Abstract
Protein carbonylation is the most commonly used measure of oxidative modification of proteins. It is frequently measured spectrophotometrically or immunochemically by derivatizing proteins with the classical carbonyl reagent, 2,4-dinitrophenylhydrazine. We developed an immunochemical dot blot method for quantitation of protein carbonylation in homogenates or purified proteins. Dimethyl sulfoxide was employed as the solvent because it very efficiently extracts proteins from tissues and keeps them soluble. It also readily dissolves 2,4-dinitrophenylhydrazine and wets PVDF membranes. The detection limit is 0.19 ± 0.04 pmol carbonyl. Sixty ng protein is sufficient to measure protein carbonyl content. This level of sensitivity allowed measurement of protein carbonylation in individual Drosophila.
Keywords: Protein carbonylation; 2,4-dinitrophenylhydrazine; Oxidative stress; Immunoblot; Dot blot
Oxidative modification of proteins, both reversible and irreversible, occurs during redox signaling and other cellular processes. It also occurs as a consequence of acute or chronic oxidative stress in many conditions. The list of diseases and processes in which oxidative damage is implicated reads like a textbook of pathology and includes atherosclerosis, cancer, neurodegenerative diseases such as Alzheimers and Parkinsons, and the aging process [1;2]. As a consequence of the oxidative stress, cells carry an increased burden of oxidatively damaged macromolecules, including nucleic acids, lipids, and proteins. A large number of the resulting modifications of proteins have been characterized and studied [3]. Protein carbonylation occurs in many of these modifications, providing an integrated assessment of oxidative damage. The National Library of Medicine’s Medical Subject Headings entry for protein carbonylation notes that, “It is a standard marker for oxidative stress.” [4]. In this context, the term protein carbonylation refers to “reactive” carbonyl groups, that is, to aldehydes and ketones.
Detection and quantitation of carbonylated proteins is accomplished after derivatization of the carbonyl groups. A variety of derivatizing reagents have been utilized for spectrophotometric, fluorimetric, and immunochemical analysis. The most commonly employed is the classical carbonyl reagent, 2,4-dinitrophenylhydrazine (DNPH)1, perhaps first applied to proteins by Dixon [5]. Spectrophotometry was initially used for quantitation of derivatized proteins. Immunochemical detection is now more commonly utilized, either as an ELISA or as a Western blot of SDS-PAGE separated proteins. When detected with anti-DNPH antibodies carrying fluorescent labels, the Western blot is quantitative and fairly sensitive. Still, in our hands, 10 to 20 μg protein per sample is needed to obtain duplicate analyses. Our goal was to develop a more sensitive dot blot method in which many samples of ~ 1 μl volume could be applied to one PVDF membrane, allowing convenient quantitation of multiple samples with replicates.
DNPH has limited solubility in water and many common solvents. It has limited, but sufficient, solubility in strong acids and that is the reason stock solutions are usually made in 2M HCl. Few proteins are soluble in HCl, which will not wet PVDF membranes. These problems were obviated by employing dimethyl sulfoxide (DMSO) as solvent. It readily dissolves DNPH, wets PVDF, and is an excellent protein solvent[6]. The resulting method for protein carbonylation requires ~60 ng protein for one analysis and as many samples as desired can be measured on a single PVDF membrane.
Materials and methods
Oxidatively modified proteins and tissue homogenates
Recombinant E. coli glutamine synthetase is produced in our laboratory for a variety of investigations, and we have used it for years to produce standards of varying carbonyl content [7]. After determination of their carbonyl content by the reference spectrophotometric method [8], the standards were aliquoted, dried in a vacuum centrifuge, and stored at −80°. Control glutamine synthetase had a carbonyl content of 0.11±0.01 mol carbonyl/mol protein, and the oxidized standard had a content of 0.68±0.01 mol carbonyl/mol protein. These values, as are those shown in the figures are the mean and standard deviation. Mixtures of control and oxidized glutamine synthetase provided standards of intermediate carbonyl content.
