Abstract
Patients with triple-negative breast cancer (TNBC) — defined by lack of estrogen receptor and progesterone receptor expression as well as lack of human epidermal growth factor receptor 2 (HER2) amplification — have a poor prognosis. There is a need for targeted therapies to treat this condition. TNBCs frequently harbor mutations in TP53, resulting in loss of the G1 checkpoint and reliance on checkpoint kinase 1 (Chk1) to arrest cells in response to DNA damage. Previous studies have shown that inhibition of Chk1 in a p53-deficient background in response to DNA damage. We therefore tested whether inhibition of Chk1 could potentiate the cytotoxicity of the DNA damaging agent irinotecan in TNBC using xenotransplant tumor models. Tumor specimens from patients with TNBC were engrafted into humanized mammary fat pads of immunodeficient mice to create 3 independent human-in-mouse TNBC lines: 1 WT (WU-BC3) and 2 mutant for TP53 (WU-BC4 and WU-BC5). These lines were tested for their response to irinotecan and a Chk1 inhibitor (either UCN-01 or AZD7762), either as single agents or in combination. The combination therapy induced checkpoint bypass and apoptosis in WU-BC4 and WU-BC5, but not WU-BC3, tumors. Moreover, combination therapy inhibited tumor growth and prolonged survival of mice bearing the WU-BC4 line, but not the WU-BC3 line. In addition, knockdown of p53 sensitized WU-BC3 tumors to the combination therapy. These results demonstrate that p53 is a major determinant of how TNBCs respond to therapies that combine DNA damage with Chk1 inhibition.
Introduction
Triple-negative breast cancer (TNBC) lacks the expression of estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2) gene amplification and carries a particularly poor prognosis due to its association with aggressive tumor characteristics and the lack of effective targeted therapies. Interestingly, TP53 mutation is observed in up to 44% of TNBC compared with 15% in the more indolent ER-positive breast cancers (1). p53 plays a key role in arresting cell-cycle progression in the presence of genotoxic stress in order to maintain genome integrity. In response to DNA damage, normal cells arrest in G1 (via p53) to allow time for DNA repair or they proceed into apoptosis if the DNA damage is too severe. In contrast, p53-deficient tumor cells rely on checkpoint kinase 1 (Chk1) to arrest cell-cycle progression in the S and G2 phases. In response to replicative or genotoxic stress, Chk1 phosphorylates its key target, the Cdc25A phosphatase (2–5). This leads to ubiquitin-mediated proteolysis of Cdc25A and cell-cycle arrest (4–9). When the S and G2 checkpoints are abrogated by inhibition of Chk1, p53-deficient cancer cells undergo mitotic catastrophe and apoptosis (10–17).
Several preclinical studies have demonstrated that Chk1 inhibitors selectively potentiate the effects of DNA-damaging agents, such as chemotherapy or radiation, in TP53-mutated cancer cells, and several Chk1 inhibitors are being tested in clinical trials (18). Since TNBC is commonly associated with TP53 mutation, we hypothesized that a potential therapeutic strategy for treating TNBC would be to inhibit Chk1 to enhance the cytotoxicity of DNA-damaging agents. We tested this hypothesis by using 2 different Chk1 inhibitors (UCN-01 and AZD7762).
UCN-01 (7-hydroxystaurosporine) is a multitarget serine-threonine protein kinase inhibitor that potently inhibits Chk1 (IC50 = 10 nM) and was the first Chk1 inhibitor to be identified (14). UCN-01 exhibits preclinical synergy with DNA-damaging agents (19). AZD7762 is a newer generation, more selective Chk1 inhibitor. AZD7762 inhibits Chk1 by reversibly binding to the ATP-binding site of Chk1, with an IC50 of 5 nM and a KI of 3.6 nM (20).
In this study, we tested the hypothesis that loss of p53 function would exhibit synthetic lethality with DNA damage and Chk1 inhibition in TNBC. We predicted that inhibition of Chk1 would enhance the antitumor effects of irinotecan (DNA-damaging agent) by eliminating checkpoint responses selectively in tumors harboring TP53 mutations. We employed early passage human-in-mouse (HIM) models (21), which are patient tumor explants engrafted into the “humanized” mammary fat pads of immunocompromised mice. We denoted these HIM models as Washington University–breast cancer (WU-BC) tumor lines. Three TNBC HIM lines were chosen, 1 WT and 2 mutant for TP53. Mice engrafted with these tumors were treated with irinotecan and 2 different Chk1 inhibitors (UCN-01 and AZD7762) either as single agents or in combination for long-term survival and tumor growth studies as well as short-term pathway analysis. In addition, isogenic lines of WU-BC3 differing only in p53 status were generated, and the response of these lines to the combination of irinotecan and AZD7762 was assessed to determine the contribution made by p53 status to tumor response.
Results
Generation of HIM tumor models of TNBC.
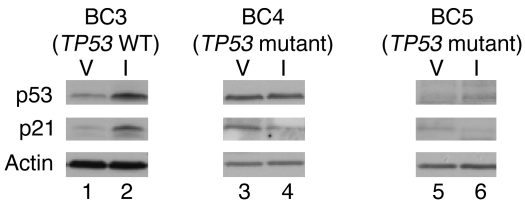
We employed methodologies first described by Kuperwasser et al. (21) to establish a panel of HIM breast cancer xenograft models in immunodeficient NOD/SCID mice. We selected 3 human TNBC HIM models (WU-BC3, WU-BC4, and WU-BC5) that differed in p53 status for our study. The WU-BC3 line was generated by engrafting the primary breast tumor of a patient with metastatic TNBC into the humanized mammary fat pad of NOD/SCID mice. DNA sequencing revealed that this tumor was WT for TP53. WU-BC4 was generated using an abdominal metastasis from a patient with TNBC. DNA sequencing revealed that this tumor encoded a homozygous R175H mutation in TP53. WU-BC5 was generated from the brain metastasis in a patient with TP53 mutant (S166S[insC]/S166S[insC]) TNBC and encoded a truncated p53 protein of approximately 18 kDa (data not shown). The functional integrity of the p53 pathway was assessed in each HIM line by determining whether DNA damage induced the accumulation of p53 and its downstream effector, p21. As seen in Figure 1, treatment of mice with irinotecan resulted in the stabilization of p53 and accumulation of p21 in WU-BC3 (TP53 WT) but not WU-BC4 (TP53 mutant) or WU-BC5 (TP53 mutant) tumor cells.
Figure 1. Functional integrity of p53 pathway in HIM models.
Mice harboring WU-BC3, WU-BC4, and WU-BC5 were treated with vehicle (V) or irinotecan (I), and tumors were isolated 24 hours later. Lysates were prepared and resolved by SDS-PAGE and analyzed for the indicated proteins by Western blotting. Note the increased baseline p53 expression due to the missense mutation in WU-BC4 (lane 3) compared with WU-BC3 (lane 1) and the absence of intact p53 protein due to the deletion mutation in WU-BC5 (lane 5).
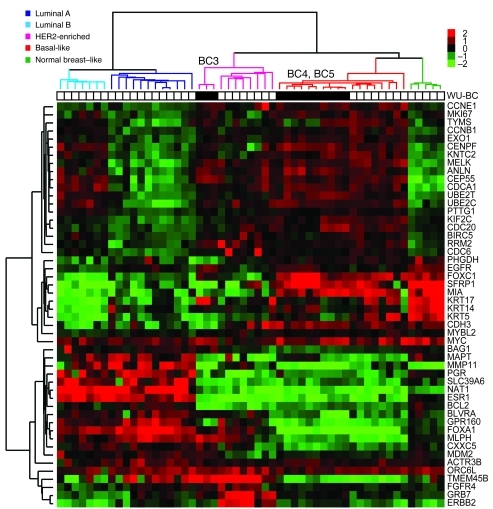
Gene-expression profiling and application of the PAM50 subtype–based predictor (22) categorized both WU-BC4 and WU-BC5 as basal like (Figure 2), the most common intrinsic molecular subtype of TNBC (23). WU-BC3 was identified as nonbasal TNBC and clustered most closely with the HER2-E subtype, but without HER2 overexpression (Figure 2). The HER2-E molecular subtype has been reported in 9% of TNBC (24). Importantly, tumors from different passages of the same HIM model clustered more closely with each other and with their original human counterpart than with any other tumor (Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI58765DS1).
Figure 2. Identification of the PAM50 intrinsic subtypes in the WU-BC models.
Hierarchical clustering of different passages of WU-BC3, WU-BC4, and WU-BC5, their human counterparts (for WU-BC4 and WU-BC5), an ovarian metastasis from a mouse harboring WU-BC5, and 40 breast samples representing all breast tumor subtypes, including the normal breast–like group (i.e., prototypic tumor samples), were analyzed. Black squares represent samples from the xenografts. White squares represent human specimens. Expression of the PAM50 genes was centered on the prototypic tumor samples. Columns, samples; rows, genes; red and green squares denote expression levels above or below the median, respectively. The magnitude of deviation from the median is represented by color saturation. The array and gene trees represent overall similarities in gene expression.
