Abstract
Malignant gliomas are the most common and the most lethal primary brain tumors in adults. Among malignant gliomas, 60%–80% show loss of P14ARF tumor suppressor activity due to somatic alterations of the INK4A/ARF genetic locus. The tumor suppressor activity of P14ARF is in part a result of its ability to prevent the degradation of P53 by binding to and sequestering HDM2. However, the subsequent finding of P14ARF loss in conjunction with TP53 gene loss in some tumors suggests the protein may have other P53-independent tumor suppressor functions. Here, we report what we believe to be a novel tumor suppressor function for P14ARF as an inhibitor of tumor-induced angiogenesis. We found that P14ARF mediates antiangiogenic effects by upregulating expression of tissue inhibitor of metalloproteinase–3 (TIMP3) in a P53-independent fashion. Mechanistically, this regulation occurred at the gene transcription level and was controlled by HDM2-SP1 interplay, where P14ARF relieved a dominant negative interaction of HDM2 with SP1. P14ARF-induced expression of TIMP3 inhibited endothelial cell migration and vessel formation in response to angiogenic stimuli produced by cancer cells. The discovery of this angiogenesis regulatory pathway may provide new insights into P53-independent P14ARF tumor-suppressive mechanisms that have implications for the development of novel therapies directed at tumors and other diseases characterized by vascular pathology.
Introduction
The CDKN2A (INK4A/ARF) genetic locus on chromosome 9p21 is among the most widely inactivated in human cancer (1) and encodes two tumor suppressors: the p16INK4A cell cycle suppressor and P14ARF (2), a regulator of P53 stability (3). Somatic alterations at this locus occur frequently in solid tumors, and germline deletion of the ARF gene predisposes to the melanoma-astrocytoma syndrome (4). The importance of P14ARF and its mouse homolog p19Arf in tumor suppression has been confirmed by numerous experimental studies (2, 3, 5–8), and the specific knockout of the p19Arf gene results in an increased frequency of diverse tumor types in mouse (5, 7, 9). It is known that p19Arf binds to and inactivates Mdm2, a negative regulator of the p53 tumor suppressor (3). P14ARF-induced stabilization of the P53 transcription factor leads to the expression of critical P53 target genes, which can mediate cell cycle arrest or induce apoptosis (6, 7, 10). Therefore, it is widely assumed that P14ARF can suppress tumor growth through P53, and that P14ARF loss or HDM2 amplification are alternative ways to inactivate the same tumor suppressor pathway (11).
Nevertheless, there are several lines of evidence suggesting that p19Arf has additional p53-independent tumor suppressor activities (12). Differences are observed in the types and frequencies of tumors that arise in p19Arf–/– versus Trp53–/– mice, including a predisposition to gliomas in the former (9).
P14ARF loss is particularly relevant to the high-grade progression of malignant astrocytomas, which are the most common and the most lethal intracranial tumors (13, 14). Initial studies had suggested that alterations in P53 or P14ARF might be mutually exclusive in human gliomas, although exceptions existed with co-alterations of both genes in some tumors (13, 15). Recent studies performed by The Cancer Genome Atlas (TCGA) Research Network have shown that glioblastoma multiforme (GBM) can be separated into four distinct subgroups with distinct genetic alterations and expression profiles. Interestingly, in the three subtypes that express mutant P53, a significant proportion of the tumors that carry TP53 mutations also harbor CDKN2A homozygous deletions (16), suggesting that selective pressure exists in some glioblastomas for the loss of both genes, raising the possibility that P14ARF might have tumor-inhibitory functions beyond P53 activation. The loss of P14ARF expression occurs with the transition to grade IV in proneural astrocytoma progression, which synchronizes with the onset of the robust angiogenesis that characterizes GBMs (for review, see refs. 14, 17). This observation led us to hypothesize a potential link between pathological vascularization and P14ARF activity. In the present study, we examined whether the disruption of P14ARF gene expression is one of the genetic events that trigger pathological angiogenesis in GBMs. We found that P14ARF upregulates the expression of the tissue inhibitor of metalloproteinase–3 (TIMP3) through a novel P53-independent HDM2/SP1 signaling pathway, which results in the negative regulation of angiogenesis.
Results
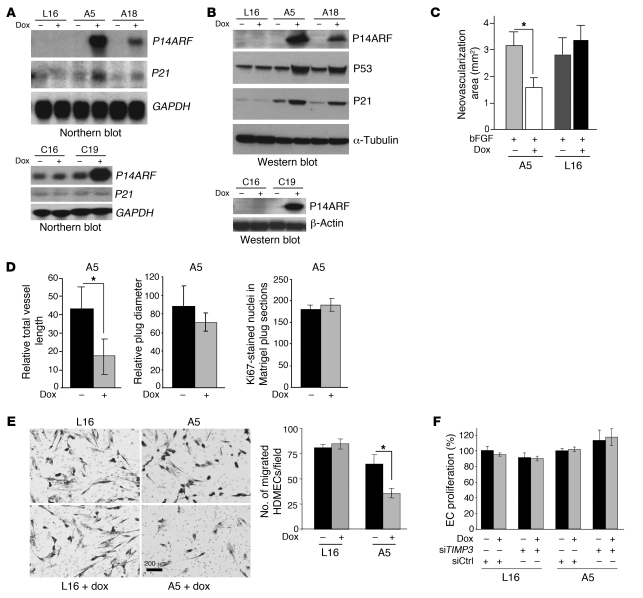
To investigate the role of P14ARF in the control of tumor-induced angiogenesis, we conditionally expressed P14ARF in malignant human glioma cells. We generated Tet-on P14ARF clones A5 and A18 from the rtTA-expressing cell line LN229-L16 (L16; WT for P53; ARF null). The Tet-on P14ARF clone C19 was similarly generated from the rtTA-expressing cell line LNZ308-C16 (C16; null for P53; ARF WT). L16 and C16 parental cells were used as controls for the nonspecific effects of doxycycline (dox) (Figure 1A). Northern and Western blot analyses confirmed that P14ARF induction was tightly regulated by dox, with concomitant stabilization of P53 and downstream induction of P21 expression in WT P53 but not in TP53-null cells, as expected (Figure 1B). To evaluate the effects of P14ARF restoration on glioma cells’ ability to induce angiogenesis in vivo, we performed corneal (Figure 1C) and Matrigel plug angiogenesis (Figure 1D) assays in mice, as previously described (18, 19). Corneas were implanted with micropellets containing human bFGF, a strong inducer of neovascularization, and serum-free conditioned medium (CM) of L16 and A5 tumor cells grown in the presence or absence of induction by dox for 48 hours. The CM of A5 cells expressing P14ARF (induced by dox) significantly inhibited corneal neovascularization when compared with CM of uninduced cells (Figure 1C). The mean area of neovascularization in corneas with pellets containing CM of P14ARF-induced cells (ARF-CM) was significantly reduced (2-fold) compared with pellets containing CM from uninduced cells (Ctrl-CM). As demonstrated with CM of parental LN229-L16 cells, the low doses of dox used in our experiments had no effect on corneal vascularization per se, excluding nonspecific effects of dox. For the Matrigel plug assay (18), A5 cells were mixed with Matrigel and implanted subcutaneously in nu/nu mice and the mice fed dox in the drinking water. The length of newly formed vessels was significantly reduced (~2.5-fold) in A5-Matrigel plugs from mice treated with dox as compared with controls (Figure 1D). This comparison was performed on Matrigel plugs of similar size, and there were no differences in rates of tumor cell proliferation in the A5 and A5 plus dox plugs, as determined by the number of nuclei positive for Ki67 factor (Figure 1D). Taken together, these in vivo studies demonstrate that P14ARF controls neoangiogenesis induced by tumor cells independently of its effects on cell proliferation.
Figure 1. P14ARF expression in tumor cells inhibits angiogenesis in vivo and EC migration in vitro.
(A) Northern blots showing dox-inducible expression of P14ARF mRNA in LN229-L16 (L16) derived clones A5 and A18 (upper panel) and LNZ308-C16 derived clone C19 (lower panel) glioma cells. (B) Western blots showing induction of P14ARF in cells from A. P14ARF stabilizes P53 and induces p21 expression in A5 and A18 cells (WT for P53) but not in TP53-null C19 cells, as expected (data not shown). (C) Mouse cornea angiogenesis assays showing that CM from P14ARF-expressing A5 cells inhibits vessel formation. The neovascularized areas (mm2) in the corneas were quantified (error bars indicate SEM; *P < 0.05, ANOVA). The experiments were repeated twice independently (n = 2) with 6 mice/group. (D) Matrigel plug angiogenesis assay showing the antiangiogenic activity of P14ARF-expressing cells. The total length of vessels per surface area (left panel), the relative plug diameter (middle panel), and the number of dividing cells (right panel) were measured and compared in groups with or without dox. Unpaired 2-tailed Student’s t test, *P < 0.05; 4 mice/group, n = 2). (E) CM from P14ARF-induced cells inhibits EC migration in a modified Boyden chamber assay. The number of HDMECs migrated to the lower side of the filter were counted in 3 randomly-selected fields and the results expressed as mean ± SD per field (right panel). *P < 0.05, unpaired 2-tailed Student’s t test; n = 3. Scale bar: 200 μm. (F) CM of P14ARF-expressing cells does not alter HDMEC proliferation expressed as percentage of untreated cells (mean ± SD; n = 4).
To establish which aspect of the angiogenic process is regulated by P14ARF restoration, we tested CM from cells with and without P14ARF induction for its effect on EC migration and proliferation. CM from P14ARF-expressing cells was able to significantly reduce (2-fold) EC migration through collagen matrix in a modified Boyden chamber assay as compared with uninduced cells (Figure 1E). CM of dox-treated parental LN229-L16 cells elicited the same cell migration as that of untreated cells, excluding nonspecific effects of dox. In contrast, the basal proliferation of ECs was not affected by CM of cells with or without dox treatment (Figure 1F).
