [105] |
| TWIST | 7p21.1 | Transcription factor, negative regulation of cellular determination | >20% | >70% | [104, 131] |
| NID2 | 14q22.1 | Cell adhesion, extracellular matrix interaction | ND | >70% | [131] |
| PDLIM4 | 5q31.1 | Protein binding | >30% | ND | [101] |
Acknowledgments
Financial Disclosure
This publication was supported in part by grant number P30 CA006927 from the National Cancer Institute. Its contents are solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health. Additional funds were provided by Fox Chase Cancer Center via institutional support of the Kidney Cancer Keystone Program. One of the authors (PC) has received financial compensation as a member of the scientific advisory board for Oncomethylome Sciences Inc.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Cohen HT, McGovern FJ. Renal-cell carcinoma. N Engl J Med. 2005;353:2477–2490. doi: 10.1056/NEJMra043172. [DOI] [PubMed] [Google Scholar]
- 3.Zambrano NR, Lubensky IA, Merino MJ, Linehan WM, Walther MM. Histopathology and molecular genetics of renal tumors: toward unification of a classification system. J Urol. 1999;162:1246–1258. [PubMed] [Google Scholar]
- 4.Herr H, Lamm DL, Denis L. Management of Superficial Bladder Cancer, Chapter 26. Principles and Practice of Genitourinary Oncology. 1997;273 [Google Scholar]
- 5.Cairns P, Sidransky D. Bladder cancer. 2. New York: McGraw-Hill; 2002. [Google Scholar]
- 6.Dalgliesh GL, Furge K, Greenman C, et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463:360–363. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomas RK, Baker AC, Debiasi RM, et al. High-throughput oncogene mutation profiling in human cancer. Nat Genet. 2007;39:347–351. doi: 10.1038/ng1975. [DOI] [PubMed] [Google Scholar]
- 8.Beroukhim R, Brunet JP, Di Napoli A, et al. Patterns of gene expression and copy-number alterations in von-hippel lindau disease-associated and sporadic clear cell carcinoma of the kidney. Cancer Res. 2009;69:4674–4681. doi: 10.1158/0008-5472.CAN-09-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoque MO, Lee CC, Cairns P, Schoenberg M, Sidransky D. Genome-wide genetic characterization of bladder cancer: a comparison of high-density single-nucleotide polymorphism arrays and PCR-based microsatellite analysis. Cancer Res. 2003;63:2216–2222. [PubMed] [Google Scholar]
- 10.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 11.Dunn BK. Hypomethylation: one side of a larger picture. Ann N Y Acad Sci. 2003;983:28–42. doi: 10.1111/j.1749-6632.2003.tb05960.x. [DOI] [PubMed] [Google Scholar]
- 12.Cui H, Niemitz EL, Ravenel JD, et al. Loss of imprinting of insulin-like growth factor-II in Wilms’ tumor commonly involves altered methylation but not mutations of CTCF or its binding site. Cancer Res. 2001;61:4947–4950. [PubMed] [Google Scholar]
- 13.Cui H, Onyango P, Brandenburg S, Wu Y, Hsieh CL, Feinberg AP. Loss of imprinting in colorectal cancer linked to hypomethylation of H19 and IGF2. Cancer Res. 2002;62:6442–6446. [PubMed] [Google Scholar]
- 14.Byun HM, Wong HL, Birnstein EA, Wolff EM, Liang G, Yang AS. Examination of IGF2 and H19 loss of imprinting in bladder cancer. Cancer Res. 2007;67:10753–10758. doi: 10.1158/0008-5472.CAN-07-0329. [DOI] [PubMed] [Google Scholar]
- 15.Herman JG, Latif F, Weng Y, et al. Silencing of the VHL tumor-suppressor gene by DNA methylation in renal carcinoma. Proc Natl Acad Sci U S A. 1994;91:9700–9704. doi: 10.1073/pnas.91.21.9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dulaimi E, Ibanez de Caceres I, Uzzo RG, et al. Promoter hypermethylation profile of kidney cancer. Clin Cancer Res. 2004;10:3972–3979. doi: 10.1158/1078-0432.CCR-04-0175. [DOI] [PubMed] [Google Scholar]
- 17.Schmidt L, Duh FM, Chen F, et al. Germline and somatic mutations in the tyrosine kinase domain of the MET proto-oncogene in papillary renal carcinomas. Nat Genet. 1997;16:68–73. doi: 10.1038/ng0597-68. [DOI] [PubMed] [Google Scholar]
- 18.Tomlinson IP, Alam NA, Rowan AJ, et al. Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat Genet. 2002;30:406–410. doi: 10.1038/ng849. [DOI] [PubMed] [Google Scholar]
- 19.Nickerson ML, Warren MB, Toro JR, et al. Mutations in a novel gene lead to kidney tumors, lung wall defects, and benign tumors of the hair follicle in patients with the Birt-Hogg-Dube syndrome. Cancer Cell. 2002;2:157–164. doi: 10.1016/s1535-6108(02)00104-6. [DOI] [PubMed] [Google Scholar]