Chk1 inhibitors potentiated the apoptosis-inducing effects of irinotecan selectively in TP53 mutant tumors.
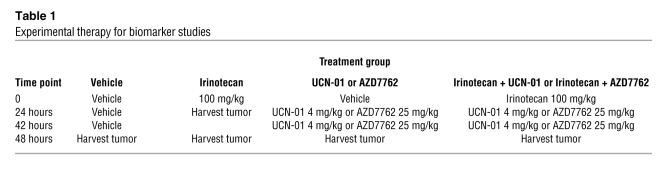
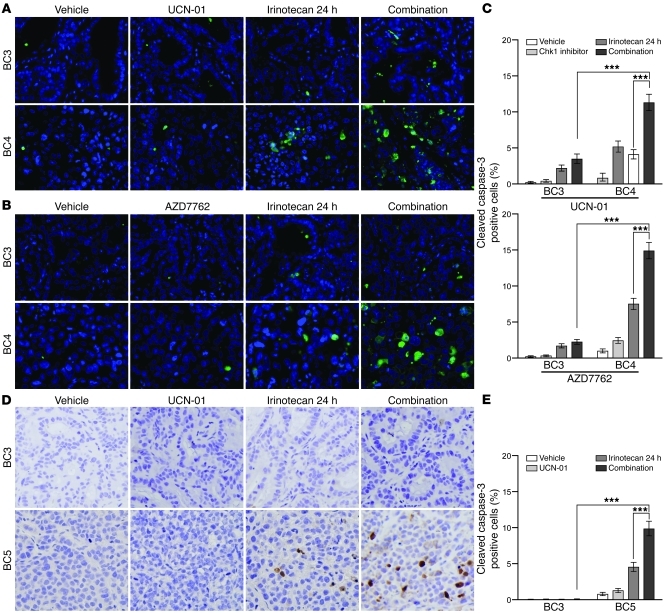
To determine how the TNBC HIM models differing in p53 status respond to DNA damage and/or Chk1 inhibition, mice bearing either WU-BC3 or WU-BC4 were randomly assigned to the treatment groups outlined in Table 1. These included vehicle (DMSO, i.p. injection at hours 0, 24, and 42), irinotecan (100 mg/kg i.p. at hour 0), Chk1 inhibitor (4 mg/kg UCN-01 i.p. or 25 mg/kg AZD7762 i.p. at hours 24 and 42), or a combination therapy of irinotecan at hour 0 followed by Chk1 inhibitor at hours 24 and 42. Two mice carrying 2 breast cancer xenografts each were subjected to 1 of these treatment regimens. One treatment group (irinotecan only) was sacrificed at hour 24, and the remaining treatment groups were sacrificed at hour 48. Tumors were processed for immunofluorescence (IF) staining and Western blotting. We first assessed the effect of therapy on the induction of apoptosis by monitoring for the appearance of cleaved caspase-3. Representative images of the IF staining for cleaved caspase-3 are shown in Figure 3, A and B, and quantitation is shown in Figure 3C. Compared with single agent alone (either irinotecan or Chk1 inhibitor), the combination therapy (irinotecan followed by either UCN-01 or AZD7762) produced a significantly greater increase in tumor cell apoptosis in TP53 mutant line WU-BC4 than in WU-BC3, the TP53 WT line (Figure 3C). The 2 Chk1 inhibitors (UCN-01 or AZD7762) were both effective at potentiating the apoptotic-inducing effects of irinotecan (Figure 3C).
Table 1 .
Experimental therapy for biomarker studies
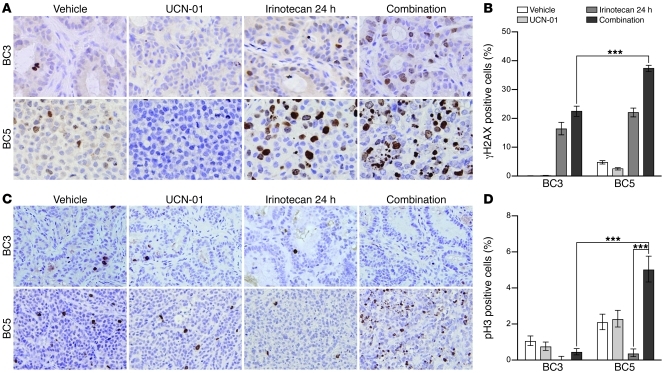
Figure 3. Chk1 inhibitors potentiate irinotecan-induced apoptosis selectively in TP53 mutant tumors.
Mice harboring WU-BC3, WU-BC4 or WU-BC5 TNBC were treated as indicated, and tumors were analyzed for cleaved caspase-3 by IF staining (A and B) or IHC staining (D). Representative images are shown in A, B, and D, with quantitations ± 95% CIs shown in C (for data in A and B) and E (for data in D), respectively. Original magnification, ×400. See Methods for experimental details. Statistics were obtained by the Wilson score method in SPSS 20. Notable comparisons are marked with ***P < 0.001.
The effect of Chk1 inhibition on irinotecan-induced apoptosis was also compared between WU-BC5 and WU-BC3 (Figure 3, D and E) following the same treatment and tumor-harvesting protocols as described above. Immunohistochemistry (IHC) of cleaved caspase-3 was performed. Tumor-bearing mice were treated with vehicle, irinotecan, UCN-01, or irinotecan followed by UCN-01. A significant induction of apoptosis following the combination therapy was observed in WU-BC5, but not in WU-BC3 (Figure 3, D and E). These results suggest that Chk1 inhibitors sensitize TP53 mutant TNBCs to the cytotoxic effects of irinotecan.
Chk1 inhibitors abrogated cell-cycle arrest and enhanced DNA-damaging effects of irinotecan selectively in the TP53 mutant tumors.
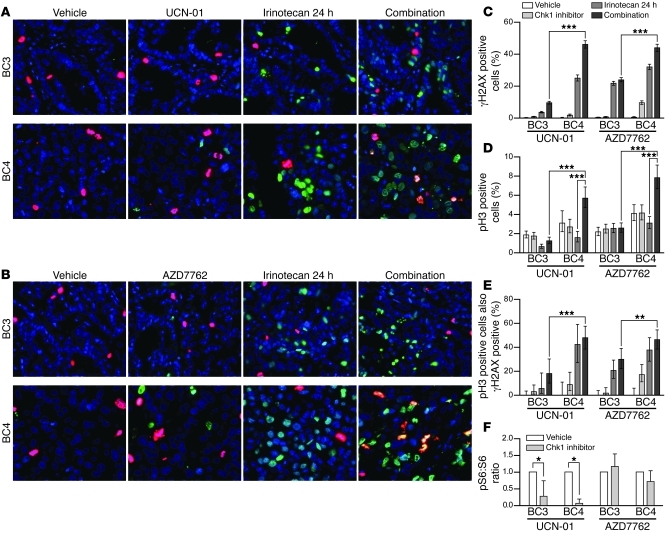
Since TP53 mutant cells rely on the function of Chk1 for S and G2 cell-cycle checkpoint regulation, the enhanced apoptotic effect of Chk1 inhibitors in combination with irinotecan in these cells could be explained by checkpoint abrogation (due to Chk1 inhibition) in the presence of DNA damage (induced by irinotecan). To test this hypothesis, we compared WU-BC3 and WU-BC4 for levels of γH2AX to assess DNA double-strand breaks (DSBs) (25) and phosphohistone H3 to identify cells in mitosis (26, 27) following the various treatment regimens. Representative IF images are shown in Figure 4, A and B, and quantitation in Figure 4, C–E. γH2AX staining was observed in approximately 5% to 30% of tumor cells from irinotecan-treated mice (Figure 4, A–C). Chk1 inhibitors alone (either UCN-01 or AZD7762) induced negligible or statistically insignificant levels of DNA DSBs in WU-BC3, and AZD7762 induced only modest DNA DSBs in WU-BC4 (Figure 4, A–C). However, combining irinotecan with either Chk1 inhibitor abrogated cell-cycle arrest selectively in the TP53 mutant tumor cells (WU-BC4), as indicated by the increase in the number of cells staining positive for phosphohistone H3 (Figure 4D). Importantly, approximately 50% of WU-BC4 staining positive for phosphohistone H3 (indicative of mitosis) also stained positive for γH2AX (Figure 4E). Thus, in the absence of a functional p53 pathway, TNBC cells under Chk1 inhibition moved into mitosis despite the fact that their genomes contained high levels of DNA DSBs.
Figure 4. Chk1 inhibitors enhance DNA damage and abrogate cell-cycle arrest induced by irinotecan selectively in p53 mutant tumors.
