Summary
We identified four PDZ domain-containing proteins, syntenin, PICK1, GRIP, and PSD95, as interactors with the kainate receptor (KAR) subunits GluR52b, GluR52c, and GluR6. Of these, we show that both GRIP and PICK1 interactions are required to maintain KAR-mediated synaptic function at mossy fiber-CA3 synapses. In addition, PKCα can phosphorylate ct-GluR52b at residues S880 and S886, and PKC activity is required to maintain KAR-mediated synaptic responses. We propose that PICK1 targets PKCα to phosphorylate KARs, causing their stabilization at the synapse by an interaction with GRIP. Importantly, this mechanism is not involved in the constitutive recycling of AMPA receptors since blockade of PDZ interactions can simultaneously increase AMPAR- and decrease KAR-mediated synaptic transmission at the same population of synapses.
Introduction
The physiological roles of AMPA and NMDA receptors in synaptic transmission and plasticity are well known (Bliss and Collingridge, 1993), and many of the molecular interactions that are involved in their targeting to, and organization at, postsynaptic sites have been identified (for reviews, see Braithwaite et al., 2000; Garner et al., 2000; Sheng, 2001). Corresponding progress has not been achieved for kainate receptors (KARs) and, despite significant advances in the characterization of some of their physiological roles, little is known about their regulation, targeting, or trafficking.
Unlike AMPA receptors (AMPARs), which are predominantly or exclusively postsynaptic, there are functional KARs at both pre- and postsynaptic sites (for reviews, see Chittajallu et al., 1999; Lerma et al., 2001). Presynaptic KARs modulate transmitter release (e.g., Chittajallu et al., 1996; Clarke et al., 1997; Contractor et al., 2000) and can be activated synaptically to both increase (Lauri et al., 2001; Schmitz et al., 2001) and decrease (Kidd et al., 2002; Schmitz et al., 2000) glutamate release. Postsynaptic KARs are involved in synaptic transmission, as first shown at hippocampal mossy fiber synapses where they mediate a slow component of synaptic transmission (Castillo et al., 1997; Vignes and Collingridge, 1997). KARs at both locations have been implicated in the induction of synaptic plasticity (Bortolotto et al., 1999; Contractor et al., 2001; Lauri et al., 2001). The expression of KAR subunit transcripts and their mRNA editing change markedly during development with high levels of mRNA present comparatively early in development (Bahn et al., 1994), suggesting that they may play a role in synaptic formation and stabilization. Furthermore, it has been reported that KARs at thalamocortical synapses are subject to developmental and activity-dependent regulation (Kidd and Isaac, 1999). These data indicate that KARs are regulated in response to developmental synaptic plasticity, but the molecular mechanisms underlying this regulation remain to be determined.
It has been shown previously that GluR6 expressed in HEK cells can be phosphorylated by cAMP-dependent protein kinase (PKA) (Raymond et al., 1993; Wang et al., 1993). SAP102, SAP97, and PSD95 also coimmunoprecipitated with GluR6, suggesting in vivo binding of the PSD95 family of proteins to KARs. Coexpression of PSD95 with GluR6 or GluR6/KA2 in HEK cells altered KAR function by reducing desensitization (Garcia et al., 1998). There have, however, been no previous reports of protein interactions with, or phosphorylation of, GluR5. Furthermore, no function for any KAR-interacting protein at synapses has been shown.
Here we identify several KAR-interacting proteins and show that, in contrast to the effects seen with AMPARs at the same population of synapses, selective blockade of either GRIP or PICK1 binding caused a rapid decrease in KAR-mediated EPSCs (EPSCK). This divergence in the regulation of two major types of glutamate receptors by the same two PDZ proteins is likely to have important functional consequences since PICK1 and GRIP interactions provide a mechanism for the rapid and differential regulation of the AMPAR and KAR complement at synapses.
Results
PICK1 and Syntenin Interact with GluR52b and GluR6 in the Yeast Two-Hybrid System
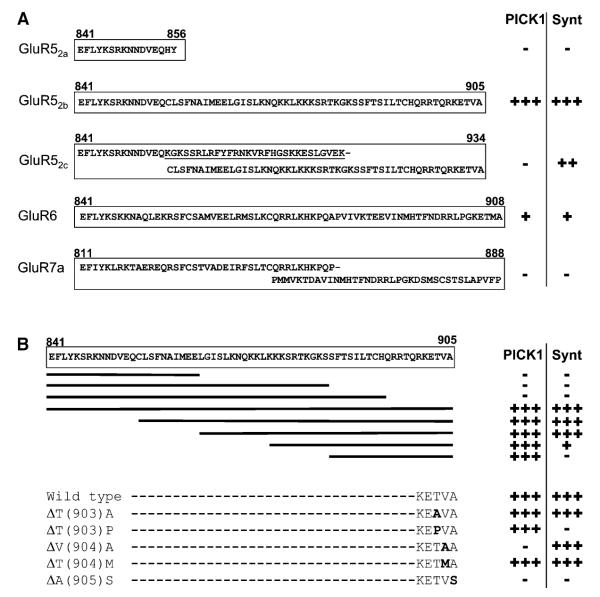
To search for proteins involved in the regulation of KARs, we performed a yeast two-hybrid screen of an adult rat brain cDNA library with KAR subunit alternative splice variant ct-GluR52b. Strong interactions were detected with two separate PDZ domain-containing proteins (Figure 1A). We isolated 84 clones encoding the entire coding sequence of syntenin, a protein with two PDZ domain repeats first reported as an interactor with syndecans, a group of cell surface proteoglycans (Grootjans et al., 1997). We also isolated ten clones encoding PICK1, a protein containing a single PDZ domain that was originally isolated for its interaction with PKCα (Staudinger et al., 1995).