- 20.da Silva NF, Gentle D, Hesson LB, Morton DG, Latif F, Maher ER. Analysis of the Birt-Hogg-Dube (BHD) tumour suppressor gene in sporadic renal cell carcinoma and colorectal cancer. J Med Genet. 2003;40:820–824. doi: 10.1136/jmg.40.11.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gad S, Lefevre SH, Khoo SK, et al. Mutations in BHD and TP53 genes, but not in HNF1beta gene, in a large series of sporadic chromophobe renal cell carcinoma. Br J Cancer. 2007;96:336–340. doi: 10.1038/sj.bjc.6603492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ricketts C, Woodward ER, Killick P, et al. Germline SDHB mutations and familial renal cell carcinoma. J Natl Cancer Inst. 2008;100:1260–1262. doi: 10.1093/jnci/djn254. [DOI] [PubMed] [Google Scholar]
- 23.Morris MR, Hesson LB, Wagner KJ, et al. Multigene methylation analysis of Wilms’ tumour and adult renal cell carcinoma. Oncogene. 2003;22:6794–6801. doi: 10.1038/sj.onc.1206914. [DOI] [PubMed] [Google Scholar]
- 24.Lynch HT, Smyrk TC, Watson P, et al. Genetics, natural history, tumor spectrum, and pathology of hereditary nonpolyposis colorectal cancer: an updated review. Gastroenterology. 1993;104:1535–1549. doi: 10.1016/0016-5085(93)90368-m. [DOI] [PubMed] [Google Scholar]
- 25.Gylling AH, Nieminen TT, Abdel-Rahman WM, et al. Differential cancer predisposition in Lynch syndrome: insights from molecular analysis of brain and urinary tract tumors. Carcinogenesis. 2008;29:1351–1359. doi: 10.1093/carcin/bgn133. [DOI] [PubMed] [Google Scholar]
- 26.Herman JG, Umar A, Polyak K, et al. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci U S A. 1998;95:6870–6875. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Catto JW, Azzouzi AR, Rehman I, et al. Promoter hypermethylation is associated with tumor location, stage, and subsequent progression in transitional cell carcinoma. J Clin Oncol. 2005;23:2903–2910. doi: 10.1200/JCO.2005.03.163. [DOI] [PubMed] [Google Scholar]
- 28.Esteller M, Levine R, Baylin SB, Ellenson LH, Herman JG. MLH1 promoter hypermethylation is associated with the microsatellite instability phenotype in sporadic endometrial carcinomas. Oncogene. 1998;17:2413–2417. doi: 10.1038/sj.onc.1202178. [DOI] [PubMed] [Google Scholar]
- 29.Esteller M, Catasus L, Matias-Guiu X, et al. hMLH1 promoter hypermethylation is an early event in human endometrial tumorigenesis. Am J Pathol. 1999;155:1767–1772. doi: 10.1016/S0002-9440(10)65492-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haibach H, Burns TW, Carlson HE, Burman KD, Deftos LJ. Multiple hamartoma syndrome (Cowden’s disease) associated with renal cell carcinoma and primary neuroendocrine carcinoma of the skin (Merkel cell carcinoma) Am J Clin Pathol. 1992;97:705–712. doi: 10.1093/ajcp/97.5.705. [DOI] [PubMed] [Google Scholar]
- 31.Pilarski R, Eng C. Will the real Cowden syndrome please stand up (again)? Expanding mutational and clinical spectra of the PTEN hamartoma tumour syndrome. J Med Genet. 2004;41:323–326. doi: 10.1136/jmg.2004.018036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liaw D, Marsh DJ, Li J, et al. Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat Genet. 1997;16:64–67. doi: 10.1038/ng0597-64. [DOI] [PubMed] [Google Scholar]
- 33.Cairns P, Evron E, Okami K, et al. Point mutation and homozygous deletion of PTEN/MMAC1 in primary bladder cancers. Oncogene. 1998;16:3215–3218. doi: 10.1038/sj.onc.1201855. [DOI] [PubMed] [Google Scholar]
- 34.Zysman MA, Chapman WB, Bapat B. Considerations when analyzing the methylation status of PTEN tumor suppressor gene. Am J Pathol. 2002;160:795–800. doi: 10.1016/S0002-9440(10)64902-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Niida Y, Stemmer-Rachamimov AO, Logrip M, et al. Survey of somatic mutations in tuberous sclerosis complex (TSC) hamartomas suggests different genetic mechanisms for pathogenesis of TSC lesions. Am J Hum Genet. 2001;69:493–503. doi: 10.1086/321972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rocco JW, Sidransky D. p16(MTS-1/CDKN2/INK4a) in cancer progression. Exp Cell Res. 2001;264:42–55. doi: 10.1006/excr.2000.5149. [DOI] [PubMed] [Google Scholar]
- 37.Herman JG, Merlo A, Mao L, et al. Inactivation of the CDKN2/p16/MTS1 gene is frequently associated with aberrant DNA methylation in all common human cancers. Cancer Res. 1995;55:4525–4530. [PubMed] [Google Scholar]
