Abstract
Multiple sclerosis (MS) is a multifocal demyelinating disease with progressive neurodegeneration caused by an autoimmune response to self-antigens in a genetically susceptible individual. While the formation and persistence of meningeal lymphoid follicles suggest persistence of antigens to drive the continuing inflammatory and humoral response, the identity of an antigen or infectious agent leading to the oligoclonal expansion of B and T cells is unknown. In this review we examine new paradigms for understanding the immunopathology of MS, present recent data defining the common genetic variants underlying disease susceptibility, and explore how improved understanding of immune pathway disruption can inform MS prognosis and treatment decisions.
Introduction
MS is a multifocal demyelinating disease with progressive neurodegeneration caused by an autoimmune response to self-antigens in a genetically susceptible individual. Clinical symptoms vary based on the site of neurologic lesions and often correlate with invasion of inflammatory cells across the blood-brain barrier with resulting demyelination and edema (1). Patients often exhibit an initial clinically isolated syndrome, followed by a series of subacute clinical events that spontaneously abate, referred to as relapsing remitting MS (RRMS). While patients generally return to near normal neurologic function at the cessation of each episode, over a variable period of time there can be irreversible progression of clinical disability termed secondary progressive MS (SPMS), although early therapeutic intervention may delay time to progression (2, 3). However, 10%–15% of MS patients will instead experience primary progressive MS, characterized by clinical progression from the initiation of disease without preceding relapses and remissions (4).
The diagnosis is made primarily on the basis of the medical history and physical exam, which was formalized as the McDonald criteria and updated to reflect the increased reliance on imaging for lesion identification (5–7). These criteria indicate that the neurologic lesions should be disseminated in both space and time to distinguish them from monophasic self-limiting diseases such as acute disseminated encephalomyelitis, and additionally they should reflect a pattern of neurological inflammation typical of MS in the absence of a more appropriate diagnosis. A diagnosis of MS reflects clinical evaluation of episodes of intermittent neurological impairment and may take into account laboratory data, such as the characteristic oligoclonal bands in the cerebrospinal fluid (CSF), which indicate intrathecal immunoglobulin production, or abnormal visual evoked responses in the absence of optic neuritis. Over the past two decades, the diagnosis has come to include the more sensitive and specific spatial identification of white matter lesions via evaluation of T2-hyperintense lesions and gadolinium-enhancing T1 lesions on MRI (8). Gadolinium enhancement serves as a marker of focal inflammation when the dye leaks through vessels due to the local permeability and breakdown of the blood-brain barrier (BBB).
In this review we examine new paradigms for understanding the immunopathology of MS as related to contributions of meningeal B cell follicles and alterations in regulatory T cell function. Additionally, we present recent data defining the common genetic variants underlying susceptibility to MS and other autoimmune diseases as the foundation for understanding the mechanisms of immune dysregulation. We conclude by exploring how improved understanding of immune pathway disruption can inform prognosis and treatment decisions in MS.
Immunopathology
White and gray matter pathology.
Circumscribed white matter plaques, typically located in the subcortical or periventricular white matter, optic nerve sheaths, brainstem, and spinal cord, have characterized classical MS pathology (9). Focal inflammatory demyelinating lesions are characterized by perivascular infiltrates that predominantly contain clonally expanded CD8+ T cells, and to a lesser degree CD4+ T cells (10, 11), γδ T cells (12), monocytes, and rare B cells and plasma cells. These active lesions contain large numbers of macrophages containing myelin debris as well as significant complement and immunoglobulin deposition (13). While early RRMS inflammatory lesions generally correlate with gadolinium-enhancing MRI lesions, the inflammation in progressive disease generally does not (14). In the progressive stage of MS, disease is characterized by gradual lesion expansion with myelin-laden macrophages typically restricted to the edge of the plaque, diffuse abnormal inflammation of the normal appearing white matter, and extensive axonal injury and transection associated with demyelinated lesions (15–18). Additionally, both neuronal loss and spinal cord axonal loss associate with the degree of meningeal inflammation (19, 20). Pathological characterization of cortical demyelination is generally limited to autopsy specimens of individuals with long-standing disease, but there have also been case reports of early RRMS presenting with inflammatory cortical demyelination before white matter lesions have formed, suggesting that some inflammation may be initiated at the subpial layer (21, 22).
Some pathologists have suggested that separate mechanisms may be responsible for the inflammatory and degenerative components of disease progression, particularly given that disability accumulation may depend more on patient age than initial disease course (23–26). Potential mechanisms of degenerative progression include Wallerian degeneration secondary to demyelination and axonal transection, damage from reactive oxygen species and nitric oxide, or energy failure from mitochondrial dysfunction (27–29). An alternative hypothesis suggests degenerative progression may be caused by disease compartmentalization behind a repaired BBB with an ongoing inflammatory response separated from peripheral regulation. Although early RRMS is mediated by the adaptive immune system, with waves of T cells trafficking into the CNS from the peripheral immune system, it is possible that CNS inflammation in SPMS is mediated by the innate immune system, with activated microglia interacting with astrocytes, leading to oligodendrocyte destruction. For example, there is specific expression of innate receptors such as TIM-3 on microglia in white matter but not gray matter in normal brain tissue. With inflammation, there is expression of the TIM-3 ligand galectin-9 on astrocytes, which can lead to TNF-α secretion by the microglia independent of T cells (30).
Meningeal B cell follicles.
Given the persistent oligoclonal bands and increases in IgG that characterize the CSF of MS patients (31) and the effectiveness of anti-CD20 antibodies such as Rituximab in decreasing relapse incidence (32), B cells and plasma cells are recognized to play an important role in MS pathology. Ectopic lymphoid follicle-like structures have been described in the meninges of SPMS patients (33). These structures contain proliferating B cells suggestive of germinal centers, as well as plasma cells, T cells, and dendritic cells. Interestingly, in the subpial cortex close to meningeal follicles there is a gradient of worsening neuronal and axonal loss, suggesting that there may be a humoral cytotoxic element partially responsible for the cortical demyelination observed in grey matter pathology (19, 20, 34). While the formation and persistence of lymphoid follicles suggest persistence of antigens to drive the continuing inflammatory and humoral response, the identity of an antigen or infectious agent that is driving this process is unknown. Epidemiologic data and enhanced EBV-specific immune reactivity in MS patients suggest that EBV may play a role in priming the immune system (35–39), and in one report, EBV infection of B cells was implicated in driving the formation of these ectopic lymph nodes (40). However, these findings have not been replicated and it has not been possible to identify an EBV footprint in parenchymal or meningeal B cells by molecular techniques.