Figure 1.
KAR Subunits Interact with PICK1 and Syntenin in the Yeast Two-Hybrid System
(A) β-galactosidase assays ranging from +++ (filters turn blue within 15 min) to negative (−). For all experiments, at least three assays were performed on three batches of independently transformed yeast.
(B) Overlapping deletion mutants and single point mutations of GluR52b.
Both interactors were tested against a range of baits comprising the ct domains of the other glutamate receptors and subunits: mGluR1-8, GluR1-4, NR1, and NR2A-D. Syntenin did not interact with any of these other baits (Hirbec et al., 2002 and data not shown). In agreement with previous reports, however, PICK1 interacted with the ct domains of GluR2 and 3 (Dev et al., 1999; Xia et al., 1999) and of mGluR7a (Dev et al., 2000; data not shown). We also tested the specificity of PICK1 and syntenin against other KAR subunits. Neither PICK1 nor syntenin interacted with GluR52a or GluR7a. Syntenin, but not PICK1, interacted with the GluR52c. The lack of detectable PICK1 binding to GluR52c in yeast was surprising since this subunit shares the same 30 extreme ct amino acids with GluR52b. In the yeast two-hybrid assay we also observed a consistent but less robust interaction between GluR6 and both PICK1 and syntenin.
Sites of Interaction
To define the exact sites of interaction between GluR52b and syntenin or PICK1, overlapping truncation mutations and a variety of point mutants of GluR52b were generated. Figure 1B shows that deletion of the last ten amino acids of GluR52b completely eliminated interaction with both syntenin and PICK1. Surprisingly, the shortest region we tested, corresponding to the last 20 residues and which contains the PDZ binding motif, interacted with PICK1 but not syntenin. Thus, the last 20 amino acids of GluR52b are necessary, but not sufficient, for the syntenin interaction.
Point mutations within the last three amino acids of GluR52b indicated the critical importance of these residues. Interestingly, syntenin and PICK1 displayed a differential tolerance to specific residue changes. Substitution of the valine at position −1 to alanine (ΔV[904]A) abolished the interaction with PICK1 but not with syntenin. Conversely, substitution of the threonine at −2 to proline (ΔT[903]P) abolished the interaction with syntenin but not with PICK1. Finally, replacing the terminal alanine with serine (ΔA[905]S) abolished the interaction with both syntenin and PICK1 (Figure 1B).
Substitution of the crucial glycine residue within the second PDZ domain of syntenin (Ponting et al., 1997) resulted in the loss of its interaction with ct-GluR52b. Similarly, substituting a glutamine for lysine in the PDZ domain of PICK1 (Dev et al., 1999) eliminated PICK1 binding to GluR52b (data not shown). These results confirm that binding to GluR52b occurs via the PDZ domain of PICK1 and the second PDZ domain of syntenin.
In Vitro Binding of GluR52b, GluR52c, and GluR6 to Recombinant Syntenin, PICK1, GRIP, and PSD95
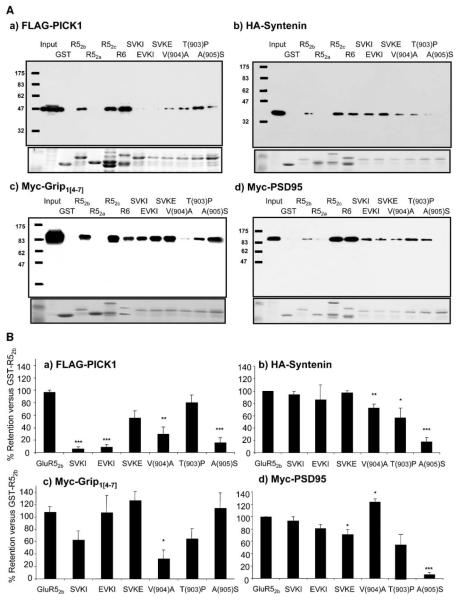
To validate and extend the yeast two-hybrid assays, we performed pull-down experiments with GST-ct-GluR52a, GST-ct-GluR52b, GST-ct-GluR52c, GST-ct-GluR6, and GST-ct-GluR7a. Although we did not detect them in the yeast two-hybrid assays, we also screened for GRIP and PSD95 retention by each of the constructs because GRIP, like PICK1, is a strong PDZ interactor with GluR2 (Dev et al., 1999; Xia et al., 1999), and PSD95 has been reported as a GluR6 interactor (Garcia et al., 1998).
As shown in Figure 2A, recombinant epitope-tagged syntenin, PICK1, PSD95, and a partial fragment of GRIP containing PDZ domains 4-7 (GRIP[4-7]; residues 430–1112) expressed in COS7 cells were all efficiently retained by GST-ct-GluR52b, GST-ct-GluR52c, and GST-ct-GluR6. Each interactor bound each of the KAR subunits in similar amounts. None of the proteins bound to GST alone, GST-ct-GluR52a, or GST-ct-GluR7a (Figure 2A and data not shown). The detection of additional interactors suggests that pull-down experiments using purified proteins can be more sensitive than yeast two-hybrid assays at detecting certain protein-protein interactions.