Mice harboring WU-BC3 and WU-BC4 TNBC were treated as indicated, and tumors were costained for phosphohistone H3 (pH3, red) and γH2AX (green). Representative IF images are shown in A and B, and quantitation ± 95% CIs are shown in C–E. Statistics were obtained by the Wilson score method in SPSS 20. **P < 0.01; ***P < 0.001. Original magnification, ×400. (F) Mice harboring WU-BC3 or WU-BC4 tumors were treated as indicated; tumors were lysed and analyzed by Western blotting with antibodies specific for S6 ribosomal protein, phosphorylated S6 ribosomal protein (pS6), and actin as a loading control. The ratio of phosphorylated S6 to total S6 protein was determined for each sample. Blots were developed using ECL detection reagent (GE Healthcare), and proteins were quantitated using ImageJ (53). Mean ratios are shown with 95% CIs. *P < 0.05.
Levels of phosphorylated ribosomal S6 protein (pS6) were also monitored, since UCN-01, but not AZD7762, is a potent 3-phosphoinositide–dependent protein kinase-1 (PDK1) inhibitor (28). As seen in Figure 4F, a significant reduction in pS6 staining was observed in UCN-01– but not AZD7762-treated HIMs, and this was independent of TP53 status. Therefore, the antitumor effect of UCN-01 is unlikely to be due to its ability to inhibit PDK1.
In a separate set of experiments, WU-BC3 and WU-BC5 were analyzed for levels of γH2AX and phosphohistone H3 by IHC staining after treating mice with either vehicle, irinotecan, UCN-01, or the combination of irinotecan and UCN-01. Abrogation of cell-cycle arrest and enhanced DNA damage were observed in TP53 mutant WU-BC5 cells, but not WU-BC3 (TP53 WT) cells in response to the combination therapy (Figure 5). These results demonstrated that the combination therapy leads to checkpoint abrogation selectively in TP53 mutant TNBCs.
Figure 5. Enhanced DNA damage and checkpoint bypass in p53-deficient TNBC treated with combination therapy.
Mice harboring WU-BC3 and WU-BC5 TNBC were treated as indicated, and tumors were stained for γH2AX (A) and phosphohistone H3 (C). Original magnification, ×400 (A); ×200 (C). Representative IHC images are shown in A and C; quantitation ± 95% CIs are shown in B and D. Statistics were obtained by the Wilson score method in SPSS 20. ***P < 0.001.
Chk1 inhibitors in combination with irinotecan improved host survival and reduced tumor growth selectively in TP53 mutant tumors.
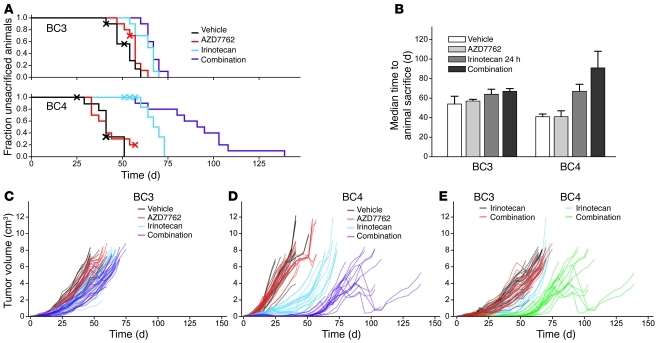
The enhanced apoptotic effect of Chk1 inhibitors in combination with irinotecan in TP53 mutant tumors was confirmed in survival studies. Mice bearing either WU-BC3 or WU-BC4 were treated with 4 cycles of vehicle, irinotecan, AZD7762, or the combination of irinotecan and AZD7762 (irinotecan on day 1 followed by AZD7762 on days 2 and 3 of a 5-day cycle). Mice were followed until death or were sacrificed if tumors reached 2 cm or mice experienced unacceptable toxicities. Most of the mice in the study were sacrificed due to tumor size reaching 2 cm. The longest survival and tumor growth suppression was observed in TP53 mutant WU-BC4–bearing mice in the combination therapy arm (Figure 6, A, B, and D). Survival and tumor growth rates observed in WU-BC3–bearing mice were similar regardless of treatment arms (Figure 6, A–C). Median time to animal sacrifice ± 95% CIs was calculated using Kaplan-Meier analysis, and pairwise significance values were calculated for the survival curves using the log-rank test. In the WU-BC4 treatment group (TP53 mutant), progression to animal death was significantly different between vehicle-treated controls and animals treated with irinotecan or the combination of irinotecan and AZD7762 (P = 0.0002 for both comparisons) (Figure 6B). These 2 treatments also varied significantly with each other in delaying tumor growth for WU-BC4 but not WU-BC3 (P = 0.006) (Figure 6, C–E). These data validate the short-term biomarker studies and suggest that combining DNA damage with Chk1 inhibition is an effective antitumor strategy for TP53 mutant TNBC.
Figure 6. Effect of combination therapy on tumor growth and host survival.
Tumor-bearing NOD/SCID mice were randomly assigned to the indicated treatment groups (vehicle, irinotecan, AZD7762, or the combination of irinotecan and AZD7762; n = 10 per group). Mice were followed until death or sacrificed if tumor size reached 2 cm or mice experienced unacceptable toxicities. (A) Kaplan-Meier survival curve following therapy initiation using animal sacrifice as the terminal event. Animals dying of other causes were right-censored from the analysis at the observed time of death and are marked on the survival curves with an X. (B) Median time to animal sacrifice for mice treated in each treatment arm. (C and D) Tumor growth following therapy initiation for individual mice in each treatment group for WU-BC3 and WU-BC4, respectively. (E) Tumor growth curve for individual mice treated with either irinotecan or the combination of irinotecan and AZD7762.
Since growth rate could potentially affect tumor response to the combination therapy, we compared tumor volume changes over time after engraftment of WU-BC3, WU-BC4, and WU-BC5 cells into the humanized mammary fat pads of mice (Supplemental Figure 2). Initially, WU-BC4 grew at a slower rate than WU-BC3 and WU-BC5, but by the time of therapeutic treatment (day 65 for WU-BC3 and WU-BC5 and day 76 for WU-BC4), the growth rate of all 3 tumor lines was similar.
Knockdown of p53 sensitizes WU-BC3 tumors to combination therapy.
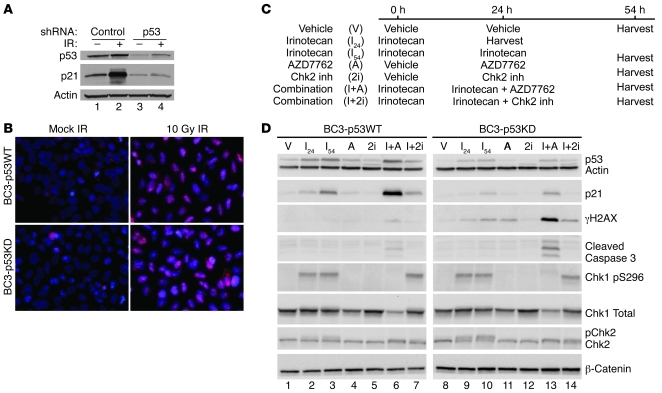
Although TP53 can be sequenced in each HIM line and the functional integrity of various pathways assessed, it is impossible to know all of the additional genetic changes present in each tumor model without a comprehensive analysis of the genome, epigenome, and transcriptome. Therefore, we generated isogenically matched HIM lines differing only in p53 status. In this way, response to therapy could be directly correlated with TP53 status. To accomplish this, WU-BC3 cells were infected with control retroviruses or retroviruses encoding p53-specific shRNAs (29) to generate BC3-p53WT and BC3-p53KD, respectively. As seen in Figure 7A, significant knockdown of p53 was achieved in BC3-p53KD cells (compare lanes 1 and 3). Cells were also exposed to 10 Gy ionizing radiation (IR) to assess functionality of the p53 DNA damage response pathway in each WU-BC3 line. A robust accumulation of p21 was observed in irradiated BC3-p53WT (lane 2) but not in BC3-p53KD cells (lane 4), verifying that p53 function was impaired in BC3-p53KD cells. Since defects in homologous recombination repair (HRR) could alter the sensitivity of TNBC cells to DNA-damaging agent, we assessed the integrity of HRR by monitoring for the appearance of RAD51 foci in response to DNA damage (Figure 7B). Both BC-p53WT and BC-p53KD cells formed RAD51 foci after exposure to 10 Gy IR, demonstrating that HRR was intact in these cells (Figure 7B). Next, WU-BC3 cells were incubated with either vehicle (DMSO), 10 nM irinotecan, 100 nM AZD7762, 10 μM Chk2 inhibitor, or a combination of irinotecan followed by AZD7762 or Chk2 inhibitor, as indicated in Figure 7C. As seen in Figure 7D, p53 and p21 levels rose in irinotecan-treated BC3-p53WT (lanes 2, 3), but increased only slightly in BC3-p53KD cells (lanes 9, 10), consistent with knockdown of p53 in BC3-p53KD cells. Treatment with irinotecan induced Chk1 autophosphorylation equally in both cell lines, but levels of γH2AX and cleaved caspase-3 were approximately 15- and 4-fold higher, respectively, in BC3-p53KD cells compared to that in BC3-p53WT cells (lane 13 versus 6) when treated with the combination of irinotecan and AZD7762. Thus, knockdown of p53 sensitized WU-BC3 TNBC cells to the combination therapy. Similar results were observed when carboplatin or gemcitabine was used in place of irinotecan (Supplemental Figure 3).