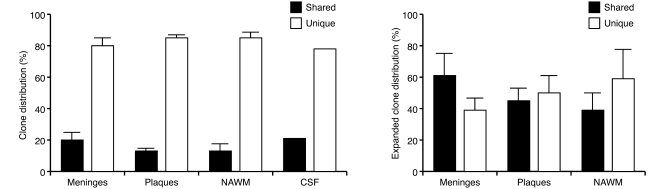
Additionally, it appears that there is significant connectivity between the populations of immune cells present in the CSF and CNS — specifically, related clonally expanded B cells have been identified in the brain parenchyma, meningeal follicles, and CSF (41) and furthermore, these B cell and plasmablast clones are responsible for the intrathecal immunoglobulin production (ref. 42 and Figure 1). Notably, however, oligoclonal bands persist with anti-CD20 monoclonal antibody treatment despite depletion of B cells in the serum and CSF, indicating that mature plasma cells are not affected by these anti-B cell treatments (43).
Figure 1. Clonally related B cells populate the brain parenchyma, plaques, normal-appearing white matter (NAWM), meninges, and CSF in MS.
Analysis of the B cell repertoires derived from the meninges, plaques, NAWM, and CSF from brains of 11 individuals with MS demonstrated that the majority of the B cells resided exclusively in one area, but a small proportion of clones were shared among different locations. Analysis of a clonally expanded B cell subset revealed that 39%–62% of these clones populated different locations within the MS CNS. Reproduced with permission from Brain (41).
Paradigm for Th17 cell pathogenesis.
There continues to be intense investigation of mechanisms leading to disease initiation in humans and how it might involve the transition from physiologic immunosurveillance to pathologic cascade. VLA-4 is the α4β7 integrin expressed on activated immune cells and is the ligand for VCAM expressed on CNS endothelial cells. Clinical studies with natalizumab, a monoclonal antibody against VLA-4, emphasize the importance of immunosurveillance. Specifically, natalizumab has proven successful in reducing relapse in RRMS patients (44) by inhibiting lymphocyte migration through the BBB to prevent pathogenic autoreactive T cells from entering the CNS. However, there have been cases of natalizumab-treated patients developing progressive multifocal leukoencephalopathy, a devastating neurological disorder associated with JC virus reactivation (45), which suggests that in addition to preventing pathogenic T cells from trafficking into the CNS, natalizumab also inhibits immune cells that are scanning for and controlling latent disease.
Although Th1 cells were previously thought to drive MS inflammation, it is now thought that pro-inflammatory Th17 cells are also critically involved in the initiation of disease pathogenesis, as pathogenic Th17 cells gain early access to the CNS (46). Immune cells may enter the CNS for surveillance via the choroid plexus, which spans much of the blood-CSF barrier and produces CSF by filtering solutes from the serum and secreting them at the ventricle-facing membrane. Investigations using the EAE model described a Th17 cell–mediated entry through the choroid plexus in the initiation of disease via a CCR6-CCL20 axis (with CCR6 expressed on Th17 cells and CCL20 expressed on the choroid plexus epithelium) (47). If this model is applicable to human disease, we can speculate that in the context of defective immune regulation (Figure 2), peripherally activated autoreactive Th17 cells may bind to adhesion molecules and chemokine receptors expressed on the choroid plexus and migrate across the blood-CSF barrier into the CSF of the ventricles. Once within the ventricles, cells might penetrate the periventricular parenchyma or continue circulating until they access the pia and subpial cortex. IL-23 and transcription factor RORγt promote GM-CSF production by Th17 cells, which has recently been shown to be a nonredundant requirement for EAE susceptibility and encephalogenicity and may promote further IL-23 production by antigen-presenting cells in a positive feedback loop that contributes to smoldering axoglial damage (48, 49). Cytokines IL-17 and IL-22 released by Th17 cells increase BBB permeability and initiate a cascade of immune cell penetration by autoreactive Th17 cells (50) as well as Th1 (IFN-γ secreting), γδ T cells, cytotoxic CD8+ cells, B cells, and immunoglobulin-secreting plasma cells, which could then form perivascular infiltrates.
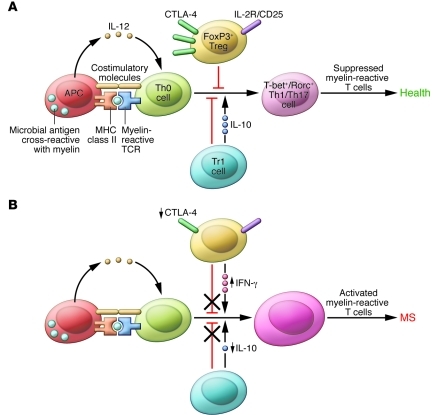
Figure 2. Defects in peripheral immune regulation lower the activation barrier for autoreactive T cells.
(A) In normal homeostasis, APCs digest microbial antigens or self proteins and present them to naive T cells in the context of co-stimulatory molecules. An appropriate cytokine milieu can drive differentiation of these naive autoreactive T cells to a Th1 or Th17 cell phenotype; however, these potentially pathogenic T cells are not activated due to the actions of peripheral regulatory immune cell populations, such as FoxP3+ Tregs and Tr1 cells. Via the actions of co-inhibitory molecules and cytokines such as IL-10 and TGF-β, autoreactive T cells become anergic and autoimmune disease is prevented. Other mechanisms, such as thymic deletion and lack of co-stimulatory molecules on APCs, are also involved in controlling autoreactive T cells. (B) MS patients have defects in peripheral immune regulation, including higher expression of co-stimulatory molecules on APCs, lower CTLA-4 levels, and lower IL-10 production. Additionally, MS patients have an increased frequency of IFN-γ–secreting Tregs relative to healthy controls. Thus, the barrier for activation of autoreactive T cells is lowered for MS patients. Activated myelin-reactive T cells can then adhere to and extravasate across the choroid plexus and BBB, where they can initiate an inflammatory milieu that gives license to further waves of inflammation and eventual epitope spreading.
Thus, we can posit a two-step model of disease for MS, which may be analogous to diabetes in NOD mice, in which peri-insulitis precedes the inflammatory invasion of the insula (51). First, autoreactive Th17 cells may be attracted to and migrate through the choroid plexus to access the CSF and periventricular and subpial CNS. Second, Th17 cells that have invaded the CNS increase BBB permeability by release of cytokines and initiate a pathological cascade of inflammation correlating with a clinical exacerbation. Interestingly, a recent paper suggested that a pathogenic Th17 cell response might be controlled by utilizing the CCR6-CCL20 axis to redirect cells to the duodenum, where they acquire a regulatory suppressive phenotype (52). Future studies may investigate whether there is a defect in this system in MS patients or whether this could be potentially harnessed as a therapeutic approach.
What are the autoantigens?