- 38*.Smiraglia DJ, Rush LJ, Fruhwald MC, et al. Excessive CpG island hypermethylation in cancer cell lines versus primary human malignancies. Hum Mol Genet. 2001;10:1413–1419. doi: 10.1093/hmg/10.13.1413. This study illustrates the point that tumor cell lines need not be representative of the extent of aberrant methylation in a primary tumor. [DOI] [PubMed] [Google Scholar]
- 39.Dulaimi E, Uzzo RG, Greenberg RE, Al-Saleem T, Cairns P. Detection of bladder cancer in urine by a tumor suppressor gene hypermethylation panel. Clin Cancer Res. 2004;10:1887–1893. doi: 10.1158/1078-0432.ccr-03-0127. [DOI] [PubMed] [Google Scholar]
- 40.Robertson KD, Jones PA. The human ARF cell cycle regulatory gene promoter is a CpG island which can be silenced by DNA methylation and down-regulated by wild-type p53. Mol Cell Biol. 1998;18:6457–6473. doi: 10.1128/mcb.18.11.6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Res. 2001;61:3225–3229. [PubMed] [Google Scholar]
- 42.Esteller M, Sparks A, Toyota M, et al. Analysis of adenomatous polyposis coli promoter hypermethylation in human cancer. Cancer Res. 2000;60:4366–4371. [PubMed] [Google Scholar]
- 43.Maruyama R, Toyooka S, Toyooka KO, et al. Aberrant promoter methylation profile of bladder cancer and its relationship to clinicopathological features. Cancer Res. 2001;61:8659–8663. [PubMed] [Google Scholar]
- 44.Neuhausen A, Florl AR, Grimm MO, Schulz WA. DNA methylation alterations in urothelial carcinoma. Cancer Biol Ther. 2006;5:993–1001. doi: 10.4161/cbt.5.8.2885. [DOI] [PubMed] [Google Scholar]
- 45.Becker KF, Atkinson MJ, Reich U, et al. E-cadherin gene mutations provide clues to diffuse type gastric carcinomas. Cancer Res. 1994;54:3845–3852. [PubMed] [Google Scholar]
- 46.Guilford P, Hopkins J, Harraway J, et al. E-cadherin germline mutations in familial gastric cancer. Nature. 1998;392:402–405. doi: 10.1038/32918. [DOI] [PubMed] [Google Scholar]
- 47.Grady WM, Willis J, Guilford PJ, et al. Methylation of the CDH1 promoter as the second genetic hit in hereditary diffuse gastric cancer. Nat Genet. 2000;26:16–17. doi: 10.1038/79120. [DOI] [PubMed] [Google Scholar]
- 48.Kawakami T, Okamoto K, Ogawa O, Okada Y. Multipoint methylation and expression analysis of tumor suppressor genes in human renal cancer cells. Urology. 2003;61:226–230. doi: 10.1016/s0090-4295(02)02110-6. [DOI] [PubMed] [Google Scholar]
- 49.Chung WB, Hong SH, Kim JA, Sohn YK, Kim BW, Kim JW. Hypermethylation of tumor-related genes in genitourinary cancer cell lines. J Korean Med Sci. 2001;16:756–761. doi: 10.3346/jkms.2001.16.6.756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ribeiro-Filho LA, Franks J, Sasaki M, et al. CpG hypermethylation of promoter region and inactivation of E-cadherin gene in human bladder cancer. Mol Carcinog. 2002;34:187–198. doi: 10.1002/mc.10064. [DOI] [PubMed] [Google Scholar]
- 51*.Reinhold WC, Reimers MA, Maunakea AK, et al. Detailed DNA methylation profiles of the E-cadherin promoter in the NCI-60 cancer cells. Mol Cancer Ther. 2007;6:391–403. doi: 10.1158/1535-7163.MCT-06-0609. A detailed study across the promoter region of E-Cadherin revealed significant differences in methylation between individual CpG sites in the same tumor cell line and between different tumor cell lines in the NCI-60 panel and defines a threshold of approximately 20% to 30% of promoter CpG sites methylated, necessary for silencing of mRNA expression. [DOI] [PubMed] [Google Scholar]
- 52.Stirzaker C, Millar DS, Paul CL, et al. Extensive DNA methylation spanning the Rb promoter in retinoblastoma tumors. Cancer Res. 1997;57:2229–2237. [PubMed] [Google Scholar]
- 53.Bachman KE, Herman JG, Corn PG, et al. Methylation-associated silencing of the tissue inhibitor of metalloproteinase-3 gene suggest a suppressor role in kidney, brain, and other human cancers. Cancer Res. 1999;59:798–802. [PubMed] [Google Scholar]
- 54.Kagan J, Srivastava S, Barker PE, Belinsky SA, Cairns P. Towards Clinical Application of Methylated DNA Sequences as Cancer Biomarkers: A Joint NCI’s EDRN and NIST Workshop on Standards, Methods, Assays, Reagents and Tools. Cancer Res. 2007;67:4545–4549. doi: 10.1158/0008-5472.CAN-06-2888. [DOI] [PubMed] [Google Scholar]
- 55.Dammann R, Li C, Yoon JH, Chin PL, Bates S, Pfeifer GP. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nat Genet. 2000;25:315–319. doi: 10.1038/77083. [DOI] [PubMed] [Google Scholar]