Figure 7. p53 status is a key determinant of how TNBCs respond to DNA damage followed by Chk1 inhibition.
WU-BC3 cells were infected with control retroviruses or retroviruses encoding p53-specific shRNAs to generate the BC3-p53WT and BC3-p53KD lines, respectively. (A and B) Cells were either mock irradiated (–) or exposed to 10 Gy IR (+) and then monitored for the integrity of the p53 pathway by observing changes in the levels of p53 and p21 by Western blotting (A) and for the integrity of HRR by monitoring for the appearance of Rad51 foci (B; pink stain, Rad 51). Original magnification, ×400. (C) Timeline of treatment of cells with either vehicle, irinotecan, AZD7762, the Chk2 inhibitor, or irinotecan in combination with AZD7762 or the Chk2 inhibitor. Cell lysates were prepared and analyzed for the indicated proteins by Western blotting (D).
Since AZD7762 inhibits both Chk1 and Chk2 (20), we tested to determine whether Chk2 inhibition contributed to the synergistic antitumor effects observed when AZD7762 was combined with chemotherapy. A selective Chk2 inhibitor (compound 2h in Arienti, et al., ref. 30) was tested alone or in combination with irinotecan in BC3-p53WT and BC3-p53KD cells (Figure 7, C and D). As expected, addition of the Chk2 inhibitor blocked autophosphorylation of Chk2 in irinotecan-treated cells, as evidenced by the loss of the slower electrophoretic form of Chk2 (lanes 7 and 14), but did not affect Chk1 autophosphorylation (Chk1 pS296, lanes 7 and 14). Unlike when AZD7762 was used, specific inhibition of Chk2 in combination with irinotecan did not enhance levels of γH2AX or cleaved caspase-3 above that of irinotecan alone in either cell type (lane 3 vs. 7 and lane 10 vs. 14). Therefore, we conclude that the enhanced DNA damage and apoptosis observed when irinotecan was combined with AZD7762 was through inhibition of Chk1, not Chk2.
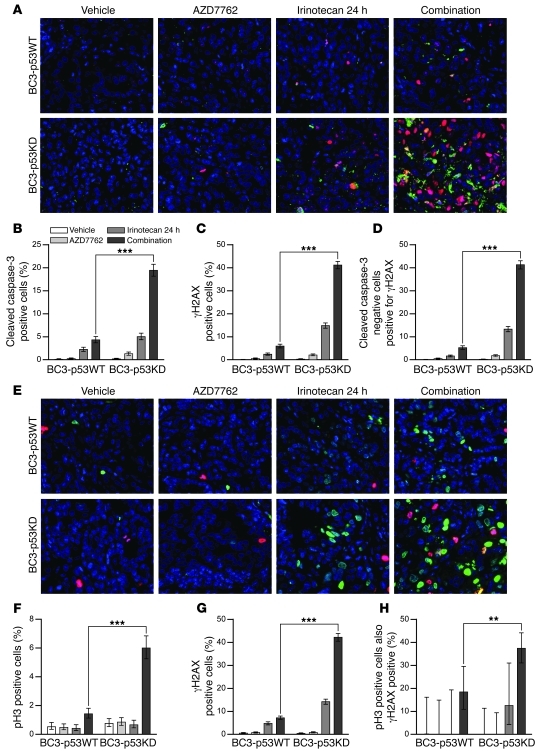
The importance of p53 deficiency in sensitizing tumors to the apoptotic-inducing effects of DNA damage followed by Chk1 inhibition was further investigated in vivo using isogenic lines BC3-p53WT and BC-p53KD. Mice bearing BC3-p53WT or BC3-p53KD tumors were treated with either vehicle, irinotecan, AZD7762, or a combination of irinotecan followed by AZD7762 using the same protocol as described for WU-BC3, WU-BC4, and WU-BC5 (Figures 3–5). Tumors were processed for costaining of cleaved caspase-3 and γH2AX (Figure 8, A–D) and for phosphohistone H3 and γH2AX (Figure 8, E–H). Irinotecan followed by AZD7762 resulted in a significant increase in apoptosis in tumor cells knocked down for p53 (BC3-p53KD) compared with control cells (BC3-p53WT) (Figure 8B). Additionally, the combination therapy led to significant increases in the number of BC3-p53KD cells staining positive for both γH2AX (indicative of DNA DSBs) (Figure 8, C and G) and phosphohistone H3 (indicative of mitosis) (Figure 8F) compared with that in BC3-p53WT cells (Figure 8, C and G). Approximately 40% of BC-p53KD cells staining positive for phosphohistone H3 also stained positive for γH2AX (Figure 8H), suggesting that the combination therapy forced p53-deficient tumor cells into mitosis despite their high content of DNA DSBs. Given that 40% of BC3-p53KD cells staining negative for cleaved caspase-3 were positive for γH2AX (Figure 8D) and that 80% of γH2AX-positive cells were negative for cleaved caspase-3 (data not shown), γH2AX staining selectively detected irinotecan-induced DNA DSBs that failed to be repaired in p53-deficient cells rather than being an indirect effect of Caspase Activated DNase-mediated (CAD-) apoptotic DNA cleavage.
Figure 8. p53 status is the key determinant of TNBC response to DNA damage followed by Chk1 inhibition in vivo.
Mice harboring BC3-p53WT and BC3-p53KD were treated as indicated, and tumors were analyzed by IF staining for cleaved caspase-3 and costaining for phosphohistone H3 or γH2AX. (A) Representative IF costaining for cleaved caspase-3 (green) and γH2AX (red). Quantitation is shown for cleaved caspase-3–positive (B), γH2AX-positive (C), and cleaved caspase-3–negative cells that are positive for γH2AX (D). (E) Representative IF costaining for pH3 (red) and γH2AX (green). Original magnification, ×400. Quantitation is shown in F (pH3), G (γH2AX), and H (pH3-positive cells also positive for γH2AX). Statistics were obtained by the Wilson score method in SPSS 20. **P < 0.01; ***P < 0.001.
Discussion
Using HIM tumor models of TNBC, we demonstrated that UCN-01 (nonselective Chk1 inhibitor) and AZD7762 (selective Chk1 inhibitor) abrogated irinotecan-induced S and G2 cell-cycle arrest, increased apoptosis, and reduced tumor growth in TP53 mutant, but not TP53 WT, TNBC. In addition, knockdown of p53 sensitized WU-BC3 TNBC cells to the combination therapy of irinotecan and a Chk1 inhibitor, either UCN-01 or AZD7762. Importantly, the combination treatment significantly (P = 0.006) extended the survival of mice harboring p53-deficient but not p53-proficient TNBC.
Most of the existing preclinical breast cancer xenograft models utilize cancer cell lines that have undergone multiple passages ex vivo prior to implantation. Not surprisingly, discordance between preclinical predictions and clinical trial data has been observed, thereby posing significant challenges in the development of novel anticancer therapeutics using these models. A major strength of the HIM model is that it uses tumors obtained directly from patients that are immediately transplanted and propagated in the context of a humanized mammary fat pad, resulting in a closer resemblance to the human tumor counterpart (21). Our data demonstrated that the established xenografts (passages 1–5) and their original human tumor clustered more closely with each other than with any other tumor (Figure 2 and Supplemental Figure 1). In a whole genomic sequencing analysis comparing a first-passage HIM tumor, the primary breast tumor from which the HIM model was established, and a brain metastasis in an African American patient with basal-like breast cancer, the HIM tumor did not gain any de novo mutations, retained all mutations present in the primary breast tumor, and displayed a mutation enrichment pattern that resembled the brain metastasis (31). The similarity between the HIM model and its human tumor counterpart in gene expression pattern and genomic mutation spectrum makes it a powerful system for functional and therapeutic studies. The difference in the gene-expression profile and molecular subclassification between WU-BC3 (HER2-E subtype) and the basal-like tumors (WU-BC4 and WU-BC5) demonstrates that HIM models are able to capture the molecular heterogeneity of TNBC.