Glutamine synthetase need not be used as the standard; almost any protein which can be oxidatively modified should suffice, and one can use metal-catalyzed oxidation [9], ionizing radiation (see results), hypochlorous acid [10], or ozone [11] to introduce carbonyl groups. Note that almost all proteins have some carbonyl groups when isolated, often at least 0.05 mol carbonyl/mol protein. These carbonyl groups can be reduced to alcohols by borohydride if one wishes to have a standard with even lower protein content[12]. Incubate the protein solution at 37° for 1 h with 20 mM NaBH4 (Sigma 480886) in 100 mM Tris, pH 8.5, 2 mM EDTA, followed by dialysis into 50 mM Hepes, 100 mM KCl, and 1 mM MnCl2. It is not essential to use this dialysis buffer; we use it because it is the preferred buffer for glutamine synthetase. Fatty acid free bovine serum albumin (BSA; Boehringer Mannheim 100069) was used as supplied or when noted, treated with borohydride to remove carbonyl groups.
Deinococcus radiodurans extracts were a gift from Michael Daly (Uniformed Services University of the Health Sciences). Cells were subjected to 10 or 20 kGray γ irradiation to carbonylate proteins [13] after which extracts were prepared. Cells were suspended in 50 mM sodium phosphate buffer, pH 7.4, 1 mM DTPA, broken in an Emulsiflex homogenizer, and centrifuged at 31,000 g for 30 min at 4° to remove debris. Protein concentration was determined by Bradford’s method with BSA as standard [14]. After dialysis to remove low molecular weight compounds, the ratio of absorbances at 280 nm to 260 nm was 0.6, indicative of substantial nucleic acid content as expected for a bacterial homogenate. Aliquots were stored at −80°.
A liver extract from 8–10 week old Swiss Webster mice (PelFreeze) was prepared on ice with a Potter-Elvehjem homogenizer and centrifuged for 5 min at 16,000 g at 4°. Aliquots of the supernatant were subjected to 5, 10, 15, and 20 kGray γ irradiation and then stored at −20°. We used samples containing 15 μg protein for SDS gel electrophoresis followed by Western blot determination of protein carbonylation. Samples with 9 μg protein were dried and utilized in the dot blot assays.
Extracts of individual Drosophila were prepared from flies stored at −80°, a gift from Stephen Helfand (Brown University). The fly was placed in a conical, 0.5 ml polypropylene tube with 30 μl homogenizing solution (92.5% DMSO/7.5% H2O (v/v) and acidified with 0.5% trifluoroacetic acid) and homogenized on ice for 30 sec using a plastic pestle (Kimble Chase KT749521-0500) and a motorized handheld homogenizer. The extracts were centrifuged for 5 min at 13,000 g, and the supernatants removed for analysis.
Dot blot assay for protein carbonylation
A stock solution of 500 mM DNPH (Aldrich D19930-3) was prepared in DMSO (Sigma-Aldrich 154938), taking into account the water content of the solid DNPH which is typically ~30%. The derivatizing solution was prepared from the stock DNPH, DMSO, and trifluoroacetic acid (Pierce 28901) to give final concentrations of 20 mM DNPH and 0.5% trifluoroacetic acid in 92.5% DMSO. Although we routinely prepared the derivatizing solution fresh daily, it is stable for several days. Samples, with or without added BSA as carrier (see results), were dried in plastic conical tubes (Eppendorf 022364111) in a vacuum centrifuge. Derivatizing solution was added to give a protein concentration of 300 ng/μl, and the tubes were agitated at room temperature for 15 min. The derivatized samples are stable in solution for at least 1 h so that the samples can be derivatized as a batch and then applied to the membrane.
We find it convenient to use Fast Green FCF as a fluorescent protein stain[7] which can be quantitated in our Odyssey infrared scanner (LI-COR, Lincoln, NE), and thus we prepare a separate blot for this purpose. However, one might be able to use visible range fluorescence to determine protein and our usual infrared tagged secondary antibody for carbonyl determination on a single blot. Although the method described below requires only about 350 ng protein per replicate, use of a more sensitive protein assay would reduce this even lower since 80% of the sample is used in the Fast Green FCF protein assay.
