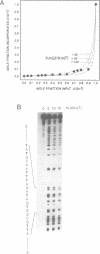
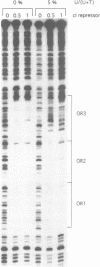
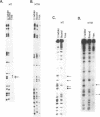
Abstract
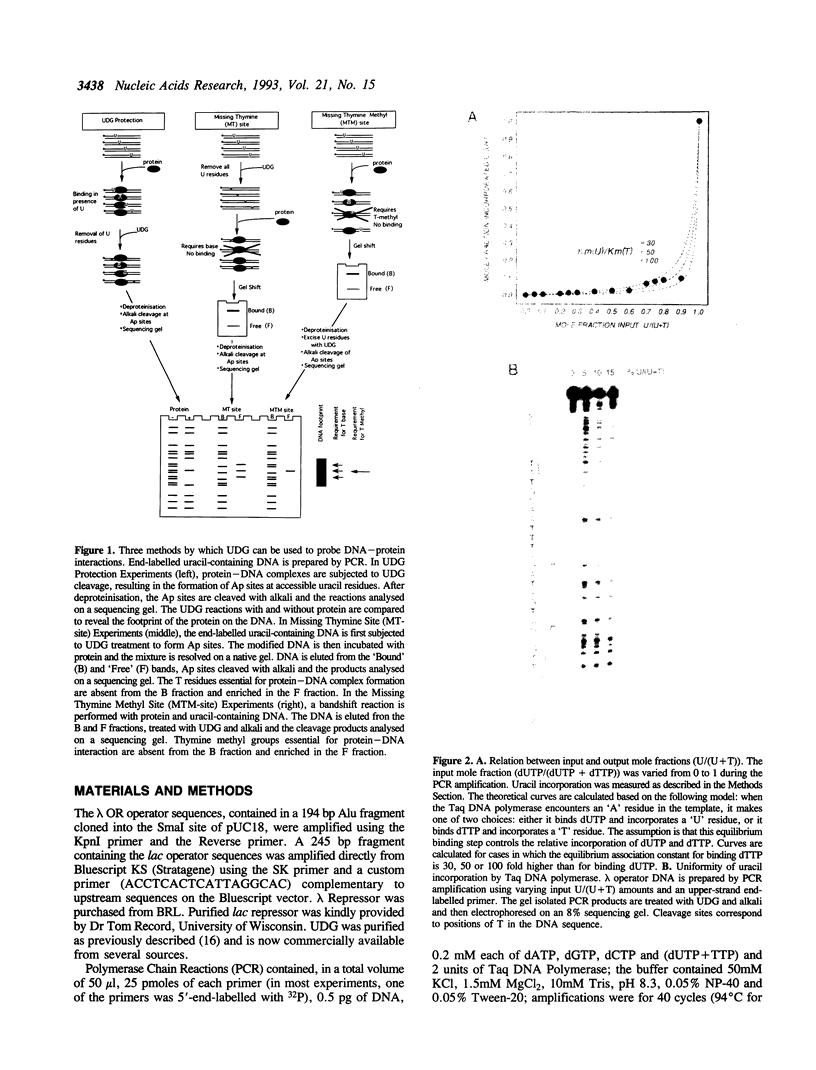
The DNA repair enzyme Uracil-DNA Glycosylase (UDG) can be used to investigate three different features of protein-DNA interactions. Complexes can be probed by simple protection experiments ('footprinting') or by two kinds of interference assays: a missing thymine site (MT-site) experiment and a missing thymine methyl site (MTM-site) experiment. The three probing methods are assessed using the well-characterized in vitro systems of lambda repressor and lac repressor binding to their respective operator sites. The results obtained with UDG probing agree well with previous probing experiments on the same systems and, in certain cases, extend previous interpretations: for example, comparison of the results obtained with the two interference assays shows that formation of the lac repressor-operator complex requires interactions with the methyl group of one particular thymine residue (T-13) in the operator but also requires interactions with other parts of the thymine base at operator positions 7, 8, 9, 21, 23 and 24. Overall, the properties of UDG recommend it as a versatile and convenient method to investigate DNA-protein interactions both in vitro and possibly in vivo.
