Abstract
Many bacterial pathogens inject into host cells effector proteins that are substrates for host tyrosine kinases such as Src and Abl family kinases. Phosphorylated effectors eventually subvert host cell signaling, aiding disease development. In the case of the gastric pathogen Helicobacter pylori, which is a major risk factor for the development of gastric cancer, the only known effector protein injected into host cells is the oncoprotein CagA. Here, we followed the hierarchic tyrosine phosphorylation of H. pylori CagA as a model system to study early effector phosphorylation processes. Translocated CagA is phosphorylated on Glu-Pro-Ile-Tyr-Ala (EPIYA) motifs EPIYA-A, EPIYA-B, and EPIYA-C in Western strains of H. pylori and EPIYA-A, EPIYA-B, and EPIYA-D in East Asian strains. We found that c-Src only phosphorylated EPIYA-C and EPIYA-D, whereas c-Abl phosphorylated EPIYA-A, EPIYA-B, EPIYA-C, and EPIYA-D. Further analysis revealed that CagA molecules were phosphorylated on 1 or 2 EPIYA motifs, but never simultaneously on 3 motifs. Furthermore, none of the phosphorylated EPIYA motifs alone was sufficient for inducing AGS cell scattering and elongation. The preferred combination of phosphorylated EPIYA motifs in Western strains was EPIYA-A and EPIYA-C, either across 2 CagA molecules or simultaneously on 1. Our study thus identifies a tightly regulated hierarchic phosphorylation model for CagA starting at EPIYA-C/D, followed by phosphorylation of EPIYA-A or EPIYA-B. These results provide insight for clinical H. pylori typing and clarify the role of phosphorylated bacterial effector proteins in pathogenesis.
Introduction
Tyrosine phosphorylation of proteins and their interaction with cognate, Src homology (SH2) domain–containing ligands play key roles in mammalian signal transduction (1). Many bacterial pathogens use type III and type IV secretion systems (T3SS and T4SS) to inject effector proteins into the cytoplasm of host target cells where they are phosphorylated by host tyrosine kinases (2–4). Phosphorylated effectors target various components of signal transduction pathways, subverting host cell functions for the benefit of the pathogen. Intriguingly, 6 bacterial pathogens, including enteropathogenic E. coli, Helicobacter pylori, Chlamydia trachomatis, Bartonella henselae, Anaplasma phagocytophilum, and Ehrlichia chaffeensis, share this infection strategy (5–9). The tyrosine phosphorylation sites of H. pylori CagA and several other bacterial effector proteins such as BepD-F (B. henseleae), Tir (enteropathogenic E. coli), Tarp (C. trachomatis), or AnkA (A. phagocytophilum and E. chaffeensis) resemble classical substrates of eukaryotic cytosolic tyrosine kinases. The phosphorylation sites share some homology, although the corresponding effector proteins are not evolutionarily related to one another (4, 8, 9). Many of these effectors are targeted by kinases of the Src and Abl families (10–14). As Src is evolutionarily old and ubiquitously expressed, these motifs may have evolved as phosphorylation substrates in a wide range of host organisms and tissues. Once phosphorylated, effector proteins usually recruit cellular binding partners with SH2 domains to perturb downstream signaling cascades and host cell functions for bacterial benefit, and are also involved in disease development (9, 14). Many host cell interaction partners have been identified so far, but little is known regarding regulation of these early phosphorylation events. Thus, it is unclear if Src and Abl kinases target the same or different phosphorylation sites within the effectors. It is also unknown if certain kinases are preferentially activated as these proteins enter a target cell.
H. pylori is a valuable model system to study effector protein functions. This Gram-negative bacterium is a highly successful pathogen, colonizing the mucus layer of the stomach in about half of the global population (15–18). Research in the last 2 decades has indicated that the disease-associated cag pathogenicity island (cagPAI) encodes functional components of a T4SS (19, 20), a syringe-like pilus protruding from the bacterial surface. Upon contact with target cells, the T4SS injects virulence molecules such as CagA (4, 19, 21). CagA’s central role in H. pylori infection has been demonstrated in Mongolian gerbils and mice as well as in cultured gastric epithelial cells in vitro (18, 19, 22–24). A hallmark of H. pylori–infected AGS cells is the “hummingbird” or “elongation” phenotype, which is fully dependent on the presence of phosphorylated CagA (CagAPY) (11, 25). This phenotype may impact pathogenesis by influencing immune responses, wound healing, metastasis, or invasive growth of cancer cells in vivo (26, 27). Numerous cellular binding partners of CagA that highjack host signal transduction events have been identified, including tyrosine phosphatase SHP-2, C-terminal Src kinase (Csk), PI3K, and others (4, 14, 28, 29).
Glu-Pro-Ile-Tyr-Ala (EPIYA) motifs in the C-terminal region of CagA protein have been identified as phosphorylation sites using site-directed mutagenesis of injected and/or transfected CagA and mass spectrometry (11, 25, 28, 30). 3 distinct EPIYA motifs are found in Western isolates (EPIYA-A, EPIYA-B, and EPIYA-C). In East Asian strains, EPIYA-C motifs are replaced by EPIYA-D motifs, each of which is conserved in its surrounding sequence (22, 31). While the majority of reported strains have 3 EPIYA motifs per CagA molecule (EPIYA-ABC or EPIYA-ABD), the number and configuration of motifs per molecule may vary from 2 to 8 (4). Interestingly, oncogenic tyrosine kinases of the Src (10, 11) and Abl (12, 13) families have been shown to phosphorylate CagA in vitro and during infection in vivo. Mutagenesis of Y-972 in the EPIYA-C of CagA from the TIGR strain 26695 has shown that this motif is a major phosphorylation site (25). However, the majority of CagA phosphorylation studies to date used phosphorylation-deficient mutants in which all tyrosines were mutated in Western-type EPIYA-A, EPIYA-B, and EPIYA-C; these were investigated using 1-dimensional protein electrophoresis (1-DE) and panphosphotyrosine antibodies (12, 13, 28).
The EPIYA motifs are widely used to type H. pylori strains in clinical studies and are significant determinants of virulence (15–18, 31). However, it is unknown whether all EPIYA motifs are phosphorylated simultaneously or in a specific sequence, or if only 1 or 2 motifs are phosphorylated per CagA molecule. Also unknown is whether differentially phosphorylated CagA populations exist in the cell, whether a regulatory mechanism controls multiple phosphorylation events during infection, and whether Src and Abl kinases phosphorylate all EPIYA sites equally or only specific EPIYAs. Here, we used 2-dimensional protein electrophoresis (2-DE) and site-directed mutagenesis to study the time course of CagA phosphorylation during infection. We showed that 1 or 2 EPIYA motifs can be phosphorylated per CagA molecule in H. pylori TIGR strain 26695 (EPIYA-ABC) and strain TN2-GF4 (EPIYA-ABD) (32, 33), and that Src and Abl kinases phosphorylates different phosphorylation sites. We also created a series of phosphorylation-deficient and phosphorylation-mimetic H. pylori strain 26695 CagA mutants in all possible combinations. Study of CagA phosphorylation and of AGS cell elongation induced by these H. pylori mutants allowed deduction of the minimum set of EPIYA motifs necessary and sufficient for phenotypical outcome.
Results
Analysis of CagAPY protein species by 2-DE.
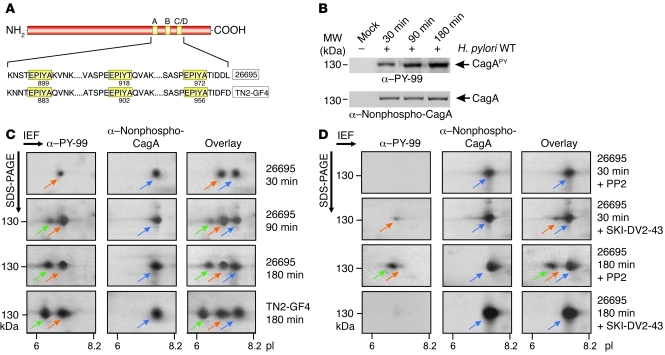
The H. pylori TIGR strain 26695 (32) cagA gene encodes 1 copy each of the Western-type motifs EPIYA-A, EPIYA-B, and EPIYA-C (Figure 1A). AGS gastric epithelial cells were infected with H. pylori for 30, 90, or 180 minutes. As reported previously (10, 25), 130-kDa phosphorylated CagA (CagAPY) signal intensity, detected by an α-phosphotyrosine antibody on 1-dimensional SDS-PAGE gels, increased time dependently (Figure 1B) and was strictly correlated with the H. pylori–induced elongation phenotype (Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI61143DS1). Similar results were observed with EPIYA-ABD of East Asian H. pylori strain TN2-GF4 (Figure 1A and data not shown). Increased CagAPY signal intensity could result from increased amounts of injected CagA molecules undergoing phosphorylation at a specific site and/or by increased phosphorylation of multiple sites per CagA molecule. To answer this question, infected AGS cell lysates were analyzed by 2-DE to separate phosphorylated and nonphosphorylated H. pylori strain 26695 CagA protein species as described (34). After 30 minutes infection, α-phosphotyrosine immunoblotting revealed a single 130-kDa CagAPY spot (spot 1) with an isoelectric point (pI) of approximately 7.0 (Figure 1C). After 90 minutes infection, the CagAPY spot 1 increased in intensity, and a second, fainter CagAPY spot appeared with pI of 6.5 (spot 2). After 180 minutes infection, spot 2 increased in intensity. No additional spots were observed after 240 minutes or longer infections, but both CagAPY spots continued to increase in intensity (data not shown). The blots were then reprobed with a monoclonal α-CagA antibody that binds exclusively to the nonphosphorylated form of CagA (25). This antibody did not label CagAPY spots 1 and 2, but in all infected samples did label a full-length CagA spot with pI of 7.5 (CagA spot 3; Figure 1C; refs. 9, 25). CagA protein species from H. pylori strain TN2-GF4 gave a similar pattern for each antibody (Figure 1C). Overlays of both the α-CagA and α-phosphotyrosine exposures yielded 3 distinct spots representing full-length CagA proteins (Figure 1C). These results were observed in both AGS and MKN-28 cells infected with 26695 or TN2-GF4 strains (Figure 1C and Supplemental Figure 2A) and are consistent with the more basic spot 3 representing nonphosphorylated CagA, and the other 2 more acidic spots representing CagAPY molecules phosphorylated at 1 or 2 positions.