TNBC is among the most difficult to treat of the breast cancer subtypes due to the lack of a specific molecular target for existing treatment strategies. It is a high priority in cancer research to develop targeted approaches to treating this aggressive form of breast cancer. Our experiments using HIM TNBC xenograft models provide proof of principle that TNBCs harboring TP53 mutations may be effectively targeted by the combination of a DNA-damaging agent followed by a Chk1 inhibitor. This synthetic lethal strategy is based on a tumor-specific mutation (TP53 mutation) and a drug, in this case a DNA-damaging agent combined with a Chk1 inhibitor, acting together to cause the tumor cell to undergo apoptosis, similar to the synthetic lethal interactions of BRCA1 mutations and poly(ADP-ribose) polymerase (PARP) inhibitors (32). WU-BC5 was derived from a brain metastasis, which harbors 50 validated point mutations, small indels, and significant copy number variations, from the same patient who was subjected to whole-genome sequencing analysis discussed above (31). Despite the complexity of the genomic background, WU-BC5 was sensitive to the combination of a DNA-damaging agent and a Chk1 inhibitor, which is likely due to TP53 mutation. Our study provides preclinical rationale for the clinical investigation of this strategy in TNBC. Our phase I trial testing the combination of irinotecan and UCN-01 in patients with advanced solid tumor malignancies showed promise in patients with TNBC (33), and the extension phase of the trial is currently being conducted in patients with metastatic TNBC.
It is interesting to note that the 2 HIM models that responded to the combination treatment (WU-BC4 and WU-BC5) were both basal like by molecular subtyping, whereas WU-BC3 is HER2-E and did not respond. Although a subtype-specific antitumor response to the combination therapy might be a possibility, the enhanced apoptotic response of WU-BC3 to the combination therapy when p53 was knocked down in these cells argues against this possibility.
In addition to Chk1, UCN-01 targets several other kinases, including PDK1 in the PI3K pathway, while AZD7706 is a more selective Chk1 inhibitor. Given that UCN-01, but not AZD7762, inhibits PDK1, yet both agents induced checkpoint bypass and apoptosis in TP53 mutant TNBC, we conclude that Chk1 inhibition, not PDK1 inhibition, is the mechanism of antitumor effect of these inhibitors. Furthermore, AZD7762, but not UCN-01, is a potent Chk2 inhibitor, arguing that Chk1, rather than Chk2, inhibition is responsible for the antitumor effects observed with these protein kinase inhibitors. In support of this conclusion, a selective Chk2 inhibitor was unable to induce checkpoint bypass or enhance the DNA damage and apoptotic effects of irinotecan in the p53 knockdown cell line BC3-p53KD (Figure 7). This is consistent with previous findings reporting that knockdown of Chk1 in the presence of endogenous Chk2 is sufficient to abrogate S and G2 checkpoints in cells with DNA damage, while Chk2 knockdown does not induce checkpoint bypass nor does Chk2 knockdown synergize with Chk1 knockdown to potentiate checkpoint bypass (34–36). Our results are also consistent with previous findings that knockdown of Chk2 fails to sensitive cells to either radiation or gemcitabine and inhibition of Chk2 by VRX0466617, a potent and selective Chk2 inhibitor, does not synergize with either doxorubicin or cisplatin in tumor cell killing (20, 37–39). Therefore, inhibition of Chk1 is the major factor responsible for mediating the antitumor effects of UCN-01 and AZD7762 in combination with irinotecan in the absence of a functional p53 pathway.
In addition to TNBC, Chk1 inhibitors may be effective in other breast cancer subtypes that are deficient in TP53. TP53 mutation is one of the most common genetic abnormalities in breast cancer (40, 41) and associates with more aggressive disease and a poor clinical outcome (40–43). Although the overall frequency of TP53 mutation is 20%–30%, the incidence is much higher in certain breast cancer subtypes. In an analysis of 330 breast cancer cases, the overall incidence of TP53 mutation was 25% (1), but was much higher in basal-like tumors (44%) and HER+ER– (43%) subtypes in contrast with luminal B (23%) and luminal A (15%) breast cancers (1). In an analysis of 543 patients with node-negative breast cancer, HER2-amplified tumors had a TP53 mutation frequency of 38.9% (44). There is also an increased incidence of TP53 mutation in cancers arising from patients with germ-line mutations of BRCA1 and BRCA2 (45, 46). In typical medullary breast carcinomas, TP53 mutation occurs in 100% of the cases (47). In addition to mutations in TP53, p53 pathway components can be inactivated by other genetic or epigenetic events. Examples include low or absent expressions of Ataxia Telangiectasia (ATM) (48) or Chk2 (49) and Murine Double Minute (MDM2) amplification (50). Therefore, it has been of great interest to develop strategies to target tumors that are defective in the p53 pathway. Several other selective Chk1 inhibitors are in preclinical and clinical development and should provide exciting new opportunities for targeting TP53 mutant tumors, including a significant fraction of breast cancer (51).
In summary, we demonstrated that the combination of a Chk1 inhibitor and a DNA-damaging agent is effective against HIM models of TNBC that bear TP53 mutations, arguing that clinical trials testing this type of strategy in human breast cancer are warranted.
Methods
Chemicals.
UCN-01 powder, Chk2 inhibitor II hydrate, and carboplatin were purchased from Sigma-Aldrich, and dissolved in DMSO at 1 mg/ml, 10 mM, and 50 mM, respectively. Irinotecan (20 mg/ml) was purchased from Hospira Inc. AZD7762 was manufactured by Axon Medcam BV. Gemcitabine was purchased from BioVision.
Establishment of orthotopic TNBC xenograft models.
The HIM xenograft models were established according to published protocols (21). Briefly, epithelium was removed from the fourth mammary glands of 3- to 4-week-old NOD/SCID mice (Charles River Laboratories). Two to four weeks following the clearance procedure, immortalized human mammary stromal fibroblasts derived from a patient undergoing a reduction mammoplasty were irradiated and then injected into the cleared mammary fat pads to create humanized mammary fat pads. Approximately 3 weeks later, human breast tumor cells were implanted into the humanized mammary fat pads. Established xenografts were then passaged in the mammary fat pads of recipient humanized NOD/SCID mice for our studies.
Preparing human breast tumors for engraftment.
Human breast tumors from needle biopsies or tumors passaged in mice were suspended in complete medium (DMEM/F12 medium [cat. no. SH30023.01; HyClone] supplemented with 10% bovine calf serum [cat. no. SH30072.03; HyClone] and antibiotics [100 U/ml penicillin, 100 U/ml streptomycin and 250 ng/ml fungizone]) on ice. Tumors were minced into approximately 1-mm pieces under sterile conditions and then transferred to 15 ml conical tubes containing 3 mg/ml collagenase (cat. no. 1088793; Roche), 250 U/ml hyaluronidase (cat. no. H-3506; Sigma-Aldrich), and antibiotics. Samples were incubated at 37°C until minced tissues dissociated into single cells. Cells were pelleted and supernatants discarded. Cells were washed in PBS. Cells were resuspended in 5–10 ml rbc lysis buffer (cat. no. R-7757; Sigma-Aldrich) and incubated for 15 minutes at 37°C. Cells were pelleted and washed in 10 ml PBS. Cells were resuspended in an equal volume of 0.05% trypsin-EDTA (Gibco; Invitrogen) and incubated for 5 minutes at 37°C. Trypsin was inactivated with complete medium, and cells were pelleted and then washed twice with PBS. All centrifugation steps were performed at 230 g for 5 minutes at 4°C. Cells were resuspended in complete medium and filtered through a sterile 40-μm filter. 1 × 106 tumor cells and 5 × 105 fibroblasts (2.5 × 105 that were exposed to 4 Gy IR and 2.5 × 105 untreated cells) were mixed and added to an equal volume of a 1:1 mixture of Matrigel (cat. no. 354234; BD Biosciences) and Collagen I (cat. no. 08-115; Upstate). The suspension was kept on ice until injection. The cell-suspension mixture was injected into the area of humanization with a 27-gauge syringe. The final volume was 35 μl per mammary gland. Established tumors implanted in the left and right humanized mammary fat pads of NOD/SCID mice were allowed to grow until their maximum diameter reached approximately 0.7 to 1.0 cm. Mice were sacrificed and single-cell suspensions were prepared from each tumor for further passaging in mice.
Microarray analysis.
Total RNA from human counterpart and xenograft tumors was purified, amplified, and labeled (Cy5-sample, Cy3-control), and microarray hybridizations were performed using Agilent 4× 44K Whole Human Genome Microarrays. For Cy3-controls, we used Stratagene Human Universal Reference (22) enriched with equal amounts of RNA from the MCF7 and ME16C cell lines. Microarrays were hybridized overnight, washed, dried, and scanned using an Agilent Scanner. The image files were analyzed and loaded into the UNC-CH Microarray Database ( https://genome.unc.edu/). Final normalized log2 ratios (Cy5-sample/Cy3-control) for each probe were obtained after removing probes with a Lowess normalized intensity value of less than 10 in the Cy5-sample and/or the Cy3-control. Platform normalization procedures were then applied as previously described (52), and intrinsic subtype classifications were identified from the PAM50 microarray-based assay described in Parker et al. (22). Microarray data for this study have been deposited in GEO Omnibus (GSE28860).