A single-antigen specificity dictating the autoreactive immune response in MS patients has been difficult to elucidate, in contrast to the higher-affinity antibodies that recognize self-antigens that define a number of neurologic diseases, such as the acetylcholine receptor in myasthenia gravis (53) and aquaporin-4 in neuromyelitis optica (54, 55). There has been intense investigation of T cell reactivity to myelin antigens including myelin basic protein (MBP), proteolipid protein, and myelin oligodendrocyte glycoprotein (MOG), as well as oligodendroglia-specific enzyme transaldolase and heat shock protein αB-crystallin (56–63). We have recently identified a significantly higher frequency of MOG-reactive T cells in MS patients using MHC tetramers loaded with antigen (64), and antibodies to this protein have been recovered from plaques of MS patients (59, 61). However, the general consensus is that there is not one, but several pathological antigens involved in this disease. Additionally, although the initial autoreactivity may be specific to a particular antigen, the process of epitope spreading may expand the pool of specificities of activated immune cells to include multiple epitopes of the target antigen, and even additional antigens (65). Autoreactive T cells have been described in both healthy controls and patients with MS (66), and these naive autoreactive T cells may enter the inflamed CNS, where they are activated by local antigen-presenting cells and may contribute to secondary antigen-specific inflammation. The environmental trigger for MS is unknown, although epidemiologic data has identified numerous infectious and non-infectious factors associated with increased MS risk and microbial antigens are strongly suspected (67, 68). Potential cross-reactivity between epitopes of MBP, MOG, and transaldolase with microbial antigens has been demonstrated, suggesting that mechanisms of molecular mimicry may be responsible for the initial trigger (69, 70).
Genetics of MS
Much of our early understanding of heritability of the disease derived from epidemiological studies and disease concordance in twins or family members. However, limited by underpowered candidate gene studies and linkage analyses based on multiple-case families or matched case-controls, there was little significant advance for 30 years in discovering new genes that were significantly and reproducibly linked to MS besides the MHC locus on chromosome 6p21 (71). Our knowledge of MS genetics has expanded dramatically in the last 5 years based on fundamental advances in human genetics such as sequencing of the human genome and the generation of the haplotype map (HapMap).
Genome-wide association studies.
Association studies are an alternative approach to classic linkage analyses and have greater statistical power to detect variants that confer a modest risk of disease (72, 73). Using this approach, genome-wide association studies (GWASs) using SNPs from the HapMap project allowed the use of an unbiased approach in scanning the whole genome and identifying SNPs associated with disease (74, 75). The initial GWAS further localized the MHC risk locus to HLA DRB*1501, clearly identified additional susceptibility loci in IL2RA and IL7RA, and identified several other gene regions that have since been replicated (76–78). IL2R and IL7R are of particular interest because they are expressed on regulatory T cells, which have been strongly implicated in the regulation of the immune response in MS (66). Eight additional GWASs and a meta-analysis were performed and, in total, identified approximately 14 regions with genome-wide significance (77, 79–85), including several previously identified associations; among these are CD58, IL2RA, EVI5, CLEC16A, IL7RA, and TYK2 (77) and TNFRSF1A, IRF8, and CD6 (78).
In August of 2011, an international collaborative GWAS with over 9,000 cases of MS replicated many of these previously suggested associations and identified an additional 29 novel susceptibility loci — identifying approximately half of the genetic risk for MS (ref. 86 and Figure 3). Moreover, these SNPs can be analyzed with respect to the likely or known function of nearby genes. There is significant enrichment of genes linked to lymphocyte function, particularly genes with a role in T cell activation and proliferation, cytokine pathways, co-stimulatory molecules, and signal transduction. The extensive genetic information gained from the GWASs of the past five years, in combination with novel methods of pathway and network analysis of risk variants, has opened the door to explaining defects in immune regulation from allelic differences.
Figure 3. Genome regions showing association with MS.
Evidence for association from linear mixed model analysis of the discovery data (thresholded at a –log10 P value of 12) is shown at left. Non-MHC regions containing associated SNPs are indicated in red and labeled with the rs number (green text for newly identified loci, black text for loci with strong evidence of association, and gray text for previously reported loci) and risk allele of the most significant SNP. Asterisks indicate that the locus contains a secondary SNP signal. Odds ratios (ORs; diamonds) and 95% confidence intervals (whiskers) are estimated from a meta-analysis of discovery and replication data (+ indicates estimates for previously known loci from discovery data only). Risk allele frequency estimates in the control populations are indicated by vertical bars (scale of 0 to 1, left to right). A candidate gene and the number of genes are reported for each region of association. Black dots indicate that the candidate gene is physically the nearest gene included in the GO immune system process term. “Tags functional SNP” indicates whether the most-significant SNP tags a SNP predicted to affect the function of the candidate gene. Where such a SNP exists, the gene is selected as the candidate gene; otherwise, the nearest gene is selected unless there are strong biological reasons for a different choice. The final column indicates whether SNPs are correlated (r2 > 0.1) with SNPs associated with other autoimmune diseases. CeD, celiac disease; CrD, Crohn’s disease; PS, psoriasis; RA, rheumatoid arthritis; T1D, type 1 diabetes; UC, ulcerative colitis. Reproduced with permission from Nature (86).
Immune dysregulation.
The next step in linking allelic variants to mechanisms of immune dysregulation is characterizing phenotypic differences associated with variants in disease states. Several allelic variants in genes for cytokine receptors and co-stimulatory molecules have been associated with defects in Treg homeostasis. Our group and others have previously described that despite normal frequency of Tregs in MS patients compared to healthy controls, there is a loss of functional suppression by Tregs in response to autoreactive T cells (66, 87). Treg functional plasticity may also contribute to the autoimmune disease, as several studies have demonstrated that Tregs can produce the inflammatory cytokine IL-17 under certain conditions, such as in the presence of IL-1β and IL-6 (88–90). When investigating patients with MS, we have observed that patients with RRMS have an increased frequency of Th1 type, IFN-γ+ secreting Tregs compared to healthy controls (Figure 4 and ref. 91). These human Tregs acquire a Th1-Treg effector phenotype when stimulated in the presence of IL-12, and this phenotype observed in the in vitro–derived Th1-Treg cells was confirmed in ex vivo Tregs from untreated RRMS patients (91). The role of IL-12 in inducing this population in vivo was suggested by the observation that IFN-β treatment in RRMS patients, which in addition to having other biologic effects can decrease IL-12 levels, normalized the frequency of IFN-γ+Foxp3+ Tregs to that of healthy controls. Finally, a similar observation was made in patients with type 1 diabetes (92). Thus Treg reprogramming to a newly described Th1 type of IFN-γ+Foxp3+ Tregs may provide a new biomarker and raises the issue as to whether this reprogramming may play a role in disease pathogenesis.
Figure 4. Tregs from individuals with RRMS secrete IFN-γ ex vivo.