Figure 1. Analysis of CagAPY protein species during infection with H. pylori by 1-DE and 2-DE.
(A) CagA proteins vary in the C-terminal EPIYA phosphorylation sites depending on geographical origin. Western CagA proteins (e.g., strain 26695) contain the EPIYA-A, EPIYA-B, and EPIYA-C segments, while East Asian strains contain the EPIYA-A, EPIYA-B, and EPIYA-D segments (e.g., strain TN2-GF4). These motifs are tyrosine phosphorylation sites that can be phosphorylated by Abl and Src tyrosine kinases (10–13). (B) AGS cells were infected for the indicated times with strain 26695. The resulting protein lysates were separated by 1-DE, and phosphorylation of injected CagA was examined using α–PY-99 and α-CagA antibodies (arrows). (C) Separation of CagA protein species from B by 2-DE. Depending on the time of infection, full-length CagAPY appeared as 1 spot (spot 1, red arrows, pI = 7.0) or 2 spots (spots 1 and 2; spot 2, green arrows, pI = 6.5) as indicated. The α-CagA antibody probe revealed a third spot (spot 3, blue arrows, pI = 7.5). Overlay of both exposures yielded 2 or 3 spots as shown. Strain TN2-GF4 exhibited the same pattern as 26695 (bottom). (D) Inhibition of Src with PP2 (10 μM) or Abl with SKI-DV2-43 (1 μM) revealed significant changes in spot intensity depending on the time of infection.
To test this hypothesis, lysates of infected cells were incubated in the presence or absence of PTP-N1, a protein tyrosine phosphatase. As expected, PTP-N1 treatment significantly attenuated the intensity of CagAPY spots 1 and 2 and increased the intensity of nonphosphorylated CagA spot 3 as labeled by the α-CagA antibody (Supplemental Figure 2B). Mock-infected or cagA-deficient H. pylori isogenic mutant-infected AGS cells showed no 2-DE gel labeling with α-CagA or α-phosphotyrosine antibodies (Supplemental Figure 2B). Also, as expected, infections with T4SS-deficient H. pylori strains ΔvirB5 (cagL–) and ΔvirB10 (cagY–), which express but do not inject CagA protein (21, 35), yielded no CagAPY spots (Supplemental Figure 2B).
Src and Abl kinases are activated differentially and have distinct effects on CagAPY spots.
Next, we assessed the role of Src and Abl kinases in the CagAPY patterns described above using activation-specific phosphoantibodies. Supplemental Figure 3 shows that c-Src was transiently activated between 30 and 90 minutes after infection, whereas Abl was constitutively activated with very high activities between 90 and 180 minutes after infection. Using the potent and relatively specific Src kinase inhibitor PP2 (36) and Abl kinase inhibitor SKI-DV2-43 (37), we studied the role of both kinases in producing the observed CagAPY spots. Having confirmed specific inhibition of c-Src or c-Abl tyrosine kinase activities and phenotypical outcome by the inhibitors (Supplemental Figure 4), we pretreated AGS cells with PP2 or SKI-DV2-43 for 30 minutes, infected them with H. pylori for 30 minutes, and analyzed cell lysates on 2-DE gels. Treatment with SKI-DV2-43 had no effect on CagAPY spot 1, but PP2 eliminated this signal completely (Figure 1D), suggesting that c-Src but not c-Abl phosphorylates CagA early in infection. Conversely, after 180 minutes of infection, PP2 moderately attenuated CagAPY spots 1 and 2, while SKI-DV2-43 eliminated both signals completely (Figure 1D). This suggests that c-Abl but not c-Src predominantly phosphorylates CagA at later time points, giving rise to the 2 CagAPY spots. The significant attenuation of CagAPY was not due to a bactericidal effect of the inhibitors because H. pylori viability was unaffected (data not shown). Interestingly, PP2 also had some effect on c-Abl activity after 180 minutes (Supplemental Figure 4), suggesting that c-Src may participate in the early signal cascade that triggers c-Abl activation.
c-Src and c-Abl kinases phosphorylate different tyrosines in CagA EPIYA motifs.
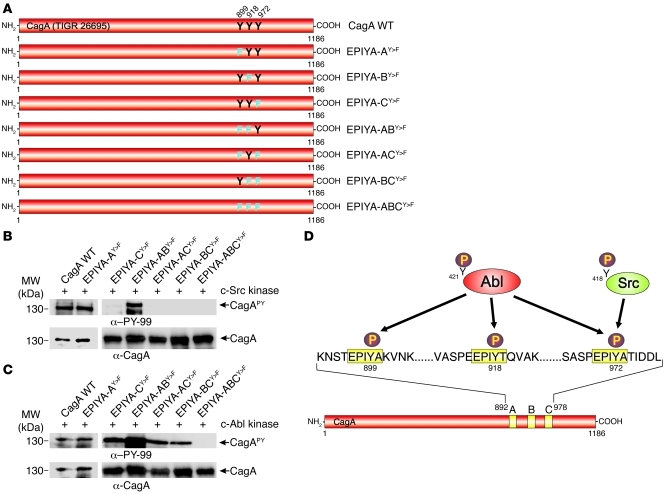
To determine whether c-Src and c-Abl phosphorylate all or only selected CagA EPIYA motifs, we generated single, double, and triple Tyr-to-Phe (Y>F) point mutations of all 3 strain 26695 EPIYA motifs in all possible combinations. These CagAs were expressed from the shuttle plasmid pSB19 and were named as shown in Figure 2A. In vitro phosphorylation analyses using recombinant c-Src or c-Abl kinases showed that c-Src failed to phosphorylate any CagAs carrying a mutation in the third EPIYA motif, suggesting that c-Src phosphorylates CagA at this site, which corresponds to EPIYA-C, but not at EPIYA-A or EPIYA-B (Figure 2B). In contrast, c-Abl phosphorylated WT CagA and all single or double mutants, but not the triple EPIYA-ABCY>F mutant (Figure 2C). This shows that c-Abl can phosphorylate all 3 EPIYA motifs, EPIYA-A, EPIYA-B, and EPIYA-C (Figure 2D). In addition, EPIYA-D in strain TN2-GF4, which is related to EPIYA-C (Figure 1A), was also phosphorylated by c-Src and c-Abl (Supplemental Figure 5). These results are consistent with the inhibitor data described above and suggest that c-Src and c-Abl phosphorylate a specific set of EPIYA motifs in a time-dependent fashion as summarized in Figure 3.
Figure 2. In vitro phosphorylation of CagA mutants by c-Abl or c-Src kinases.
(A) Site-directed Y>F mutagenesis of CagA EPIYA-A, EPIYA-B, and EPIYA-C in strain 26695. Single, double, and triple mutants were named as indicated. (B and C) Lysates of H. pylori expressing the mutated CagA EPIYA motifs were subjected to in vitro phosphorylation assays using recombinant c-Src kinase (B) or c-Abl kinase (C). Immunoblotting using α–PY-99 and α-CagA antibodies (arrows) indicated that both c-Src and c-Abl phosphorylated CagA in a different fashion. (D) Schematic diagram of the data, showing that c-Src only phosphorylated Y-972 in EPIYA-C, while Abl can phosphorylate Y-899, Y-918, and Y-972 in EPIYA-A, EPIYA-B, and EPIYA-C, respectively.
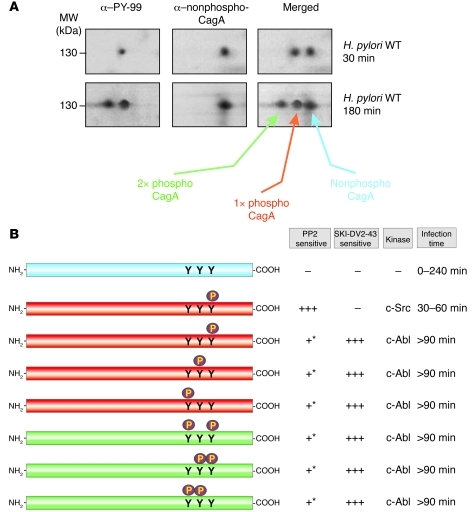
Figure 3. Modeling of phosphorylated CagA protein species detected in Figures 1 and 2.
(A) Representative 2-DE pictures of CagA protein species produced after 30 and 180 minutes of H. pylori infection. (B) Different possible phosphorylated CagA protein species induced during infection and their time-dependent sensitivity to pharmacological inhibition of Src and Abl kinases. Colors indicate the confirmed or proposed phosphorylation status of CagA motifs in each 2-DE spot. +++, strong effect; + moderate effect; –, no effect. Asterisks indicate that the slight inhibitory effect of PP2 on CagA protein species phosphorylated by c-Abl (>90 minutes) may be due to c-Src being an upstream component of the signal cascade leading to full c-Abl activation during infection.
Role of EPIYA motifs in CagA phosphorylation and AGS cell elongation.
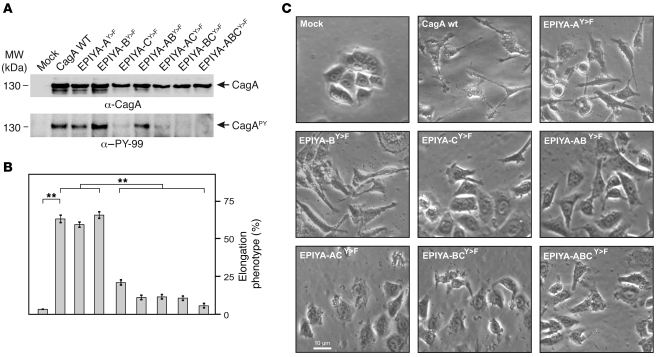
To determine which EPIYA motifs are important for CagA phosphorylation and phenotypic outcome during infection, we transformed nonpolar H. pylori ΔcagA deletion mutants (25) with pSB19 plasmids expressing the different strain 26695 WT and CagA mutants. Expression of equal amounts of CagA variants was confirmed by immunoblotting (Figure 4A). AGS cells were infected with these strains, and CagAPY levels were determined (Figure 4A). Supplemental Figure 6B shows that WT CagA and the EPIYA-AY>F mutant infections induced CagAPY signals of equal intensity, while the EPIYA-BY>F mutant induced a slightly more intense CagAPY signal. In agreement with previous reports (25), the EPIYA-CY>F mutant induced very faint CagAPY signals. Interestingly, CagA variants carrying 2 mutated motifs such as EPIYA-ACY>F and EPIYA-BCY>F also induced very faint CagAPY signals, while the EPIYA-ABY>F mutant induced slightly attenuated CagAPY signals compared with WT infection (Supplemental Figure 6B). The CagA triple mutant EPIYA-ABCY>F induced no CagAPY signal, as expected. These data show that CagA mutants with a phosphorylatable tyrosine in EPIYA-C (AY>F, BY>F, and ABY>F) produced the strongest CagAPY signals, indicating that EPIYA-C has a central role in CagA phosphorylation and that EPIYA-A and EPIYA-B can also be phosphorylated during infection.