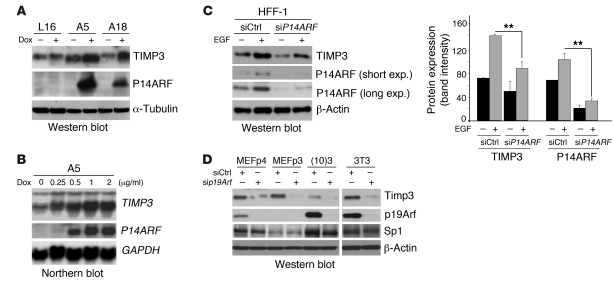
To gain insight into how P14ARF regulates vessel formation in vivo and EC migration in vitro, we used microarray analyses to examine the expression patterns of angiogenesis-related genes following P14ARF expression in A5 cells (Supplemental Table 1; supplemental material available online with this article; doi: 10.1172/JCI38596DS1). Gene expression analysis showed that TIMP3 mRNA was significantly upregulated up to 3-fold by P14ARF, while other angiogenic regulators were either barely detected or not significantly changed by P14ARF. We confirmed that the mRNA and protein expression levels of TIMP3 were increased in P14ARF-induced cells A5 and A18 by Northern and Western blotting (Figure 2A and Supplemental Figure 1), and this effect was dox dose–dependent (Figure 2B). The mRNA levels of other factors known to modulate angiogenesis and EC migration, such as VEGF, TIMP3 substrates (MMP-2 and MMP-9), and thrombospondin (TSP1) remained unchanged (Supplemental Figure 2A and Supplemental Figure 3C). Quantification of VEGF in the CM of A5, A18, and C19 glioma cells by ELISA (Supplemental Figure 2B) indicated that P14ARF’s effect on glioma-mediated angiogenesis is not dependent on VEGF production.
Figure 2. P14ARF induces TIMP3 expression.
(A) Western blot showing induction of TIMP3 protein expression by P14ARF in A5 and A18 cells. Dox induction was as in Figure 1. α-Tubulin was used as a loading control. (B) Northern blot showing that P14ARF induces TIMP3 mRNA expression in a dose-dependent manner (0.25 to 2 μg/ml dox induction for 48 hours). GAPDH mRNA was used as a loading control. (C) Left panel: Western blot showing the effect of endogenous P14ARF gene activation by EGF on the expression of TIMP3. HFF-1 cells were cultured in serum-free medium for 12 hours and then treated with EGF (100 ng/ml) for 24 hours. Short and long exposures (exp.) are shown for P14ARF. Right panel: Densitometry analysis of P14ARF and TIMP3 expression was performed with ImageJ software ( http://rsbweb.nih.gov/ij/) (right panel). Combined data of 2 independent experiments (n = 2) are presented with SD. **P < 0.01, unpaired 2-tailed Student’s t test. (D) Western blot showing silencing of p19Arf reduces TIMP3 expression in mouse fibroblasts. MEFs (passages 3 and 4 [p3 and p4]), (10)3 , and 3T3 cells were transfected in serum-free conditions with either 40 nM siCtrl or sip19Arf with Lipofectamine RNAiMax for 48 hours. Serum (3%) was added to medium following 20 hours of transfection. β-Actin was used as loading control.
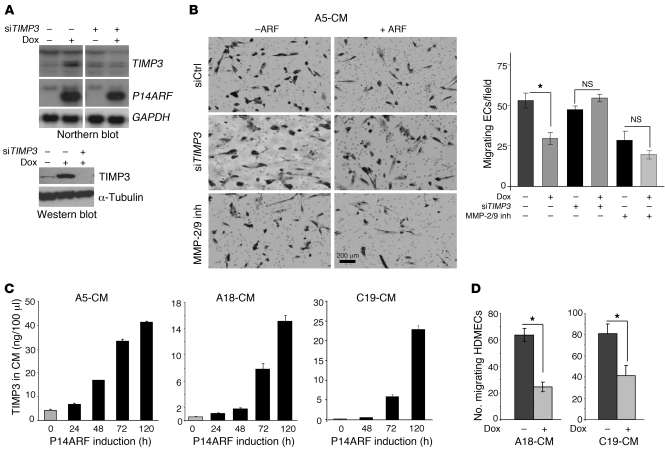
To determine whether TIMP3 induction would also occur under physiological conditions of endogenous P14ARF modulation, we treated an immortalized human foreskin fibroblast cell line (HFF-1) with EGF. Oncogenic stimulation by EGF signaling is known to trigger a negative feedback loop augmenting P14ARF expression (20). EGF-mediated increases in P14ARF expression led to enhanced TIMP3 expression, which was partially decreased by silencing of P14ARF expression (Figure 2C and Supplemental Figure 4). Similarly, the silencing of Arf in different mouse embryonic fibroblasts (MEFs) resulted in a decrease in Timp3 expression (Figure 2D). To determine whether TIMP3 was the downstream effector of the P14ARF-mediated inhibition of EC migration, we knocked down TIMP3 using specific siRNA (Figure 3A). Quantification of migrated ECs in response to CM from TIMP3-silenced cells showed that the inhibitory effect of P14ARF was largely prevented (Figure 3B). Since TIMP3 is an inhibitor of MMP-2 and -9, which are critical MMPs for EC invasion (21), we examined whether their pharmacological inhibition would mimic the inhibitory effects of ARF-CM. Indeed, inhibiting MMP-2/9 did significantly reduce EC migration. The combination of MMP-2/9 inhibition with the inhibitory effect of ARF-CM was less than additive, suggesting that their mechanisms of action on EC migration might be redundant. The inhibitory effect of ARF-CM was specific to EC migration, as their proliferation was unaffected (Figure 1F). The ARF-CM of A18 and C19 cells were also able to inhibit EC migration, but because these cells secrete less TIMP3 than do A5 cells (Figure 3C), 5 days of culture were needed to accumulate enough TIMP3 in the CM. The levels of TIMP3 had to reach approximately 15–20 ng per 100 μl CM to inhibit EC migration (Figure 3D). Combined, these results indicate that the inhibitory effects of P14ARF on EC migration are mediated in a large part by the secreted TIMP3.
Figure 3. Induction of TIMP3 expression by P14ARF inhibits EC migration.
(A) Northern blot (upper panel) and Western blot (lower panel) demonstrating the silencing of TIMP3 with siRNA in A5 glioma cells. (B) Modified Boyden chamber migration assay showing that TIMP3 is required for the inhibition of EC migration by the CM of P14ARF-expressing cells (2 μg/ml dox for 48 hours). Prior to CM collection, the A5 cells were transfected with negative control or TIMP3 siRNAs. HDMECs were seeded in the upper chamber with medium containing 1× CM (diluted from 30× concentrated), and the migration across gelatin type B–coated filters was performed. The migrated cells were photographed (left panel) and counted (right panel). The MMP-2/9 inhibitor I (inh) was used at 650 nM. Scale bar: 200 μm. (C) ELISA quantification of TIMP3 production in serum-free culture medium of A5, A18, and C19 cells with or without P14ARF induction by dox for up to 5 days. Data are expressed in ng/100 μl of CM and as mean ± SD. Each experiment was performed in duplicate. (D) Modified Boyden chamber migration assay showing the effect of 5-day CM of A18 and C19 cells with or without dox on EC (HDMEC) migration. The assay was performed as described in B. The unpaired 2-tailed Student’s t test was used for B and D to assess the statistical differences between experimental groups, *P <0.05; n = 3.
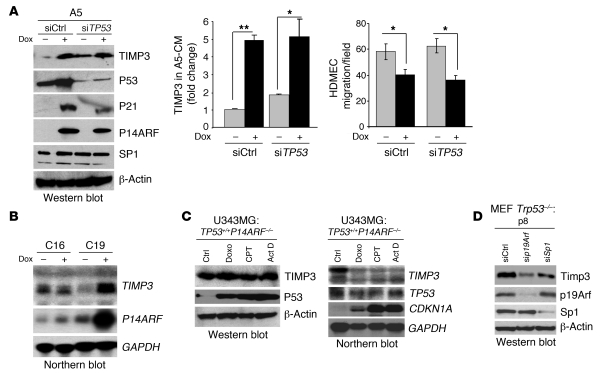
Given the known role of P14ARF protein in stabilizing P53 (3, 6) and the relevance of the latter in the control of angiogenesis in gliomas (22), we examined whether P14ARF regulation of TIMP3 expression was dependent upon P53. First, the silencing of TP53 expression in A5 cells caused a significant reduction in levels of P53 and its downstream target CDKN1A/P21, yet P14ARF’s ability to induce TIMP3 mRNA (Supplemental Figure 3A) and protein expression levels (Figure 4A and Supplemental Figure 3B) remained unchanged. Furthermore, transfection of A5 cells with siTP53 did not reduce the inhibitory effect of ARF-CM on EC migration (Figure 4A, right panel. In fact, TP53 silencing even slightly increased the basal TIMP3 mRNA and protein expression levels (Figure 4A and Supplemental Figure 3A). Second, P14ARF was able to induce TIMP3 mRNA in TP53-null C19 cells (Figure 4B), while it was unaffected in parental LNZ308-C16 cells, which were used as controls for nonspecific effects of dox. Third, the physiological activation of endogenous WT P53 by genotoxic agents in INK4A/ARF-null U343MG human glioma cells increased P53 protein levels, as well as the mRNA levels of its target gene CDKN1A/p21, yet failed to affect TIMP3 mRNA and protein levels (Figure 4C). Fourth, the silencing of endogenous p19Arf with siRNA in Trp53-null MEFs led to a decrease in Timp3 levels (Figure 4D). Taken together, these results confirm that the regulation of TIMP3 by P14ARF is not dependent on P53 stabilization by P14ARF and suggest the existence of another cellular pathway coupling P14ARF (p19Arf) to TIMP3.
Figure 4. The upregulation of TIMP3 gene expression by P14ARF is independent of P53.