(A) The frequency of FACS-sorted IFN-γ+ and IL-17+ Tregs in healthy control individuals (left) and untreated individuals with RRMS (middle, n = 17) gated on Foxp3+ Tregs. Results are shown at right from a purity analysis of the sorted IFN-γ+Foxp3+ and IFN-γ–Foxp3+ populations from subjects with RRMS used for the methylation analysis described in C. (B) Percentage of IFN-γ+Foxp3+ and IL-17+Foxp3+ Tregs (n = 17) as a proportion of total Foxp3+ Tregs. (C) Representative example of methylation analysis of the Treg cell-specific demethylated region of the FOXP3 locus in sorted IFN-γ+Foxp3+ and IFN-γ–Foxp3+ Tregs from subjects with RRMS. An analysis of IFN-γ+Foxp3– memory T cells from subjects with RRMS is shown as a control. (D) Proliferation of responder T cells (Tresp) cultured with ex vivo FACS-sorted Tregs from healthy control subjects and untreated subjects with MS (Treg/Tresp ratio of 1:2) in the presence or absence of an IFN-γ–specific antibody (n = 4). (E) The frequency of IFN-γ+ and IL-17+ Tregs in healthy control subjects (left) or IFN-β–treated patients with RRMS (right) as assessed by intracellular cytokine staining and FACS analysis. The bar diagram (right) shows the percentage of IFN-γ+Foxp3+ and IL-17+Foxp3+ cells as a proportion of total Foxp3+ Tregs in healthy controls or IFN-β–treated patients with RRMS (n = 12). Reproduced with permission from Nature Medicine (91).
Another aspect of immune dysregulation relates to defects in co-stimulatory molecules acting as negative regulators, including the CD226/T cell Ig and ITIM domain (CD226/TIGIT) and CD58/CD2 pathways and TIM-3, a Th1-associated surface molecule. CD226 and co-inhibitory molecule TIGIT may behave similarly to the well-known CD28/CTLA4 pathway (93), in which both CD28 and CTLA4 can bind the same B7 molecules on antigen-presenting cells but transmit excitatory and inhibitory signals, respectively. CD226 has been genetically linked to MS susceptibility, although it is not yet known how the risk allele may affect modulation of T cell activity via interaction with TIGIT (94). The co-stimulatory molecule CD58/LFA-3 engages CD2 to co-stimulate T cell receptor signaling, including Treg activation (95). Increased levels of CD58 appear to be protective for MS (96), likely because the increased CD2 co-stimulation promotes FoxP3 expression and improves Treg suppressive activity (97, 98). Defects in CD58 expression with reduced CD2 co-stimulation might explain the low FoxP3 expression levels observed in MS patients (99). Finally, TIM-3 also acts as a negative regulator by modulating Th1 and Th17 cytokine secretion, and loss of TIM-3 T cell function regulation has been described in MS patients (100, 101).
Comparison among autoimmune diseases.
As GWASs began to identify risk alleles in other autoimmune diseases, it became clear that there were genes shared in common with MS, and that many of these were strongly associated with immune pathways. Given their shared genetic architecture, it is perhaps not surprising that MS and other autoimmune diseases share common defects in immune function and regulation, such as defective Treg suppression, which has been described in type 1 diabetes, systemic lupus erythematosus, psoriasis, and rheumatoid arthritis (102–105). A recent study confirmed the commonality of genetic associations across seven autoimmune diseases and identified clusters of genes that associated with multiple, but not necessarily all, of the diseases (106). These gene clusters can be linked to pathways and networks based on published protein-protein interactions (107, 108). Some of these networks seem to be affected in multiple diseases, while others are specific to just one or two. A feasible model of the interconnected mechanisms of autoimmune disease pathogenesis may be on the horizon.
The future of MS
How can physicians make the best use of this “periodic table of MS” derived from genetic analyses to better understand the disease? First, we should consider the likelihood of predicting disease development based on a panel of risk alleles, particularly in families known to have a history of MS or autoimmune disorders. So far, studies using a genetic profile of SNPs to estimate disease risk have demonstrated a modest, but incomplete, ability to predict disease (109–111). While using a larger gene set can improve discriminatory accuracy, the application does not yet allow for distinguishing between similar disease processes. Resolution of accuracy may improve with a more strongly powered study, as with 10,000 cases and 10,000 controls, yet it is likely that the predictive accuracy of the model will be insufficient without also including potential environmental triggers, such as vitamin D levels or EBV serostatus, to better understand how these factors modulate risk (36, 112). Currently one of the best models of predicting a patient’s risk of developing MS is simply taking a family history and noting whether family members have also suffered from MS or other autoimmune diseases. Other possibilities, such as predicting disease course or severity, have so far not been successful using datasets from previous GWASs (113).
Second, we should consider how the identification of pathways associated with autoimmune disease risk alleles will affect our treatment decisions for MS as a disease, and for patients as individuals based on their particular pattern of risk alleles. It may finally be feasible to predict drug response based on genetic variants with our expanding knowledge of individual allelic variants. Two recent studies have already examined how genotypes can relate to treatment response. Specific HLA-DRB1 genotypes were linked to the risk of developing neutralizing antibodies to recombinant IFN-β during therapy, and thus to ineffective treatment (114). SNPs have also been examined in IRF5, a master regulator of IFN activity, and one SNP was linked to poor IFN-β therapy response as measured by increased T2 lesions relative to other genotypes (115). Further studies are likely to uncover more associations between genetic variants and treatment response. The future MS physician will be able to use this information to personalize the treatment plan to the patient and choose the most appropriate immunomodulatory treatment to minimize relapses and delay progressive degeneration early in the disease course.
Finally, we should continue to pursue the allelic variation that accounts for the final portion of MS disease risk. Total genome sequencing of patients and controls will prove to be the next major technique in elucidating the remaining allelic variants, and the next major challenge will be developing better analytic methods to improve discrimination between neutral and disease-contributing variants. Sequencing can act in lieu of fine-mapping to elucidate many of the remaining common variants, and in the appropriate populations (i.e., families with a strong history of MS and other autoimmune diseases) may also bring to light rare variants that have been theorized to contribute to disease risk (116). Success will depend on international collaborations and consortia, exemplified by the International Multiple Sclerosis Genetics Consortium, automation of robotics systems to reduce intersystem variability, and interdisciplinary studies creating genotype-phenotype maps to better understand MS pathogenesis, prognosis, and potential therapeutic approaches.
Acknowledgments
We would like to thank Kevin O’Connor and Chris Cotsapas for helpful discussions and critical reading of the manuscript. This work was supported by NIH grants U19 AI07035, P01 AI07374, P01 AI04575, RC2 NS07034, CA 1061-A-18, U19 AI08999, R01 AI09156, P01 AI03967, the Nancy Taylor Foundation for Chronic Diseases, and NIH Medical Scientist Training Program grant TG T32GM07205.
Footnotes
Conflict of interest: David A. Hafler has served as a consultant for Biogen Idec Inc., Sanofi-Aventis Inc., Novartis Pharmaceuticals, Teva Neuroscience, and Pfizer.
Citation for this article: J Clin Invest. 2012;122(4):1180–1188. doi:10.1172/JCI58649.