Figure 4. Role of EPIYA motifs in CagA phosphorylation and AGS cell elongation during H. pylori infection.
(A) AGS cells were infected for 4 hours with CagA-expressing H. pylori strains as indicated. Phosphorylation of CagA was examined using α–PY-99 and α-CagA antibodies (arrows). (B) The number of elongated cells in each experiment was quantitated in triplicate in 10 different 0.25-mm2 fields. (C) Phase-contrast micrographs of AGS cells infected with the different strains as indicated. **P ≤ 0.001.
These phosphorylation data were correlated with the elongation phenotype by quantitating the number of elongated AGS cells in the same experiments. Interestingly, all constructs inducing strong CagAPY signals (WT CagA, EPIYA-AY>F, and EPIYA-BY>F) exhibited a strong elongation phenotype in more than 60% of cells, with only 1 exception. The EPIYA-ABY>F mutant induced strong CagAPY signals, but the elongation phenotype was significantly suppressed (Figure 4, B and C), suggesting that EPIYA-C phosphorylation is necessary but not sufficient for triggering the phenotypical outcome. All other CagA constructs (EPIYA-CY>F, EPIYA-ACY>F, EPIYA-BCY>F, and EPIYA-ABCY>F) were unable to induce the elongation phenotype. Taken together, these data suggest that none of the 3 EPIYA motifs alone can induce the elongation phenotype, which appears to require CagA phosphorylation of at least 2 different EPIYA motifs.
Interaction of phosphomimetic CagA mutants with host signaling factors.
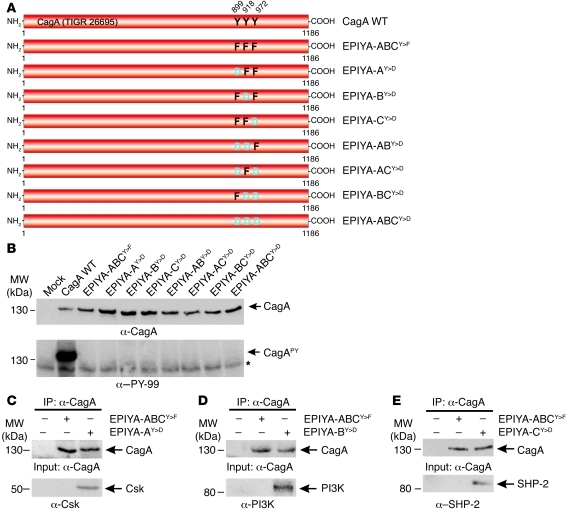
To determine the minimum number of phosphorylated EPIYA motifs needed for phenotypic outcome during infection, we generated phosphorylation-mimetic strain 26695 CagA mutants with EPIYA tyrosines replaced by aspartic acid (Y>D), which serves as a phosphorylation substrate in other proteins (38, 39). To avoid additional phosphorylation of the constructs by c-Src or c-Abl, all nontargeted tyrosines in adjacent EPIYA motifs were replaced by phenylalanines. The resulting constructs were transferred into the ΔcagA deletion mutant and named as shown in Figure 5A. Comparable expression of each CagA mutant was verified by immunoblotting, and infection of AGS cells confirmed that none of the CagA mutants induced a tyrosine phosphorylation signal (Figure 5B). To ensure that each construct was injected and functional, protein lysates of infected AGS cells were subjected to α-CagA immunoprecipitation and immunoblotting using antibodies against previously reported host interaction partners of CagA phosphorylated at EPIYA-A (Csk), EPIYA-B (PI3K), or EPIYA-C (SHP-2) (4, 14, 28, 29). Figure 5, C–E, and Supplemental Figure 7, A–C, show that each of these factors indeed formed a complex with the respective phosphomimetic CagA EPIYAY>D mutant. As control, immunoprecipitation of the nonphosphorylatable triple EPIYA-ABCY>F mutant revealed no signal for any of the above factors (Figure 5, C–E). In addition, we observed that Csk also bound to EPIYA-BY>D but only very poorly to EPIYA-CY>D, PI3K bound weakly to EPIYA-AY>D or EPIYA-CY>D, while SHP-2 also bound to EPIYA-BY>D but only weakly to EPIYA-AY>D (Supplemental Figure 7, A–C). These data indicate that phosphomimetic mutants are injected into AGS cells where they bind functionally to known host interaction partners of CagAPY.
Figure 5. Generation of phosphomimetic CagA mutants and their interaction with host signaling factors during H. pylori infection.
(A) Site-directed Y>D mutagenesis of CagA EPIYA-A, EPIYA-B, and EPIYA-C to generate phosphomimetic mutants. Nontargeted tyrosines in adjacent EPIYA motifs were replaced by phenylalanines to avoid additional phosphorylation events per molecule. The resulting single, double, and triple mutants were named as indicated. (B) AGS cells were infected for 4 hours with CagA-expressing H. pylori strains as indicated. CagA phosphorylation was examined using α–PY-99 and α-CagA antibodies (arrows). All strains expressed similar amounts of CagA, but only H. pylori expressing WT CagA revealed a phosphorylation signal. The asterisk in the lower panel indicates antibody cross-reactivity with an unknown phosphorylated host cell protein. (C–E) AGS cells were infected for 4 hours with CagA-expressing H. pylori strains as indicated, and cell lysates were subjected to IP with α-CagA antibodies. Western blotting using α-Csk (C), α-PI3K (D), or α–SHP-2 (E) indicated that each of these factors formed a complex with the respective phosphomimetic CagA EPIYAY>D mutant, but not with the nonphosphorylatable triple EPIYA-ABCY>F mutant as control.
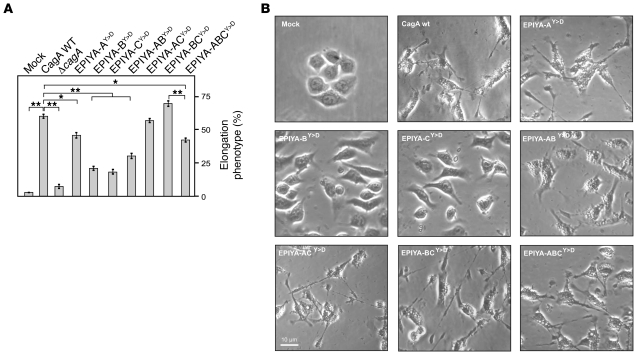
Phosphomimetic CagA mutants induce the AGS cell elongation phenotype.
We next assessed the extent of AGS cell elongation following infection with phosphomimetic Y>D mutants. Interestingly, infection with a CagA EPIYA-ACY>D mutant expressing the phosphomimetic residues for 2 EPIYA motifs (EPIYA-A and EPIYA-C) induced an elongation phenotype indistinguishable from WT CagA infection (Figure 6, A and B). Another double phosphomimetic mutant, EPIYA-BCY>D, also induced high numbers of elongated AGS cells compared with WT or ACY>D strains, while a ABY>D mutant induced intermediate levels (Figure 6, A and B). Furthermore, infection with a AY>D mutant expressing a single phosphomimetic EPIYA motif induced intermediate elongation data, as did a triple phosphomimetic ABCY>D mutant, while infection with BY>D or CY>D mutants significantly suppressed the number of elongated cells (Figure 6, A and B). These data are consistent with the results obtained by 2-DE, suggesting that 2 (but not 3) phosphorylated EPIYA motifs per CagA molecule induce signaling that leads to profound AGS cell elongation and that EPIYA-A and EPIYA-C (ACY>D) and EPIYA-B and EPIYA-C (BCY>D) are preferred over EPIYA-A and EPIYA-B (ABY>D) in this context.
Figure 6. H. pylori expressing phosphomimetic CagA mutants induce AGS cell elongation during infection.
(A) AGS cells were infected for 4 hours with CagA-expressing H. pylori strains as indicated. The number of elongated cells in each experiment was quantitated in triplicate in 10 different 0.25-mm2 fields. (B) Representative phase-contrast micrographs of infected AGS cells as indicated. *P ≤ 0.01; **P ≤ 0.001.
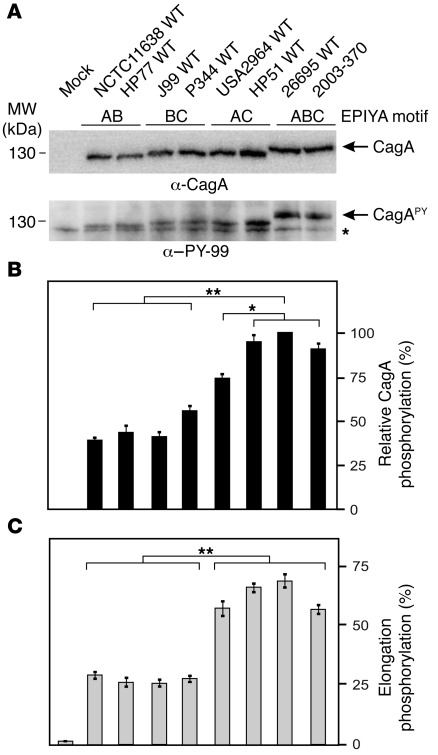
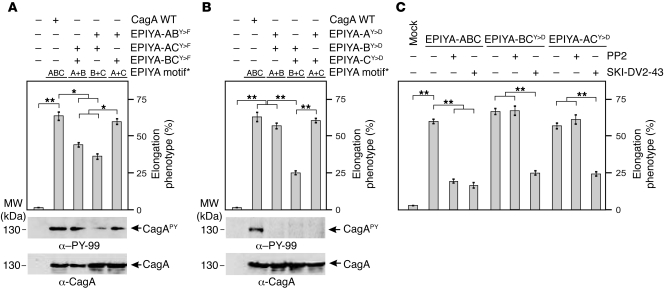
Western H. pylori clinical strains expressing CagA EPIYA-AC, but not EPIYA-AB or EPIYA-BC, induce profound AGS cell elongation.