(A) Effect of TP53 silencing on the induction of TIMP3 by P14ARF and on EC migration. Left panel: Western blot showing that the silencing of TP53 expression by siRNA in A5 cells does not affect the induction of TIMP3 expression by P14ARF. Note that the silencing of TP53 has no effect on SP1 protein levels. Middle panel: ELISA quantification of TIMP3 levels in CM of A5 cells previously transfected with either negative control siRNA (siCtrl) or TP53 siRNAs (siTP53) and cultured in medium with or without dox without serum for 48 hours. The assay was carried out in triplicate, and results are expressed as mean ± SD. Right panel: Boyden chamber HDMEC migration assay in the presence of CM from control and TP53 siRNA–transfected A5 cells with or without dox. Results are expressed as mean (±SD) cell number per field. *P < 0.05, **P < 0.01, unpaired 2-tailed Student’s t test. (B) Northern blot showing that P14ARF strongly activates TIMP3 mRNA expression in TP53-null C19 cells. (C) Western blot (left) and Northern blot (right) showing that activation of endogenous WT P53 with genotoxic agents does not induce TIMP3 protein or mRNA expression. U343MG cells were treated for 24 hours with doxorubicin (Doxo; 1 μg/ml), camptothecin (CPT; 5 μM), or actinomycin D (Act D; 10 nM) to activate P53. Note that P53 activation by genotoxic agents strongly increases its stability, yet TIMP3 protein levels remain unchanged. (D) Western blot analysis showing that silencing p19Arf or Sp1 in Trp53-null MEFs (passage 8) decreases Timp3 expression.
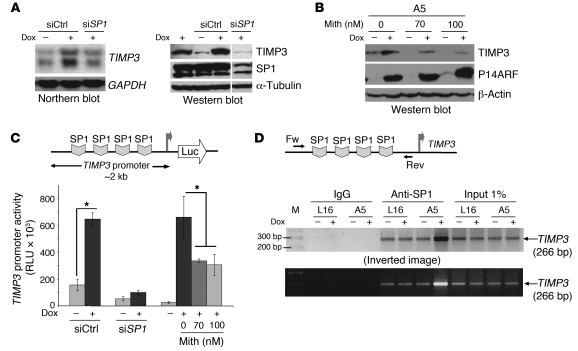
Since the mechanism underlying P14ARF control over TIMP3 expression is unknown, we hypothesized that P14ARF may activate the transcription of the TIMP3 gene through the activation of a specific transcription factor. The human TIMP3 upstream sequence (–390 to +1) contains a number of binding motifs for known transcription factors, including SP1 and potential NF1 and c/EBP sites (23). Because SP1 has 4 binding sites located in the region (–112 and +1) proximal to the transcription start site, we examined whether SP1 is the downstream mediator of P14ARF’s effects on TIMP3. The siRNA-mediated silencing of Sp1 expression in Trp53-null MEFs caused a decrease in Timp3 protein levels (Figure 4D) and prevented the induction of TIMP3 mRNA and protein levels by P14ARF in A5 glioma cells (Figure 5A). The pharmacological inhibition of SP1-DNA binding by mithramycin (24) led to a dose-response inhibition of TIMP3 induction by P14ARF at doses of 70 nM or greater (Figure 5B). Furthermore, P14ARF increased the activity of the human TIMP3 promoter by 3-fold in luciferase reporter assays (Figure 5C), an effect that was prevented by the silencing of SP1, suggesting that TIMP3 promoter activation by P14ARF is SP1 dependent. Since P14ARF activates TIMP3 transcription without affecting SP1 protein levels (Figure 5A, right panel), we investigated whether P14ARF increases the binding activity of SP1 to the TIMP3 promoter. ChIP assays demonstrated that the induction of P14ARF increased the binding of SP1 on the TIMP3 promoter; IgG served as negative control (Figure 5D). The activation of SP1 was P14ARF specific, since dox did not modify the promoter-binding activity of SP1 in P14ARF-deficient L16 parental cells. As a whole, these results demonstrate that SP1 plays a critical role in P14ARF-mediated activation of the transcription of the TIMP3 gene.
Figure 5. The upregulation of TIMP3 gene expression by P14ARF is SP1 dependent.
(A) The silencing of SP1 (siSP1) abrogates upregulation of TIMP3 mRNA (Northern) and protein (Western) expression by P14ARF in A5 cells. The noncontiguous lane of the composite blot separated by a thin white line was run on the same gel and exposed similarly. (B) Western blot showing the pharmacological inhibition of SP1 activity with mithramycin (Mith; 70 and 100 nM, 36 hours) prevents TIMP3 induction by P14ARF. (C) Luciferase reporter assay showing SP1 is required for TIMP3 promoter activation by P14ARF. A5 cells were transfected with a TIMP3 promoter–luciferase reporter and 24 hours later treated with siRNAs (siCtrl or siSP1). Twenty-four hours later, P14ARF expression was induced with dox with or without mithramycin (70 and 100 nM) for 36 hours. Cell lysates were then analyzed for luciferase activity. The values of firefly luciferase normalized to protein content are presented as the mean ± SD of triplicates; *P < 0.05, paired Student’s t test; n = 2. (D) ChIP assay demonstrating P14ARF-dependent binding of SP1 to the TIMP3 promoter. Chromatin fragments prepared from A5 cells with or without P14ARF expression were immunoprecipitated with rabbit anti–human SP1 (PEP2) or rabbit IgGs. Fw and Rev primers framing the SP1 binding sites in the TIMP3 promoter were used for PCR. The binding of rabbit IgG to the TIMP3 promoter served as negative control. Note: Dox treatment did not alter the basal level of SP1 binding in parental LN229-L16 cells, excluding nonspecific effects of dox. Three independent repeats (n = 3) gave similar results.
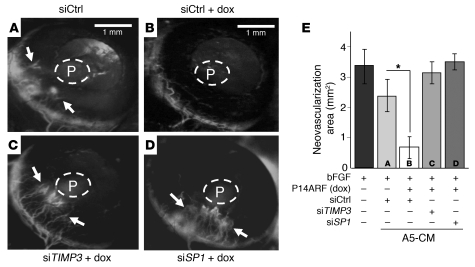
To determine whether SP1 and TIMP3 are also important for the negative regulation of angiogenesis by P14ARF in vivo, we performed corneal angiogenesis assays with micropellets containing human bFGF and CM of tumor cells transfected with different siRNAs (siCtrl, siTIMP3, and siSP1) and grown in the presence or absence of P14ARF induction. The CM of cells expressing P14ARF (ARF-CM) and transfected with negative control siRNA (siCtrl) exhibited a strong inhibitory effect on corneal neovascularization as compared with CM of uninduced cells (Figure 6, A and B). The mean area of neovascularization in corneas with pellets containing ARF-CM was significantly reduced, approximately 2-fold, compared with Ctrl-CM pellets (Figure 6E). The effect of ARF-CM on corneal neovascularization was strongly reversed by silencing of TIMP3 or SP1 expression in A5 cells (Figure 6, C and D, versus Figure 6B). These findings confirm that the antiangiogenic effect of P14ARF in vivo requires SP1 activation and TIMP3 production.
Figure 6. P14ARF regulates angiogenesis in a TIMP3-dependent manner.
Mouse cornea angiogenesis assay showing that P14ARF inhibits angiogenesis in a TIMP3- and SP1-dependent manner. Pellets containing 25 ng bFGF alone or mixed with concentrated CM of A5 cells with or without P14ARF induction for 48 hours were implanted in the cornea (see Methods). CM from cells pretreated with siRNAs against SP1, TIMP3, or control (siSP1, siTIMP3, or siCtrl, respectively) were also tested. Representative photographs (a–d) of mouse cornea at 5 days after implantation show FITC-dextran–labeled capillaries (arrows) progressing toward the insertion site of the pellet in the cornea (P). Note that in A, vessels are blurred by extensive vascular leakiness of the FITC-dextran associated with the strong angiogenic response. (E) The neovascularized areas (mm2) in the corneas were quantified as described in Methods (error bars indicate SEM). Note: siRNA against TIMP3 or SP1 prevented P14ARF-mediated inhibition of corneal angiogenesis. *P < 0.05, ANOVA. The experiments were repeated twice independently with 6 mice/group. Scale bars: 1 mm.
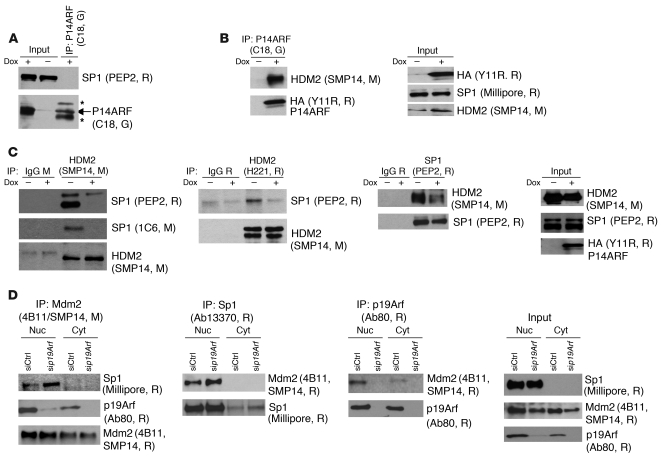
We next explored how P14ARF might activate SP1 to upregulate the TIMP3 gene. Our initial coimmunoprecipitation (co-IP) studies did not suggest any physical interaction between P14ARF and SP1 (Figure 7A), while confirming the known P14ARF-HDM2 interaction (Figure 7B). Since we showed that P53 is dispensable for TIMP3 regulation by P14ARF (Figure 4 and Supplemental Figure 3, A and B) and that P14ARF does not affect SP1 protein levels (Figure 4A and Figure 5A, right panel), we excluded also that SP1 activation may result from its known interaction with P53 (25). We then reasoned that the mechanism by which P14ARF could activate SP1 might be antagonization of HDM2 and prevention of its physical interaction with SP1. To test this hypothesis, we carried out co-IP experiments in A5 cells with HDM2 antibodies (SMP14 and H221) and SP1 antibody (PEP2) and found that HDM2-SP1 binding is reduced upon P14ARF activation (Figure 7C). To further validate this mechanism in the physiological setting, we performed co-IP studies on nuclear and cytosolic extracts of mouse 3T3 fibroblasts and found that the Mdm2-Sp1 interaction was increased by the knockdown of endogenous p19Arf (Figure 7D, first and second panels). As a control, we also verified that p19Arf interacts with Mdm2 (Figure 7D, third panel).