References
- 1.Hafler DA. Multiple sclerosis. J Clin Invest. 2004;113(6):788–794. doi: 10.1172/JCI21357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones JL, et al. Improvement in disability after alemtuzumab treatment of multiple sclerosis is associated with neuroprotective autoimmunity. Brain. 2010;133(pt 8):2232–2247. doi: 10.1093/brain/awq176. [DOI] [PubMed] [Google Scholar]
- 3.Gunnarsson M, et al. Axonal damage in relapsing multiple sclerosis is markedly reduced by natalizumab. Ann Neurol. 2011;69(1):83–89. doi: 10.1002/ana.22247. [DOI] [PubMed] [Google Scholar]
- 4.Miller DH, Leary SM. Primary-progressive multiple sclerosis. Lancet Neurol. 2007;6(10):903–912. doi: 10.1016/S1474-4422(07)70243-0. [DOI] [PubMed] [Google Scholar]
- 5.McDonald WI, et al. Recommended diagnostic criteria for multiple sclerosis: guidelines from the International Panel on the diagnosis of multiple sclerosis. Ann Neurol. 2001;50(1):121–127. doi: 10.1002/ana.1032. [DOI] [PubMed] [Google Scholar]
- 6.Polman CH, et al. Diagnostic criteria for multiple sclerosis:2005 revisions to the “McDonald Criteria”. Ann Neurol. 2005;58(6):840–846. doi: 10.1002/ana.20703. [DOI] [PubMed] [Google Scholar]
- 7.Polman CH, et al. Diagnostic criteria for multiple sclerosis:2010 revisions to the McDonald criteria. Ann Neurol. 2011;69(2):292–302. doi: 10.1002/ana.22366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Filippi M, Rocca MA. MR imaging of multiple sclerosis. Radiology. 2011;259(3):659–681. doi: 10.1148/radiol.11101362. [DOI] [PubMed] [Google Scholar]
- 9.Lassmann H, Bruck W, Lucchinetti CF. The immunopathology of multiple sclerosis: an overview. Brain Pathol. 2007;17(2):210–218. doi: 10.1111/j.1750-3639.2007.00064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Babbe H, et al. Clonal expansions of CD8(+) T cells dominate the T cell infiltrate in active multiple sclerosis lesions as shown by micromanipulation and single cell polymerase chain reaction. J Exp Med. 2000;192(3):393–404. doi: 10.1084/jem.192.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Booss J, Esiri MM, Tourtellotte WW, Mason DY. Immunohistological analysis of T lymphocyte subsets in the central nervous system in chronic progressive multiple sclerosis. J Neurol Sci. 1983;62(1–3):219–232. doi: 10.1016/0022-510X(83)90201-0. [DOI] [PubMed] [Google Scholar]
- 12.Wucherpfennig KW, Newcombe J, Li H, Keddy C, Cuzner ML, Hafler DA. Gamma delta T-cell receptor repertoire in acute multiple sclerosis lesions. Proc Natl Acad Sci U S A. 1992;89(10):4588–4592. doi: 10.1073/pnas.89.10.4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prineas JW, Graham JS. Multiple sclerosis: capping of surface immunoglobulin G on macrophages engaged in myelin breakdown. Ann Neurol. 1981;10(2):149–158. doi: 10.1002/ana.410100205. [DOI] [PubMed] [Google Scholar]
- 14.Filippi M, Rossi P, Campi A, Colombo B, Pereira C, Comi G. Serial contrast-enhanced MR in patients with multiple sclerosis and varying levels of disability. AJNR Am J Neuroradiol. 1997;18(8):1549–1556. [PMC free article] [PubMed] [Google Scholar]
- 15.Bo L, Vedeler CA, Nyland HI, Trapp BD, Mork SJ. Subpial demyelination in the cerebral cortex of multiple sclerosis patients. J Neuropathol Exp Neurol. 2003;62(7):723–732. doi: 10.1093/jnen/62.7.723. [DOI] [PubMed] [Google Scholar]
- 16.Kutzelnigg A, et al. Cortical demyelination and diffuse white matter injury in multiple sclerosis. Brain. 2005;128(pt 11):2705–2712. doi: 10.1093/brain/awh641. [DOI] [PubMed] [Google Scholar]
- 17.Trapp BD, Peterson J, Ransohoff RM, Rudick R, Mork S, Bo L. Axonal transection in the lesions of multiple sclerosis. N Engl J Med. 1998;338(5):278–285. doi: 10.1056/NEJM199801293380502. [DOI] [PubMed] [Google Scholar]
- 18.Peterson JW, Bo L, Mork S, Chang A, Trapp BD. Transected neurites, apoptotic neurons, and reduced inflammation in cortical multiple sclerosis lesions. Ann Neurol. 2001;50(3):389–400. doi: 10.1002/ana.1123. [DOI] [PubMed] [Google Scholar]
- 19.Magliozzi R, et al. Meningeal B-cell follicles in secondary progressive multiple sclerosis associate with early onset of disease and severe cortical pathology. Brain. 2007;130(pt 4):1089–1104. doi: 10.1093/brain/awm038. [DOI] [PubMed] [Google Scholar]
- 20.Androdias G, Reynolds R, Chanal M, Ritleng C, Confavreux C, Nataf S. Meningeal T cells associate with diffuse axonal loss in multiple sclerosis spinal cords. Ann Neurol. 2010;68(4):465–476. doi: 10.1002/ana.22054. [DOI] [PubMed] [Google Scholar]
- 21.Popescu BF, Bunyan RF, Parisi JE, Ransohoff RM, Lucchinetti CF. A case of multiple sclerosis presenting with inflammatory cortical demyelination. Neurology. 2011;76(20):1705–1710. doi: 10.1212/WNL.0b013e31821a44f1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Calabrese M, Gallo P. Magnetic resonance evidence of cortical onset of multiple sclerosis. Mult Scler. 2009;15(8):933–941. doi: 10.1177/1352458509106510. [DOI] [PubMed] [Google Scholar]
- 23.Compston A, Coles A. Multiple sclerosis. Lancet. 2008;372(9648):1502–1517. doi: 10.1016/S0140-6736(08)61620-7. [DOI] [PubMed] [Google Scholar]
- 24.Bo L, Geurts JJ, van der Valk P, Polman C, Barkhof F. Lack of correlation between cortical demyelination and white matter pathologic changes in multiple sclerosis. Arch Neurol. 2007;64(1):76–80. doi: 10.1001/archneur.64.1.76. [DOI] [PubMed] [Google Scholar]
- 25.Lucchinetti C, Bruck W, Parisi J, Scheithauer B, Rodriguez M, Lassmann H. Heterogeneity of multiple sclerosis lesions: implications for the pathogenesis of demyelination. Ann Neurol. 2000;47(6):707–717. doi: 10.1002/1531-8249(200006)47:6<707::AID-ANA3>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 26.Confavreux C, Vukusic S. Age at disability milestones in multiple sclerosis. Brain. 2006;129(pt 3):595–605. doi: 10.1093/brain/awh714. [DOI] [PubMed] [Google Scholar]
- 27.Dziedzic T, et al. Wallerian degeneration: a major component of early axonal pathology in multiple sclerosis. Brain Pathol. 2010;20(5):976–985. doi: 10.1111/j.1750-3639.2010.00401.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith KJ, Lassmann H. The role of nitric oxide in multiple sclerosis. Lancet Neurol. 2002;1(4):232–241. doi: 10.1016/S1474-4422(02)00102-3. [DOI] [PubMed] [Google Scholar]
- 29.Campbell GR, et al. Mitochondrial DNA deletions and neurodegeneration in multiple sclerosis. Ann Neurol. 2011;69(3):481–492. doi: 10.1002/ana.22109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anderson AC, et al. Promotion of tissue inflammation by the immune receptor Tim-3 expressed on innate immune cells. Science. 2007;318(5853):1141–1143. doi: 10.1126/science.1148536. [DOI] [PubMed] [Google Scholar]