We sought to confirm the foregoing CagA mutagenesis data by infecting AGS cells with Western H. pylori clinical strains expressing combinations of 2 EPIYA motifs, specifically EPIYA-AB, EPIYA-BC, or EPIYA-AC (Table 1). The respective EPIYA motifs and flanking sequences are shown in Supplemental Figure 8. AGS cells were infected with these strains or, as a control, with 2 isolates expressing 3 EPIYA repeats (EPIYA-ABC). AGS cell infection with EPIYA-AB, EPIYA-BC, or EPIYA-AC expressed similar amounts of CagA, but the intensity of CagAPY signals varied significantly (Figure 7A). While the clinical strains HP51 and USA2964 expressing the EPIYA-AC induced CagAPY levels comparable to strain 26695 or 2003-370 EPIYA-ABC, CagAPY levels induced by AB or BC strains (NCTC11638, HP77, J99, and P344) were significantly reduced by about 40%–60% (Figure 7, A and B). Quantitation of the induced elongation phenotypes revealed that strains expressing EPIYA-AC induced profound AGS cell elongation, but strains expressing EPIYA-AB or EPIYA-BC did not (Figure 7C).
Table 1 .
Characteristics of H. pylori strains used in the present study
Figure 7. Clinical Western H. pylori strains expressing CagA EPIYA-AC, but not EPIYA-AB or EPIYA-BC, induce profound AGS cell elongation.
(A) AGS cells were infected for 4 hours with strains expressing the combinations EPIYA-AB, EPIYA-BC, EPIYA-AC, and EPIYA-ABC (control). See Supplemental Figure 8 for specific EPIYA motifs and flanking sequences. CagA phosphorylation was examined using α–PY-CagA and α-CagA antibodies (arrows). The asterisk in the lower panel indicates antibody cross-reactivity with an unknown phosphorylated host cell protein. (B) Quantification of CagA phosphorylation signals using the luminescence image analyzer. (C) The number of elongated cells in each experiment was quantitated in triplicate in 10 different 0.25-mm2 fields. *P ≤ 0.01; **P ≤ 0.001.
East Asian H. pylori clinical strains expressing CagA EPIYA-AD or EPIYA-BD, but not EPIYA-AB, induce moderate AGS cell elongation.
AGS cells were also infected with clinical East Asian H. pylori strains expressing combinations of 2 EPIYA motifs, specifically EPIYA-AB, EPIYA-BD, or EPIYA-AD (Table 1), or, as controls, with strain TN2-GF4 or 35A clinical isolates expressing 3 motifs (EPIYA-ABD). AGS cells infected with strains carrying EPIYA-AB, EPIYA-BD, or EPIYA-AD expressed comparable amounts of CagA (with slightly reduced CagA in the case of EPIYA-BD strain Ca19), but the intensity of CagAPY signals varied significantly (Supplemental Figure 9A). While the ABD strains TN2-GF4 and 35A induced CagAPY levels comparable to their ABC strain counterparts (26695 or 2003-370), CagAPY levels of strains Oki105 and Oki149 (expressing EPIYA-AB), strains Ca19 and 3461 (EPIYA-BD), and strains Oki326 and Oki388 (EPIYA-AD) were significantly reduced by about 40%–60% (Supplemental Figure 9, A and B). Quantitation of the induced elongation phenotypes revealed that compared with EPIYA-ABD, EPIYA-BD induced strong AGS cell elongation, whereas induction by EPIYA-AD was moderate and by EPIYA-AB very weak (Supplemental Figure 9C). Thus, while similar weak activities were seen for EPIYA-AB of both Western and East Asian strains, the activities of EPIYA-BC/BD and EPIYA-AC/AD varied with geographical origin, suggesting that even small sequence variations observed in the EPIYA flanking region may impact phenotypical outcome (Supplemental Figure 8).
Dual infection with strains expressing different single phosphorylatable or phosphomimetic EPIYA motifs also induces AGS cell elongation.
The above results showed that Western H. pylori strains expressing CagA with only 1 phosphorylatable or phosphomimetic EPIYA motif failed to induce the AGS cell elongation phenotype, while strains with 2 specific phosphorylatable or phosphomimetic EPIYA motifs efficiently induced elongation. However, 2-DE data had revealed that populations of CagAPY proteins expressing either 1 or 2 phosphorylated EPIYA motifs were generated during infection (Figure 1). Accordingly, using a dual infection approach, we sought to determine whether a mixed population of CagA proteins expressing 1 phosphorylated tyrosine residue (at different positions) may also induce elongation. AGS cells were coinfected with 2 strains expressing mutations allowing phosphorylation of only 1 EPIYA motif each, either EPIYA-A+B, EPIYA-B+C, or EPIYA-A+C (Figure 8A and Supplemental Figure 10A). Cell infection with any 1 of these strains alone failed to induce the elongation phenotype (Figure 4), but coinfection with 1 of these strains enhanced elongation (Figure 8A). Notably, coinfection with strains expressing CagA EPIYA-BCY>F and EPIYA-ABY>F (representing the single phosphorylatable EPIYA-A+C) efficiently induced cell elongation that was almost indistinguishable from an infection with WT strain 26695 expressing EPIYA-ABC (Figure 8A). In addition, coinfection with strains expressing single phosphorylatable EPIYA-A+B or EPIYA-B+C slightly enhanced induction of AGS cell elongation, but did not reach WT CagA levels.
Figure 8. Dual infection of H. pylori strains expressing different single phosphorylatable or phosphomimetic EPIYA motifs induces AGS cell elongation.
AGS cells were infected for 4 hours with CagA EPIYAY>F (A) or EPIYAY>D (B) mutant strains as indicated. The available single phosphorylatable or phosphomimetic EPIYA motifs for each double infection are indicated. The number of elongated cells in each experiment was quantitated in triplicate in 10 different 0.25-mm2 fields, and CagA phosphorylation was examined using α–PY-99 and α-CagA antibodies (arrows). (C) AGS infection for 4 hours with the indicated CagA-expressing strains in the presence or absence of c-Src inhibitor PP2 (10 μM) or c-Abl inhibitor SKI-DV2-43 (1 μM) revealed significant changes in AGS cell elongation as quantitated in triplicate in 10 different 0.25-mm2 fields. *P ≤ 0.01; **P ≤ 0.001.
We also sought to determine if AGS cell elongation could be induced in coinfections with H. pylori strains expressing only 1 phosphomimetic EPIYA motif. AGS cells were coinfected with 2 strains expressing either AY>D, BY>D, or CY>D CagA proteins in all possible combinations (Figure 8B and Supplemental Figure 10B). The elongation results confirmed data shown in Figure 8A, and indicated that 2 different CagA proteins expressing either phosphomimetic EPIYA-A or EPIYA-C efficiently induce AGS cell elongation. Coinfection with 2 strains, one expressing phosphomimetic EPIYA-A and the other expressing phosphomimetic EPIYA-B, induced AGS cell elongation more efficiently than the corresponding Y>F mutants, while the phosphomimetic B and C mutants failed to induce AGS cell elongation. We conclude that the cell elongation phenotype in Western strains can be induced by single CagA proteins with 2 phosphorylatable EPIYA motifs, or by a combination of 2 different CagA proteins each with a single phosphorylatable EPIYA motif or by the corresponding phosphomimetic variants.
Different roles for c-Src and c-Abl kinases in CagA-induced AGS cell elongation.
Having shown that c-Src and c-Abl phosphorylated different EPIYA sites and that inhibiting either kinase abrogated AGS cell elongation (Figure 3 and Supplemental Figure 4), we asked if inhibited AGS cell elongation is due to impaired CagA phosphorylation and/or kinase-impaired downstream signaling. We infected AGS cells with the phosphomimetic EPIYA-BCY>D or EPIYA-ACY>D mutants in the presence of PP2 or SKI-DV2-43, respectively. Figure 8C shows that while both inhibitors suppressed the elongation phenotype in the WT ABD control, c-Abl or c-Src inhibition had different effects during infection by phosphomimetic EPIYA mutants. SKI-DV2-43 inhibition of c-Abl downregulated the phenotype induced by EPIYA-BCY>D or EPIYA-ACY>D, and PP2 inhibition of c-Src was unable to block AGS cell elongation by these mutants (Figure 8C). This suggests that the role of c-Src in phenotypical outcome is restricted to initial CagA phosphorylation and that c-Abl also promotes other downstream signaling functions.
Discussion
Posttranslational phosphorylation is the most common mechanism by which protein functions can be significantly altered and plays a key role in regulating intracellular signaling cascades (40–42). During their lifetime in the cell, about one-third of all eukaryotic proteins are modified by serine, threonine, or tyrosine phosphorylation (42). Signaling is commonly propagated by processive phosphorylation, whereby a protein kinase binds to a substrate and phosphorylates all available sites before dissociating. Multisite modification of such proteins is known as the “molecular barcode” hypothesis, in which each combination of specific protein modifications conveys a unique meaning in a given cell (41, 42). Interestingly, several bacterial pathogens, including enteropathogenic E. coli, C. trachomatis, B. henselae, A. phagocytophilum, E. chaffeensis, and H. pylori, use T3SSs and T4SSs to inject effector proteins that perturb host cell phosphorylation mechanisms (4–8). The respective Tir, Tarp, BepD-F, AnkA, and CagA effectors are tyrosine phosphorylated by host cell kinases and target various SH2 domain–containing components of eukaryotic signaling pathways, subverting host cell functions to the pathogen’s benefit and promoting disease development (14). Infection studies using knockout cell lines, inhibitors, or in vitro phosphorylation assays have shown that Src and Abl family kinases phosphorylate at least some of these effectors (10–13). However, little is known about the regulation of multiple phosphorylation events per effector molecule. Here, using 2-DE and site-directed mutagenesis, we demonstrated that only 1 or 2 H. pylori EPIYA motifs were concurrently phosphorylated per CagA molecule and that c-Src and c-Abl kinases phosphorylated different EPIYA sites. We propose a model for hierarchic phosphorylation by both kinases, allowing tight control of effector protein phosphorylation during infection.