- 56.Dreijerink K, Braga E, Kuzmin I, et al. The candidate tumor suppressor gene, RASSF1A, from human chromosome 3p21.3 is involved in kidney tumorigenesis. Proc Natl Acad Sci U S A. 2001;98:7504–7509. doi: 10.1073/pnas.131216298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Morrissey C, Martinez A, Zatyka M, et al. Epigenetic inactivation of the RASSF1A 3p21.3 tumor suppressor gene in both clear cell and papillary renal cell carcinoma. Cancer Res. 2001;61:7277–7281. [PubMed] [Google Scholar]
- 58*.Yoon JH, Dammann R, Pfeifer GP. Hypermethylation of the CpG island of the RASSF1A gene in ovarian and renal cell carcinomas. Int J Cancer. 2001;94:212–217. doi: 10.1002/ijc.1466. The above four publications are important because they reported aberrant hyperemethylation of a gene to be frequent and across different tumor types. [DOI] [PubMed] [Google Scholar]
- 59.Chan MW, Chan LW, Tang NL, et al. Frequent hypermethylation of promoter region of RASSF1A in tumor tissues and voided urine of urinary bladder cancer patients. Int J Cancer. 2003;104:611–616. doi: 10.1002/ijc.10971. [DOI] [PubMed] [Google Scholar]
- 60.Lee MG, Kim HY, Byun DS, et al. Frequent epigenetic inactivation of RASSF1A in human bladder carcinoma. Cancer Res. 2001;61:6688–6692. [PubMed] [Google Scholar]
- 61.Lee WH, Morton RA, Epstein JI, et al. Cytidine methylation of regulatory sequences near the pi-class glutathione S-transferase gene accompanies human prostatic carcinogenesis. Proc Natl Acad Sci U S A. 1994;91:11733–11737. doi: 10.1073/pnas.91.24.11733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chan MW, Chan LW, Tang NL, et al. Hypermethylation of multiple genes in tumor tissues and voided urine in urinary bladder cancer patients. Clin Cancer Res. 2002;8:464–470. [PubMed] [Google Scholar]
- 63.Esteller M, Corn PG, Urena JM, Gabrielson E, Baylin SB, Herman JG. Inactivation of glutathione S-transferase P1 gene by promoter hypermethylation in human neoplasia. Cancer Res. 1998;58:4515–4518. [PubMed] [Google Scholar]
- 64.Esteller M, Hamilton SR, Burger PC, Baylin SB, Herman JG. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res. 1999;59:793–797. [PubMed] [Google Scholar]
- 65.Hoque MO, Begum S, Topaloglu O, et al. Quantitative detection of promoter hypermethylation of multiple genes in the tumor, urine, and serum DNA of patients with renal cancer. Cancer Res. 2004;64:5511–5517. doi: 10.1158/0008-5472.CAN-04-0799. [DOI] [PubMed] [Google Scholar]
- 66.Kissil JL, Feinstein E, Cohen O, et al. DAP-kinase loss of expression in various carcinoma and B-cell lymphoma cell lines: possible implications for role as tumor suppressor gene. Oncogene. 1997;15:403–407. doi: 10.1038/sj.onc.1201172. [DOI] [PubMed] [Google Scholar]
- 67.Katzenellenbogen RA, Baylin SB, Herman JG. Hypermethylation of the DAP-kinase CpG island is a common alteration in B-cell malignancies. Blood. 1999;93:4347–4353. [PubMed] [Google Scholar]
- 68.Gonzalgo ML, Yegnasubramanian S, Yan G, et al. Molecular profiling and classification of sporadic renal cell carcinoma by quantitative methylation analysis. Clin Cancer Res. 2004;10:7276–7283. doi: 10.1158/1078-0432.CCR-03-0692. [DOI] [PubMed] [Google Scholar]
- 69.Wethkamp N, Ramp U, Geddert H, et al. Expression of death-associated protein kinase during tumour progression of human renal cell carcinomas: hypermethylation-independent mechanisms of inactivation. Eur J Cancer. 2006;42:264–274. doi: 10.1016/j.ejca.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 70.Tada Y, Wada M, Taguchi K, et al. The association of death-associated protein kinase hypermethylation with early recurrence in superficial bladder cancers. Cancer Res. 2002;62:4048–4053. [PubMed] [Google Scholar]
- 71.Christoph F, Kempkensteffen C, Weikert S, et al. Methylation of tumour suppressor genes APAF-1 and DAPK-1 and in vitro effects of demethylating agents in bladder and kidney cancer. Br J Cancer. 2006;95:1701–1707. doi: 10.1038/sj.bjc.6603482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Marsit CJ, Houseman EA, Christensen BC, et al. Examination of a CpG island methylator phenotype and implications of methylation profiles in solid tumors. Cancer Res. 2006;66:10621–10629. doi: 10.1158/0008-5472.CAN-06-1687. [DOI] [PubMed] [Google Scholar]
- 73.Sathyanarayana UG, Maruyama R, Padar A, et al. Molecular detection of noninvasive and invasive bladder tumor tissues and exfoliated cells by aberrant promoter methylation of laminin-5 encoding genes. Cancer Res. 2004;64:1425–1430. doi: 10.1158/0008-5472.can-03-0701. [DOI] [PubMed] [Google Scholar]