- 31.Kabat EA, Glusman M, Knaub V. Quantitative estimation of the albumin and gamma globulin in normal and pathologic cerebrospinal fluid by immunochemical methods. Am J Med. 1948;4(5):653–662. doi: 10.1016/S0002-9343(48)90389-1. [DOI] [PubMed] [Google Scholar]
- 32.Hauser SL, et al. B-cell depletion with rituximab in relapsing-remitting multiple sclerosis. N Engl J Med. 2008;358(7):676–688. doi: 10.1056/NEJMoa0706383. [DOI] [PubMed] [Google Scholar]
- 33.Serafini B, Rosicarelli B, Magliozzi R, Stigliano E, Aloisi F. Detection of ectopic B-cell follicles with germinal centers in the meninges of patients with secondary progressive multiple sclerosis. Brain Pathol. 2004;14(2):164–174. doi: 10.1111/j.1750-3639.2004.tb00049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Magliozzi R, et al. A Gradient of neuronal loss and meningeal inflammation in multiple sclerosis. Ann Neurol. 2010;68(4):477–493. doi: 10.1002/ana.22230. [DOI] [PubMed] [Google Scholar]
- 35.Willis SN, et al. Epstein-Barr virus infection is not a characteristic feature of multiple sclerosis brain. Brain. 2009;132(pt 12):3318–3328. doi: 10.1093/brain/awp200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thacker EL, Mirzaei F, Ascherio A. Infectious mononucleosis and risk for multiple sclerosis: a meta-analysis. Ann Neurol. 2006;59(3):499–503. doi: 10.1002/ana.20820. [DOI] [PubMed] [Google Scholar]
- 37.Lassmann H, Niedobitek G, Aloisi F, Middeldorp JM. Epstein-Barr virus in the multiple sclerosis brain: a controversial issue--report on a focused workshop held in the Centre for Brain Research of the Medical University of Vienna, Austria. Brain. 2011;134(pt 9):2772–2786. doi: 10.1093/brain/awr197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lunemann JD, et al. Increased frequency and broadened specificity of latent EBV nuclear antigen-1-specific T cells in multiple sclerosis. Brain. 2006;129(pt 6):1493–1506. doi: 10.1093/brain/awl067. [DOI] [PubMed] [Google Scholar]
- 39.Lunemann JD, et al. EBNA1-specific T cells from patients with multiple sclerosis cross react with myelin antigens and co-produce IFN-gamma and IL-2. J Exp Med. 2008;205(8):1763–1773. doi: 10.1084/jem.20072397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Serafini B, et al. Dysregulated Epstein-Barr virus infection in the multiple sclerosis brain. J Exp Med. 2007;204(12):2899–2912. doi: 10.1084/jem.20071030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lovato L, et al. Related B cell clones populate the meninges and parenchyma of patients with multiple sclerosis. Brain. 2011;134(pt 2):534–541. doi: 10.1093/brain/awq350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Obermeier B, et al. Related B cell clones that populate the CSF and CNS of patients with multiple sclerosis produce CSF immunoglobulin. J Neuroimmunol. 2011;233(1–2):245–248. doi: 10.1016/j.jneuroim.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Piccio L, et al. Changes in B- and T-lymphocyte and chemokine levels with rituximab treatment in multiple sclerosis. Arch Neurol. 2010;67(6):707–714. doi: 10.1001/archneurol.2010.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Polman CH, et al. A randomized, placebo-controlled trial of natalizumab for relapsing multiple sclerosis. N Engl J Med. 2006;354(9):899–910. doi: 10.1056/NEJMoa044397. [DOI] [PubMed] [Google Scholar]
- 45.Yousry TA, et al. Evaluation of patients treated with natalizumab for progressive multifocal leukoencephalopathy. N Engl J Med. 2006;354(9):924–933. doi: 10.1056/NEJMoa054693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Steinman L. A brief history of T(H)17, the first major revision in the T(H)1/T(H)2 hypothesis of T cell-mediated tissue damage. Nat Med. 2007;13(2):139–145. doi: 10.1038/nm1551. [DOI] [PubMed] [Google Scholar]
- 47.Reboldi A, et al. C-C chemokine receptor 6-regulated entry of TH-17 cells into the CNS through the choroid plexus is required for the initiation of EAE. Nat Immunol. 2009;10(5):514–523. doi: 10.1038/ni.1716. [DOI] [PubMed] [Google Scholar]
- 48.Codarri L, et al. RORgammat drives production of the cytokine GM-CSF in helper T cells, which is essential for the effector phase of autoimmune neuroinflammation. Nat Immunol. 2011;12(6):560–567. doi: 10.1038/ni.2027. [DOI] [PubMed] [Google Scholar]
- 49.El-Behi M, et al. The encephalitogenicity of T(H)17 cells is dependent on IL-1- and IL-23-induced production of the cytokine GM-CSF. Nat Immunol. 2011;12(6):568–575. doi: 10.1038/ni.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kebir H, et al. Human TH17 lymphocytes promote blood-brain barrier disruption and central nervous system inflammation. Nat Med. 2007;13(10):1173–1175. doi: 10.1038/nm1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Toyoda H, Formby B. Contribution of T cells to the development of autoimmune diabetes in the NOD mouse model. Bioessays. 1998;20(9):750–757. doi: 10.1002/(SICI)1521-1878(199809)20:9<750::AID-BIES8>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 52.Esplugues E, et al. Control of TH17 cells occurs in the small intestine. Nature. 2011;475(7357):514–518. doi: 10.1038/nature10228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vincent A. Unravelling the pathogenesis of myasthenia gravis. Nat Rev Immunol. 2002;2(10):797–804. doi: 10.1038/nri916. [DOI] [PubMed] [Google Scholar]
- 54.Lennon VA, et al. A serum autoantibody marker of neuromyelitis optica: distinction from multiple sclerosis. Lancet. 2004;364(9451):2106–2112. doi: 10.1016/S0140-6736(04)17551-X. [DOI] [PubMed] [Google Scholar]
- 55.Lennon VA, Kryzer TJ, Pittock SJ, Verkman AS, Hinson SR. IgG marker of optic-spinal multiple sclerosis binds to the aquaporin-4 water channel. J Exp Med. 2005;202(4):473–477. doi: 10.1084/jem.20050304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cruz M, Olsson T, Ernerudh J, Hojeberg B, Link H. Immunoblot detection of oligoclonal anti-myelin basic protein IgG antibodies in cerebrospinal fluid in multiple sclerosis. Neurology. 1987;37(9):1515–1519. doi: 10.1212/wnl.37.9.1515. [DOI] [PubMed] [Google Scholar]
- 57.Warren KG, Catz I. Autoantibodies to myelin basic protein within multiple sclerosis central nervous system tissue. J Neurol Sci. 1993;115(2):169–176. doi: 10.1016/0022-510X(93)90221-J. [DOI] [PubMed] [Google Scholar]
- 58.Greer JM, Pender MP. Myelin proteolipid protein: an effective autoantigen and target of autoimmunity in multiple sclerosis. J Autoimmun. 2008;31(3):281–287. doi: 10.1016/j.jaut.2008.04.018. [DOI] [PubMed] [Google Scholar]