Previous phosphorylation studies of CagA and other bacterial effector proteins relied on conventional 1-DE gels, which preclude quantitation of individual phosphorylation events. We separated phosphorylated and nonphosphorylated CagA proteins by 2-DE. Overlays of CagA and phosphotyrosine immunoblots consistently yielded 3 discrete full-length CagA signals at pI values of 7.5, 7.0, and 6.5. Mass spectrometry had previously confirmed these 3 spots to be CagA, but phosphorylated peptides were not observed, probably due to their low abundance (34). Our data identify spot 3 of pI 7.5 as nonphosphorylated CagA, and the other 2 spots (pI of 7.0 and 6.5) as CagAPY molecules phosphorylated at 1 or 2 tyrosines, respectively, consistent with known acidic shifts of increasingly phosphorylated eukaryotic proteins (43, 44). Confirmatory evidence came from phosphatase treatment, which led to a significant attenuation of CagAPY spots 1 and 2 and an increase of the nonphosphorylated CagA spot 3. As another control, infections with isogenic T4SS-defective strains yielded no CagAPY spots, as expected. These data suggest that in typical Western H. pylori strains carrying the 3 motifs EPIYA-ABC (e.g., TIGR 26695) or East Asian strains with EPIYA-ABD (e.g., TN2-GF4), only 1 or 2 EPIYA sites, but not 3, can be simultaneously phosphorylated per CagA molecule.
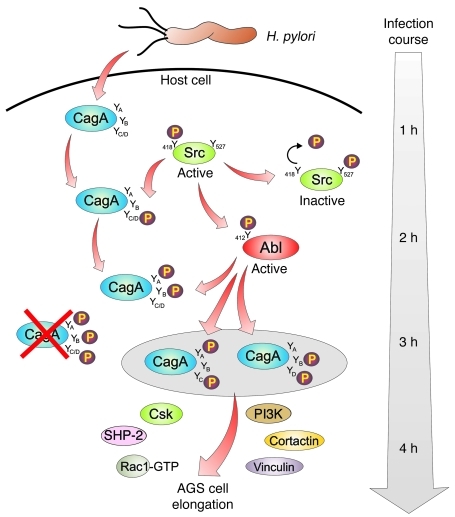
The role of c-Src and c-Abl kinases in CagAPY expression during infection was investigated using activation-specific antibodies, and Figure 9 presents a model for time-dependent c-Src– and c-Abl–mediated CagA phosphorylation and related signaling. In agreement with previous reports (12, 13, 45), our data indicated that c-Src was transiently activated between 30 and 90 minutes, whereas c-Abl was constitutively highly activated between 90 and 180 minutes. Interestingly, our inhibitor studies by 2-DE showed that c-Src but not c-Abl phosphorylated CagA at 1 site (spot 1) after 30 minutes of infection. Conversely, after 180 minutes of infection, inhibition of c-Abl eliminated CagAPY spots 1 and 2 completely, while PP2 treatment had only slight effects, indicating that c-Abl but not c-Src predominantly phosphorylates CagA proteins on 1 or 2 sites at later time points (Figure 9). Another important question was whether c-Src and c-Abl phosphorylate all or only selected EPIYA motifs in CagA. In vitro phosphorylation assays using single, double, and triple EPIYA Y>F point mutations and phosphospecific antibodies for EPIYA-C/D surprisingly showed that c-Src phosphorylates only EPIYA-C/D, but not EPIYA-A or EPIYA-B. Thus, CagAPY spot 1 on 2-DE gels at 30 minutes represents EPIYA-C/D phosphorylated by c-Src. In contrast, c-Abl phosphorylated all 4 motifs, EPIYA-A, EPIYA-B, EPIYA-C, and EPIYA-D, and these data are consistent with time-dependent CagAPY spot development and with inhibitor experiments. Thus, after 90–180 minutes of infection, 2-DE gel CagAPY spots 1 and 2 represented a mixed population of CagAPY phosphorylated by c-Abl on various EPIYAs (Figure 3).
Figure 9. Model showing successive CagA phosphorylation, production of specific CagAPY protein species during infection, and requirements of CagA phosphorylation–dependent signaling leading to AGS cell elongation.
H. pylori injects CagA by a T4SS-dependent process. Early in infection (1–2 hours), c-Src is activated by Y-418 phosphorylation, resulting in rapid phosphorylation of EPIYA-C or EPIYA-D in translocated CagA. CagAPY then inactivates c-Src in a negative feedback loop whereby direct binding of CagAPY to c-Src and Csk phosphorylates the negative regulatory site Y-527 and dephosphorylates Y-418 in c-Src. The c-Abl kinase is also activated by H. pylori initially involving c-Src. In contrast to c-Src, c-Abl kinase is continuously activated at later times of infection (2–4 hours). Activated c-Abl (phosphorylated at Y-412) continues phosphorylating CagA at the indicated EPIYAs when c-Src is inactive. We propose that the indicated phosphorylated forms of CagA, SHP-2, Csk, PI3K, cortactin, vinculin, and Rac1 create discrete protein complexes to activate downstream signaling that leads to cytoskeletal rearrangements and AGS cell elongation.
In eukaryotic proteins, phosphorylation of one residue may affect phosphorylation of other residues. In several cases of multisite phosphorylation, a protein kinase activity is required to “prime” phosphorylation of nearby site(s) by a second kinase (40, 42, 46, 47). Proteins known to require priming are glycogen synthase kinase (GSK3), a critical enzyme in the insulin and Wnt signaling pathways (42), and vasodilator-stimulated phosphoprotein (VASP), a regulator of actin dynamics with key roles in morphogenesis, axon guidance, endothelial barrier function, and cancer cell invasion (48). Both GSK3 and VASP are regulated by serine/threonine and not tyrosine phosphorylation. In this study, we found that CagA mutants with a phosphorylatable tyrosine in EPIYA-C (AY>F, BY>F, and ABY>F) produced the strongest CagAPY signals, while the single CY>F mutant clearly inhibited phosphorylation of other EPIYAs, indicating that EPIYA-C (and probably EPIYA-D) is critical in initiating further CagA phosphorylation. We conclude that the major phospho-CagA populations induced by Western H. pylori strain 26695 infection are EPIYA-CPY, EPIYA-ACPY, and EPIYA-BCPY, and those induced by the East Asian strain TN2-GF4 are EPIYA-DPY, EPIYA-ADPY and EPIYA-BDPY.
This CagAPY scenario is reminiscent of 2 other eukaryotic signaling proteins controlled by tyrosine phosphorylation. The focal adhesion multidomain “docking protein” 130-kDa Crk-associated substrate (p130-Cas) contains multiple protein-protein interaction domains with numerous Tyr-X-X-Pro (YXXP) motifs that are major sites of tyrosine phosphorylation. Like CagA, p130-Cas can be phosphorylated by Src and Abl family kinases, creating binding sites for SH2-containing signaling effectors (49–51). Most notably, recruitment of Crk adaptor proteins has been strongly implicated in promoting GTPase Rac-1 activation and cell motility, events that are also induced by CagAPY (13, 52, 53). Grab2-associated binder (Gab), another docking protein family member, is phosphorylated on 2 tyrosine residues by the c-Met receptor tyrosine kinase (54), creating Gab1 binding motifs for SHP-2, stimulating SHP-2 phosphatase, and activating MAP kinase cascades (55). Genetic studies have suggested that CagA may show Gab-like adapter protein activity in Drosophila (56). Thus, CagAPY has striking functional similarities to p130-Cas and Gab adapter proteins, in terms of both tyrosine phosphorylation and recruitment of SH2 factors that induce downstream signaling cascades.
This study also provided several insights into which EPIYA motifs are important for the H. pylori–induced cell elongation phenotype. First, we confirmed using a triple CagA EPIYA-ABCY>F mutant that CagA phosphorylation was necessary for phenotypical outcome, as reported previously (11, 13, 28). Second, using single and double CagAY>F mutants, we showed that AGS cell elongation could not be induced by any single phosphorylated EPIYA motif. Third, phosphorylation of EPIYA-C was necessary but not sufficient for inducing elongation. Lastly, at least 2 phosphorylated EPIYAs were necessary for the elongation phenotype. These findings are in good agreement with results of infections with clinical Western and East Asian H. pylori strains encoding only 2 EPIYA motifs. Quantitation of the induced elongation phenotypes revealed that strains expressing EPIYA-AC or EPIYA-BD induced profound AGS cell elongation, while EPIYA-AB, EPIYA-BC, or EPIYA-AD strains were significantly impaired in this respect.
Additional insights were gained by creation of phosphorylation-mimetic CagA EPIYAY>D mutants with aspartic acids replacing tyrosines, as described previously for several eukaryotic signaling molecules (38, 39). Although lacking tyrosine phosphorylation (Figure 5B), these constructs were functional because immunoprecipitation of CagA identified known host interaction partners of EPIYA-A (Csk), EPIYA-B (PI3K), and EPIYA-C (SHP-2) (4, 14, 28, 29) for the corresponding phosphomimetic EPIYAY>D motifs. The injected CagAY>D mutant proteins allowed further study of the requirements of the elongation phenotype. Interestingly, infection with an EPIYA-ACY>D mutant induced an elongation phenotype indistinguishable from WT H. pylori infection, in good agreement with foregoing results. All other phosphomimetic EPIYAY>D mutants, including the triple EPIYA-ABCY>D mutant strain, induced marginal or intermediate cell elongation. A provocative finding was that single EPIYA-AY>F and double phosphomimetic EPIYA-BCY>D mutants induced many elongated AGS cells, as did East Asian clinical strains expressing phosphorylatable EPIYA-BD, while some EPIYA-BC strains did not. The reason for this discrepancy is as yet unclear. We propose that the elongation phenotype is primarily induced by simultaneous phosphorylation of EPIYA-AC or EPIYA-BD motifs, when present, and that the arrangement of specific amino acids surrounding the EPIYA motifs may significantly influence phenotypical outcome.