Dry Immobilon-FL PVDF membranes (Millipore, IPFL00010) were placed on two 96-well polypropylene plates (Thermo 267245), one for protein determination and one for carbonyl determination. Each sample was spotted in triplicate on each membrane. To provide a guide for spotting samples, a flat 96-well plastic grid was placed on top of the membrane and taped to the plate. We used the grid from a pipet tip rack (Rainin), but any similar template should work. Samples were vortexed vigorously before withdrawing a 1 μl aliquot with a 2 μl pipette (Rainin P-2) and applying it to the first membrane for protein determination. BSA standards ranging from 0 to 600 ng/μl were also applied. Samples were then diluted five-fold with the DNPH derivatizing solution to give a protein concentration of about 60 ng/μl. These and the carbonyl standards ranging from 0.1 to 0.8 pmol carbonyl/μl were applied to the second membrane. After air drying for at least 15 min, blots were rinsed with acetic acid to remove excess DNPH. After placing the membrane in an acid resistant polystyrene (BD Biosciences 35112) or polypropylene dish, the membrane was washed twice in acetic acid for 2 min with agitation. After removal of the second wash, another 5 ml acetic acid was added to keep the membrane wet, and water was gradually added to cover the membrane. This solution was removed and replaced with water. After 5 min, the membrane was ready for protein or carbonyl determination. The membrane can be dried and stored in the dark at room temperature or at −80° for 2 days if desired. (We tested these storage conditions to determine whether membranes prepared in one laboratory could be shipped to another for scanning.)
For carbonyl quantitation, membranes were blocked for 1 h in Odyssey blocking buffer (LI-COR 927-40010), then incubated for 2 h with a 1:10,000 dilution of goat anti-DNPH primary antibody (Bethyl A150-117A) in blocking buffer with 0.1% Tween 20. They were then washed 3 times with 0.1% Tween-PBS. The secondary antibody, donkey anti-goat IRDye 800CW (LI-COR 926-32214) was also diluted 1:10,000 with blocking buffer with 0.1% Tween 20 and incubated for 1 h in the dark. Washing was repeated as for the primary antibody after which the blots were equilibrated with water and scanned in the 800-nm channel. For quantitation, a circle large enough to encompass the largest spot on the blot was drawn with the software. The circle included the dense central portion of each spot and the more diffuse border. Automatic background subtraction with a 2 mm border width was utilized. Carbonyl content was calculated from the standards included on every blot.
Protein determination by Fast Green FCF [7] was modified slightly for rapid staining of the PVDF membrane. Wet membranes were stained for 45 sec with 0.01% Fast Green FCF (Sigma F7252) in 30% methanol with 7% acetic acid. They were destained in water, scanned in the 700 nm channel of the Odyssey scanner, and again integrated with automatic background subtraction with a 2 mm border width. Protein content was calculated from the BSA standards on the blot.
Western blot assay for protein carbonylation
Samples were dissolved with a final concentration of 6% SDS and derivatized by addition of 20 mM DNPH in 10% trifluoroacetic acid as described [15]. Samples were neutralized and made 2% in β-mercaptoethanol. Gel electrophoresis was performed on duplicate gels, either 14% or 8–16% acrylamide (Invitrogen EC64855 or EC60455). Separations were performed in 25 mM Tris, 192 mM glycine, pH 8.3 buffer with 0.1% SDS at 180 V. One gel was stained for protein quantitation by Coomassie Blue fluorescence [7] and the other was transferred onto an Immobilon-FL PVDF membrane for carbonyl determination as in the dot blot assay. Transfers were performed overnight at 4° at 15 V in 12.5 mM Tris, 96 mM glycine, 20% methanol, 0.005% SDS, pH 8.0. After transfer, the gel was stained with Coomassie Blue to determine efficiency of transfer. Visualized bands were integrated by the Odyssey scanner software using its “right-left” background subtraction.