Figure 7. P14ARF regulates SP1/HDM2 interaction.
Protein extracts from dox-treated A5 cells were immunoprecipitated with the indicated antibodies, followed by Western blotting. R, G, and M indicate rabbit, goat, and mouse, respectively, which were the source for the indicated antibodies. Asterisks indicate detection of IgGs by secondary antibody. (A) Co-IP showing the absence of P14ARF-SP1 interaction. (B) Co-IP verifying that P14ARF interacts with HDM2 in A5 glioma cells. P14ARF was immunoprecipitated with an anti-P14ARF antibody (C18). Input levels (right panels) for P14ARF were detected with an anti-HA tag (Y11R, R) antibody. (C) Co-IP assays demonstrating that P14ARF reduces HDM2-SP1 interaction. In the two left panels, immunoprecipitations with anti-HDM2 antibodies show that HDM2 interacts with SP1 and this interaction decreases when P14ARF is induced. This regulation is confirmed by reverse immunoprecipitation of HDM2-SP1 complexes with anti-SP1 antibody in the third panel. Western blotting of HDM2 and SP1 in the cell extracts (input) shows their expression is not affected by P14ARF induction (far right panel). Species-matched IgGs were used as controls. (D) Co-IP experiments showing that p19Arf silencing increases the endogenous Sp1-Mdm2 interaction. After siRNA silencing of p19Arf for 48 hours in mouse 3T3 fibroblasts, nuclear (Nuc) and cytosolic (Cyt) extracts were used for co-IP. First and second panels: Co-IPs using anti-Mdm2 and anti-Sp1 antibodies demonstrate that Mdm2 binding to Sp1 is increased in the nuclear fraction upon p19Arf silencing. Third panel: The co-IP with anti-p19Arf antibody confirms p19Arf interaction with Mdm2 in the nucleus. Fourth panel: Input levels of Sp1, Mdm2, and p19Arf in nuclear and cytoplasmic extracts.
Because P14ARF and SP1 bind to the same central region of HDM2 (26), these data are consistent with the interpretation that the P14ARF-HDM2 binding relieves SP1 from an inhibitory interaction with HDM2 so it can transactivate TIMP3. These findings are consistent with our observation by ChIP assay of enhanced SP1 availability and binding to the TIMP3 promoter following P14ARF induction.
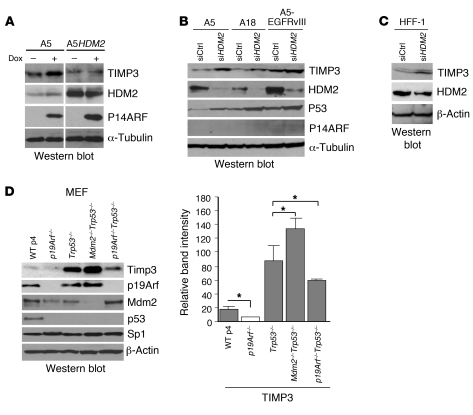
To confirm that TIMP3 expression is indeed regulated by HDM2, we examined the effect of HDM2 overexpression and silencing. Stable overexpression of HDM2 in A5 cells led to the abrogation of the increase in TIMP3 expression induced by P14ARF (Figure 8A, lane 4 vs. lane 2). The silencing of HDM2 in 3 different P14ARF-inducible glioma cell lines — including one expressing the ligand-independent EGFRvIII, which characterizes the most aggressive form of gliomas — induced the expression of TIMP3 and stabilized P53, as expected (Figure 8B). Similar results were obtained in normal HFF-1 cells (Figure 8C). Furthermore, genetic ablation of p19Arf reduced Timp3 levels in MEFs, irrespective of Trp53 status (Figure 8D and Supplemental Figure 5). Conversely, double knockout of Mdm2 and Trp53 increased the levels of Timp3 expression in comparison to Trp53-null and WT MEFs (Figure 8D, lane 4 versus lanes 3 and 1). These findings demonstrate that p19Arf loss decreases, while Mdm2 loss increases Timp3 levels independently of Trp53 status.
Figure 8. Role of HDM2 in the regulation of TIMP3.
(A) Western blot showing that overexpression of HDM2 abrogates the increase in TIMP3 induced by P14ARF. A5HDM2 is a clone of A5 cells stably transfected with an HDM2 expression vector (pCMV-MDM2-Neo). The noncontiguous lanes of the composite blot separated by a white line were run on the same gel and exposed similarly. (B) Western blot showing that HDM2 silencing increases TIMP3 expression in the absence of P14ARF induction (no dox). P14ARF-null human glioblastoma cell lines A5, A18, and A5-EGFRvIII were treated with specific siRNA for HDM2 (30 nM) for 48 hours without P14ARF induction. The induction of P53 protein levels was used as an independent measure of the silencing and the functional inactivation of HDM2. (C) Western blot showing that silencing of HDM2 increases physiological levels of TIMP3 in HFF-1 cells. (D) Left panel: Western blot showing that knockout of p19Arf or Mdm2 affects the basal levels of Timp3 in MEFs. Note that Mdm2–/– MEFs are not available due to the embryonic lethality of Mdm2–/– mice. WT MEFs were passage 4 cultures. p19Arf-, Trp53-, double Mdm2/p53-, and double p19Arf/Trp53-null MEFs were passage 8 cultures. The experiment was repeated 2 times independently with 2 different batches of MEFs. Right panel: Quantification of Western blot band intensity of Timp3 from 2 different batches of MEFs from D. Results are expressed as mean ± SD. *P < 0.05, 2-tailed Student’s t test.
In summary, using silencing, overexpression, and knockout strategies, we demonstrate that SP1 and HDM2 (Mdm2) are critical in the mechanism of P14ARF’s control over TIMP3 expression.
Discussion
Loss of P14ARF gene expression, as occurs in many malignancies, can abrogate tumor surveillance mechanisms and increase cancer susceptibility. Mice lacking p19Arf are highly prone to tumor development (5, 9), and in humans, P14ARF deficiency leads to the astrocytoma-melanoma syndrome (4). Conversely, increased p19Arf gene dosage confers cancer resistance to transgenic mice (27). p19Arf has previously been shown to function as a sensor of growth factor–mediated oncogenic signals and to exert its tumor suppressor function by stabilizing P53 and, thereby, negatively regulating cell proliferation or inducing apoptosis (28). Nevertheless, the finding of co-alterations in TP53 and P14ARF in many tumors suggests that P14ARF might have additional P53-independent tumor-suppressive functions.
We now report that P14ARF regulates another hallmark of solid cancer growth, the process of tumor-induced neovascularization, inhibition of which can prevent tumor growth by limiting the necessary supply of nutrients and oxygen to cancer cells (29). We have discovered a HDM2/SP1/TIMP3 signaling network controlled by P14ARF that leads to the inhibition of angiogenesis. We showed that P14ARF exerts this activity through the inhibition of EC migration, an essential component of new vessel formation. The motile process of ECs is directionally regulated by chemotactic stimuli released by the tumor (30) and involves the degradation of the extracellular matrix by specific proteases (31). We found that P14ARF counterbalances the effects of proangiogenic signals released by the tumor cells by stimulating the expression of TIMP3, a secreted factor that can inhibit EC migration in vitro and vessel formation in vivo. TIMP3 is a member of a family of secreted proteins that inhibit the activity of MMPs, which are important vascular remodeling factors. TIMP3 is a physiological antagonist for MMP-2 and -9, which are critical for the degradation of the extracellular matrix associated with the progression of migrating ECs (32). Additional antiangiogenic mechanisms of action of TIMP3 include the inhibition of VEGFR2 signaling through the trapping of the proangiogenic factor VEGF and blockage of VE-cadherin expression on ECs (33, 34). That TIMP3’s angio-suppressive functions are a barrier to tumor development is underscored by the finding that TIMP3 is epigenetically silenced in 40%–80% cases of glioblastoma (35) and in primary cancers of the kidney, colon, breast, and lung (36). Furthermore, restoration of TIMP3 expression blocks tumor growth (37).
The regulation of TIMP3 by P14ARF is new, and the underlying mechanisms are unknown. Given that P14ARF counteracts excessive mitogenic stimuli by raising the levels of P53, we considered involvement of P53 transcriptional activity (6). We found that P14ARF upregulated the expression of TIMP3 at the transcriptional level, but in a P53-independent fashion, consistent with the absence of P53 binding sites in the human TIMP3 gene. Other P53-independent functions for P14ARF have been reported. P14ARF can trigger growth arrest or apoptosis of TP53-deficient human tumor cell lines in culture and inhibit their growth as xenografts in nu/nu mice (38–40). p19Arf can also block the expression of Pdgfβ by pericytes independent of Mdm2 and p53 and thereby control the extent of perivascular cell proliferation in the hyaloid vascular system of the developing mouse eye (41). Recent reports have also suggested a role for P14ARF in the modulation of VEGF and the activity of its transactivator HIF (42–45). We did not observe an effect of P14ARF on VEGF levels in human glioma cells, which may reflect differences in cell types and experimental conditions.
TIMP3 transcription can be triggered by the activation of growth factor pathways. Indeed, our data have shown that EGF stimulates TIMP3 expression and this was inhibited by P14ARF silencing. To further examine whether additional oncogenic signals such as E1A or Ras would do the same, we infected Trp53–/– and Trp53–/–p19Arf–/– MEFs with retroviral expression vectors and found that Timp3 was reduced regardless of p19Arf expression (data not shown); this suggests that some oncogenic pathways (E1A, Ras) directly downregulate Timp3 expression, while others (EGF) activate Timp3 through p19Arf upregulation, likely providing selective pressure to abolish p19Arf or Timp3 function.