Figure 2.
Ct-GluR52b and GluR6 Bind to Recombinant Syntenin, PICK1, GRIP, and PSD95 In Vitro
(A) Representative Western blots of GST-ct-fusion pull-downs of recombinant epitope-tagged interacting proteins. The lower insert shows the corresponding Coomassie blue-stained gels.
(B) Quantification of GST pull-downs. The data are the mean ± SEM (n > 3) expressed as a percentage of the amount retained by wild-type GST-ct-GluR52b.
Specificity of In Vitro Interactions between GluR52b and the PDZ Proteins
Point Mutations
The yeast two-hybrid experiments suggested that the extreme C-terminal residues of GluR52b are critical for the interaction with syntenin and PICK1. Three point mutations, namely – EPVA (ct-GluR52bΔT[903]P), –ETAA (ct-GluR52bΔV[904]A) and –ETVS (ct-GluR52bΔA[905]S) were of particular interest since they differentiated between these two interactors (Figure 1). We therefore quantified the effects of these point mutations in pull-down assays (Figure 2B). Consistent with the yeast data, GST-ct-GluR52bΔT(903)P bound PICK1 at levels similar to the wild-type GST-ct-GluR52b, but bound to syntenin, GRIP, and PSD95 at reduced levels. GST-ct-GluR52bΔV(904)A bound strongly to PSD95 and to a lesser extent syntenin but had much reduced binding to PICK1 and GRIP. Interestingly, GST-ct-GluR52bΔA(905)S showed very robust binding to GRIP but little or no binding to PICK1, syntenin, or PSD95. Thus, in neurons, infusion of GST-ct-GluR52bΔV(904)A would be predicted to act as an inhibitor of syntenin and PSD95 binding, whereas GST-ct-GluR52bΔA(905)S would selectively prevent GRIP binding to KARs.
Peptide Blockers
In a complementary strategy, we included peptides corresponding to the wild-type or point mutants of C-terminal 11 amino acids of GluR2 in the GST-ct-GluR52b pull-down assays. We, and others, have used these peptides to discriminate between GRIP and PICK1 interactions with GluR2 (Li et al., 1999; Daw et al., 2000; Kim et al., 2001). Inclusion of the wild-type GluR2 sequence peptide (pep2-SVKI) (Dev et al., 1999; Xia et al., 1999) prevented the interaction between GST-ct-GluR52b and PICK1 (6% ± 4%, p < 0.001; Figure 2). This peptide had no effect on the binding to syntenin or PSD95 (94% ± 6% and 93% ± 7%, respectively). Surprisingly, pep2-SVKI had only an intermediate, and statistically nonsignificant, effect on GRIP binding to GST-ct-GluR52b (52% ± 15%, p > 0.05). Since pep2-SVKI effectively inhibits the binding of GRIP to GluR2, this latter result raised the possibility that GluR52b and GluR2 bind to different PDZ domains of GRIP. To test this idea, we investigated the binding of a more restricted truncation of GRIP that included only PDZ domains 4 and 5 (GRIP[4-5]). GRIP[4-5] bound efficiently to GluR2, GluR52b, and GluR6 in pull-down assays (data not shown). These data suggest that GluR52b and GluR2 bind differentially to PDZ domains 4 and 5 on GRIP.
Consistent with its actions on GluR2 (Daw et al., 2000; Li et al., 1999), pep2-EVKI prevented the binding of PICK1 to GST-ct-GluR52b (8% ± 5%, p < 0.001), but it had no significant effect on the interaction with syntenin, GRIP, or PSD95 (86% ± 24%, 90% ± 28%, and 81% ± 6%, respectively). The pep2-SVKE peptide had little effect on the binding of GST-ct-GluR52b to any of the PDZ proteins. These results demonstrate that pep2-SVKI and pep2-EVKI can selectively discriminate between PICK1 and the other identified PDZ interactors at KARs.
These biochemical tools allowed us to distinguish between the effects of PICK1 and GRIP by selectively blocking their binding. However, we were unable to selectively prevent binding of syntenin or PSD95. Therefore, we focused mainly on defining the roles of PICK1 and GRIP in neurons using PSD95 as a control where appropriate.
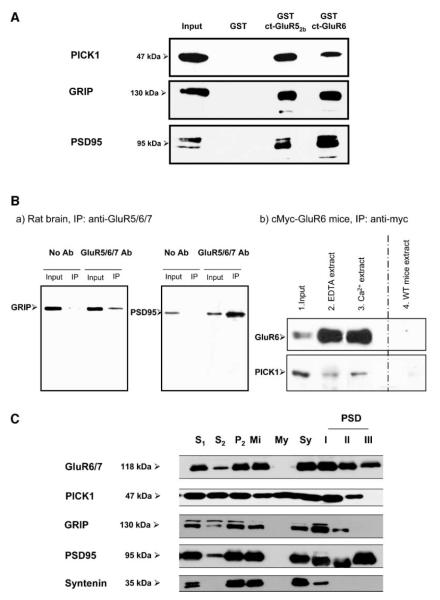
Interaction between KARs and PICK1, GRIP, and PSD95 in Brain
GST pull-downs from solubilized rat brain extracts were consistent with the results with recombinant proteins expressed in COS7 cells. An identical pattern of interaction was obtained using native PICK1, GRIP, and PSD95 from P2 membranes. Each of the proteins was retained by both GST-ct-GluR52b and GST-ct-GluR6, but not by GST alone (Figure 3A).