Results and discussion
Comparison of the Western blot and dot blot methods
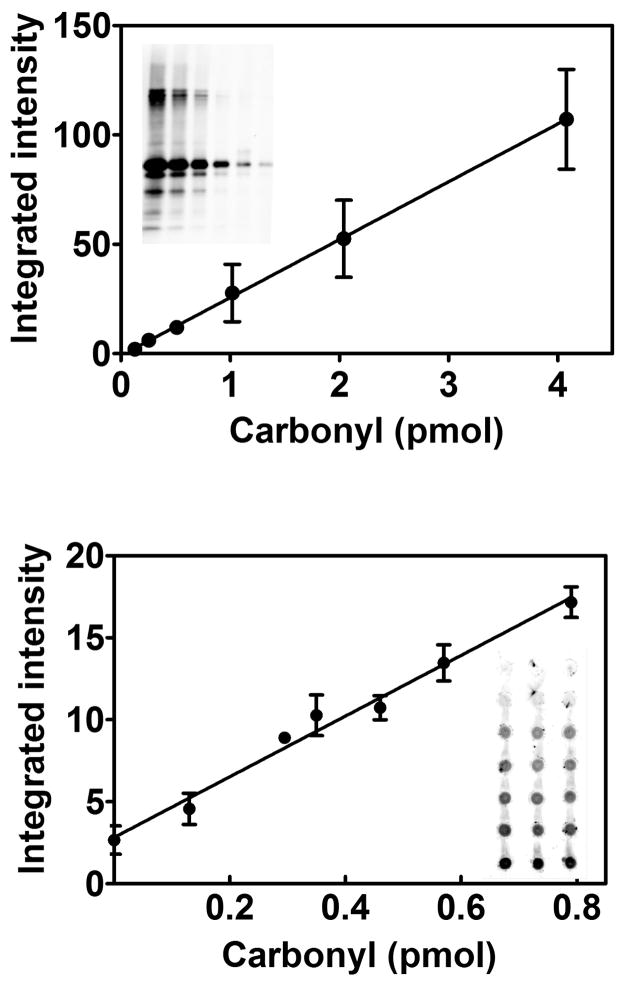
We compared the results from the Western blot and the dot blot methods with samples of glutamine synthetase of varying carbonyl content. The resulting responses were linear over the ranges tested, using a lower range for the dot blot because of its greater sensitivity (Fig. 1). These calibration curves were constructed by applying 60 ng protein, with the carbonyl content varied by mixing control and oxidized glutamine synthetase standards. To investigate whether total protein per dot affected the results, we constructed a calibration standard with only oxidized glutamine synthetase standard, applying from 10 to 200 ng protein (0 to 2.6 pmol carbonyl). We constructed a second calibration standard with the same amount of glutamine synthetase, but with addition of 100 ng BSA. The two calibration standards were identical, indicating that a variation in total protein from 10 to 300 ng has no effect. As indicated in the methods section, we treated a portion of the BSA with borohydride to reduce any carbonyl groups present in the protein. The addition experiment was performed with both the untreated and the reduced BSA with no difference in the results. The reason was that the carbonyl content of this preparation of BSA was extremely low, and thus borohydride treatment was not required. If BSA is added as a carrier protein, it would be prudent to measure its carbonyl content to determine whether borohydride treatment is needed.
Fig. 1.
Comparison of the calibration curves for the Western blot method (upper) and the dot blot method (lower). Lanes of the 14% SDS-PAGE reducing gel were loaded with varying amounts DNPH derivatized oxidized glutamine synthetase standard. From left to right, the load was 4.1, 2.0, 1.0, 0.50, 0.25, and 0.13 pmol carbonyl. The graph shows the averaged results from 2 separate blots. The regression line was fit to all points, giving y=26.6x −1.4, r2>.99. The dot blot samples were spotted in triplicate, with the top row containing no protein. All other rows were loaded with 60 ng glutamine synthetase. The carbonyl content from top to bottom was 0.13, 0.30, 0.35, 0.46, 0.57, and 0.79 pmol. The equation of the fit line is y = 16.3x + 7.7, r2>.99 with 3 measurements for each point.
The accuracy and precision of the method were assessed by analysis of a glutamine synthetase sample containing 0.46 pmol carbonyl, calibrated by the spectrophotometric method. Within day accuracy and precision was determined on 6 separate membranes. The carbonyl content was 0.50 ± 0.05 pmol. Between day accuracy and precision was determined on 6 different blots over 3 ½ months. It was 0.49 ± 0.04 pmol. A signal twice that of background was taken to be the sensitivity limit of the method and was 0.19 ± 0.04 pmol carbonyl.