Experimental therapy for biomarker analysis.
1 × 106 breast cancer cells were implanted into each humanized mammary fat pad of recipient mice for experimental therapies. Tumors were allowed to grow to approximately 0.5 cm prior to treatment. To assess the functional integrity of the p53 pathway, NOD/SCID mice bearing either WU-BC3, WU-BC4, or WU-BC5 tumors were treated with vehicle (DMSO) or irinotecan (100 mg/kg i.p.). Tumors were harvested 24 hours later and analyzed for p53 and p21 by Western blotting. To assess the effects of Chk1 inhibitors (UCN-01 or AZD7762) on irinotecan-induced DNA damage, cell-cycle arrest, and apoptosis, 2 mice (each harboring 2 xenograft tumors, 1 at each mammary fat pad) were allocated to each treatment group for each HIM model (DMSO only, irinotecan only, Chk1 inhibitor only, and the combination of irinotecan and Chk1 inhibitor). Table 1 outlines the experimental procedure used for therapy and tumor harvesting. Irinotecan (or vehicle) was administered (100 mg/kg) i.p. at hour 0, followed by UCN-01 (4 mg/kg) or AZD7762 (25 mg/kg) or vehicle i.p. at hours 24 and 42. Mice were then euthanized and tumors harvested at hour 48, with the exception of 1 mouse in the irinotecan-only treated group, which was sacrificed at hour 24. Each xenograft tumor was cut into 2 pieces with 1 piece fresh frozen for Western blotting and the other piece fixed and embedded in paraffin blocks for IHC or IF staining.
Experimental therapy for tumor growth and survival studies.
Approximately 1 × 106 breast cancer cells derived from WU-BC3 and WU-BC4 tumors were implanted into left and right humanized mammary fat pads of recipient NOD/SCID mice for treatment. Ten mice (20 xenograft tumors) were allocated to each group for each HIM model. Treatment began when tumors reached approximately 0.5 cm. DMSO or irinotecan (50 mg/kg i.p.) was administered on day 1, followed by AZD7762 (25 mg/kg i.p.) or vehicle on days 2 and 3 of a 5-day cycle. Mice were subjected to a total of 4 cycles of therapy. Tumors were measured by calipers prior to drug treatment, every 2–3 days following the initiation of drug therapy, and at the termination of the experiment. Tumor volume was calculated using the following equation: V = 0.5 × ([greatest diameter] × [shortest diameter]2). Mice were followed until death or were sacrificed earlier if tumors reached 2 cm in size or if mice experienced intolerable toxicities.
Xenograft tumor processing for Western blotting.
Tumors were lysed in RIPA buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% Triton X-100, 0.1% sodium dodecyl sulfate, 0.5% deoxycholic acid) containing 5 μg/ml aprotinin, 10 μg/ml leupeptin, 1 μM PMSF, 1 mM sodium fluoride, and 10 mM β-glycerophosphate. Samples were subjected to 3 rounds of freeze/thawing at –80°C and 37°C, respectively. Samples were then incubated on ice for 10 to 15 minutes, followed by sonication on ice for 7 seconds using a small tip sonicator. Samples were placed on ice for 20 seconds, followed by 2 rounds of sonication, and then placed on ice for an additional 30 minutes. Samples were centrifuged at 10,000 g at 4°C for 5 minutes. Supernatants were removed to fresh tubes, and protein concentrations were determined using Bio-Rad Protein Assay Dye Reagent Concentrate.
Western blotting.
Samples (50–100 μg) were run on Bio-Rad Criterion gels and transferred to PVDF for Western blotting using standard procedures. Antibodies used for Western blotting include those recognizing S6 ribosomal protein (#2217; Cell Signaling Technology), phospho-S6 ribosomal protein Ser240/244 (#4838; Cell Signaling Technology), p21 (sc-6246;, Santa Cruz Biotechnology Inc.), p53 (sc-126; Santa Cruz Biotechnology Inc.), cleaved caspase-3 (#9961; Cell Signaling Technology), γH2A.X (#05-636; Millipore), Chk1 (#2344; Santa Cruz Biotechnology Inc.), Chk1 pS296 (#2580-1; Epitomics), Chk2 (#05-649; Millipore), actin (A-4200; Sigma-Aldrich), and β-catenin (#610154; BD Biosciences). Secondary antibodies used were HRP goat anti-rabbit (#81-1620; Invitrogen) and HRP goat anti-mouse (#115-035-062l; Jackson ImmunoResearch). Protein was detected using ECL (#RPN 2106; GE Healthcare).
Quantitation of pS6 levels.
Blots were developed using ECL Detection Reagent (GE Healthcare), and proteins were quantitated using ImageJ (53). The ratio of phosphorylated S6 to total S6 protein was determined for each sample. GraphPad Prism software was used for graphing and statistical analysis.
IF staining.
Each mammary tumor was excised, half was rapidly frozen on dry ice (to use for Western blotting), and the remainder was fixed in 10% neutral buffered formalin (Fisher Scientific) overnight. Tumor samples were embedded in paraffin, and 5-μm sections were cut and then baked in an oven at 70°C for 20 minutes. Sections were deparaffinized by immersing in xylene (Fisher Scientific) 3 times for 5 minutes each and rehydrated by immersing twice through a decreasing gradient of alcohol (100%, 95%, 70%, 50%, and distilled H20) for 2 minutes each. Antigen retrieval was carried out by boiling samples in rodent decloaker agent (Biocare Medical, RD913M) for 30 minutes followed by cooling at room temperature for an additional 30 minutes. Endogenous peroxidase activity was blocked by incubating sections in Peroxidase Blocking Reagent (Dako) for 10 minutes. Nonspecific interactions were blocked by incubating sections in Protein Block (Dako) for 1 hour at room temperature. Primary and secondary antibodies were diluted in Antibody Diluent (Dako). Sections were then incubated with either cleaved caspase-3 (Asp175) antibody (1:100; Cell Signaling Technology) or costained by incubating with phosphohistone H3 (Ser28) antibody (1:1000; Sigma-Aldrich) and γH2AX (Ser139) antibody (1:100; Cell Signaling Technology) overnight at 4°C. The following secondary antibodies (Molecular Probes) were applied for 1 hour: FITC-conjugated anti-rabbit antibody (Alexa Fluor 488) for cleaved caspase-3 or γH2AX and TRITC-conjugated anti-rat antibody (Alexa Fluor 594) for phosphohistone H3 staining. All fluorescently labeled sections were counterstained with DAPI using the ProLong Gold Antifade–DAPI reagent (Invitrogen).
IHC.
For IHC staining, tumor sections were processed as described for IF staining except that endogenous peroxidase activity was blocked by incubating in 3% hydrogen peroxide dissolved in methanol for 15 minutes. Antigen retrieval and blocking were carried out as described for IF staining. Sections were incubated with phosphohistone H3 (Ser10) antibody (1:400; Millipore), γH2AX (Ser139) antibody (1:200; Millipore), and cleaved caspase-3 (Asp175) antibody overnight at 4°C followed by incubation with secondary EnVision + Single Reagents HRP anti-rabbit antibody (Dako) for 30 minutes at room temperature. Positive staining was visualized using HRP substrate diaminobenzidine diluted in REAL substrate buffer (Dako) applied for 10 minutes, followed by intermediate rinses in PBS. Sections were counterstained with Meyer’s hematoxylin for 1 minute, dehydrated in an ascending ethanol series (70%, 95%, and 100%), and cleared with xylene. Sections were mounted using Cytoseal XYL Mounting Medium (Richard-Allan Scientific).
Infection of WU-BC3 cells.
WU-BC3 cells were grown in McCoy’s 5A (16600; Invitrogen) supplemented with 10% FBS, 1% glutamine, and 1% penicillin-streptomycin. When logarithmically growing cells reached approximately 50% confluency, they were infected with control retroviruses (pMKOPuro, Addgene plasmid 8452) (29) or pMKOPuro expressing p53-specific shRNA using a lift infection protocol. Briefly, cells were trypsinized and then rinsed with fresh medium to inactivate the trypsin. Cells were infected in suspension at an MOI of 20 in the presence of 8 μg/ml polybrene. Infected cells were then replated and incubated for 4 hours. The next day, cells were reinfected for an additional 4 hours. After the second infection, the medium was changed and the cells were cultured for 24 hours followed by drug selection with 1 μg/ml puromycin (#A11138-03; Invitrogen). Drug-resistant colonies were pooled and expanded. These cells were then treated with vehicle (DMSO), 10 nm irinotecan, 100 μM carboplatin, 30 nM gemcitabine, 100 nM AZD7762, and 10 μM Chk2 inhibitor II hydrate as described in the figure legends. Cells were lysed and proteins were resolved on Criterion gels (Bio-Rad) and transferred to nitrocellulose for Western blotting.