The human TIMP3 promoter contains 4 potential binding sites for transcription factor SP1 close to the transcription initiation site, and we demonstrated that SP1 was the key modulator of TIMP3 regulation by P14ARF. SP1 has also been implicated in the activation of the TIMP1 (46), TIMP2 (47), and Timp4 (48) genes, raising the possibility that P14ARF and SP1 might exert coordinated antiangiogenic effects through other members of the TIMP family. This was not the case, as our data show that TIMP1 and TIMP2 levels remained unchanged by P14ARF. The expression of TIMPs hinders tumor progression, as suggested by studies in astrocytoma and melanoma (33, 37, 49). These findings contrast with the more established role for SP1 as a factor that upregulates growth-related genes and, thereby, promotes many aspects of cancer biology, including cell proliferation, survival, invasion, and angiogenesis (50). The stimulation of SP1 constitutive activity can be achieved by alterations in tumor suppressor genes. Tumor suppressors such as P53 (51), p73 (52), von Hippel–Landau (53), and retinoblastoma protein (Rb) (54) can physically interact with SP1, form a complex, and block binding of SP1 to growth-promoting genes. It is currently unknown what precisely determines the activation state of SP1-regulated genes and whether specific SP1 regulators determine coordinated activation of pro- or anti-tumorigenic target genes. The transcriptional activity of SP1 is regulated by many cofactors (50, 55), and the response of SP1 target genes is under epigenetic control (56). Tumor suppressors such as P14ARF can exert multi-modal actions, and while stimulating SP1 activation on TIMPs, may still retain control over cell cycle progression through the HDM2/P53/P21 cell cycle control axis. Clearly, these multiple biological effects of SP1 are to be integrated in an overall response, which will be cell type specific and context dependent.
The mechanism underlying the activation of SP1 by P14ARF revealed in our study remains unknown. We did not observe a difference in the levels of SP1 protein upon P14ARF induction. We show that P14ARF relieves transcription factor SP1 from negative regulation by HDM2, which allows its binding to the promoter of the TIMP3 gene. The specific mechanism underlying the suppressive effect of HDM2 on SP1 remains to be further defined. The function of transcription factors can be modified through binding of cofactors and regulatory proteins, which may activate, neutralize, or even turn them into repressors (50). Because HDM2 binds to SP1 in the region that contains the zinc finger domain (54), one potential mechanism by which HDM2 could inhibit SP1 activity is through the inhibition of DNA binding (57), as supported by our ChIP data. Because P14ARF and SP1 bind to HDM2 in the same central region of HDM2 (54, 58), it is plausible that P14ARF reverses HDM2 inhibition of SP1 by displacing the latter from HDM2, as seen in our co-IP experiments. HDM2 binding to P14ARF (59, 60) will free SP1 and activate it. In this model, the DNA-binding activity of SP1 is regulated by the competing activities of HDM2 and P14ARF. Consistent with this notion, our data support that the balance of P14ARF and HDM2 will maintain basal P14ARF-induced TIMP3 levels. Our study is the first to our knowledge to suggest that HDM2 has oncogenic activity by blocking antiangiogenic signals, independent of its P53-inactivating function. This may explain why in certain tumors, HDM2 amplification may be selected in lieu of TP53 mutations. Our data and previous findings suggesting that Mdm2 might also activate proangiogenic responses (61) support the targeting of HDM2 for controlling tumor angiogenesis, regardless of the P53 status of the tumors.
In summary, we report tumor-suppressive activity for P14ARF as an inhibitor of tumor-induced angiogenesis. This activity is achieved through a hitherto unknown P14ARF/HDM2/SP1/TIMP3 signaling axis that regulates TIMP3 expression through a balance between P14ARF and HDM2 levels, independent of P53. P14ARF enhances the transcriptional activity of SP1 on TIMP3 by antagonizing the inhibitory interaction of HDM2 with SP1. Thus, anticancer strategies designed to restore WT P53 function instead of P14ARF may be insufficient to counter tumors in which P14ARF exerts additional P53-independent tumor suppressor activities. Our data suggest that therapies directed toward restoring P14ARF activity, preventing HDM2-SP1 interaction, inactivating HDM2, or enhancing TIMP3 activities should all lead to the abrogation of tumor angiogenesis and could be widely applicable for the treatment of cancer and other vasculopathies.
Methods
Cell lines and transfections.
Human glioblastoma cell lines LN229 (polymorphism in TP53 at codon 98 and deleted for the P14ARF gene) and LNZ308 (devoid of endogenous P53 protein) (62) were used to generate clones with tetracycline-inducible expression of P14ARF. Clones LN229-L16 (derived from LN229) (18) and LNZ308-C16 (derived from LNZ308) (63) expressing stably the reverse tetracycline-controlled transactivator (rtTA) were stably transfected with pTRE-HA-ARF, an expression vector containing an HA-tagged P14ARF cDNA under the control of the tetracycline-responsive element (TRE). Hygromycin-resistant clones A5 and A18 (derived from LN229-L16) and C19 (derived from LNZ308-C16) exhibited tightly regulated P14ARF cDNA expression upon dox (2 μg/ml) induction with minimal background and were retained for the subsequent studies. A5-EGFRvIII human malignant glioma cells were generated by stably transfecting an expression vector for the ligand-independent EGFRvIII receptor (14) in the A5 clone. A5HDM2 cells were generated by stable transfection of A5 cells with an HDM2 expression vector (pCMVMDM2-Neo), provided by Bert Vogelstein (The Johns Hopkins Oncology Center, Baltimore, Maryland, USA). Human U343MG glioma cells (TP53 WT, P14ARF deleted), human HFF-1 fibroblasts, and mouse 3T3 fibroblasts were obtained from ATCC. WT, p19Arf-null, Trp53-null, and Trp53/Mdm2 and p19Arf/Trp53 double-knockout MEFs were provided by Martine Roussel and Charles Sherr (St. Jude Children’s Research Hospital, Memphis, Tennessee, USA) and were cultured as described previously (12). (10)3 mouse fibroblasts were provided by Arnold Levine (Institute for Advanced Study, Princeton, New Jersey, USA). Primary cultures of human dermal microvascular ECs (HDMECs) were obtained from the Emory Core Facility, Department of Dermatology. HDMECs were cultured in complete medium MCDB131 (Invitrogen), supplemented with 10% FBS, which was supplemented with 10 ng/ml bFGF, 0.5 μg/ml hydrocortisone, and antibiotics.
Glioma cells were routinely cultured in DMEM containing 5% tetracycline-free calf serum (Invitrogen). HFF-1 cells and all MEFs were cultured in DMEM with 10% FBS (HyClone Laboratories), nonessential amino acids, and 100 μg/ml of penicillin and streptomycin (Invitrogen) at 37°C in a humidified atmosphere of 5% CO2. All transfections with plasmids were performed using GenePORTER as recommended (Gene Therapy Systems). For gene silencing experiments, cells were transfected with 20–100 nM final concentration of TIMP3 siRNA (Santa Cruz Biotechnology Inc.), SP1, TP53, and HDM2 siRNAs (Ambion) or negative control siRNA using Lipofectamine 2000, Lipofectamine RNAiMax (Invitrogen), or HiPerFect (QIAGEN) in serum-free medium for 5–24 hours. The silencing of human P14ARF was performed with human CDKN2A siRNA (Ambion) and of mouse p19Arf with duplex siRNA oligonucleotides: Cdkn2a-MSS10, 5′-GACAUCAAGACAUCGUGCGAUAUUU-3′ and Cdkn2a-MSS12, 5′-UUAGCUCUGCUCUUGGGAUUGGCCG-3′.
Western blot analysis.
Whole-cell extracts were obtained by lysing cells in Laemmli buffer or RIPA buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 0.5 mM EDTA, 0.5% sodium deoxycholate, 0.1% SDS, 1% Triton X-100, and 1× mini Complete anti-proteases, Roche), and protein concentrations were determined using the DC protein assay (Bio-Rad). Nuclear and cytosolic extractions were performed with an NE-PER Nuclear and Cytoplasmic extraction reagents kit (Pierce). The protein samples (50–100 μg per well) were then boiled for 5 minutes after addition of DTT to a final concentration of 50 mM and resolved by SDS-PAGE on either 12.5% Tris-HCl gels or 4%–20% gradient gels (Bio-Rad) and transferred to NitroBind pure nitrocellulose membranes (GE Water and Process Technologies). The membranes were immunoblotted using polyclonal rabbit anti-HA antibodies to detect P14ARF (1:2,000, Covance), mouse anti-P53 (clone DO-7; 1:1,000 dilution, Dako), mouse anti-P21 (Ab-11; 1:500 dilution, NeoMarkers), rabbit and mouse anti-SP1 (PEP2, 1C6; 1:500), mouse anti-HDM2 (SMP14, 1:500), rabbit anti-HDM2 (H221; 1:500), rabbit anti-P53/p53 (FL393; 1:500), goat anti–β-actin, and mouse anti–α-tubulin (1:1,000) (Santa Cruz Biotechnology Inc.), rabbit anti-TIMP3 (1:400, Chemicon, Millipore), rabbit anti-SP1 and rabbit anti-p19Arf (1:1,000, Abcam) and mouse anti-HDM2 (2A10, 4B11, 1:300, Calbiochem) antibodies.
Immunodetection was performed using the corresponding secondary horseradish peroxidase–conjugated antibodies. Horseradish peroxidase activity was detected using a SuperSignal West Pico chemiluminescence kit (Pierce).
Northern blot analysis.
To generate probes specific for each human gene of interest, RT-PCR amplification was performed. The sequences of the PCR forward (Fw) and reverse (Rev) primers were as follows: P14ARF-Fw, 5′-AAACCATGGATGGTCCGCAGGTTCTTGGTG-3′, P14ARF-Rev, 5′-AGCTGGATCCCATCATCATTGACCTGGTCTTCTA-3′; TIMP3 Fw, 5′-AGGTCGCGTCTATGATGGCAAGAT-3′, TIMP3 Rev, 5′-AGCCAGGGTAACCGAAATTGGAGA-3′; CDKN1A/P21-Fw, 5′-GCAGTGTGTCGGGTGAAG-3′, CDKN1A/P21 Rev, 5′-CTCAGCAAGCAACGAAGTG-3′. The annealing temperatures used in PCR reactions were either 60°C or 62°C. Northern blots were generated as previously described (64).
EC assays.