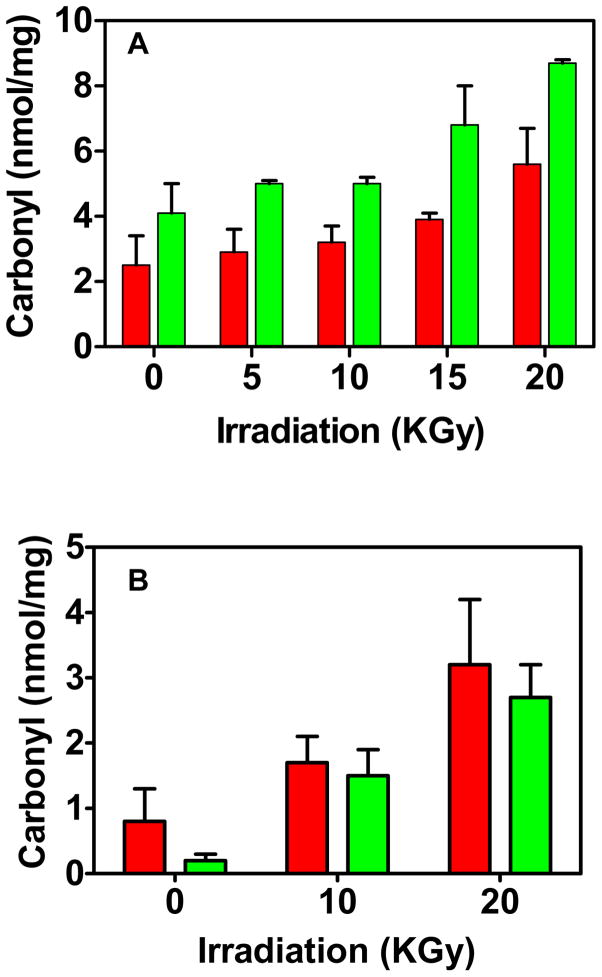
We then compared results for a protein mixture, namely liver extracts subjected to irradiation to oxidize proteins. The increase in carbonylated protein content induced by irradiation is evident for both the Western and dot blot methods. The absolute values are ~2 nmol/mg protein higher for the dot blot (Fig. 2).
Fig. 2.
Comparison of the Western blot (red) and dot blot (green) methods applied to extracts exposed to varying γ irradiation. (A) Mouse liver homogenate. Four blots were averaged for the Western blot and two for the dot blot, each with triplicate analyses (B). Deinococcus radiodurans homogenate. Four blots were averaged for the Western blot, each with duplicate samples. Two blots were also averaged for the dot blot, each with sextuplicate analyses.
Nucleic acids have carbonyl groups and can artifactually elevate the protein carbonyl measurement in the spectrophotometric assay but do not interfere in the Western blot assay [8;16]. DNA has been reported to artifactually elevate protein carbonyl in a slot blot assay in which DNPH derivatization is performed on the membrane after applying the sample [17]. We therefore tested the effect of DNA added to the glutamine synthetase standard. We tested both sheared herring sperm DNA (Phoenix Biotechnologies 2006) and mouse genomic DNA, a gift from Geumsoo Kim (National Heart, Lung, and Blood Institute). After derivatization with DNPH, the usual 60 ng protein was spotted onto the PVDF membrane with DNA from 0 to 2.0 ng. Even at the highest DNA level, no elevation of signal occurred. In our experience, artifactual elevation of protein carbonylation by nucleic acids has not been observed in assays of animal tissues, but it is a serious problem with bacterial extracts [8]. To provide additional experimental investigation of the issue and also to provide another comparison of the Western and dot blot methods, we analyzed extracts of Deinococcus radiodurans subjected to irradiation to increase carbonylation. D. radiodurans is a large Gram positive bacterium with relatively high DNA content. After disrupting the cells we did not perform the usual streptomycin sulfate precipitation of nucleic acids. As a consequence, even after dialysis to remove low molecular weight compounds, the A280/260 ratio of the extract was 0.6 due to substantial DNA contamination. From this measurement we calculated that 20 ng DNA would be applied with each 60 ng protein blot[18]. Nevertheless, the Western blot and dot blot carbonyl contents were very similar.
From these experiments we conclude that DNA does not interfere in the dot blot assay. While the Western blot method is widely used, there is no basis for considering it to be the reference method, particularly given the variability introduced by differing transfer efficiencies in moving proteins from the gel to the membrane. The 2 nmol/mg higher carbonyl content detected by the dot blot assay of liver homogenate suggests the presence of a carbonyl pool which is retained in the dot blot but not in the Western blot.