- 74.Takahashi T, Suzuki M, Shigematsu H, et al. Aberrant methylation of Reprimo in human malignancies. Int J Cancer. 2005;115:503–510. doi: 10.1002/ijc.20910. [DOI] [PubMed] [Google Scholar]
- 75.Iliopoulos D, Guler G, Han SY, et al. Fragile genes as biomarkers: epigenetic control of WWOX and FHIT in lung, breast and bladder cancer. Oncogene. 2005;24:1625–1633. doi: 10.1038/sj.onc.1208398. [DOI] [PubMed] [Google Scholar]
- 76.Kvasha S, Gordiyuk V, Kondratov A, et al. Hypermethylation of the 5’CpG island of the FHIT gene in clear cell renal carcinomas. Cancer Lett. 2008;265:250–257. doi: 10.1016/j.canlet.2008.02.036. [DOI] [PubMed] [Google Scholar]
- 77.Morris MR, Gentle D, Abdulrahman M, et al. Tumor suppressor activity and epigenetic inactivation of hepatocyte growth factor activator inhibitor type 2/SPINT2 in papillary and clear cell renal cell carcinoma. Cancer Res. 2005;65:4598–4606. doi: 10.1158/0008-5472.CAN-04-3371. [DOI] [PubMed] [Google Scholar]
- 78.Agathanggelou A, Dallol A, Zochbauer-Muller S, et al. Epigenetic inactivation of the candidate 3p21.3 suppressor gene BLU in human cancers. Oncogene. 2003;22:1580–1588. doi: 10.1038/sj.onc.1206243. [DOI] [PubMed] [Google Scholar]
- 79.Stoehr R, Wissmann C, Suzuki H, et al. Deletions of chromosome 8p and loss of sFRP1 expression are progression markers of papillary bladder cancer. Lab Invest. 2004;84:465–478. doi: 10.1038/labinvest.3700068. [DOI] [PubMed] [Google Scholar]
- 80.Marsit CJ, Karagas MR, Andrew A, et al. Epigenetic inactivation of SFRP genes and TP53 alteration act jointly as markers of invasive bladder cancer. Cancer Res. 2005;65:7081–7085. doi: 10.1158/0008-5472.CAN-05-0267. [DOI] [PubMed] [Google Scholar]
- 81.Dahl E, Wiesmann F, Woenckhaus M, et al. Frequent loss of SFRP1 expression in multiple human solid tumours: association with aberrant promoter methylation in renal cell carcinoma. Oncogene. 2007;26:5680–5691. doi: 10.1038/sj.onc.1210345. [DOI] [PubMed] [Google Scholar]
- 82.Kagara I, Enokida H, Kawakami K, et al. CpG hypermethylation of the UCHL1 gene promoter is associated with pathogenesis and poor prognosis in renal cell carcinoma. J Urol. 2008;180:343–351. doi: 10.1016/j.juro.2008.02.044. [DOI] [PubMed] [Google Scholar]
- 83.Urakami S, Shiina H, Enokida H, et al. Combination analysis of hypermethylated Wnt-antagonist family genes as a novel epigenetic biomarker panel for bladder cancer detection. Clin Cancer Res. 2006;12:2109–2116. doi: 10.1158/1078-0432.CCR-05-2468. [DOI] [PubMed] [Google Scholar]
- 84.de Caestecker MP, Piek E, Roberts AB. Role of transforming growth factor-beta signaling in cancer. J Natl Cancer Inst. 2000;92:1388–1402. doi: 10.1093/jnci/92.17.1388. [DOI] [PubMed] [Google Scholar]
- 85.Roberts AB, Wakefield LM. The two faces of transforming growth factor beta in carcinogenesis. Proc Natl Acad Sci U S A. 2003;100:8621–8623. doi: 10.1073/pnas.1633291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Teicher BA. Malignant cells, directors of the malignant process: role of transforming growth factor-beta. Cancer Metastasis Rev. 2001;20:133–143. doi: 10.1023/a:1013177011767. [DOI] [PubMed] [Google Scholar]
- 87.Suzuki M, Shigematsu H, Shames DS, et al. DNA methylation-associated inactivation of TGFbeta-related genes DRM/Gremlin, RUNX3, and HPP1 in human cancers. Br J Cancer. 2005;93:1029–1037. doi: 10.1038/sj.bjc.6602837. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 88.Okuda H, Toyota M, Ishida W, et al. Epigenetic inactivation of the candidate tumor suppressor gene HOXB13 in human renal cell carcinoma. Oncogene. 2006;25:1733–1742. doi: 10.1038/sj.onc.1209200. [DOI] [PubMed] [Google Scholar]
- 89.Yamada D, Kikuchi S, Williams YN, et al. Promoter hypermethylation of the potential tumor suppressor DAL-1/4.1B gene in renal clear cell carcinoma. Int J Cancer. 2006;118:916–923. doi: 10.1002/ijc.21450. [DOI] [PubMed] [Google Scholar]
- 90.To KK, Zhan Z, Bates SE. Aberrant promoter methylation of the ABCG2 gene in renal carcinoma. Mol Cell Biol. 2006;26:8572–8585. doi: 10.1128/MCB.00650-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kawakami T, Chano T, Minami K, Okabe H, Okada Y, Okamoto K. Imprinted DLK1 is a putative tumor suppressor gene and inactivated by epimutation at the region upstream of GTL2 in human renal cell carcinoma. Hum Mol Genet. 2006;15:821–830. doi: 10.1093/hmg/ddl001. [DOI] [PubMed] [Google Scholar]