The CM used in the migration/invasion assays was collected from the indicated human glioma cells grown in 100-mm-diameter Petri dishes at 70% confluence in the presence or absence of dox (2 μg/ml) in 4 ml serum-free medium for 48–120 hours. The cells had been previously transfected with either negative control siRNA, TIMP3, or TP53 siRNAs or treated with 650 nM MMP-2 and -9 inhibitor I (Calbiochem). CM was concentrated up to 30-fold using ultrafiltration concentrators with 3-kDa-cutoff Millipore membranes (Amicon Ultra). This pore size is permeable for dox; therefore, upon re-dilution to 1×, the concentration of dox was reduced by 30-fold (lower than 0.07 μg/ml).
EC migration assays were performed with HDMECs for 24 hours using 24-well modified Boyden chambers with an 8-μm pore size (Transwell, BD) and coated with 0.1% hydrolyzed collagen (gelatin, type B). To minimize any effects by growth factors, we decreased the FBS in the culture medium by switching it from 10% to 0.5% serum overnight. Subsequently, cells were trypsinized and plated on the gelatin-coated filter of the upper chamber in 700 μl of medium containing 0.25% serum and 1× CM (diluted from 30× serum-free concentrated CM). Cells were left to adhere to the Transwell filter for 5 hours, and the migration assay was then initiated by adding 500 μl of medium containing 10% serum to the lower chamber. After 24 hours of incubation, the insert was removed, and unmigrated cells (upper side) were scraped off using a cotton swab. The migrated cells (lower side) were fixed, stained using the DiffQuik kit (Dade Behring Inc.), and counted under a light microscope. Pictures were taken from 3 random fields per slide, and cells were counted. The results are expressed as mean (SD) number per field.
EC proliferation assays were performed by plating HDMECs at 60% confluence in a 6-well plate in MCDB131 medium and supplemented with CM (diluted to 1× concentration from 30× concentrated CM of glioma cells treated or untreated with dox) in the presence or absence of bFGF (5 ng/ml). The cells were incubated at 37°C for 72 hours, fixed, and stained with 0.1% crystal violet to evaluate cell density as described previously (18).
Matrigel plug angiogenesis assay.
We used a Matrigel plug assay (18) to assess the regulation of angiogenesis in vivo by glioma cells in the presence or absence of P14ARF expression. Briefly, growth factor–reduced Matrigel (BD Biosciences — Discovery Labware) mixed with A5 cells (2 × 106) in a 0.5-ml total volume was injected subcutaneously into nu/nu mice. Eight mice were injected and subsequently divided into two groups. One group was given 2 mg/ml dox in drinking water with 5% sucrose for 10 days, while the other group received sucrose water only. After 10 days, Matrigel plugs were harvested, fixed in 10% formalin, and embedded in paraffin. A section across the largest dimension of each Matrigel plug was performed and stained with either H&E or hematoxylin and immunostained for Ki67 (mouse anti-Ki67 antibody, Biocare Medical). The total length of vessels in each cross-section was determined as previously described (18). Relative total length was defined as total length of all vessels counted divided by the surface area of each plug section.
Mouse corneal angiogenesis assay.
One hundred pellets were made as previously described (65) by mixing sterile bFGF (Research Diagnostics Inc.) at a final concentration of 25 ng/pellet with sucralfate (Teva Pharmaceuticals) and 7.5 μl of 80× concentrated CM. The CM was collected from A5 or L16 glioma cells cultured in 100-mm-diamter Petri dishes at 70% confluence with or without dox (2 μg/ml) for 48 hours. Mice were anesthetized with ketamine (100 mg/kg) and xylazine (10 mg/kg), the eyes were topically anesthetized with 0.5% proparacaine and 2% Alocril, and the globes proptosed with a forceps. Pellets were implanted surgically in the cornea approximately 1 mm from the limbus as previously described (65).
On day 5 after implantation, mice were anesthetized as before. Fifty microliters of a 2.5-mg/ml solution of sterile FITC-dextran (MW, ~70,000 Da, Sigma-Aldrich) were injected into the retro-orbital sinus. The eyes were proptosed and digital images captured under a fluorescence dissecting microscope (Leica). Images were transferred to Adobe Photoshop for measurements. The maximum vessel length and the neovascularization zone (in clock hours) were used to calculate the area of neovascularization, using the following formula: area (mm2) = 0.2 × π × VL (mm) × CH, where VL is vessel length and CH is clock hours (one clock hour = 30° of an arc).
Luciferase reporter assay of TIMP3 promoter activity.
Transcription factor binding sites in the TIMP3 gene promoter were identified using PROSCAN version 7.1 ( http://www-bimas.cit.nih.gov/molbio/proscan/). A5 cells were transfected with pGL3-TIMP3 (66) either alone or cotransfected with CMV-Sp1 plasmid (67). Twenty-four hours later, cells were trypsinized and plated to be transfected with SP1 or negative control siRNAs (Ambion) for an additional 24 hours. Empty vector pcDNA3 and negative control siRNA were used as negative controls. Plasmid transfections were carried out with GenePORTER, and siRNA transfections were performed with Lipofectamine 2000 (Invitrogen). Twenty-four hours after transfection with siRNAs, cells were left untreated or treated with 2 μg/ml dox for 36 hours. Cells were lysed, and the relative firefly luciferase activities were measured using a Luciferase Reporter Assay System (Promega) and a luminometer (Sirius model, Berthold Detection Systems). The firefly luciferase activity was normalized to protein content. All luciferase assays were carried out in triplicate, and values represent RLU of luciferase activity/μg protein present in whole-cell lysate.
ChIP assays.
The ChIP assays were performed with a commercial kit (Upstate, Millipore) using the manufacturer’s protocol with minor adjustments. LN229-L16 and A5 cells were grown in 150-mm dishes at 3 × 106 cells/plate in medium with 2% serum in the presence or absence of dox (2 μg/ml) for 48 hours. Subsequently, the cells were fixed for 10 minutes at 37°C by adding formaldehyde directly to the culture medium to a final concentration of 1%. The cells were washed with cold PBS, lysed for 10 minutes with 1% SDS, 10 mM Tris HCl, pH 8.0, and sonicated 4 times for 10 seconds each on ice (Sonic Dismembrator model 100, Fisher Scientific), and then cell debris was removed by centrifugation. Aliquots were taken to control for DNA input. The remaining lysate was diluted 10 times in 0.01% SDS, 1% Triton X-100, 1 mM EDTA, 10 mM Tris-HCl, pH 8.0, 150 mM NaCl, and phosphatase/protease inhibitors; precleared with agarose beads/salmon sperm DNA; and incubated overnight (4°C) with antibodies against SP1 (PEP2) or with non-specific rabbit IgG (Santa Cruz Biotechnology Inc.). Formaldehyde-fixed DNA-protein complexes were pulled down with protein A–conjugated agarose beads and extracted with 1% SDS, 0.1 M NaHCO3. The DNA-protein crosslinking was reversed at 65°C for 5 hours, and the released proteins were eliminated through digestion with proteinase K. The co-immunoprecipitated genomic DNA was purified using Mini columns (QIAGEN) and eluted with 10 mM Tris-HCl, pH 8.0. The primers used for PCR to amplify the TIMP3 promoter encompassing the SP1 binding sites were TIMP3 Fw, 5′-CCACGGCGGCATTATTCCCTATAA-3′, TIMP3 Rev, 5′-AGGAGCAAGAGGAGGAGGAGAA-3′. The expected size of the PCR product is 266 bp.
Co-IP assays.
Cells were cultured in DMEM supplemented with 2% serum and were treated with dox (2 μg/ml) to induce P14ARF expression. Thereafter, the cells were washed with cold PBS and lysed in ice-cold RIPA buffer without deoxycholate and containing anti-proteases (Roche Applied Science) for 30 minutes, and the lysates were cleared by centrifugation at 14,000 g at 4°C for 15 minutes. Total A5 protein extracts (500 μg) and nuclear and cytosolic 3T3 protein extracts were precleared, then incubated overnight with 3–5 μg antibodies. Anti-SP1 (PEP2, rabbit), anti-P14ARF (C-18, goat; Santa Cruz Biotechnology Inc.), anti-HDM2 (SMP14 mouse, Sigma-Aldrich; H221, rabbit, Santa Cruz Biotechnology Inc.), anti-HDM2 2A10 (Calbiochem, mouse), anti-Sp1 or anti-p19Arf (Abcam, rabbit), and protein A/G–conjugated agarose beads (Roche) for trapping immune complexes. Rabbit and mouse IgG (Santa Cruz Biotechnology Inc.) were used as negative controls for nonspecific interactions. Then, beads were spun down at 460 g, pellets were washed 3 times with cold lysis buffer containing anti-proteases, and the immune complexes were released by boiling for 5 minutes in Laemmli buffer. Proteins were then resolved by SDS-PAGE and immunoblotted with antibodies against Sp1, Mdm2, P14ARF, and p19Arf.
ELISAs.
Concentrations of TIMP3 and VEGF in cell CM were measured using ELISA kits (R&D Systems). Measurements were performed in duplicate, and results, normalized to protein extracts, are reported as mean ± SD.
Microarray analysis.
A5 cells were cultured in 2% serum-supplemented medium in the presence and absence of 2 μg/ml dox during 48 hours. Total RNA from two independent experiments was prepared using TRIzol (Invitrogen), according to the manufacturer’s protocol. Integrity of the purified RNA was first monitored by agarose gel electrophoresis and then reverse transcribed to DNA, followed by in vitro transcription using a BioArray HighYield RNA transcript labeling kit (Enzo). Biotin-labeled cRNA was then fragmented according to Affymetrix standard protocols. cRNA probes were hybridized overnight at 45°C to the Human Genome U133A arrays (Affymetrix), washed in a Fluidics workstation, and scanned according to the manufacturer’s protocols. The Affymetrix CEL files were normalized using the robust multi-chip analysis (RMA) method. After data normalization, GeneTraffic analysis (Iobion Informatics) was performed as 2-class unpaired data (group of experimental samples with P14ARF induction for 48 hours versus group of untreated samples). The list of genes up- and downregulated by P14ARF was generated using the cutoff at log ratio greater than 0.4 and P value less than 0.05. The probe sets estimated absent using Affymetrix Microarray Suite 5.0 (MAS 5.0) software, in both baseline and experimental samples, were excluded from the final list. Hierarchical clustering of genes was next performed using GeneTraffic software. Microarray gene expression data were deposited at a MIAME-compliant public database, EMBL-EBI ArrayExpress, and can be accessed with E-MEXP-3476.