Monitoring integrity of HRR.
BC3-p53WT or BC3-p53KD cells were exposed to 10 Gy IR for 1 hour and then fixed in 3.7% paraformaldehyde for 20 minutes. Cells were permeabilized in 0.5% Triton X-100 for 2 minutes. followed by blocking in Dako Protein Block (Dako) for 60 minutes. Permeabilized cells were incubated in the presence of rabbit anti-Rad51 antibody (0.5 μg/ml; cat. no. ABE257, Millipore) overnight at 4°C. Alexa Fluor 594–conjugated goat anti-rabbit IgG antibody (5 μg/ml; cat. no. A-11012, Millipore) was used for IF detection. Slides were covered with ProLong Gold Antifade-DAPI reagent (Invitrogen).
Statistics.
Tumor cells staining positive for phosphohistone H3 and/or γH2AX or cleaved caspase-3 were counted in 5 randomly selected fields per tumor at ×400 magnification. Approximately 1,382 to 2,182 cells were counted. Proportions were calculated in SPSS 20 using the Wilson score method with continuity correct described by Newcombe (54). All proportions are presented with 95% CIs. Survival and tumor growth data were analyzed and graphed in SPSS 19 (IBM) using animal sacrifice as the terminal event. Animals dying of other causes were right-censored from the analysis at the observed time of death and are marked on the survival curves with an X (Figure 6A). Median times to animal sacrifice ± 95% CIs were calculated using Kaplan-Meier analysis, and pairwise significance values were calculated using the log-rank test. P < 0.05 was used for significance for all reported statistics. Additionally, lower P value thresholds are noted in figures to emphasize important statistical tests.
Study approval.
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the NIH. The protocol was approved by the Committee on the Ethics of Animal Experiments of Washington University. Human breast tumor samples for establishing the HIM models were obtained under the auspices of an Institutional Review Board–approved protocol at the Siteman Cancer Center and Washington University School of Medicine. Informed consent was obtained from all patients involved.
Supplementary Material
Acknowledgments
We thank the patients and their families for donating the tumor tissues to generate the HIM lines described in this study. We thank Diana Schorry for help with IHC. This work was supported by grants from the Komen Foundation (KG081551to H. Piwnica-Worms, C.X. Ma, and S. Li), from the National Heart, Lung, and Blood Institute (F30HL107009 to R.J. Goiffon) and by (NIH grant P50 CA94056 to the Molecular Imaging Center at Washington University). The generation and characterization of HIM models was funded by grants from the Komen Foundation (BCTR0707808 and KG090422 to M.J. Ellis and S. Li), the Siteman Cancer Center at Barnes-Jewish Hospital, and Washington University School of Medicine through the NIH/NCI (P30 CA091842) and NIH/NCRR Washington University-ICTS (UL1 RR024992), and from the Fashion Footwear Association of New York (FFANY) (to M.J. Ellis). H. Piwnica-Worms is a Research Professor of the American Cancer Society.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2012;122(4):1541–1552. doi:10.1172/JCI58765.
See the related Commentary beginning on page 1202.
References
- 1.Carey LA, et al. Race, breast cancer subtypes, and survival in the carolina breast cancer study. JAMA. 2006;295(21):2492–2502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 2.Kim SH, Li C, Maller JL. A maternal form of the phosphatase Cdc25A regulates early embryonic cell cycles in Xenopus laevis. Dev Biol. 1999;212(2):381–391. doi: 10.1006/dbio.1999.9361. [DOI] [PubMed] [Google Scholar]
- 3.Shimuta K, Nakajo N, Uto K, Hayano Y, Okazaki K, Sagata N. Chk1 is activated transiently and targets Cdc25A for degradation at the Xenopus midblastula transition. Embo J. 2002;21(14):3694–3703. doi: 10.1093/emboj/cdf357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sorensen CS, et al. Chk1 regulates the S phase checkpoint by coupling the physiological turnover and ionizing radiation-induced accelerated proteolysis of Cdc25A. Cancer Cell. 2003;3(3):247–258. doi: 10.1016/S1535-6108(03)00048-5. [DOI] [PubMed] [Google Scholar]
- 5.Zhao H, Watkins JL, Piwnica-Worms H. Disruption of the checkpoint kinase 1/cell division cycle 25A pathway abrogates ionizing radiation-induced S and G2 checkpoints. Proc Natl Acad Sci U S A. 2002;99(23):14795–14800. doi: 10.1073/pnas.182557299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen MS, Ryan CE, Piwnica-Worms H. Chk1 kinase negatively regulates mitotic function of Cdc25A phosphatase through 14-3-3 binding. Mol Cell Biol. 2003;23(21):7488–7497. doi: 10.1128/MCB.23.21.7488-7497.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Falck J, Mailand N, Syljuasen RG, Bartek J, Lukas J. The ATM-Chk2-Cdc25A checkpoint pathway guards against radioresistant DNA synthesis. Nature. 2001;410(6830):842–847. doi: 10.1038/35071124. [DOI] [PubMed] [Google Scholar]
- 8.Goloudina A, Yamaguchi H, Chervyakova DB, Appella E, Fornace AJ, Jr, Bulavin DV. Regulation of human Cdc25A stability by Serine 75 phosphorylation is not sufficient to activate a S phase checkpoint. Cell Cycle. 2003;2(5):473–478. [PubMed] [Google Scholar]
- 9.Hassepass I, Voit R, Hoffmann I. Phosphorylation at serine 75 is required for UV-mediated degradation of human Cdc25A phosphatase at the S-phase checkpoint. J Biol Chem. 2003;278(32):29824–29829. doi: 10.1074/jbc.M302704200. [DOI] [PubMed] [Google Scholar]
- 10.Levesque AA, Kohn EA, Bresnick E, Eastman A. Distinct roles for p53 transactivation and repression in preventing UCN-01-mediated abrogation of DNA damage-induced arrest at S and G2 cell cycle checkpoints. Oncogene. 2005;24(23):3786–3796. doi: 10.1038/sj.onc.1208451. [DOI] [PubMed] [Google Scholar]
- 11.Bunch RT, Eastman A. Enhancement of cisplatin-induced cytotoxicity by 7-hydroxystaurosporine (UCN-01), a new G2-checkpoint inhibitor. Clin Cancer Res. 1996;2(5):791–797. [PubMed] [Google Scholar]
- 12.Fuse E, et al. Unpredicted clinical pharmacology of UCN-01 caused by specific binding to human alpha1-acid glycoprotein. Cancer Res. 1998;58(15):3248–3253. [PubMed] [Google Scholar]
- 13.Yu L, et al. UCN-01 abrogates G2 arrest through a Cdc2-dependent pathway that is associated with inactivation of the Wee1Hu kinase and activation of the Cdc25C phosphatase. J Biol Chem. 1998;273(50):33455–33464. doi: 10.1074/jbc.273.50.33455. [DOI] [PubMed] [Google Scholar]
- 14.Graves PR, et al. The Chk1 protein kinase and the Cdc25C regulatory pathways are targets of the anticancer agent UCN-01. J Biol Chem. 2000;275(8):5600–5605. doi: 10.1074/jbc.275.8.5600. [DOI] [PubMed] [Google Scholar]
- 15.Busby EC, Leistritz DF, Abraham RT, Karnitz LM, Sarkaria JN. The radiosensitizing agent 7-hydroxystaurosporine (UCN-01) inhibits the DNA damage checkpoint kinase hChk1. Cancer Res. 2000;60(8):2108–2112. [PubMed] [Google Scholar]
- 16.Kohn EA, Yoo CJ, Eastman A. The protein kinase C inhibitor Go6976 is a potent inhibitor of DNA damage-induced S and G2 cell cycle checkpoints. Cancer Res. 2003;63(1):31–35. [PubMed] [Google Scholar]
- 17.Wang Q, Fan S, Eastman A, Worland PJ, Sausville EA, O’Connor PM. UCN-01: a potent abrogator of G2 checkpoint function in cancer cells with disrupted p53. J Natl Cancer Inst. 1996;88(14):956–965. doi: 10.1093/jnci/88.14.956. [DOI] [PubMed] [Google Scholar]
- 18.Ma CX, Janetka JW, Piwnica-Worms H. Death by releasing the breaks: CHK1 inhibitors as cancer therapeutics. Trends Mol Med. 2010;17(2):88–96. doi: 10.1016/j.molmed.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tse AN, Carvajal R, Schwartz GK. Targeting Checkpoint Kinase 1 in Cancer Therapeutics. Clin Cancer Res. 2007;13(7):1955–1960. doi: 10.1158/1078-0432.CCR-06-2793. [DOI] [PubMed] [Google Scholar]