As noted above, H. pylori strains expressing CagA with only 1 phosphorylatable or phosphomimetic EPIYA motif failed to induce the elongation phenotype, while strains with 2 phosphorylatable or phosphomimetic EPIYA motifs efficiently induced elongation in single infections. Interestingly, AGS cell coinfection with 2 strains expressing mutations allowing phosphorylation of 1 EPIYA motif each, or the corresponding phosphomimetic mutants for EPIYA-AC and, to a lesser extent, EPIYA-AB (but not EPIYA-BC), efficiently restored the elongation phenotype. This phenotype can therefore be induced by a single CagA protein with 2 phosphorylatable EPIYA motifs (preferably EPIYA-AC), or by 2 CagA proteins, each with a single phosphorylatable EPIYA motif, which may form dimers to trigger this response. In this context, recent studies have identified a CagA multimerization sequence (CM) that interacts with the Par1 kinase independently of CagA tyrosine phosphorylation (57). Mutation of the CM sequence also caused reduced SHP-2 binding activity and attenuated induction of the AGS cell elongation phenotype (57).
EPIYA-ABC and EPIYA-ABD are the predominant combination of CagA phosphorylation sites in Western and East Asian H. pylori strains, respectively (15–18, 31). Thus, the present study carries important clinical implications. East Asian H. pylori strains expressing EPIYA-D show higher binding affinities for SHP-2, induce a more pronounced cell elongation phenotype, and provoke more gastric epithelial cell IL-8 secretion than Western H. pylori strains expressing EPIYA-ABC (58–61). Mutated SHP-2 is associated with human malignancies, and mouse studies have shown that perturbations of SHP-2 signaling promote development of gastric adenocarcinoma (62, 63). The in vitro cell elongation phenotype reflects the infection-induced pathophysiological contraction of epithelial cells that compromises the integrity of the epithelial barrier, providing bacterial access to subepithelial compartments and facilitating cell migration and metastasis. Lastly, higher levels of H. pylori–induced IL-8 secretion provoke greater recruitment of infiltrating phagocytic cells that potentially exacerbate inflammation-related epithelial damage, promoting eventual mucosal progression to dysplasia and carcinoma. Thus, to the extent that H. pylori EPIYA motif strain differences contribute to clinical virulence, understanding the mechanistic basis of EPIYA motif–driven cellular pathophysiology may inform genomic typing of clinical H. pylori strains as an important factor in therapeutic decision making.
In summary, this study defined a hierarchic CagA tyrosine phosphorylation process in which c-Src phosphorylated EPIYA-C/D early in infection (but not EPIYA-A or EPIYA-B), while c-Abl phosphorylated EPIYA-A, EPIYA-B, EPIYA-C, and EPIYA-D later in infection. Importantly, CagA phosphorylation was tightly regulated over the entire course of infection. We showed that CagA was phosphorylated on 1 or 2, but never simultaneously on 3, EPIYA motifs per CagA in 26695 or TN2-GF4 strains. EPIYA-C/D was the first motif to be phosphorylated, and coexisted later with phosphorylated EPIYA-A or EPIYA-B. This study also showed that none of the phosphorylated EPIYA motifs alone was sufficient to induce AGS cell elongation. The preferred combination of phosphorylated EPIYA motifs in Western strains was EPIYA-A and EPIYA-C, induced either as individual phosphorylation of 2 CagA proteins or simultaneously of 1 CagA protein. These data were verified using phosphomimetic CagA variants in H. pylori. Future studies will investigate strains expressing different EPIYA motif combinations or strains in which EPIYA-C or EPIYA-D are amplified up to 6 times (4). Finally, since CagA phosphorylation sites show significant homology with bacterial effector proteins in other pathogens (4, 64), further study of other bacterial tyrosine-phosphorylated effector proteins will undoubtedly augment our understanding of molecular mimicry and elucidate mechanisms underlying infection-associated pathophysiology.
Methods
H. pylori isolates.
WT strains were virulent type I isolates, with a functional T4SS encoded by the cagPAI (Table 1). The cagA deletion mutant in strain P1 was recently described (13, 65). H. pylori was grown in thin layers on horse serum GC agar plates supplemented with vancomycin (10 μg/ml), nystatin (1 μg/ml), and trimethoprim (5 μg/ml) as described previously (66–68). Antibiotics were obtained from Sigma-Aldrich. Bacteria were grown at 37°C for 2 days in an anaerobic jar containing a Campygen gas mix of 5% O2, 10% CO2, and 85% N2 (Oxoid).
Cloning, complementation, and site-directed mutagenesis of CagA.
To analyze the EPIYA motifs in different CagA proteins, cagA gene subfragments from H. pylori strains were amplified by PCR cloned into the pCR2.1 vector (Invitrogen) and sequenced (25). For construction of a complementation vector, cagA containing its own promoter was amplified from strain 26695 (accession no. AAD07614) and cloned in the E. coli/H. pylori shuttle vector pHel3 containing the oriT of RP4 and a kanamycin resistance gene cassette (Aph-A3) as a selectable marker (69), resulting in vector pSB19. Site-directed mutagenesis of tyrosines Y-899, Y-918, and Y-972 in the CagA sequence was done using the Sculptor mutagenesis kit as directed (Amersham Pharmacia Biotech). All resulting plasmids were transformed into H. pylori (69).
Eukaryotic cell culture, inhibitors, infection, and elongation phenotype quantitation assays.
MKN-28 or AGS cells were grown in 6-well plates containing RPMI 1640 medium (Invitrogen) supplemented with 25 mM HEPES buffer and 10% heat-inactivated FBS (Biochrom) for 2 days to approximately 70% confluence. Cells were serum-deprived overnight and infected at a MOI of 50 for the indicated times per experiment (see figure legends). The c-Abl tyrosine kinase inhibitor SKI-DV2-43 (37) and c-Src inhibitor PP2 (Calbiochem) were dissolved in Me2SO and added to the cells 30 minutes before infection. After infection, the cells were harvested in ice-cold PBS containing 1 mmol/l Na3VO4 (Sigma-Aldrich). Elongated AGS cells in each experiment were quantitated in 10 different 0.25-mm2 fields using an Olympus IX50 phase contrast microscope. All experiments were performed in triplicate and subjected to statistical analysis as described below.
Protein tyrosine phosphatase treatment of protein lysates.
Pellets of 2 × 106 infected and noninfected AGS cells were collected in PBS and washed twice (600 g, room temperature) in 100 μl 1× phosphatase reaction buffer (Biorbyt). The final pellet was resuspended in 50 μl 1× reaction buffer containing 0.1% Triton X-100 for mild cell lysis. The phosphatase reactions were initiated by adding 10 U protein tyrosine phosphatase PTP-N1 (Biorbyt). Reactions continued for 3 hours at 37°C.
In vitro phosphorylation of CagA with recombinant c-Src or c-Abl.
1010 H. pylori cells expressing either WT CagAs or phosphorylation-deficient CagAs were harvested in 1 ml of kinase buffer as described previously (10). A total of 5 U of recombinant human c-Src (Upstate) or c-Abl (NEB) and 1 μmol/l of ATP were mixed with 30 μl of the H. pylori lysate and incubated for 30 minutes at 30°C as described previously (13).
2-DE.
AGS cells with attached bacteria were harvested and washed by centrifugation as described above to ensure that only intracellular proteins were analyzed, and then dissolved in 9 M urea, 70 mM DTT, and 2% Triton X-100. Fully denatured and reduced cellular proteins were analyzed using a 2-DE gel system (23 cm × 30 cm × 0.75 mm) with a resolving power of about 5,000 protein species (9). Proteins (150 μg) were loaded to the anodic side of the IEF gel (1 mm diameter). Resolved proteins in the second-dimension gel were detected by Coomassie Blue staining and immunoblot analysis and identified by mass spectrometry as described previously (34).
Conventional SDS-PAGE and immunoblot analysis.
Cell pellets with attached bacteria were mixed with equal amounts of 2× SDS-PAGE buffer and boiled for 5 minutes. Proteins were separated by SDS-PAGE on 6%–12% polyacrylamide gels and blotted onto PVDF membranes (Immobilon-P; Millipore). Membranes were blocked in TBS-T (140 mM NaCl; 25 mM Tris-HCl, pH 7.4; 0.1% Tween-20) with 3% BSA or 5% skim milk for 1 hour at room temperature. Membranes were incubated with mouse monoclonal α-phosphotyrosine antibody PY-99 (Santa Cruz) or monoclonal anti-CagA antibody (Austral Biologicals). Monoclonal antibodies recognizing SHP-2, c-Src, c-Abl, Csk, and PI3K were purchased from Santa Cruz. Activation-specific anti–c-Abl–PY-412 and anti–c-Src–PY-418 antibodies were purchased from NEB. We also produced a phosphospecific rabbit α–PY–EPIYA-C/D antibody using a peptide-based antigen (H-VGLSASPEPIpYAT; Jerini), as described previously (21), which recognizes both phosphorylated EPIYA-C and EPIYA-D. Phosphorylated and nonphosphorylated CagA proteins were detected using horseradish peroxidase–conjugated anti-mouse or anti-rabbit polyvalent sheep immunoglobulin secondary antibodies in the ECL Plus chemoluminescence Western Blot system (Amersham Pharmacia Biotech). Band intensities were quantitated with the Lumi-Imager F1 (Roche Diagnostics).
Statistics.
All data were evaluated using 1-tailed Student’s t test with SigmaStat statistical software (version 2.0). A P value of 0.01 or less was considered significant. Data are presented as mean ± SD.