Statistics.
ANOVA was used to compare corneal angiogenesis in mice groups that had received pellets with bFGF and CM of A5 with or without dox cells. Unpaired 2-tailed Student’s t test was used in all other studies, unless stated otherwise. A P value less than 0.05 was considered statistically significant.
Study approval.
The in vivo Matrigel plug and cornea assays conducted in mice were reviewed and approved by the IACUC of Emory University and by the IACUC of the Cleveland Clinic, respectively.
Supplementary Material
Acknowledgments
This work was supported by grants from the NIH (CA86335 and CA116804 to E.G. Van Meir, P30 CA138292 to the Winship Cancer Institute, and AR42687 to Emory Diseases Research Core Center), the Pediatric Brain Tumor Foundation and the Southeastern Brain Tumor Foundation (to E.G. Van Meir and A. Zerrouqi), the American Brain Tumor Association (to B. Pyrzynska). We thank Martine Roussel, Charles Sherr, and Gerard Zambetti (St. Jude Children’s Research Hospital) for providing knockout MEFs, Arnold Levine (Institute for Advanced Study, Princeton, New Jersey, USA) for providing (10)3 fibroblasts, Yue Xiong (University of North Carolina, Chapel Hill, North Carolina, USA) for the pTRE-HA-ARF plasmid, Stephan Smale (University of California, Berkeley, California, USA) for the CMVSp1 construct, Bert Vogelstein (The Johns Hopkins Oncology Center) for pCMV-MDM2-NEO, and Paula Vertino for helpful discussions.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2012;122(4):1283–1295. doi:10.1172/JCI38596.
References
- 1.Sharpless NE. INK4a/ARF: a multifunctional tumor suppressor locus. Mutat Res. 2005;576(1–2):22–38. doi: 10.1016/j.mrfmmm.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 2.Quelle DE, Zindy F, Ashmun RA, Sherr CJ. Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell. 1995;83(6):993–1000. doi: 10.1016/0092-8674(95)90214-7. [DOI] [PubMed] [Google Scholar]
- 3.Pomerantz J, et al. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2’s inhibition of p53. Cell. 1998;92(6):713–723. doi: 10.1016/S0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- 4.Randerson-Moor JA, et al. A germline deletion of p14(ARF) but not CDKN2A in a melanoma-neural system tumour syndrome family. Hum Mol Genet. 2001;10(1):55–62. doi: 10.1093/hmg/10.1.55. [DOI] [PubMed] [Google Scholar]
- 5.Kamijo T, et al. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 1997;91(5):649–659. doi: 10.1016/S0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- 6.Weber HO, Samuel T, Rauch P, Funk JO. Human p14(ARF)-mediated cell cycle arrest strictly depends on intact p53 signaling pathways. Oncogene. 2002;21(20):3207–3212. doi: 10.1038/sj.onc.1205429. [DOI] [PubMed] [Google Scholar]
- 7.Kelly-Spratt KS, Gurley KE, Yasui Y, Kemp CJ. p19Arf suppresses growth, progression, and metastasis of Hras-driven carcinomas through p53-dependent and -independent pathways. PLoS Biol. 2004;2(8):E242. doi: 10.1371/journal.pbio.0020242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kamijo T, et al. Loss of the ARF tumor suppressor reverses premature replicative arrest but not radiation hypersensitivity arising from disabled atm function. Cancer Res. 1999;59(10):2464–2469. [PubMed] [Google Scholar]
- 9.Kamijo T, Bodner S, van de Kamp E, Randle DH, Sherr CJ. Tumor spectrum in ARF-deficient mice. Cancer Res. 1999;59(9):2217–2222. [PubMed] [Google Scholar]
- 10.Radfar A, Unnikrishnan I, Lee HW, DePinho RA, Rosenberg N. p19(Arf) induces p53-dependent apoptosis during abelson virus-mediated pre-B cell transformation. Proc Natl Acad Sci U S A. 1998;95(22):13194–13199. doi: 10.1073/pnas.95.22.13194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ichimura K, Bolin MB, Goike HM, Schmidt EE, Moshref A, Collins VP. Deregulation of the p14ARF/MDM2/p53 pathway is a prerequisite for human astrocytic gliomas with G1-S transition control gene abnormalities. Cancer Res. 2000;60(2):417–424. [PubMed] [Google Scholar]
- 12.Weber JD, et al. p53-independent functions of the p19(ARF) tumor suppressor. Genes Dev. 2000;14(18):2358–2365. doi: 10.1101/gad.827300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watanabe T, Katayama Y, Yoshino A, Komine C, Yokoyama T. Deregulation of the TP53/p14ARF tumor suppressor pathway in low-grade diffuse astrocytomas and its influence on clinical course. Clin Cancer Res. 2003;9(13):4884–4890. [PubMed] [Google Scholar]
- 14.Van Meir EG, Hadjipanayis CG, Norden AD, Shu HK, Wen PY, Olson JJ. Exciting new advances in neuro-oncology: the avenue to a cure for malignant glioma. CA Cancer J Clin. 2010;60(3):166–193. doi: 10.3322/caac.20069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fulci G, et al. p53 gene mutation and ink4a-arf deletion appear to be two mutually exclusive events in human glioblastoma. Oncogene. 2000;19(33):3816–3822. doi: 10.1038/sj.onc.1203700. [DOI] [PubMed] [Google Scholar]
- 16.Verhaak RG, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brat DJ, Van Meir EG. Vaso-occlusive and prothrombotic mechanisms associated with tumor hypoxia, necrosis, and accelerated growth in glioblastoma. Lab Invest. 2004;84(4):397–405. doi: 10.1038/labinvest.3700070. [DOI] [PubMed] [Google Scholar]
- 18.Kaur B, Brat DJ, Devi NS, Van Meir EG. Vasculostatin, a proteolytic fragment of brain angiogenesis inhibitor 1, is an antiangiogenic and antitumorigenic factor. Oncogene. 2005;24(22):3632–3642. doi: 10.1038/sj.onc.1208317. [DOI] [PubMed] [Google Scholar]
- 19.Kaur B, et al. Vasculostatin inhibits intracranial glioma growth and negatively regulates in vivo angiogenesis through a CD36-dependent mechanism. Cancer Res. 2009;69(3):1212–1220. doi: 10.1158/0008-5472.CAN-08-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Groth A, Weber JD, Willumsen BM, Sherr CJ, Roussel MF. Oncogenic Ras induces p19ARF and growth arrest in mouse embryo fibroblasts lacking p21Cip1 and p27Kip1 without activating cyclin D-dependent kinases. J Biol Chem. 2000;275(35):27473–27480. doi: 10.1074/jbc.M003417200. [DOI] [PubMed] [Google Scholar]
- 21.Birkedal-Hansen H, et al. Matrix metalloproteinases: a review. Crit Rev Oral Biol Med. 1993;4(2):197–250. doi: 10.1177/10454411930040020401. [DOI] [PubMed] [Google Scholar]
- 22.Van Meir EG, Polverini PJ, Chazin VR, Su Huang HJ, de Tribolet N, Cavenee WK. Release of an inhibitor of angiogenesis upon induction of wild type p53 expression in glioblastoma cells. Nat Genet. 1994;8(2):171–176. doi: 10.1038/ng1094-171. [DOI] [PubMed] [Google Scholar]
- 23.Wick M, et al. Structure of the human TIMP-3 gene and its cell cycle-regulated promoter. Biochem J. 1995;311(pt 2):549–554. doi: 10.1042/bj3110549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blume SW, Snyder RC, Ray R, Thomas S, Koller CA, Miller DM. Mithramycin inhibits SP1 binding and selectively inhibits transcriptional activity of the dihydrofolate reductase gene in vitro and in vivo. J Clin Invest. 1991;88(5):1613–1621. doi: 10.1172/JCI115474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gualberto A, Baldwin AS., Jr p53 and Sp1 interact and cooperate in the tumor necrosis factor-induced transcriptional activation of the HIV-1 long terminal repeat. J Biol Chem. 1995;270(34):19680–19683. doi: 10.1074/jbc.270.34.19680. [DOI] [PubMed] [Google Scholar]
- 26.Daujat S, Neel H, Piette J. MDM2: life without p53. Trends Genet. 2001;17(8):459–464. doi: 10.1016/S0168-9525(01)02369-1. [DOI] [PubMed] [Google Scholar]
- 27.Matheu A, et al. Increased gene dosage of Ink4a/Arf results in cancer resistance and normal aging. Genes Dev. 2004;18(22):2736–2746. doi: 10.1101/gad.310304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Serrano M. The INK4a/ARF locus in murine tumorigenesis. Carcinogenesis. 2000;21(5):865–869. doi: 10.1093/carcin/21.5.865. [DOI] [PubMed] [Google Scholar]
- 29.Folkman J. The role of angiogenesis in tumor growth. Semin Cancer Biol. 1992;3(2):65–71. [PubMed] [Google Scholar]
- 30.Yassini PR, Stickler DL, Bloomfield SM, Wiggins RC, Konat GW. Glioma-stimulated chemoattraction of endothelial cells and fibroblasts in vitro: a model for the study of glioma-induced angiogenesis. Metab Brain Dis. 1994;9(4):391–399. doi: 10.1007/BF02098885. [DOI] [PubMed] [Google Scholar]
- 31.Cornelius LA, Nehring LC, Roby JD, Parks WC, Welgus HG. Human dermal microvascular endothelial cells produce matrix metalloproteinases in response to angiogenic factors and migration. J Invest Dermatol. 1995;105(2):170–176. doi: 10.1111/1523-1747.ep12317080. [DOI] [PubMed] [Google Scholar]