Figure 3.
Binding of Ct-GluR52b and Ct-GluR6 to Native PICK1, GRIP, and PSD95
(A) GST pull-downs of native proteins obtained from solubilized rat brain P2 sonicates. The data are representative of at least three separate blots for each antibody.
(B) Coimmunoprecipitation from 14-day-old rats (a). PSD95 and GRIP were coimmunopreci-pitated with a monoclonal anti-GluR5/6/7 antibody. Coimmunoprecipitation of PICK1 from myc-GluR6 transgenic mice (b). Mouse brain tissue expressing myc-tagged GluR6 was precipitated using a monoclonal anti-myc antibody.
(C) Subcellular distributions in adult rat brain. 1000 × g supernatant from crude homogenate (S1); cytosol (S2); crude membrane fraction (P2); mitochondria (Mi); myelin fraction (My); synaptosomes (Sy) and fractions resistable to single Triton (PSD I), double Triton (PSD II), or triton and sarcosyl (PSD III) extraction. 20 μg protein per lane.
We next performed coimmunoprecipitation experiments to demonstrate the interaction between the native proteins in brain. Consistent with a previous study (Garcia et al., 1998), we successfully coprecipitated native PSD95 from rat brain using anti-GluR6/7 antibody (data not shown). However, we were unable to coimmunoprecipitate any of the other PDZ interactors with this antibody. The most likely reason is that the anti-GluR6/7 antibody recognizes an epitope within the C-terminal domain of GluR6 that occludes interacting proteins other than PSD95. In an attempt to overcome this technical problem, we used a monoclonal anti-GluR5/6/7 antibody that recognizes the N-terminal domain of the KAR subunits to immunoprecipitate native KARs from rat brain. With this antibody we were able to coimmunoprecipitate native PSD95 and GRIP (Figure 3Ba). However, as often observed for monoclonal compared to polyclonal antibodies, this antibody gave consistently lower yields of KAR immunoprecipitation than the C-terminal-directed polyclonal anti-GluR6/7 antibody (data not shown). This lower efficiency, combined with the fact that the antibodies to PICK1 are less sensitive than those for GRIP or PSD95, prevented detection of PICK1 coimmunoprecipitation from rat brain.
Because of the limitations of the available antibodies against native receptors, we pursued the alternative strategy of using transgenic mice in which the GluR6 subunit is myc tagged on its N-terminal extracellular domain (Coussen et al., 2002). Using a monoclonal anti-myc antibody, both GRIP and PICK1 (Figure 3Bb) were successfully coimmunoprecipitated with myc-GluR6. The presence or the absence of calcium in the extraction medium did not have any significant effect on the amount of interacting protein precipitated. No immunoprecipitate was obtained in parallel control experiments using anti-myc antibody in wild-type (i.e., not expressing myc-GluR6) mice.
Subcellular Localization of GluR6/7 and PDZ-Containing Interactors
We compared the subcellular distribution profiles of GluR6/7 and the PDZ-interacting proteins in the rat brain. There is no effective antibody available for GluR5, so direct comparison with this subunit was not possible. Immunoreactivity for GluR6/7 and the interacting proteins was detected in various fractions including synaptosomes (Figure 3C). Progressive detergent extraction of the postsynaptic density (PSD) fraction of synaptosomes showed GluR6/7 in all fractions, with relatively high, but diminishing levels the more harsh the extraction procedure. PSD95 was the only interactor enriched in the PSDIII, indicating its very tight association with the postsynaptic density. GRIP and PICK1 were abundant in PSDI, present in PSDII, but absent from PSDIII. Syntenin was abundant in the synaptosomal fraction, but within the postsynaptic fraction was only present in PSDI at lower levels. These data demonstrate that KARs and each of the interactors are present at synapses but that they have differing levels of attachment to the postsynaptic density.