- 92.Reu FJ, Bae SI, Cherkassky L, et al. Overcoming resistance to interferon-induced apoptosis of renal carcinoma and melanoma cells by DNA demethylation. J Clin Oncol. 2006;24:3771–3779. doi: 10.1200/JCO.2005.03.4074. [DOI] [PubMed] [Google Scholar]
- 93.Lee MG, Huh JS, Chung SK, et al. Promoter CpG hypermethylation and downregulation of XAF1 expression in human urogenital malignancies: implication for attenuated p53 response to apoptotic stresses. Oncogene. 2006;25:5807–5822. doi: 10.1038/sj.onc.1209867. [DOI] [PubMed] [Google Scholar]
- 94.Kempkensteffen C, Hinz S, Schrader M, et al. Gene expression and promoter methylation of the XIAP-associated Factor 1 in renal cell carcinomas: correlations with pathology and outcome. Cancer Lett. 2007;254:227–235. doi: 10.1016/j.canlet.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 95.Ibanez de Caceres I, Dulaimi E, Hoffman AM, Al-Saleem T, Uzzo RG, Cairns P. Identification of novel target genes by an epigenetic reactivation screen of renal cancer. Cancer Res. 2006;66:5021–5028. doi: 10.1158/0008-5472.CAN-05-3365. [DOI] [PubMed] [Google Scholar]
- 96.Agathanggelou A, Bieche I, Ahmed-Choudhury J, et al. Identification of novel gene expression targets for the Ras association domain family 1 (RASSF1A) tumor suppressor gene in non-small cell lung cancer and neuroblastoma. Cancer Res. 2003;63:5344–5351. [PMC free article] [PubMed] [Google Scholar]
- 97.Mori K, Enokida H, Kagara I, et al. CpG hypermethylation of collagen type I alpha 2 contributes to proliferation and migration activity of human bladder cancer. Int J Oncol. 2009;34:1593–1602. doi: 10.3892/ijo_00000289. [DOI] [PubMed] [Google Scholar]
- 98*.Liang G, Gonzales FA, Jones PA, Orntoft TF, Thykjaer T. Analysis of gene induction in human fibroblasts and bladder cancer cells exposed to the methylation inhibitor 5-aza-2’-deoxycytidine. Cancer Res. 2002;62:961–966. This was the first use of a genome-wide expresssion array after demethylating drug treatment of a tumor cell line as a global discovery tool. [PubMed] [Google Scholar]
- 99.Suzuki H, Gabrielson E, Chen W, et al. A genomic screen for genes upregulated by demethylation and histone deacetylase inhibition in human colorectal cancer. Nat Genet. 2002;31:141–149. doi: 10.1038/ng892. [DOI] [PubMed] [Google Scholar]
- 100.Morris MR, Gentle D, Abdulrahman M, et al. Functional epigenomics approach to identify methylated candidate tumour suppressor genes in renal cell carcinoma. Br J Cancer. 2008;98:496–501. doi: 10.1038/sj.bjc.6604180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Morris MR, Ricketts C, Gentle D, et al. Identification of candidate tumour suppressor genes frequently methylated in renal cell carcinoma. Oncogene. 2010;29:2104–2117. doi: 10.1038/onc.2009.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Seliger B, Handke D, Schabel E, Bukur J, Lichtenfels R, Dammann R. Epigenetic control of the ubiquitin carboxyl terminal hydrolase 1 in renal cell carcinoma. J Transl Med. 2009;7:90. doi: 10.1186/1479-5876-7-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dalgin GS, Drever M, Williams T, King T, DeLisi C, Liou LS. Identification of novel epigenetic markers for clear cell renal cell carcinoma. J Urol. 2008;180:1126–1130. doi: 10.1016/j.juro.2008.04.137. [DOI] [PubMed] [Google Scholar]
- 104*.McRonald FE, Morris MR, Gentle D, et al. CpG methylation profiling in VHL related and VHL unrelated renal cell carcinoma. Mol Cancer. 2009;8:31. doi: 10.1186/1476-4598-8-31. This is the first report of Illumina beadchip methylation technology in renal cancer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wu G, Guo Z, Chang X, et al. LOXL1 and LOXL4 are epigenetically silenced and can inhibit ras/extracellular signal-regulated kinase signaling pathway in human bladder cancer. Cancer Res. 2007;67:4123–4129. doi: 10.1158/0008-5472.CAN-07-0012. [DOI] [PubMed] [Google Scholar]
- 106.Veerla S, Panagopoulos I, Jin Y, Lindgren D, Hoglund M. Promoter analysis of epigenetically controlled genes in bladder cancer. Genes Chromosomes Cancer. 2008;47:368–378. doi: 10.1002/gcc.20542. [DOI] [PubMed] [Google Scholar]
- 107*.Cairns P. Gene methylation and early detection of genitourinary cancer: the road ahead. Nat Rev Cancer. 2007;7:531–543. doi: 10.1038/nrc2170. A discussion of the promise and challenges of aberrant methylation for early detection of cancer. [DOI] [PubMed] [Google Scholar]