- 59.McLaughlin KA, et al. Age-dependent B cell autoimmunity to a myelin surface antigen in pediatric multiple sclerosis. J Immunol. 2009;183(6):4067–4076. doi: 10.4049/jimmunol.0801888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Raine CS, Cannella B, Hauser SL, Genain CP. Demyelination in primate autoimmune encephalomyelitis and acute multiple sclerosis lesions: a case for antigen-specific antibody mediation. Ann Neurol. 1999;46(2):144–160. doi: 10.1002/1531-8249(199908)46:2<144::AID-ANA3>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 61.O’Connor KC, et al. Antibodies from inflamed central nervous system tissue recognize myelin oligodendrocyte glycoprotein. J Immunol. 2005;175(3):1974–1982. doi: 10.4049/jimmunol.175.3.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Banki K, et al. Oligodendrocyte-specific expression and autoantigenicity of transaldolase in multiple sclerosis. J Exp Med. 1994;180(5):1649–1663. doi: 10.1084/jem.180.5.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van Noort JM, et al. The small heat-shock protein alpha B-crystallin as candidate autoantigen in multiple sclerosis. Nature. 1995;375(6534):798–801. doi: 10.1038/375798a0. [DOI] [PubMed] [Google Scholar]
- 64.O’Connor KC, et al. Self-antigen tetramers discriminate between myelin autoantibodies to native or denatured protein. Nat Med. 2007;13(2):211–217. doi: 10.1038/nm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McMahon EJ, Bailey SL, Castenada CV, Waldner H, Miller SD. Epitope spreading initiates in the CNS in two mouse models of multiple sclerosis. Nat Med. 2005;11(3):335–339. doi: 10.1038/nm1202. [DOI] [PubMed] [Google Scholar]
- 66.Viglietta V, Baecher-Allan C, Weiner HL, Hafler DA. Loss of functional suppression by CD4+CD25+ regulatory T cells in patients with multiple sclerosis. J Exp Med. 2004;199(7):971–979. doi: 10.1084/jem.20031579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ascherio A, Munger KL. Environmental risk factors for multiple sclerosis. Part I: the role of infection. Ann Neurol. 2007;61(4):288–299. doi: 10.1002/ana.21117. [DOI] [PubMed] [Google Scholar]
- 68.Ascherio A, Munger KL. Environmental risk factors for multiple sclerosis. Part II: Noninfectious factors. Ann Neurol. 2007;61(6):504–513. doi: 10.1002/ana.21141. [DOI] [PubMed] [Google Scholar]
- 69.Esposito M, et al. Human transaldolase and cross-reactive viral epitopes identified by autoantibodies of multiple sclerosis patients. J Immunol. 1999;163(7):4027–4032. [PubMed] [Google Scholar]
- 70.Wucherpfennig KW, Catz I, Hausmann S, Strominger JL, Steinman L, Warren KG. Recognition of the immunodominant myelin basic protein peptide by autoantibodies and HLA-DR2-restricted T cell clones from multiple sclerosis patients. Identity of key contact residues in the B-cell and T-cell epitopes. J Clin Invest. 1997;100(5):1114–1122. doi: 10.1172/JCI119622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sawcer S, et al. A high-density screen for linkage in multiple sclerosis. Am J Hum Genet. 2005;77(3):454–467. doi: 10.1086/444547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273(5281):1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 73.Yang Q, Khoury MJ, Friedman J, Little J, Flanders WD. How many genes underlie the occurrence of common complex diseases in the population? Int J Epidemiol. 2005;34(5):1129–1137. doi: 10.1093/ije/dyi130. [DOI] [PubMed] [Google Scholar]
- 74.Tishkoff SA, Verrelli BC. Role of evolutionary history on haplotype block structure in the human genome: implications for disease mapping. Curr Opin Genet Dev. 2003;13(6):569–575. doi: 10.1016/j.gde.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 75.International HapMap Consortium et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449(7164):851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.International Multiple Sclerosis Genetics Consortium et al. Risk alleles for multiple sclerosis identified by a genomewide study. N Engl J Med. 2007;357(9):851–862. doi: 10.1056/NEJMoa073493. [DOI] [PubMed] [Google Scholar]
- 77.Australia and New Zealand Multiple Sclerosis Genetics Consortium (ANZgene) Genome-wide association study identifies new multiple sclerosis susceptibility loci on chromosomes 12 and 20. Nat Genet. 2009;41(7):824–828. doi: 10.1038/ng.396. [DOI] [PubMed] [Google Scholar]
- 78.De Jager PL, et al. Meta-analysis of genome scans and replication identify CD6, IRF8 and TNFRSF1A as new multiple sclerosis susceptibility loci. Nat Genet. 2009;41(7):776–782. doi: 10.1038/ng.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wellcome Trust Case Control Consortium et al. Association scan of 14,500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nat Genet. 2007;39(11):1329–1337. doi: 10.1038/ng.2007.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Comabella M, et al. Identification of a novel risk locus for multiple sclerosis at 13q31.3 by a pooled genome-wide scan of 500,000 single nucleotide polymorphisms. PLoS One. 2008;3(10):e3490. doi: 10.1371/journal.pone.0003490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Baranzini SE, et al. Genome-wide association analysis of susceptibility and clinical phenotype in multiple sclerosis. Hum Mol Genet. 2009;18(4):767–778. doi: 10.1093/hmg/ddn388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jakkula E, et al. Genome-wide association study in a high-risk isolate for multiple sclerosis reveals associated variants in STAT3 gene. Am J Hum Genet. 2010;86(2):285–291. doi: 10.1016/j.ajhg.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sanna S, et al. Variants within the immunoregulatory CBLB gene are associated with multiple sclerosis. Nat Genet. 2010;42(6):495–497. doi: 10.1038/ng.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Aulchenko YS, et al. Genetic variation in the KIF1B locus influences susceptibility to multiple sclerosis. Nat Genet. 2008;40(12):1402–1403. doi: 10.1038/ng.251. [DOI] [PubMed] [Google Scholar]
- 85.Nischwitz S, et al. Evidence for VAV2 and ZNF433 as susceptibility genes for multiple sclerosis. J Neuroimmunol. 2010;227(1–2):162–166. doi: 10.1016/j.jneuroim.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 86.International Multiple Sclerosis Genetics Consortium et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476(7359):214–219. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Haas J, et al. Reduced suppressive effect of CD4+CD25high regulatory T cells on the T cell immune response against myelin oligodendrocyte glycoprotein in patients with multiple sclerosis. Eur J Immunol. 2005;35(11):3343–3352. doi: 10.1002/eji.200526065. [DOI] [PubMed] [Google Scholar]