Full text
PDF
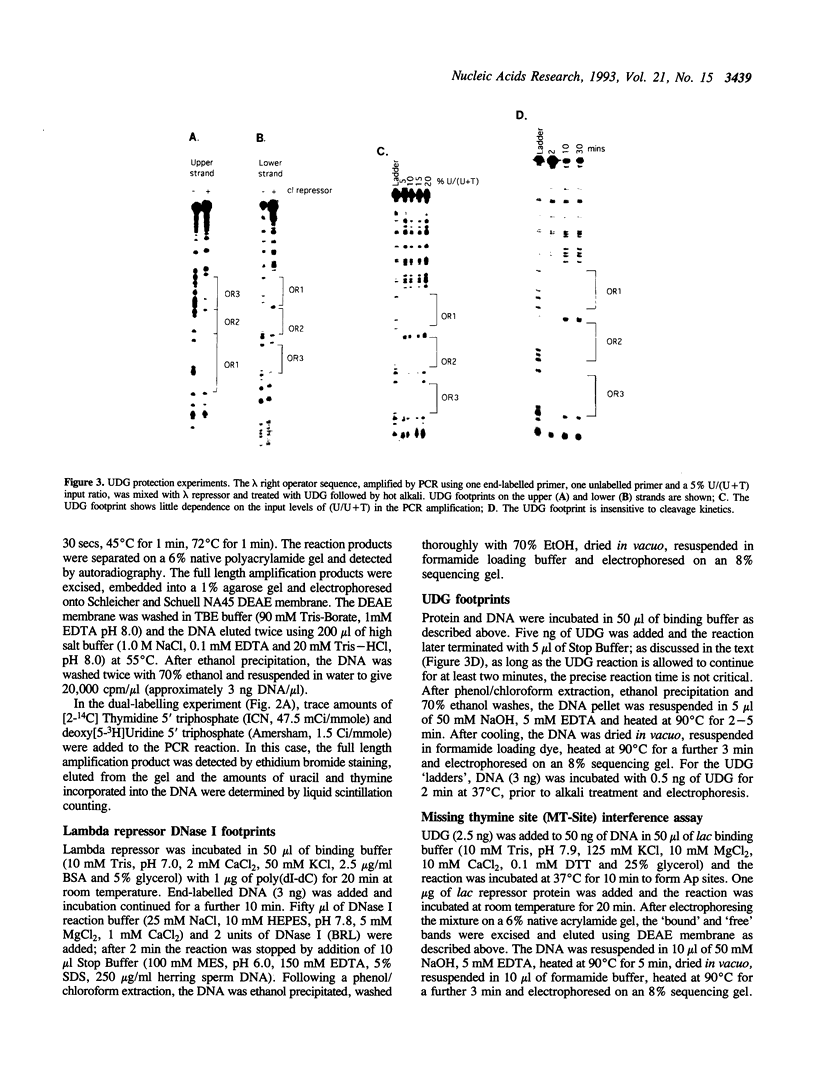

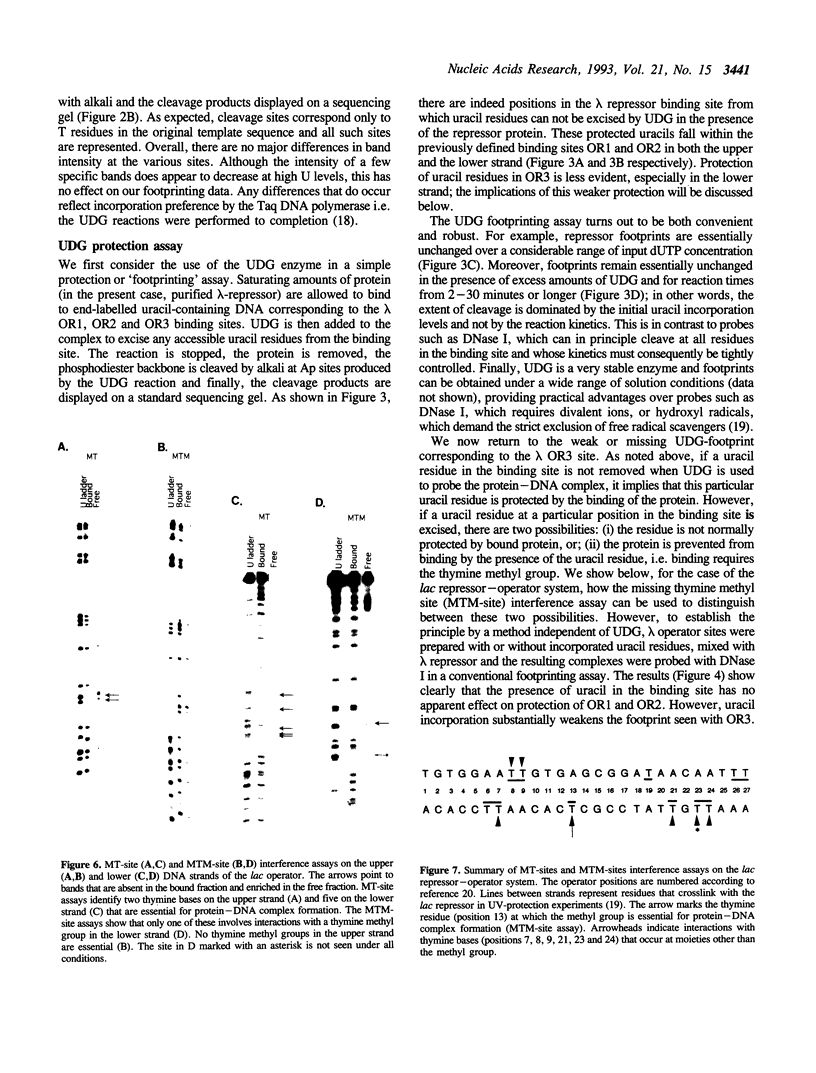


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson J. E., Ptashne M., Harrison S. C. Structure of the repressor-operator complex of bacteriophage 434. 1987 Apr 30-May 6Nature. 326(6116):846–852. doi: 10.1038/326846a0. [DOI] [PubMed] [Google Scholar]
- Beamer L. J., Pabo C. O. Refined 1.8 A crystal structure of the lambda repressor-operator complex. J Mol Biol. 1992 Sep 5;227(1):177–196. doi: 10.1016/0022-2836(92)90690-l. [DOI] [PubMed] [Google Scholar]
- Delort A. M., Duplaa A. M., Molko D., Teoule R., Leblanc J. P., Laval J. Excision of uracil residues in DNA: mechanism of action of Escherichia coli and Micrococcus luteus uracil-DNA glycosylases. Nucleic Acids Res. 1985 Jan 25;13(2):319–335. doi: 10.1093/nar/13.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeddel D. V., Yansura D. G., Winston C., Caruthers M. H. Studies on gene control regions. VII. Effect of 5-bromuracil-substituted lac operators on the lac operator-lac repressor interaction. J Mol Biol. 1978 Aug 25;123(4):661–687. doi: 10.1016/0022-2836(78)90211-5. [DOI] [PubMed] [Google Scholar]
- Gottesfeld J. M., Bloomer L. S. Nonrandom alignment of nucleosomes on 5S RNA genes of X. laevis. Cell. 1980 Oct;21(3):751–760. doi: 10.1016/0092-8674(80)90438-9. [DOI] [PubMed] [Google Scholar]
- Hillen W., Schollmeier K., Gatz C. Control of expression of the Tn10-encoded tetracycline resistance operon. II. Interaction of RNA polymerase and TET repressor with the tet operon regulatory region. J Mol Biol. 1984 Jan 15;172(2):185–201. doi: 10.1016/s0022-2836(84)80037-6. [DOI] [PubMed] [Google Scholar]
- Lamerichs R. M., Boelens R., van der Marel G. A., van Boom J. H., Kaptein R., Buck F., Fera B., Rüterjans H. H NMR study of a complex between the lac repressor headpiece and a 22 base pair symmetric lac operator. Biochemistry. 1989 Apr 4;28(7):2985–2991. doi: 10.1021/bi00433a037. [DOI] [PubMed] [Google Scholar]