- 108.Teodoridis JM, Hardie C, Brown R. CpG island methylator phenotype (CIMP) in cancer: causes and implications. Cancer Lett. 2008;268:177–186. doi: 10.1016/j.canlet.2008.03.022. [DOI] [PubMed] [Google Scholar]
- 109.Zhang L, Yu J, Willson JK, Markowitz SD, Kinzler KW, Vogelstein B. Short mononucleotide repeat sequence variability in mismatch repair-deficient cancers. Cancer Res. 2001;61:3801–3805. [PubMed] [Google Scholar]
- 110.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 111.Nguyen CT, Weisenberger DJ, Velicescu M, et al. Histone H3-lysine 9 methylation is associated with aberrant gene silencing in cancer cells and is rapidly reversed by 5-aza-2’-deoxycytidine. Cancer Res. 2002;62:6456–6461. [PubMed] [Google Scholar]
- 112.Fraga MF, Ballestar E, Villar-Garea A, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet. 2005;37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- 113.Ozdag H, Teschendorff AE, Ahmed AA, et al. Differential expression of selected histone modifier genes in human solid cancers. BMC Genomics. 2006;7:90. doi: 10.1186/1471-2164-7-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Stransky N, Vallot C, Reyal F, et al. Regional copy number-independent deregulation of transcription in cancer. Nat Genet. 2006;38:1386–1396. doi: 10.1038/ng1923. [DOI] [PubMed] [Google Scholar]
- 115.Seligson DB, Horvath S, McBrian MA, et al. Global levels of histone modifications predict prognosis in different cancers. Am J Pathol. 2009;174:1619–1628. doi: 10.2353/ajpath.2009.080874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ellinger J, Kahl P, Mertens C, et al. Prognostic relevance of global histone H3 lysine 4 (H3K4) methylation in renal cell carcinoma. Int J Cancer. 2010;127:2360–2366. doi: 10.1002/ijc.25250. [DOI] [PubMed] [Google Scholar]
- 117.Lamy P, Andersen CL, Dyrskjot L, Torring N, Orntoft T, Wiuf C. Are microRNAs located in genomic regions associated with cancer? Br J Cancer. 2006;95:1415–1418. doi: 10.1038/sj.bjc.6603381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Saito Y, Jones PA. Epigenetic activation of tumor suppressor microRNAs in human cancer cells. Cell Cycle. 2006;5:2220–2222. doi: 10.4161/cc.5.19.3340. [DOI] [PubMed] [Google Scholar]
- 119.Friedman JM, Liang G, Liu CC, et al. The putative tumor suppressor microRNA-101 modulates the cancer epigenome by repressing the polycomb group protein EZH2. Cancer Res. 2009;69:2623–2629. doi: 10.1158/0008-5472.CAN-08-3114. [DOI] [PubMed] [Google Scholar]
- 120.Catto JW, Miah S, Owen HC, et al. Distinct microRNA alterations characterize high- and low-grade bladder cancer. Cancer Res. 2009;69:8472–8481. doi: 10.1158/0008-5472.CAN-09-0744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Dyrskjot L, Ostenfeld MS, Bramsen JB, et al. Genomic profiling of microRNAs in bladder cancer: miR-129 is associated with poor outcome and promotes cell death in vitro. Cancer Res. 2009;69:4851–4860. doi: 10.1158/0008-5472.CAN-08-4043. [DOI] [PubMed] [Google Scholar]
- 122.Wiklund ED, Bramsen JB, Hulf T, et al. Coordinated epigenetic repression of the miR-200 family and miR-205 in invasive bladder cancer. Int J Cancer. 2010 doi: 10.1002/ijc.25461. [DOI] [PubMed] [Google Scholar]
- 123.Jung M, Mollenkopf HJ, Grimm C, et al. MicroRNA profiling of clear cell renal cell cancer identifies a robust signature to define renal malignancy. J Cell Mol Med. 2009;13:3918–3928. doi: 10.1111/j.1582-4934.2009.00705.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Nakada C, Matsuura K, Tsukamoto Y, et al. Genome-wide microRNA expression profiling in renal cell carcinoma: significant down-regulation of miR-141 and miR-200c. J Pathol. 2008;216:418–427. doi: 10.1002/path.2437. [DOI] [PubMed] [Google Scholar]
- 125.Juan D, Alexe G, Antes T, et al. Identification of a MicroRNA Panel for Clear-cell Kidney Cancer. Urology. 2009 doi: 10.1016/j.urology.2009.10.033. [DOI] [PubMed] [Google Scholar]
- 126.Banumathy G, Cairns P. Signaling pathways in renal cell carcinoma. Cancer Biol Ther. 2010;10 doi: 10.4161/cbt.10.7.13247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Battagli C, Uzzo RG, Dulaimi E, et al. Promoter hypermethylation of tumor suppressor genes in urine from kidney cancer patients. Cancer Res. 2003;63:8695–8699. [PubMed] [Google Scholar]
- 128.Friedrich MG, Weisenberger DJ, Cheng JC, et al. Detection of methylated apoptosis-associated genes in urine sediments of bladder cancer patients. Clin Cancer Res. 2004;10:7457–7465. doi: 10.1158/1078-0432.CCR-04-0930. [DOI] [PubMed] [Google Scholar]