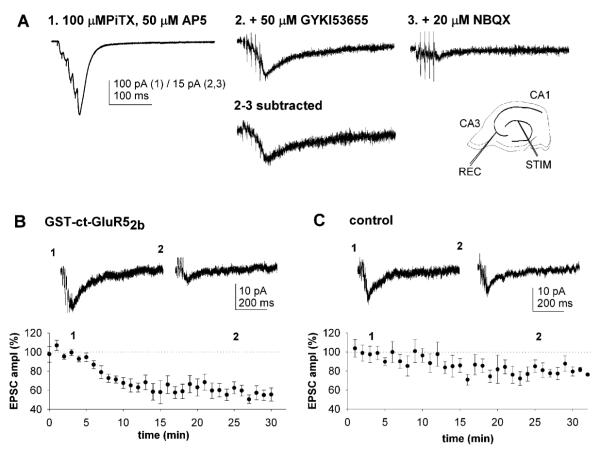
Effects on KAR-Mediated EPSCs of Blocking the PDZ Interactions
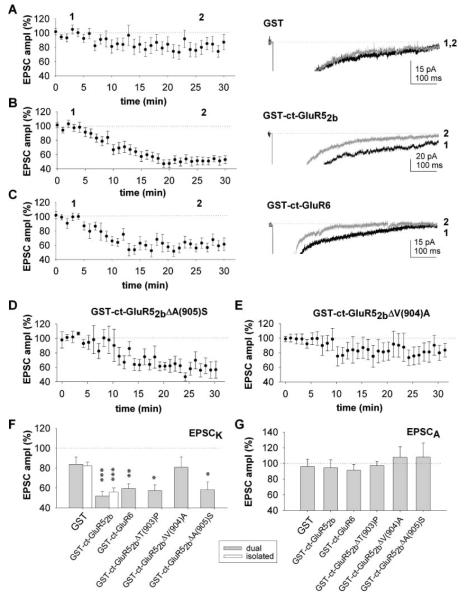
We next investigated the functional roles of these interacting proteins in regulating postsynaptic KARs at mossy fiber-CA3 synapses where there is a well-characterized KAR-mediated component to transmission (Castillo et al., 1997; Vignes and Collingridge, 1997). Whole-cell patch-clamp recordings were obtained from CA3 neurons in hippocampal slices with electrodes containing GST fusion proteins (100 nM) comprising the entire ct-GluR52b or ct-GluR6. KAR-mediated synaptic responses were evoked by stimulating mossy fibers with brief high-frequency trains (five stimuli at 100 Hz) (Castillo et al., 1997; Lauri et al., 2001; Vignes and Collingridge, 1997).
The effects of blocking KAR protein-protein interactions were investigated on pharmacologically isolated KAR-mediated EPSCs (Figure 4A) (Lauri et al., 2001). Inclusion of GST-ct-GluR52b in the whole-cell recording solution caused a rapid reduction in the size of KAR-mediated EPSCs (EPSCK; Figure 4B). The EPSCK amplitude decreased to about 50% of control within ~15 min of first obtaining whole-cell access and remained depressed for as long as recordings were maintained. In interleaved experiments with GST alone in the electrode solution, there was a small, slow reduction in EPSCK (Figure 4C) similar to that also reported recently in the absence of GST and attributed to a slow run down in KAR-mediated currents (Lauri et al., 2001). However, this reduction in EPSCK observed in control experiments was much smaller and slower than that observed for GST-ct-GluR52b.
Figure 4.
Rapid Regulation of KAR-Mediated EPSCs by Intracellular Protein Interactions
(A) Pharmacological isolation of EPSCK. The traces show a dual component EPSC evoked by five shocks delivered at 100 Hz under control conditions (1), in the presence of GYKI53655 (50 μM) (2), and following addition of NBQX (20 μM) (3). The subtraction (2 and 3) shows EPSCK. Inset: stimulating and recording positions.
(B) Pooled data (n = 12) of the amplitude of EPSCK (evoked by five stimuli at 100 Hz in the presence of GYKI53655) during recordings with electrodes containing GST-ct-GluR52b (100 nM). Data in this and the following figures are normalized to the first three points and are presented as mean ± SEM.
(C) Controls with solution containing no protein (n = 8; 83% ± 6% of control at 30 min) or GST alone (100 nM; n = 6; 82% ± 6% of control at 30 min), showing the effect on EPSCK amplitude in experiments interleaved with those in (B). Insets in (B) and (C) are representative EPSC traces, of four to six successive sweeps, taken at the times indicated (1 and 2).
To monitor the effects on both AMPARs and KARs within the same experiments, we collected dual component AMPAR- and KAR-mediated EPSCs, evoked by five stimuli at 100 Hz in the absence of GYKI53655 (Figure 5). The amplitude of the slow KAR-mediated tail was measured 110–120 ms after the last stimulus throughout the recordings. Effects on AMPAR-mediated EPSC (EPSCA) were determined by measuring the peak amplitude of EPSCs evoked by single shock stimuli at the start and at the end of the each experiment. Under these conditions, GST-ct-GluR52b (Figures 5B and 5F), but not GST alone (Figures 5A–5F), caused ~50% reduction in EPSCK, similar to that observed for the pharmacologically isolated EPSCK (Figure 5F). However, neither GST-ct-GluR52b nor GST alone had any effect on EPSCA (Figure 5G). We also found that GST-ct-GluR6 caused a similar depression in EPSCK and was similarly selective for KARs over AMPARs (Figures 5C, 5F, and 5G).
Figure 5.
GRIP Acutely Regulates KAR-Mediated EPSCs
(A) Pooled data (n = 21) for the effects of GST on EPSCK, measured from dual component EPSCs. The example traces were obtained by the times indicated (1 and 2).
(B) Pooled data (n = 19) and corresponding example traces for the effects of GST-ct-GluR52b.
(C) Pooled data (n = 17) and corresponding example traces for the effects of GST-ct-GluR6.
(D) Pooled data (n = 11) for the effects of GST-ct-GluR52bΔA(905)S.
(E) Pooled data (n = 16) for the effects of GST-ct-GluR52bΔV(904)A.
(F) Summary data showing the effects at 30 min of the GST constructs on EPSCK (the open bars are the isolated EPSCK data from Figure 4). *p < 0.05, **p < 0.01, ***p < 0.005.
(G) Summary data for EPSCA from the same experiments, taken at 30 min.