- Lindahl T. An N-glycosidase from Escherichia coli that releases free uracil from DNA containing deaminated cytosine residues. Proc Natl Acad Sci U S A. 1974 Sep;71(9):3649–3653. doi: 10.1073/pnas.71.9.3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T., Andersson A. Rate of chain breakage at apurinic sites in double-stranded deoxyribonucleic acid. Biochemistry. 1972 Sep 12;11(19):3618–3623. doi: 10.1021/bi00769a019. [DOI] [PubMed] [Google Scholar]
- Lo Y. M., Mehal W. Z., Fleming K. A. Rapid production of vector-free biotinylated probes using the polymerase chain reaction. Nucleic Acids Res. 1988 Sep 12;16(17):8719–8719. doi: 10.1093/nar/16.17.8719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConlogue L., Brow M. A., Innis M. A. Structure-independent DNA amplification by PCR using 7-deaza-2'-deoxyguanosine. Nucleic Acids Res. 1988 Oct 25;16(20):9869–9869. doi: 10.1093/nar/16.20.9869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller P. R., Wold B. In vivo footprinting of a muscle specific enhancer by ligation mediated PCR. Science. 1989 Nov 10;246(4931):780–786. doi: 10.1126/science.2814500. [DOI] [PubMed] [Google Scholar]
- Ogata R., Gilbert W. Contacts between the lac repressor and the thymines in the lac operator. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4973–4976. doi: 10.1073/pnas.74.11.4973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu W. T., Struhl K. Uracil interference, a rapid and general method for defining protein-DNA interactions involving the 5-methyl group of thymines: the GCN4-DNA complex. Nucleic Acids Res. 1992 Feb 25;20(4):771–775. doi: 10.1093/nar/20.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasse-Dwight S., Gralla J. D. KMnO4 as a probe for lac promoter DNA melting and mechanism in vivo. J Biol Chem. 1989 May 15;264(14):8074–8081. [PubMed] [Google Scholar]