- 20.Zabludoff SD, et al. AZD7762, a novel checkpoint kinase inhibitor, drives checkpoint abrogation and potentiates DNA-targeted therapies. Mol Cancer Ther. 2008;7(9):2955–2966. doi: 10.1158/1535-7163.MCT-08-0492. [DOI] [PubMed] [Google Scholar]
- 21.Kuperwasser C, et al. Reconstruction of functionally normal and malignant human breast tissues in mice. Proc Natl Acad Sci U S A. 2004;101(14):4966–4971. doi: 10.1073/pnas.0401064101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parker JS, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27(8):1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perou CM, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 24.Prat A, Perou CM. Deconstructing the molecular portraits of breast cancer. Mol Oncol. 2011;5(1):5–23. doi: 10.1016/j.molonc.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sedelnikova OA, Pilch DR, Redon C, Bonner WM. Histone H2AX in DNA damage and repair. Cancer Biol Ther. 2003;2(3):233–235. doi: 10.4161/cbt.2.3.373. [DOI] [PubMed] [Google Scholar]
- 26.Xu B, Kim S-T, Kastan MB. Involvement of Brca1 in S-phase and G2-phase Checkpoints after Ionizing Radiation. Mol Cell Biol. 2001;21(10):3445–3450. doi: 10.1128/MCB.21.10.3445-3450.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tapia C, Kutzner H, Mentzel T, Savic S, Baumhoer D, Glatz K. Two mitosis-specific antibodies, MPM-2 and phospho-histone H3 (Ser28), allow rapid and precise determination of mitotic activity. Am J Surg Pathol. 2006;30(1):83–89. doi: 10.1097/01.pas.0000183572.94140.43. [DOI] [PubMed] [Google Scholar]
- 28.Sato S, Fujita N, Tsuruo T. Interference with PDK1-Akt survival signaling pathway by UCN-01 (7-hydroxystaurosporine). Oncogene. 2002;21(11):1727–1738. doi: 10.1038/sj.onc.1205225. [DOI] [PubMed] [Google Scholar]
- 29.Stewart SA, et al. Lentivirus-delivered stable gene silencing by RNAi in primary cells. Rna. 2003;9(4):493–501. doi: 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arienti KL, et al. Checkpoint kinase inhibitors: SAR and radioprotective properties of a series of 2-arylbenzimidazoles. J Med Chem. 2005;48(6):1873–1885. doi: 10.1021/jm0495935. [DOI] [PubMed] [Google Scholar]
- 31.Ding L, et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464(7291):999–1005. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bryant HE, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434(7035):913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 33.Fracasso PM, et al. A Phase 1 study of UCN-01 in combination with irinotecan in patients with resistant solid tumor malignancies. Cancer Chemother Pharmacol. 2011;67(6):1225–1237. doi: 10.1007/s00280-010-1410-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao H, Watkins JL, Piwnica-Worms H. Disruption of the checkpoint kinase 1/cell division cycle 25A pathway abrogates ionizing radiation-induced S and G2 checkpoints. Proc Natl Acad Sci U S A. 2002;99(23):14795–14800. doi: 10.1073/pnas.182557299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiao Z, Xue J, Sowin TJ, Zhang H. Differential roles of checkpoint kinase 1, checkpoint kinase 2, and mitogen-activated protein kinase-activated protein kinase 2 in mediating DNA damage-induced cell cycle arrest: implications for cancer therapy. Mol Cancer Ther. 2006;5(8):1935–1943. doi: 10.1158/1535-7163.MCT-06-0077. [DOI] [PubMed] [Google Scholar]
- 36.Cho SH, Toouli CD, Fujii GH, Crain C, Parry D. Chk1 is essential for tumor cell viability following activation of the replication checkpoint. Cell Cycle. 2005;4(1):131–139. doi: 10.4161/cc.4.1.1299. [DOI] [PubMed] [Google Scholar]
- 37.Carlessi L, Buscemi G, Larson G, Hong Z, Wu JZ, Delia D. Biochemical and cellular characterization of VRX0466617, a novel and selective inhibitor for the checkpoint kinase Chk2. Mol Cancer Ther. 2007;6(3):935–944. doi: 10.1158/1535-7163.MCT-06-0567. [DOI] [PubMed] [Google Scholar]
- 38.Karnitz LM, et al. Gemcitabine-induced activation of checkpoint signaling pathways that affect tumor cell survival. Mol Pharmacol. 2005;68(6):1636–1644. doi: 10.1124/mol.105.012716. [DOI] [PubMed] [Google Scholar]
- 39.Carrassa L, Broggini M, Erba E, Damia G. Chk1, but not Chk2, is involved in the cellular response to DNA damaging agents: differential activity in cells expressing or not p53. Cell Cycle. 2004;3(9):1177–1181. [PubMed] [Google Scholar]
- 40.Gasco M Shami S, Crook T. The p53 pathway in breast cancer. Breast Cancer Res. 2002;4(2):70–76. doi: 10.1186/bcr426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lacroix M, Toillon RA, Leclercq G. p53 and breast cancer, an update. Endocr Relat Cancer. 2006;13(2):293–325. doi: 10.1677/erc.1.01172. [DOI] [PubMed] [Google Scholar]
- 42.Malamou-Mitsi V, et al. Evaluation of the prognostic and predictive value of p53 and Bcl-2 in breast cancer patients participating in a randomized study with dose-dense sequential adjuvant chemotherapy. Ann Oncol. 2006;17(10):1504–1511. doi: 10.1093/annonc/mdl147. [DOI] [PubMed] [Google Scholar]
- 43.Andersson J, et al. Worse survival for TP53 (p53)-mutated breast cancer patients receiving adjuvant CMF. Ann Oncol. 2005;16(5):743–748. doi: 10.1093/annonc/mdi150. [DOI] [PubMed] [Google Scholar]
- 44.Bull SB, et al. The combination of p53 mutation and neu/erbB-2 amplification is associated with poor survival in node-negative breast cancer. J Clin Oncol. 2004;22(1):86–96. doi: 10.1200/JCO.2004.09.128. [DOI] [PubMed] [Google Scholar]
- 45.Smith PD, et al. Novel p53 mutants selected in BRCA-associated tumours which dissociate transformation suppression from other wild-type p53 functions. Oncogene. 1999;18(15):2451–2459. doi: 10.1038/sj.onc.1202565. [DOI] [PubMed] [Google Scholar]
- 46.Phillips KA, et al. Frequency of p53 mutations in breast carcinomas from Ashkenazi Jewish carriers of BRCA1 mutations. J Natl Cancer Inst. 1999;91(5):469–473. doi: 10.1093/jnci/91.5.469. [DOI] [PubMed] [Google Scholar]
- 47.De Cremoux P, et al. p53 mutation as a genetic trait of typical medullary breast carcinoma. J Natl Cancer Inst. 1999;91(7):641–643. doi: 10.1093/jnci/91.7.641. [DOI] [PubMed] [Google Scholar]
- 48.Angele S, et al. Abnormal expression of the ATM and TP53 genes in sporadic breast carcinomas. Clin Cancer Res. 2000;6(9):3536–3544. [PubMed] [Google Scholar]
- 49.Sullivan A, et al. Concomitant inactivation of p53 and Chk2 in breast cancer. Oncogene. 2002;21(9):1316–1324. doi: 10.1038/sj.onc.1205207. [DOI] [PubMed] [Google Scholar]
- 50.Ho GH, et al. Genetic alterations of the p14ARF-hdm2-p53 regulatory pathway in breast carcinoma. Breast Cancer Res Treat. 2001;65(3):225–232. doi: 10.1023/A:1010686518990. [DOI] [PubMed] [Google Scholar]
- 51.Ye XS, Fincher RR, Tang A, Osmani SA. The G2/M DNA damage checkpoint inhibits mitosis through Tyr15 phosphorylation of p34cdc2 in Aspergillus nidulins. . EMBO J. 1997;16(1):182–192. doi: 10.1093/emboj/16.1.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ellis MJ, et al. Randomized phase II neoadjuvant comparison between letrozole, anastrozole, and exemestane for postmenopausal women with estrogen receptor-rich stage 2 to 3 breast cancer: clinical and biomarker outcomes and predictive value of the baseline PAM50-based intrinsic subtype--ACOSOG Z1031. J Clin Oncol. 2011;29(17):2342–2349. doi: 10.1200/JCO.2010.31.6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with Image J. Biophotonics Int. 2004;11(7):36–42. [Google Scholar]
- 54.Newcombe RG. Interval estimation for the difference between independent proportions: comparison of eleven methods. Stat Med. 1998;17(8):873–890. doi: 10.1002/(SICI)1097-0258(19980430)17:8<873::AID-SIM779>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.