To determine which of the PDZ interactors is important for acutely regulating synaptic KARs, we investigated the effects of infusing the specific point mutants of GST-ct-GluR52b and correlated their actions with their binding profiles in the GST pull-down assays. Substituting a serine for the alanine at the 0 position (ΔA[905]S) or a proline for threonine at the −2 position (ΔT[903]P) had no affect on the ability of GST-ct-GluR52b to depress EPSCK (Figures 5D and 5F). In contrast, substitution of an alanine for the valine at the −1 position (ΔV[904]A) greatly reduced the ability of GST-ct-GluR52b to block EPSCK (Figures 5E and 5F). From the biochemical analyses of the interaction profiles (Figure 2), these results suggest that the binding of GRIP but not PSD95 or syntenin is important in acutely maintaining the function of synaptic KARs. However, these data do not exclude a role for PICK1.
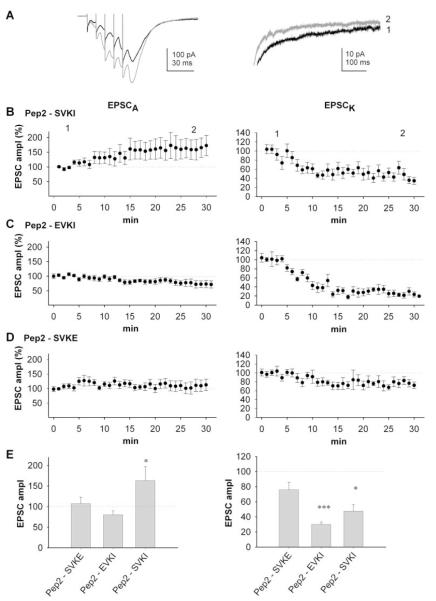
We also used the ct-GluR2 peptides “pep2-SVKI,” “pep2-EVKI,” and “pep2-SVKE” that contain a PDZ binding motif and point mutants thereof. These peptides have previously been used in biochemical and electrophysiological studies to disrupt the binding of PDZ domain-containing proteins to GluR2 (Li et al., 1999; Daw et al., 2000; Kim et al., 2001). Infusion of either pep2-SVKI or pep2-EVKI (100 μM) caused a marked reduction in EPSCK measured as the amplitude of the slow tail of the dual component EPSC (Figures 6A–6C and 6E). This effect was very similar to that observed with GST-ct-GluR52b. In contrast, pep2-SVKE (100 μM) had little effect (Figures 6D and 6E). In the biochemical experiments, pep2-SVKI and pep2-EVKI selectively blocked the PICK1-GluR52b interaction, while pep2-SVKE had no strong inhibition of any interaction. Therefore, these results suggest that, in addition to GRIP, PICK1 also has a role in the acute regulation of synaptic KARs.
Figure 6.
Rapid, Differential Regulation of AMPAR- and KAR-Mediated Synaptic Transmission by PDZ Proteins
(A) EPSCs evoked by five shocks at 100 Hz obtained 3 min (black traces) and 25 min (gray traces) following obtaining whole-cell access with intracellular pep2-SVKI. Note the increase in EPSCA and decrease in EPSCK, which is more clearly seen in the expansion of the EPSC decay (right-hand trace).
(B) Pooled data (n = 14) of amplitude of EPSCA and EPSCK for neurons infused with pep2-SVKI.
(C) Pooled data (n = 9) of amplitude of EPSCA and EPSCK for neurons infused with pep2-EVKI.
(D) Pooled data (n = 9) of amplitude of EPSCA and EPSCK for neurons infused with pep2-SVKE.
(E) Summary data for EPSCA (left panel) and EPSCK (right panel) from the same experiments, taken at 30 min.
Differential Regulation of Synaptic AMPARs and KARs
We simultaneously investigated the effects of the peptides on AMPAR-mediated EPSCs by measuring the peak amplitude of the fifth EPSC evoked by the five shock stimulation train. Consistent with our previous results for AMPAR-mediated EPSCs at CA1 synapses (Daw et al., 2000), pep2-SVKI caused an increase in EPSCA amplitude in approximately one-third of cells (5/14 cells). When the effects on all 14 cells were averaged, pep2-SVKI caused a significant increase in EPSCA of 63% ± 34% (Figures 6A, 6B, and 6E), together with a concomitant decrease in EPSCK of 52% ± 8%. Also in agreement with our previous study at CA1 synapses (Daw et al., 2000; but see Kim et al., 2001) infusion of the pep2-EVKI did not affect EPSCA amplitude (Figures 6C and 6E); however, as described above, pep2-EVKI caused a large reduction in EPSCK in the same neurons. The inactive control peptide pep2-SVKE had no effect on EPSCA (Figures 6D and 6E).
Regulation of Synaptic KARs by PKC
The electrophysiological data suggest that disruption of either GRIP or PICK1 binding causes a loss of functional synaptic KARs. To investigate the mechanism for such rapid and selective regulation of KARs, we focused on the role of PKC since PICK1 is an interactor with PKCα (Staudinger et al., 1995, 1997). Ct-GluR52b has at least five candidate serines and one threonine for PKC phosphorylation as predicted by the NetPhos database. To test if these residues can be phosphorylated by PKCα, we performed in vitro 32P assays using GST-ct-GluR52a, GST-ct-GluR52b, and GST-ct-GluR6. In all experiments, GST-ct-GluR2 was used as a positive control since this has been previously reported to be phosphorylated by PKCα (Chung et al., 2000b; Perez et al., 2001). GluR52b and GluR6, but not GluR52a, were efficiently phosphorylated by PKC in vitro (data not shown).