Supplementary Material
Acknowledgments
We thank Bastian Thaa for technical assistance. This study was supported by Deutsche Forschungsgemeinschaft (Ba1671/3 and Ba1671/8-1 to S. Backert), the National Institute of Diabetes, Digestive, and Kidney Diseases (R56DK064371 to A. Smolka and R01DK62813 to Y. Yamaoka), and University College Dublin (R11408 to S. Backert). J. Torres is a recipient of a scholarship from Fundacion IMSS, Mexico.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2012;122(4):1553–1566. doi:10.1172/JCI61143.
Sabine Brandt’s present address is: University of Magdeburg, Department of Nephrology and Hypertension, Diabetes and Endocrinology, Magdeburg, Germany.
See the related Commentary beginning on page 1192.
References
- 1.Schlessinger J, Lemmon MA. SH2 and PTB domains in tyrosine kinase signaling. Sci STKE. 2003;2003(191):RE12. doi: 10.1126/stke.2003.191.re12. [DOI] [PubMed] [Google Scholar]
- 2.Covacci A, Rappuoli R. Tyrosine-phosphorylated bacterial proteins: Trojan horses for the host cell. J Exp Med. 2000;191(4):587–592. doi: 10.1084/jem.191.4.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dean P, Kenny B. The effector repertoire of enteropathogenic E. coli: ganging up on the host cell. . Curr Opin Microbiol. 2009;12(1):101–109. doi: 10.1016/j.mib.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Backert S, Tegtmeyer N, Selbach M. The versatility of Helicobacter pylori CagA effector protein functions: The master key hypothesis. . Helicobacter. 2010;15(3):163–176. doi: 10.1111/j.1523-5378.2010.00759.x. [DOI] [PubMed] [Google Scholar]
- 5.Clifton DR, et al. A chlamydial type III translocated protein is tyrosine-phosphorylated at the site of entry and associated with recruitment of actin. Proc Natl Acad Sci U S A. 2004;101(27):10166–10171. doi: 10.1073/pnas.0402829101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kenny B, DeVinney R, Stein M, Reinscheid DJ, Frey EA, Finlay BB. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. . Cell. 1997;91(4):511–520. doi: 10.1016/S0092-8674(00)80437-7. [DOI] [PubMed] [Google Scholar]
- 7.Schulein R, et al. A bipartite signal mediates the transfer of type IV secretion substrates of Bartonella henselae into human cells. Proc Natl Acad Sci U S A. 2005;102(3):856–861. doi: 10.1073/pnas.0406796102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rikihisa Y, Lin M. Anaplasma phagocytophilum and Ehrlichia chaffeensis type IV secretion and Ank proteins. Curr Opin Microbiol. 2010;13(1):59–66. doi: 10.1016/j.mib.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Backert S, Selbach M. Tyrosine-phosphorylated bacterial effector proteins: the enemies within. Trends Microbiol. 2005;13(10):476–484. doi: 10.1016/j.tim.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 10.Selbach M, Moese S, Hauck CR, Meyer TF, Backert S. Src is the kinase of the Helicobacter pylori CagA protein in vitro and in vivo. . J Biol Chem. 2002;277(9):6775–6778. doi: 10.1074/jbc.C100754200. [DOI] [PubMed] [Google Scholar]
- 11.Stein M, Bagnoli F, Halenbeck R, Rappuoli R, Fantl WJ, Covacci A. c-Src/Lyn kinases activate Helicobacter pylori CagA through tyrosine phosphorylation of the EPIYA-motifs. . Mol Microbiol. 2002;43(4):971–980. doi: 10.1046/j.1365-2958.2002.02781.x. [DOI] [PubMed] [Google Scholar]
- 12.Poppe M, Feller SM, Römer G, Wessler S. Phosphorylation of Helicobacter pylori CagA by c-Abl leads to cell motility. . Oncogene. 2007;26(24):3462–3472. doi: 10.1038/sj.onc.1210139. [DOI] [PubMed] [Google Scholar]
- 13.Tammer I, Brandt S, Hartig R, König W, Backert S. Activation of Abl by Helicobacter pylori: a novel kinase for CagA and crucial mediator of host cell scattering. . Gastroenterology. 2007;132(4):1309–1319. doi: 10.1053/j.gastro.2007.01.050. [DOI] [PubMed] [Google Scholar]
- 14.Selbach M, et al. Host cell interactome of tyrosine-phosphorylated bacterial proteins. Cell Host Microbe. 2009;5(4):397–403. doi: 10.1016/j.chom.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 15.Blaser MJ, Atherton JC. Helicobacter pylori persistence: biology and disease. . J Clin Invest. 2004;113(3):321–333. doi: 10.1172/JCI20925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamaoka Y, Kato M, Asaka M. Geographic differences in gastric cancer incidence can be explained by differences between Helicobacter pylori strains. . Intern Med. 2008;47(12):1077–1083. doi: 10.2169/internalmedicine.47.0975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amieva MR, El-Omar EM. Host-bacterial interactions in Helicobacter pylori infection. . Gastroenterology. 2008;134(1):306–323. doi: 10.1053/j.gastro.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 18.Polk DB, Peek RM., Jr Helicobacter pylori: gastric cancer and beyond. . Nat Rev Cancer. 2010;10(6):403–414. doi: 10.1038/nrc2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shaffer CL, et al. Helicobacter pylori exploits a unique repertoire of type IV secretion system components for pilus assembly at the bacteria-host cell interface. . PLoS Pathog. 2011;7(9):e1002237. doi: 10.1371/journal.ppat.1002237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Backert S, Meyer TF. Type IV secretion systems and their effectors in bacterial pathogenesis. Curr Opin Microbiol. 2006;9(2):207–217. doi: 10.1016/j.mib.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 21.Kwok T, et al. Helicobacter exploits integrin for type IV secretion and kinase activation. . Nature. 2007;449(7164):862–866. doi: 10.1038/nature06187. [DOI] [PubMed] [Google Scholar]
- 22.Hatakeyama M. SagA of CagA in Helicobacter pylori pathogenesis. . Curr Opin Microbiol. 2008;11(1):30–37. doi: 10.1016/j.mib.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 23.Backert S, et al. Functional analysis of the cag pathogenicity island in Helicobacter pylori isolates from patients with gastritis, peptic ulcer, and gastric cancer. . Infect Immun. 2004;72(2):1043–1056. doi: 10.1128/IAI.72.2.1043-1056.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohnishi N, et al. Transgenic expression of Helicobacter pylori CagA induces gastrointestinal and hematopoietic neoplasms in mouse. . Proc Natl Acad Sci U S A. 2008;105(3):1003–1008. doi: 10.1073/pnas.0711183105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Backert S, Moese S, Selbach M, Brinkmann V, Meyer TF. Phosphorylation of tyrosine 972 of the Helicobacter pylori CagA protein is essential for induction of a scattering phenotype in gastric epithelial cells. . Mol Microbiol. 2001;42(3):631–644. doi: 10.1046/j.1365-2958.2001.02649.x. [DOI] [PubMed] [Google Scholar]
- 26.Ridley AJ, et al. Cell migration: integrating signals from front to back. Science. 2003;302(5651):1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 27.Schneider S, Weydig C, Wessler S. Targeting focal adhesions: Helicobacter pylori–host communication in cell migration. . Cell Commun Signal. 2008;6:2. doi: 10.1186/1478-811X-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Higashi H, et al. SHP2 tyrosine phosphatase as an intracellular target of Helicobacter pylori CagA protein. . Science. 2002;295(5555):683–686. doi: 10.1126/science.1067147. [DOI] [PubMed] [Google Scholar]
- 29.Tsutsumi R, Higashi H, Higuchi M, Okada M, Hatakeyama M. Attenuation of Helicobacter pylori CagA x SHP2 signaling by interaction between CagA and C-terminal Src kinase. . J Biol Chem. 2003;278(6):3664–3670. doi: 10.1074/jbc.M208155200. [DOI] [PubMed] [Google Scholar]
- 30.Mimuro H, Suzuki T, Tanaka J, Asahi M, Haas R. Grb2 is a key mediator of Helicobacter pylori CagA protein activities. . Mol Cell. 2002;10(4):745–755. doi: 10.1016/S1097-2765(02)00681-0. [DOI] [PubMed] [Google Scholar]
- 31.Xia Y, Yamaoka Y, Zhu Q, Matha I, Gao X. A comprehensive sequence and disease correlation analyses for the C-terminal region of CagA protein of Helicobacter pylori. . PLoS One. 2009;4(11):e7736. doi: 10.1371/journal.pone.0007736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tomb JF, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. . Nature. 1997;388(6642):539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 33.Ogura K, et al. Virulence factors of Helicobacter pylori responsible for gastric diseases in Mongolian gerbil. . J Exp Med. 2000;192(11):1601–1610. doi: 10.1084/jem.192.11.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Backert S, Müller EC, Jungblut PR, Meyer TF. Tyrosine phosphorylation patterns and size modification of the Helicobacter pylori CagA protein after translocation into gastric epithelial cells. . Proteomics. 2001;1(4):608–617. doi: 10.1002/1615-9861(200104)1:4<608::AID-PROT608>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 35.Fischer W, Püls J, Buhrdorf R, Gebert B, Odenbreit S, Haas R. Systematic mutagenesis of the Helicobacter pylori cag pathogenicity island: essential genes for CagA translocation in host cells and induction of interleukin-8. . Mol Microbiol. 2001;42(5):1337–1348. doi: 10.1046/j.1365-2958.2001.02714.x. [DOI] [PubMed] [Google Scholar]
- 36.Nagao M, Yamauchi J, Kaziro Y, Itoh H. Involvement of protein kinase C and Src family tyrosine kinase in Galphaq/11-induced activation of c-Jun N-terminal kinase and p38 mitogen-activated protein kinase. . J Biol Chem. 1998;273(36):22892–22898. doi: 10.1074/jbc.273.36.22892. [DOI] [PubMed] [Google Scholar]
- 37.von Bubnoff N, et al. Inhibition of wild-type and mutant Bcr-Abl by pyrido-pyrimidine-type small molecule kinase inhibitors. Cancer Res. 2003;63(19):6395–6404. [PubMed] [Google Scholar]