- 88.Koenen HJ, Smeets RL, Vink PM, van Rijssen E, Boots AM, Joosten I. Human CD25highFoxp3pos regulatory T cells differentiate into IL-17-producing cells. Blood. 2008;112(6):2340–2352. doi: 10.1182/blood-2008-01-133967. [DOI] [PubMed] [Google Scholar]
- 89.Beriou G, et al. IL-17-producing human peripheral regulatory T cells retain suppressive function. Blood. 2009;113(18):4240–4249. doi: 10.1182/blood-2008-10-183251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ayyoub M, et al. Human memory FOXP3+ Tregs secrete IL-17 ex vivo and constitutively express the T(H)17 lineage-specific transcription factor RORgamma t. Proc Natl Acad Sci U S A. 2009;106(21):8635–8640. doi: 10.1073/pnas.0900621106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dominguez-Villar M, Baecher-Allan CM, Hafler DA. Identification of T helper type 1-like, Foxp3(+) regulatory T cells in human autoimmune disease. Nat Med. 2011;17(6):673–675. doi: 10.1038/nm.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.McClymont SA, et al. Plasticity of human regulatory T cells in healthy subjects and patients with type 1 diabetes. J Immunol. 2011;186(7):3918–3926. doi: 10.4049/jimmunol.1003099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Joller N, et al. Cutting edge: TIGIT has T cell-intrinsic inhibitory functions. J Immunol. 2011;186(3):1338–1342. doi: 10.4049/jimmunol.1003081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hafler JP, et al. CD226 Gly307Ser association with multiple autoimmune diseases. Genes Immun. 2009;10(1):5–10. doi: 10.1038/gene.2008.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Baecher-Allan CM, et al. CD2 costimulation reveals defective activity by human CD4+CD25(hi) regulatory cells in patients with multiple sclerosis. J Immunol. 2011;186(6):3317–3326. doi: 10.4049/jimmunol.1002502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.De Jager PL, et al. The role of the CD58 locus in multiple sclerosis. Proc Natl Acad Sci U S A. 2009;106(13):5264–5269. doi: 10.1073/pnas.0813310106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zheng Y, Josefowicz SZ, Kas A, Chu TT, Gavin MA, Rudensky AY. Genome-wide analysis of Foxp3 target genes in developing and mature regulatory T cells. Nature. 2007;445(7130):936–940. doi: 10.1038/nature05563. [DOI] [PubMed] [Google Scholar]
- 98.Marson A, et al. Foxp3 occupancy and regulation of key target genes during T-cell stimulation. Nature. 2007;445(7130):931–935. doi: 10.1038/nature05478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Huan J, et al. Decreased FOXP3 levels in multiple sclerosis patients. J Neurosci Res. 2005;81(1):45–52. doi: 10.1002/jnr.20522. [DOI] [PubMed] [Google Scholar]
- 100.Yang L, Anderson DE, Kuchroo J, Hafler DA. Lack of TIM-3 immunoregulation in multiple sclerosis. J Immunol. 2008;180(7):4409–4414. doi: 10.4049/jimmunol.180.7.4409. [DOI] [PubMed] [Google Scholar]
- 101.Koguchi K, Anderson DE, Yang L, O’Connor KC, Kuchroo VK, Hafler DA. Dysregulated T cell expression of TIM3 in multiple sclerosis. J Exp Med. 2006;203(6):1413–1418. doi: 10.1084/jem.20060210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.International Multiple Sclerosis Genetics Consortium (IMSGC) The expanding genetic overlap between multiple sclerosis and type I diabetes. Genes Immun. 2009;10(1):11–14. doi: 10.1038/gene.2008.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhernakova A, van Diemen CC, Wijmenga C. Detecting shared pathogenesis from the shared genetics of immune-related diseases. Nat Rev Genet. 2009;10(1):43–55. doi: 10.1038/nrg2489. [DOI] [PubMed] [Google Scholar]
- 104.Xavier RJ, Rioux JD. Genome-wide association studies: a new window into immune-mediated diseases. Nat Rev Immunol. 2008;8(8):631–643. doi: 10.1038/nri2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Torkamani A, Topol EJ, Schork NJ. Pathway analysis of seven common diseases assessed by genome-wide association. Genomics. 2008;92(5):265–272. doi: 10.1016/j.ygeno.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cotsapas C, et al. Pervasive sharing of genetic effects in autoimmune disease. PLoS Genet. 2011;7(8):e1002254. doi: 10.1371/journal.pgen.1002254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Baranzini SE, et al. Pathway and network-based analysis of genome-wide association studies in multiple sclerosis. Hum Mol Genet. 2009;18(11):2078–2090. doi: 10.1093/hmg/ddp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rossin EJ, et al. Proteins encoded in genomic regions associated with immune-mediated disease physically interact and suggest underlying biology. PLoS Genet. 2011;7(1):e1001273. doi: 10.1371/journal.pgen.1001273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sawcer S, Ban M, Wason J, Dudbridge F. What role for genetics in the prediction of multiple sclerosis? Annal Neurol. 2010;67(1):3–10. doi: 10.1002/ana.21911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang JH, et al. Modeling the cumulative genetic risk for multiple sclerosis from genome-wide association data. Genome Med. 2011;3(1):3. doi: 10.1186/gm217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.De Jager PL, et al. Integration of genetic risk factors into a clinical algorithm for multiple sclerosis susceptibility: a weighted genetic risk score. Lancet Neurol. 2009;8(12):1111–1119. doi: 10.1016/S1474-4422(09)70275-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nashold FE, Hoag KA, Goverman J, Hayes CE. Rag-1-dependent cells are necessary for 1,25-dihydroxyvitamin D(3) prevention of experimental autoimmune encephalomyelitis. J Neuroimmunol. 2001;119(1):16–29. doi: 10.1016/S0165-5728(01)00360-5. [DOI] [PubMed] [Google Scholar]
- 113.International Multiple Sclerosis Genetics Consortium Genome-wide association study of severity in multiple sclerosis. Genes Immun. 2011;12(8):615–625. doi: 10.1038/gene.2011.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Buck D, et al. Influence of the HLA-DRB1 genotype on antibody development to interferon beta in multiple sclerosis. Arch Neurol. 2011;68(4):480–487. doi: 10.1001/archneurol.2011.65. [DOI] [PubMed] [Google Scholar]
- 115.Vosslamber S, et al. Interferon regulatory factor 5 gene variants and pharmacological and clinical outcome of Interferonbeta therapy in multiple sclerosis. Genes Immun. 2011;12(6):466–472. doi: 10.1038/gene.2011.18. [DOI] [PubMed] [Google Scholar]
- 116.McClellan J, King MC. Genetic heterogeneity in human disease. Cell. 2010;141(2):210–217. doi: 10.1016/j.cell.2010.03.032. [DOI] [PubMed] [Google Scholar]