- Sasse-Dwight S., Gralla J. D. Probing the Escherichia coli glnALG upstream activation mechanism in vivo. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8934–8938. doi: 10.1073/pnas.85.23.8934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlomai J., Kornberg A. Deoxyuridine triphosphatase of Escherichia coli. Purification, properties, and use as a reagent to reduce uracil incorporation into DNA. J Biol Chem. 1978 May 10;253(9):3305–3312. [PubMed] [Google Scholar]
- Siebenlist U., Gilbert W. Contacts between Escherichia coli RNA polymerase and an early promoter of phage T7. Proc Natl Acad Sci U S A. 1980 Jan;77(1):122–126. doi: 10.1073/pnas.77.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J., Klar A. J. Active genes in budding yeast display enhanced in vivo accessibility to foreign DNA methylases: a novel in vivo probe for chromatin structure of yeast. Genes Dev. 1992 Feb;6(2):186–196. doi: 10.1101/gad.6.2.186. [DOI] [PubMed] [Google Scholar]
- Tullius T. D. DNA footprinting with hydroxyl radical. Nature. 1988 Apr 14;332(6165):663–664. doi: 10.1038/332663a0. [DOI] [PubMed] [Google Scholar]
- Tullius T. D., Dombroski B. A., Churchill M. E., Kam L. Hydroxyl radical footprinting: a high-resolution method for mapping protein-DNA contacts. Methods Enzymol. 1987;155:537–558. doi: 10.1016/0076-6879(87)55035-2. [DOI] [PubMed] [Google Scholar]
- Varshney U., Hutcheon T., van de Sande J. H. Sequence analysis, expression, and conservation of Escherichia coli uracil DNA glycosylase and its gene (ung). J Biol Chem. 1988 Jun 5;263(16):7776–7784. [PubMed] [Google Scholar]
- Warner H. R., Duncan B. K., Garrett C., Neuhard J. Synthesis and metabolism of uracil-containing deoxyribonucleic acid in Escherichia coli. J Bacteriol. 1981 Feb;145(2):687–695. doi: 10.1128/jb.145.2.687-695.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vlieg J., Berendsen H. J., van Gunsteren W. F. An NMR-based molecular dynamics simulation of the interaction of the lac repressor headpiece and its operator in aqueous solution. Proteins. 1989;6(2):104–127. doi: 10.1002/prot.340060203. [DOI] [PubMed] [Google Scholar]