Homogenization and analysis of single Drosophila
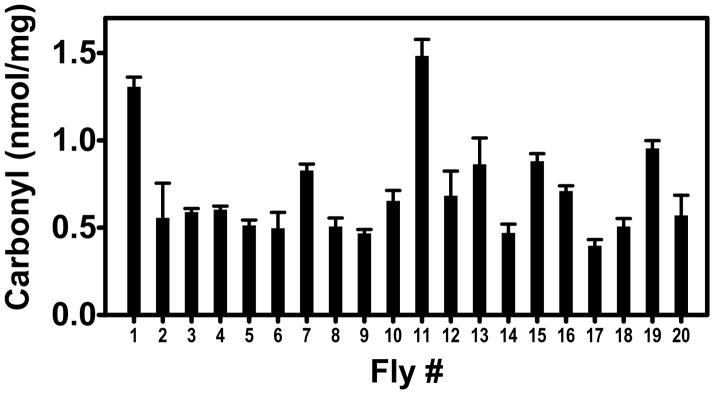
In the course of this work we observed that DMSO acidified with 0.5% trifluoroacetic acid allowed facile homogenization of liver tissue with recovery of protein at least as great as with an aqueous buffer at neutral pH. If this were also true of model organisms such as Drosophila, then the dot blot assay might allow determination of protein carbonyl levels on individual flies. This capability could be useful in studying the well-known phenotypic variations observed in colonies of genetically identical animals, for example, the rate of aging[20]. We extracted 20 individual Drosophila in 30 μl 92.5% DMSO/0.5% trifluoroacetic acid, the same solvent used for the DNPH derivatizing solution. These were taken through the dot blot procedure with determination of both protein and carbonyl content. For comparison, 30 flies were homogenized in 30 μl 50 mM sodium phosphate, pH 7.4, 1 mM DTPA. The protein extracted per fly was 30.3 ± 9.3 μg with the phosphate buffer and 63.6 ± 7.7 μg with the DMSO solution, an impressive improvement in efficiency. Carbonyl measurements were readily obtained on the individual flies and revealed the expected heterogeneity in these genetically identical organisms (Fig. 3). The assay is sufficiently sensitive that one could assay the head and thorax separately if desired.
Fig. 3.
Carbonyl content of individual Drosophila melanogaster. The average and standard deviation for triplicate analyses are shown.
Given the ease of homogenizing tissues in DMSO and the convenience of applying the extracts directly onto dry PVDF membranes, the DMSO homogenization should be useful for other membrane based chemical and immunochemical assays. We conclude by noting that the DMSO carbonyl dot blot assay decreases the sample requirement by at least an order of magnitude from that needed for a Western blot assay.
Acknowledgments
This research was supported by the Intramural Research Program of the National Heart, Lung, and Blood Institute. We thank Barbara Berlett and Elena Gaidamakova for performing the γ irradiations.
Footnotes
Abbreviations used: DNPH, 2,4-dinitrophenylhydrazine; BSA, bovine serum albumin; DMSO, dimethyl sulfoxide; DPTA, diethylenetriamine pentaacetic acid; PVDF, polyvinylidene fluoride.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Levine RL, Stadtman ER. Carbonylated proteins and their implication in physiology and pathology. In: Dalle-Donne I, Scaloni A, Butterfield DA, editors. Redox Proteomics: from Protein Modifications to Cellular Dysfunction and Diseases. Wiley Interscience; New York: 2006. pp. 123–168. [Google Scholar]
- 2.Giustarini D, Dalle-Donne I, Tsikas D, Rossi R. Oxidative stress and human diseases: Origin, link, measurement, mechanisms, and biomarkers. Crit Rev Clin Lab Sci. 2009;46:241–281. doi: 10.3109/10408360903142326. [DOI] [PubMed] [Google Scholar]
- 3.Stadtman ER, Levine RL. Chemical modification of proteins by reactive oxygen species. In: Dalle-Donne I, Scaloni A, Butterfield DA, editors. Redox Proteomics: from Protein Modifications to Cellular Dysfunction and Diseases. Wiley Interscience; New York: 2006. pp. 3–23. [Google Scholar]
- 4.Medical Subject Headings. National Library of Medicine; [Accessed: 12/11/11]. http://www.nlm.nih.gov/cgi/mesh/2011/MB_cgi. [Google Scholar]