- 32.van Hinsbergh VW, Koolwijk P. Endothelial sprouting and angiogenesis: matrix metalloproteinases in the lead. Cardiovasc Res. 2008;78(2):203–212. doi: 10.1093/cvr/cvm102. [DOI] [PubMed] [Google Scholar]
- 33.Spurbeck WW, Ng CY, Strom TS, Vanin EF, Davidoff AM. Enforced expression of tissue inhibitor of matrix metalloproteinase-3 affects functional capillary morphogenesis and inhibits tumor growth in a murine tumor model. Blood. 2002;100(9):3361–3368. doi: 10.1182/blood.V100.9.3361. [DOI] [PubMed] [Google Scholar]
- 34.Qi JH, et al. A novel function for tissue inhibitor of metalloproteinases-3 (TIMP3): inhibition of angiogenesis by blockage of VEGF binding to VEGF receptor-2. Nat Med. 2003;9(4):407–415. doi: 10.1038/nm846. [DOI] [PubMed] [Google Scholar]
- 35.Nakamura M, et al. Frequent LOH on 22q12.3 and TIMP-3 inactivation occur in the progression to secondary glioblastomas. Lab Invest. 2005;85(2):165–175. doi: 10.1038/labinvest.3700223. [DOI] [PubMed] [Google Scholar]
- 36.Bachman KE, et al. Methylation-associated silencing of the tissue inhibitor of metalloproteinase-3 gene suggest a suppressor role in kidney, brain, and other human cancers. Cancer Res. 1999;59(4):798–802. [PubMed] [Google Scholar]
- 37.Mahller YY, et al. Tissue inhibitor of metalloproteinase-3 via oncolytic herpesvirus inhibits tumor growth and vascular progenitors. Cancer Res. 2008;68(4):1170–1179. doi: 10.1158/0008-5472.CAN-07-2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eymin B, et al. Human ARF binds E2F1 and inhibits its transcriptional activity. Oncogene. 2001;20(9):1033–1041. doi: 10.1038/sj.onc.1204220. [DOI] [PubMed] [Google Scholar]
- 39.Eymin B, Leduc C, Coll JL, Brambilla E, Gazzeri S. p14ARF induces G2 arrest and apoptosis independently of p53 leading to regression of tumours established in nude mice. Oncogene. 2003;22(12):1822–1835. doi: 10.1038/sj.onc.1206303. [DOI] [PubMed] [Google Scholar]
- 40.Hemmati PG, et al. Adenovirus-mediated overexpression of p14(ARF) induces p53 and Bax-independent apoptosis. Oncogene. 2002;21(20):3149–3161. doi: 10.1038/sj.onc.1205458. [DOI] [PubMed] [Google Scholar]
- 41.Silva RL, et al. Arf-dependent regulation of Pdgf signaling in perivascular cells in the developing mouse eye. Embo J. 2005;24(15):2803–2814. doi: 10.1038/sj.emboj.7600751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gibson SL, et al. Inhibition of colon tumor progression and angiogenesis by the Ink4a/Arf locus. Cancer Res. 2003;63(4):742–746. [PubMed] [Google Scholar]
- 43.Kawagishi H, et al. ARF suppresses tumor angiogenesis through translational control of VEGFA mRNA. Cancer Res. 2010;70(11):4749–4758. doi: 10.1158/0008-5472.CAN-10-0368. [DOI] [PubMed] [Google Scholar]
- 44.Rizos H, Woodruff S, Kefford RF. p14ARF interacts with the SUMO-conjugating enzyme Ubc9 and promotes the sumoylation of its binding partners. Cell Cycle. 2005;4(4):597–603. doi: 10.4161/cc.4.4.1597. [DOI] [PubMed] [Google Scholar]
- 45.Fatyol K, Szalay AA. The p14ARF tumor suppressor protein facilitates nucleolar sequestration of hypoxia-inducible factor-1alpha (HIF-1alpha ) and inhibits HIF-1-mediated transcription. J Biol Chem. 2001;276(30):28421–28429. doi: 10.1074/jbc.M102847200. [DOI] [PubMed] [Google Scholar]
- 46.Lee M, Song SU, Ryu JK, Suh JK. Sp1-dependent regulation of the tissue inhibitor of metalloproteinases-1 promoter. J Cell Biochem. 2004;91(6):1260–1268. doi: 10.1002/jcb.20021. [DOI] [PubMed] [Google Scholar]
- 47.Zhong ZD, Hammani K, Bae WS, DeClerck YA. NF-Y and Sp1 cooperate for the transcriptional activation and cAMP response of human tissue inhibitor of metalloproteinases-2. J Biol Chem. 2000;275(24):18602–18610. doi: 10.1074/jbc.M001389200. [DOI] [PubMed] [Google Scholar]
- 48.Young DA, et al. Identification of an initiator-like element essential for the expression of the tissue inhibitor of metalloproteinases-4 (Timp-4) gene. Biochem J. 2002;364(pt 1):89–99. doi: 10.1042/bj3640089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ahonen M, Baker AH, Kahari VM. Adenovirus-mediated gene delivery of tissue inhibitor of metalloproteinases-3 inhibits invasion and induces apoptosis in melanoma cells. Cancer Res. 1998;58(11):2310–2315. [PubMed] [Google Scholar]
- 50.Black AR, Black JD, Azizkhan-Clifford J. Sp1 and kruppel-like factor family of transcription factors in cell growth regulation and cancer. J Cell Physiol. 2001;188(2):143–160. doi: 10.1002/jcp.1111. [DOI] [PubMed] [Google Scholar]
- 51.Borellini F, Glazer RI. Induction of Sp1-p53 DNA-binding heterocomplexes during granulocyte/macrophage colony-stimulating factor-dependent proliferation in human erythroleukemia cell line TF-1. J Biol Chem. 1993;268(11):7923–7928. [PubMed] [Google Scholar]
- 52.Stros M, Ozaki T, Bacikova A, Kageyama H, Nakagawara A. HMGB1 and HMGB2 cell-specifically down-regulate the p53- and p73-dependent sequence-specific transactivation from the human Bax gene promoter. J Biol Chem. 2002;277(9):7157–7164. doi: 10.1074/jbc.M110233200. [DOI] [PubMed] [Google Scholar]
- 53.Mukhopadhyay D, Knebelmann B, Cohen HT, Ananth S, Sukhatme VP. The von Hippel-Lindau tumor suppressor gene product interacts with Sp1 to repress vascular endothelial growth factor promoter activity. Mol Cell Biol. 1997;17(9):5629–5639. doi: 10.1128/mcb.17.9.5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Johnson-Pais T, Degnin C, Thayer MJ. pRB induces Sp1 activity by relieving inhibition mediated by MDM2. Proc Natl Acad Sci U S A. 2001;98(5):2211–2216. doi: 10.1073/pnas.051415898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Udvadia AJ, et al. Sp-1 binds promoter elements regulated by the RB protein and Sp-1-mediated transcription is stimulated by RB coexpression. Proc Natl Acad Sci U S A. 1993;90(8):3265–3269. doi: 10.1073/pnas.90.8.3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Clark SJ, Harrison J, Molloy PL. Sp1 binding is inhibited by (m)Cp(m)CpG methylation. Gene. 1997;195(1):67–71. doi: 10.1016/S0378-1119(97)00164-9. [DOI] [PubMed] [Google Scholar]
- 57.Guo CS, Degnin C, Fiddler TA, Stauffer D, Thayer MJ. Regulation of MyoD activity and muscle cell differentiation by MDM2, pRb, and Sp1. J Biol Chem. 2003;278(25):22615–22622. doi: 10.1074/jbc.M301943200. [DOI] [PubMed] [Google Scholar]
- 58.Bothner B, Lewis WS, DiGiammarino EL, Weber JD, Bothner SJ, Kriwacki RW. Defining the molecular basis of Arf and Hdm2 interactions. J Mol Biol. 2001;314(2):263–277. doi: 10.1006/jmbi.2001.5110. [DOI] [PubMed] [Google Scholar]
- 59.Zhang Y, Xiong Y, Yarbrough WG. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell. 1998;92(6):725–734. doi: 10.1016/S0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
- 60.Tao W, Levine AJ. P19(ARF) stabilizes p53 by blocking nucleo-cytoplasmic shuttling of Mdm2. Proc Natl Acad Sci U S A. 1999;96(12):6937–6941. doi: 10.1073/pnas.96.12.6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kondo S, et al. The transforming activities of MDM2 in cultured neonatal rat astrocytes. Oncogene. 1996;13(8):1773–1779. [PubMed] [Google Scholar]
- 62.Ishii N, et al. Frequent co-alterations of TP53, p16/CDKN2A, p14ARF, PTEN tumor suppressor genes in human glioma cell lines. Brain Pathol. 1999;9(3):469–479. doi: 10.1111/j.1750-3639.1999.tb00536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Albertoni M, et al. Anoxia induces macrophage inhibitory cytokine-1 (MIC-1) in glioblastoma cells independently of p53 and HIF-1. Oncogene. 2002;21(27):4212–4219. doi: 10.1038/sj.onc.1205610. [DOI] [PubMed] [Google Scholar]
- 64.Tan C, et al. Identification of a novel small-molecule inhibitor of the hypoxia-inducible factor 1 pathway. Cancer Res. 2005;65(2):605–612. [PubMed] [Google Scholar]
- 65.Kenyon BM, Voest EE, Chen CC, Flynn E, Folkman J, D’Amato RJ. A model of angiogenesis in the mouse cornea. Invest Ophthalmol Vis Sci. 1996;37(8):1625–1632. [PubMed] [Google Scholar]
- 66.Zeng Y, Rosborough RC, Li Y, Gupta AR, Bennett J. Temporal and spatial regulation of gene expression mediated by the promoter for the human tissue inhibitor of metalloproteinases-3 (TIMP-3)-encoding gene. Dev Dyn. 1998;211(3):228–237. doi: 10.1002/(SICI)1097-0177(199803)211:3<228::AID-AJA4>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 67.Kadonaga JT, Courey AJ, Ladika J, Tjian R. Distinct regions of Sp1 modulate DNA binding and transcriptional activation. Science. 1988;242(4885):1566–1570. doi: 10.1126/science.3059495. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.