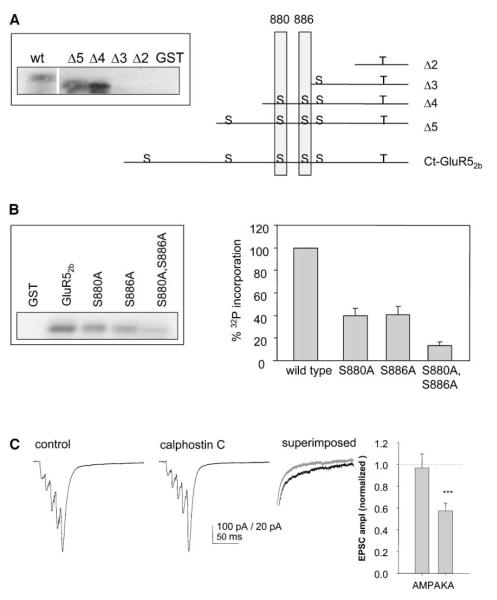
We next constructed a series of overlapping truncation mutants to determine the phosphorylation sites on GST-ct-GluR52b. As shown in Figure 7A, the Δ4 truncation was strongly phosphorylated by PKC, whereas Δ3 showed very little phosphorylation. From the predicted PKC phosphorylation sites, these data suggest that S880 and/or S886 are phosphorylated by PKCα in vitro. To better identify the phosphorylation sites, we mutated S880, S886, or both to alanines in the full-length ct-GluR52b. Each of the mutants displayed significantly decreased levels of PKC phosphorylation compared to wild-type ct-GluR52b, with the double mutant showing markedly decreased phosphorylation compared to either of the single point mutants. These results demonstrate that both S880 and S886 of GluR52b can be phosphorylated by PKCα.
Figure 7.
PKC Phosphorylates GluR52b and Inhibition of PKC Activity Reduces KAR-Mediated EPSCs
(A) Schematic of the truncation mutants generated and consensus PKC phosphorylation sites (S) on ct-GluR52b. The insert panel shows in vitro phosphorylation of each of the truncations by PKC.
(B) Representative autoradiograph showing in vitro phosphorylation of point mutants by PKCα (left panel). Quantification of data from at least three separate experiments for each mutant (right panel).
(C) Example traces (left) and summary bar graph (right) from 12 experiments in which calphostin-C (1 μM) was applied while monitoring mossy fiber synaptic transmission with five shocks delivered at 50 Hz.
To determine whether PKC activity regulates KAR function at synapses, we investigated the effects of blocking endogenous PKC activity on EPSCK in CA3 neurons. Bath application of the selective PKC inhibitor calphostin-C (1 μM) caused a rapid reduction in the amplitude of EPSCK, measured as the slow component of the dual component EPSC evoked by five shocks at 50 Hz (Figure 7B). PKC inhibition, however, had no effect on the amplitude of EPSCA simultaneously measured as the peak of the fifth EPSC in the same experiments (Figure 7C). This selective reduction in EPSCK amplitude was very similar to that observed with pep2-EVKI, which selectively blocks PICK1 binding to GluR52b. These results show that PKC can differentially regulate AMPAR and KAR function at the same population of synapses. Furthermore, this provides a mechanism whereby PICK1 can maintain KAR function by targeting PKC to KARs, and is consistent with a model in which PICK1 and GRIP differentially regulate AMPARs and KARs.
Discussion
Here we show that, like AMPAR-mediated synaptic transmission (for reviews, see Braithwaite et al., 2000; Scannevin and Huganir, 2000; Malinow and Malenka, 2002), KAR-mediated synaptic transmission is rapidly regulated by PDZ-interacting proteins. However, the consequences of PDZ-protein binding are different for the two glutamate receptor subtypes. Blocking the interaction of GluR2 with GRIP causes a rapid increase in AMPAR-mediated synaptic transmission in a subset of neurons, whereas blocking GluR2 interactions with PICK1 has no acute effect. In contrast, blocking either GRIP or PICK1 interactions causes a rapid decrease in KAR-mediated synaptic transmission. Thus, our data indicate that both GRIP and PICK1 are required to maintain KAR synaptic function. A particularly striking finding was that blocking PDZ interactions with pep2-SVKI caused a simultaneous, differential regulation of the AMPAR- and KAR-mediated components of mossy fiber synaptic transmission. Therefore, PDZ protein interactions with glutamate receptors may be involved in rapid switching of the AMPAR/KAR composition at synapses. Furthermore, we found that PKCα can phosphorylate S880 and S886 of GluR52b and that inhibition of PKC mimics the effect of blocking the PICK1 interaction with synaptic KARs. This suggests that PKC phosphorylation of GluR52b at S880/S886 is involved in the underlying mechanism for this rapid regulation of synaptic KARs by PICK1.