- 38.Martinez-Quiles N, Ho HY, Kirschner MW, Ramesh N, Geha RS. Erk/Src phosphorylation of cortactin acts as a switch on-switch off mechanism that controls its ability to activate N-WASP. Mol Cell Biol. 2004;24(12):5269–5280. doi: 10.1128/MCB.24.12.5269-5280.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tegtmeyer N, Wittelsberger R, Hartig R, Wessler S, Martinez-Quiles N, Backert S. Serine phosphorylation of cortactin controls focal adhesion kinase activity and cell scattering induced by Helicobacter pylori. . Cell Host Microbe. 2011;9(6):520–531. doi: 10.1016/j.chom.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 40.Songyang Z, et al. Catalytic specificity of protein-tyrosine kinases is critical for selective signalling. Nature. 1995;373(6514):536–539. doi: 10.1038/373536a0. [DOI] [PubMed] [Google Scholar]
- 41.Yang XJ. Multisite protein modification and intramolecular signaling. Oncogene. 2005;24(10):1653–1662. doi: 10.1038/sj.onc.1208173. [DOI] [PubMed] [Google Scholar]
- 42.Patwardhan P, Miller WT. Processive phosphorylation: mechanism and biological importance. Cell Signal. 2007;19(11):2218–2226. doi: 10.1016/j.cellsig.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scheler C, Müller EC, Stahl J, Müller-Werdan U, Salnikow J, Jungblut P. Identification and characterization of heat shock protein 27 protein species in human myocardial two-dimensional electrophoresis patterns. Electrophoresis. 1997;18(15):2823–2831. doi: 10.1002/elps.1150181518. [DOI] [PubMed] [Google Scholar]
- 44.Holland C, et al. Quantitative phosphoproteomics reveals link between Helicobacter pylori infection and RNA splicing modulation in host cells. . Proteomics. 2011;11(14):2798–2811. doi: 10.1002/pmic.201000793. [DOI] [PubMed] [Google Scholar]
- 45.Selbach M, Moese S, Hurwitz R, Hauck CR, Meyer TF, Backert S. The Helicobacter pylori CagA protein induces cortactin dephosphorylation and actin rearrangement by c-Src inactivation. . EMBO J. 2003;22(3):515–528. doi: 10.1093/emboj/cdg050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cohen P. The regulation of protein function by multisite phosphorylation--a 25 year update. Trends Biochem Sci. 2000;25(12):596–601. doi: 10.1016/S0968-0004(00)01712-6. [DOI] [PubMed] [Google Scholar]
- 47.Pawson T, Scott JD. Protein phosphorylation in signaling--50 years and counting. Trends Biochem Sci. 2005;30(6):286–90. doi: 10.1016/j.tibs.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 48.Knauer O, et al. Differential phosphoproteome profiling reveals a functional role for VASP in Helicobacter pylori-induced cytoskeleton turnover in gastric epithelial cells. . Cell Microbiol. 2008;10(11):2285–2296. doi: 10.1111/j.1462-5822.2008.01207.x. [DOI] [PubMed] [Google Scholar]
- 49.Mayer BJ, Hirai H, Sakai R. Evidence that SH2 domains promote processive phosphorylation by protein-tyrosine kinases. Curr Biol. 1995;5(3):296–305. doi: 10.1016/s0960-9822(95)00060-1. [DOI] [PubMed] [Google Scholar]
- 50.Bouton AH, Riggins RB, Bruce-Staskal PJ. Functions of the adapter protein Cas: signal convergence and the determination of cellular responses. Oncogene. 2001;20(44):6448–6458. doi: 10.1038/sj.onc.1204785. [DOI] [PubMed] [Google Scholar]
- 51.Ruest PJ, Shin NY, Polte TR, Zhang X, Hanks SK. Mechanisms of CAS substrate domain tyrosine phosphorylation by FAK and Src. Mol Cell Biol. 2001;21(22):7641–7652. doi: 10.1128/MCB.21.22.7641-7652.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suzuki M, Mimuro H, Suzuki T, Park M, Yamamoto T, Sasakawa C. Interaction of CagA with Crk plays an important role in Helicobacter pylori-induced loss of gastric epithelial cell adhesion. . J Exp Med. 2005;202(9):1235–1247. doi: 10.1084/jem.20051027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brandt S, et al. Use of a novel coinfection system reveals a role for Rac1, H-Ras, and CrkII phosphorylation in Helicobacter pylori-induced host cell actin cytoskeletal rearrangements. . FEMS Immunol Med Microbiol. 2007;50(2):190–205. doi: 10.1111/j.1574-695X.2007.00234.x. [DOI] [PubMed] [Google Scholar]
- 54.Schaeper U, Gehring NH, Fuchs KP, Sachs M, Kempkes B, Birchmeier W. Coupling of Gab1 to c-Met, Grb2, and SHP-2 mediates biological responses. . J Cell Biol. 2000;149(7):1419–1432. doi: 10.1083/jcb.149.7.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hatakeyama M. Helicobacter pylori CagA--a potential bacterial oncoprotein that functionally mimics the mammalian Gab family of adaptor proteins. . Microbes Infect. 2003;5(2):143–150. doi: 10.1016/S1286-4579(02)00085-0. [DOI] [PubMed] [Google Scholar]
- 56.Botham CM, Wandler AM, Guillemin K. A transgenic Drosophila model demonstrates that the Helicobacter pylori CagA protein functions as a eukaryotic Gab adaptor. . PLoS Pathog. 2008;4(5):e1000064. doi: 10.1371/journal.ppat.1000064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nagase L, Murata-Kamiya N, Hatakeyama M. Potentiation of Helicobacter pylori CagA virulence through homodimerization. . J Biol Chem. 2011;286(38):33622–33631. doi: 10.1074/jbc.M111.258673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Higashi H, et al. Biological activity of the Helicobacter pylori virulence factor CagA is determined by variation in the tyrosine phosphorylation sites. . Proc Nat Acad Sci USA. 2002;99(22):14428–14433. doi: 10.1073/pnas.222375399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hatakeyama M. Oncogenic mechanisms of the Helicobacter pylori CagA protein. . Nat Rev Cancer. 2004;4(9):688–694. doi: 10.1038/nrc1433. [DOI] [PubMed] [Google Scholar]
- 60.Naito M, et al. Influence of EPIYA-repeat polymorphism on the phosphorylation-dependent biological activity of Helicobacter pylori CagA. . Gastroenterology. 2006;130(4):1181–1190. doi: 10.1053/j.gastro.2005.12.038. [DOI] [PubMed] [Google Scholar]
- 61.Argent RH, Hale JL, El-Omar EM, Atherton JC. Differences in Helicobacter pylori CagA tyrosine phosphorylation motif patterns between western and East Asian strains, and influences on interleukin-8 secretion. . J Med Microbiol. 2008;57(pt 9):1062–1067. doi: 10.1099/jmm.0.2008/001818-0. [DOI] [PubMed] [Google Scholar]
- 62.Judd LM, et al. Gastric cancer development in mice lacking the SHP2 binding site on the IL-6 family co-receptor gp130. Gastroenterology. 2004;126(1):196–207. doi: 10.1053/j.gastro.2003.10.066. [DOI] [PubMed] [Google Scholar]
- 63.Tebbutt NC, et al. Reciprocal regulation of gastrointestinal homeostasis by SHP2 and STAT-mediated trefoil gene activation in gp130 mutant mice. Nat Med. 2002;8(10):1089–1097. doi: 10.1038/nm763. [DOI] [PubMed] [Google Scholar]
- 64.Backert S, Selbach M. Tyrosine-phosphorylated bacterial effector proteins: the enemies within. Trends Microbiol. 2005;13(10):476–484. doi: 10.1016/j.tim.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 65.Brandt S, Kwok T, Hartig R, König W, Backert S. NF-kappaB activation and potentiation of proinflammatory responses by the Helicobacter pylori CagA protein. . Proc Natl Acad Sci U S A. 2005;102(26):9300–9355. doi: 10.1073/pnas.0409873102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Backert S, et al. Translocation of the Helicobacter pylori CagA protein in gastric epithelial cells by a type IV secretion apparatus. . Cell Microbiol. 2000;2:155–164. doi: 10.1046/j.1462-5822.2000.00043.x. [DOI] [PubMed] [Google Scholar]
- 67.Moese S, Selbach M, Meyer TF, Backert S. cag+ Helicobacter pylori induces homotypic aggregation of macrophage-like cells by up-regulation and recruitment of intracellular adhesion molecule 1 to the cell surface. . Infect Immun. 2002;70(8):4687–4691. doi: 10.1128/IAI.70.8.4687-4691.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Selbach M, Moese S, Meyer TF, Backert S. Functional analysis of the Helicobacter pylori cag pathogenicity island reveals both VirD4-CagA-dependent and VirD4-CagA-independent mechanisms. . Infect Immun. 2002;70(2):665–671. doi: 10.1128/IAI.70.2.665-671.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Heuermann D, Haas R. A stable shuttle vector system for efficient genetic complementation of Helicobacter pylori strains by transformation and conjugation. . Mol Gen Genet. 1998;257(5):519–528. doi: 10.1007/s004380050677. [DOI] [PubMed] [Google Scholar]
- 70.Tegtmeyer N, Zabler D, Schmidt D, Hartig R, Brandt S, Backert S. Importance of EGF receptor, HER2/Neu and Erk1/2 kinase signalling for host cell elongation and scattering induced by the Helicobacter pylori CagA protein: antagonistic effects of the vacuolating cytotoxin VacA. Cell Microbiol. 2009;11(3):488–505. doi: 10.1111/j.1462-5822.2008.01269.x. [DOI] [PubMed] [Google Scholar]
- 71.Bukanov NO, Berg DE. Ordered cosmid library and high-resolution physical-genetic map of Helicobacter pylori strain NCTC11638. Mol Microbiol. 1994;11(3):509–23. doi: 10.1111/j.1365-2958.1994.tb00332.x. [DOI] [PubMed] [Google Scholar]
- 72.Panayotopoulou EG, et al. Strategy to characterize the number and type of repeating EPIYA phosphorylation motifs in the carboxyl terminus of CagA protein in Helicobacter pylori clinical isolates. . J Clin Microbiol. 2007;45(2):488–495. doi: 10.1128/JCM.01616-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Alm RA, et al. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. . Nature. 1999;397(6715):176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.