- 5.Dixon HB. Transamination of peptides. Biochem J. 1964;92:661–666. doi: 10.1042/bj0920661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singer SJ. The Properties of Proteins in Nonaqueous Solvents. In: Anfinsen CB, editor. Advances in Protein Chemistry. Academic Press; New York: 1963. pp. 1–68. [Google Scholar]
- 7.Luo S, Wehr NB, Levine RL. Quantitation of protein on gels and blots by infrared fluorescence of Coomassie blue and Fast Green. Anal Biochem. 2006;350:233–238. doi: 10.1016/j.ab.2005.10.048. [DOI] [PubMed] [Google Scholar]
- 8.Levine RL, Garland D, Oliver CN, Amici A, Climent I, Lenz AG, Ahn BW, Shaltiel S, Stadtman ER. Determination of carbonyl content in oxidatively modified proteins. Meth Enzymol. 1990;186:464–478. doi: 10.1016/0076-6879(90)86141-h. [DOI] [PubMed] [Google Scholar]
- 9.Rivett AJ, Levine RL. Metal-catalyzed oxidation of Escherichia coli glutamine synthetase: Correlation of structural and functional changes. Arch Biochem Biophys. 1990;278:26–34. doi: 10.1016/0003-9861(90)90226-o. [DOI] [PubMed] [Google Scholar]
- 10.Reddy VY, Desrochers PE, Pizzo SV, Gonias SL, Sahakian JA, Levine RL, Weiss SJ. Oxidative dissociation of human α2 macroglobulin tetramers into dysfunctional dimers. J Biol Chem. 1994;269:4683–4691. [PubMed] [Google Scholar]
- 11.Berlett BS, Levine RL, Stadtman ER. Comparison of the effects of ozone on the modification of amino acid residues in glutamine synthetase and bovine serum albumin. J Biol Chem. 1996;271:4177–4182. doi: 10.1074/jbc.271.8.4177. [DOI] [PubMed] [Google Scholar]
- 12.Climent I, Tsai L, Levine RL. Derivatization of gamma-glutamyl semialdehyde residues in oxidized proteins by fluoresceinamine. Anal Biochem. 1989;182:226–232. doi: 10.1016/0003-2697(89)90584-8. [DOI] [PubMed] [Google Scholar]
- 13.Daly MJ, Gaidamakova EK, Matrosova VY, Kiang JG, Fukumoto R, Lee DY, Wehr NB, Viteri GA, Berlett BS, Levine RL. Small-molecule antioxidant proteome-shields in Deinococcus radiodurans. PLoS One. 2010;5:e12570. doi: 10.1371/journal.pone.0012570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 15.Levine RL, Williams JA, Stadtman ER, Shacter E. Carbonyl assays for determination of oxidatively modified proteins. Meth Enzymol. 1994;233:346–357. doi: 10.1016/s0076-6879(94)33040-9. [DOI] [PubMed] [Google Scholar]
- 16.Luo S, Wehr NB. Protein carbonylation: avoiding pitfalls in the 2,4-dinitrophenylhydrazine assay. Redox Rep. 2009;14:159–166. doi: 10.1179/135100009X392601. [DOI] [PubMed] [Google Scholar]
- 17.Robinson CE, Keshavarzian A, Pasco DS, Frommel TO, Winship DH, Holmes EW. Determination of protein carbonyl groups by immunoblotting. Anal Biochem. 1999;266:48–57. doi: 10.1006/abio.1998.2932. [DOI] [PubMed] [Google Scholar]
- 18.Warburg O, Christian W. Isolierung und kristallisation des garungsferments enolase. Biochemisches Zeitschrift. 1942;310:384–421. [Google Scholar]
- 19.Dirmeier R, O’Brien KM, Engle M, Dodd A, Spears E, Poyton RO. Exposure of Yeast Cells to Anoxia Induces Transient Oxidative Stress. J Biol Chem. 2002;277:34773–34784. doi: 10.1074/jbc.M203902200. [DOI] [PubMed] [Google Scholar]
- 20.Kirkwood TB, Feder M, Finch CE, Franceschi C, Globerson A, Klingenberg CP, LaMarco K, Omholt S, Westendorp RG. What accounts for the wide variation in life span of genetically identical organisms reared in a constant environment? Mech Ageing Dev. 2005;126:439–443. doi: 10.1016/j.mad.2004.09.008. [DOI] [PubMed] [Google Scholar]