- 129.Hoque MO, Begum S, Topaloglu O, et al. Quantitation of promoter methylation of multiple genes in urine DNA and bladder cancer detection. J Natl Cancer Inst. 2006;98:996–1004. doi: 10.1093/jnci/djj265. [DOI] [PubMed] [Google Scholar]
- 130.Yates DR, Rehman I, Meuth M, Cross SS, Hamdy FC, Catto JW. Methylational urinalysis: a prospective study of bladder cancer patients and age stratified benign controls. Oncogene. 2006;25:1984–1988. doi: 10.1038/sj.onc.1209209. [DOI] [PubMed] [Google Scholar]
- 131.Renard I, Joniau S, van Cleynenbreugel B, et al. Identification and Validation of the Methylated TWIST1 and NID2 Genes through Real-Time Methylation-Specific Polymerase Chain Reaction Assays for the Noninvasive Detection of Primary Bladder Cancer in Urine Samples. Eur Urol. 2009 doi: 10.1016/j.eururo.2009.07.041. [DOI] [PubMed] [Google Scholar]
- 132.Costa VL, Henrique R, Ribeiro FR, et al. Quantitative promoter methylation analysis of multiple cancer-related genes in renal cell tumors. BMC Cancer. 2007;7:133. doi: 10.1186/1471-2407-7-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gutierrez MI, Siraj AK, Khaled H, Koon N, El-Rifai W, Bhatia K. CpG island methylation in Schistosoma- and non-Schistosoma-associated bladder cancer. Mod Pathol. 2004;17:1268–1274. doi: 10.1038/modpathol.3800177. [DOI] [PubMed] [Google Scholar]
- 134.Breault JE, Shiina H, Igawa M, et al. Methylation of the gamma-catenin gene is associated with poor prognosis of renal cell carcinoma. Clin Cancer Res. 2005;11:557–564. [PubMed] [Google Scholar]
- 135.Yates DR, Rehman I, Abbod MF, et al. Promoter hypermethylation identifies progression risk in bladder cancer. Clin Cancer Res. 2007;13:2046–2053. doi: 10.1158/1078-0432.CCR-06-2476. [DOI] [PubMed] [Google Scholar]
- 136.Gollob JA, Sciambi CJ, Peterson BL, et al. Phase I trial of sequential low-dose 5-aza-2’-deoxycytidine plus high-dose intravenous bolus interleukin-2 in patients with melanoma or renal cell carcinoma. Clin Cancer Res. 2006;12:4619–4627. doi: 10.1158/1078-0432.CCR-06-0883. [DOI] [PubMed] [Google Scholar]
- 137.Winquist E, Knox J, Ayoub JP, et al. Phase II trial of DNA methyltransferase 1 inhibition with the antisense oligonucleotide MG98 in patients with metastatic renal carcinoma: a National Cancer Institute of Canada Clinical Trials Group investigational new drug study. Invest New Drugs. 2006;24:159–167. doi: 10.1007/s10637-006-5938-1. [DOI] [PubMed] [Google Scholar]
- 138.Hammers HJ, Verheul H, Wilky B, et al. Phase I safety and pharmacokinetic/pharmacodynamic results of the histone deacetylase inhibitor vorinostat in combination with bevacizumab in patients with kidney cancer. Journal of Clinical Oncology, 2008 ASCO Annual Meeting Proceedings. 2008;26 [Google Scholar]
- 139.Verheul HM, Salumbides B, Van Erp K, et al. Combination strategy targeting the hypoxia inducible factor-1 alpha with mammalian target of rapamycin and histone deacetylase inhibitors. Clin Cancer Res. 2008;14:3589–3597. doi: 10.1158/1078-0432.CCR-07-4306. [DOI] [PubMed] [Google Scholar]
- 140.Shang D, Liu Y, Matsui Y, et al. Demethylating agent 5-aza-2’-deoxycytidine enhances susceptibility of bladder transitional cell carcinoma to Cisplatin. Urology. 2008;71:1220–1225. doi: 10.1016/j.urology.2007.11.029. [DOI] [PubMed] [Google Scholar]
- 141.Costello JF, Frühwald MC, Smiraglia DJ, et al. Aberrant CpG-island methylation has non-random and tumour-type–specific patterns. Nature Genet. 2000;25:132–138. doi: 10.1038/72785. [DOI] [PubMed] [Google Scholar]
- 142.Sjoblom T, Jones S, Wood LD, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 143.Aleman A, Cebrian V, Alvarez M, et al. Identification of PMF1 methylation in association with bladder cancer progression. Clin Cancer Res. 2008;14:8236–8243. doi: 10.1158/1078-0432.CCR-08-0778. [DOI] [PubMed] [Google Scholar]
- 144.Kwabi-Addo B, Chung W, Shen L, et al. Age-related DNA methylation changes in normal human prostate tissues. Clin Cancer Res. 2007;13:3796–3802. doi: 10.1158/1078-0432.CCR-07-0085. [DOI] [PubMed] [Google Scholar]
- 145.Wolff EM, Liang G, Cortez CC, et al. RUNX3 methylation reveals that bladder tumors are older in patients with a history of smoking. Cancer Res. 2008;68:6208–6214. doi: 10.1158/0008-5472.CAN-07-6616. [DOI] [PMC free article] [PubMed] [Google Scholar]