Molecular Characterization of the Interaction between KARs and PICK1, Syntenin, PSD95, and GRIP
We used a combination of the best available anti-KAR antibodies and tissue from transgenic myc-tagged GluR6 mice (Coussen et al., 2002) to demonstrate interactions between KARs and the PDZ proteins PSD95, PICK1, and GRIP in neurons. We also demonstrate the interaction of syntenin, another PDZ domain-containing protein, both in yeast two-hybrid and GST pull-downs. However because of the lack of suitable tools, the functional role of this protein was not investigated in the present study.
Deletion and point mutant constructs established that the interactions occur via PDZ domains. The lack of significant effect of pep2-SVKI on the GluR52b-GRIP interaction, despite the fact that it effectively blocks the interaction between the GluR2 AMPAR subunit and GRIP (Daw et al., 2000; Li et al., 1999), suggests that KARs and AMPARs might bind to different PDZ domains of GRIP. Furthermore, since both GluR2 and GluR52b bind to a reduced GRIP truncation containing only PDZ domains 4 and 5, it is possible that the differential interaction occurs within these two domains. These data show that peptide inhibitors, as well as the ct mutants, can selectively inhibit interactions between PDZ proteins and specific glutamate receptor subunits and therefore provide useful tools to investigate the functional relevance of these interactions.
An unexpected result was the lack of interaction between the C terminus of GluR52c and PICK1 in the yeast two-hybrid assay. A recent study has demonstrated that residues within the NSF binding domain of GluR2, a nonadjacent region upstream of the PDZ binding domain, are important for the GluR2-PICK1 interaction (Hanley et al., 2002). By analogy, one explanation may be that the presence of the insert within ct-GluR52c negatively influences the binding of PICK1 to the –ETVA PDZ binding domain. The fact that PICK1 bound to ct-GluR52c in pull-down experiments may reflect the greater sensitivity of this assay and/or the presence of comparatively high concentration of interacting proteins. An alternative and/or contributory explanation for the absence of interaction between ct-GluR52c and PICK1 in the two-hybrid assays could be misfolding of the longer ct-GluR52c protein in the yeast nucleus (see also Hirbec et al., 2002).
Subcellular Localization of KARs and Interacting Proteins
In agreement with functional studies (for review, see Lerma et al., 2001), our subcellular fractionation results show KAR subunits in the synaptosomal and PSD fractions consistent with their presence on both presynaptic terminals and postsynaptic membranes. Each of the interacting proteins was also expressed in the synaptosomal fraction. Further analysis of synaptosomal PSD fractions by differential detergent extraction revealed that PSD95 was most tightly associated with the postsynaptic density since it was enriched in PSDIII. GluR6/7 was also present in the PSDIII but at reduced levels compared to PSDII and PSDI. GRIP and PICK1 were enriched in the PSDI, present in PSDII, but were not detected in PSDIII, suggesting that these proteins are less tightly anchored at the postsynaptic density and may be more mobile (Figure 3C). This is consistent with the known actions of GRIP and PICK1 on AMPARs and suggests that these proteins may be involved in KAR trafficking and surface expression. For example, it has recently been shown that GRIP directly associates to kinesin motor proteins, thus providing a transport mechanism for GluR2-containing AMPARs (Setou et al., 2002). Since the kinesin binding domain on GRIP does not involve the PDZ domains, it is possible that the GRIP-kinesin complex may also be involved in the targeting and transport of KARs. Interestingly, syntenin showed a distinctly different subcellular distribution profile to the other interactors. Syntenin was abundant in the synaptosomal fraction, was present at a lower level in PSDI, but was completely absent from PSDII and PSDIII. These data suggest that syntenin may have a largely presynaptic localization, where it could play a role in the targeting of presynaptic KARs.
Functional Significance of PDZ Interactions at KARs
The only previous report investigating the role of PDZ-interacting proteins on KAR function was performed in cell lines where coexpression of PSD95 with GluR6 or GluR6/KA2 receptors reduced desensitization of the recombinant KARs (Garcia et al., 1998). Here we studied the regulation of native KARs in the mossy fiber pathway where their role in synaptic transmission has been particularly well established (Castillo et al., 1997; Contractor et al., 2001; Lauri et al., 2001; Mulle et al., 1998; Vignes et al., 1997). We reasoned that infusion of ct-GluR52b or ct-GluR6 would chelate endogenous interactors and thereby prevent their binding to native KARs. Infusion of either ct-GluR52b or ct-GluR6, but not GST alone, caused a rapid depression of KAR-mediated EPSCs. These results demonstrate that KARs, like AMPARs, are under dynamic regulation at synapses.
Analysis of the effects of point mutants of ct-GluR52b and of inhibitory peptides indicated that neither syntenin nor PSD95 are likely to be involved in the acute dynamic regulation of postsynaptic KARs at mossy fiber-CA3 synapses. PSD95 is thought to be important for the clustering of NMDARs (Kim and Sheng, 1996), AMPARs (Chen et al., 2000; Chetkovich et al., 2002), and KARs (Garcia et al., 1998) and also binds a range of other cytoskeletal and signaling proteins (Brenman et al., 1996; El-Husseini et al., 2000; Tezuka et al., 1999). Although we have recently shown that syntenin can interact with a variety of glutamate receptors in vitro (Hirbec et al., 2002), very little is known concerning the role of syntenin in neurons. Its interaction with syndecans and neurofascin (Grootjans et al., 1997; Koroll et al., 2001; Zimmermann et al., 2001), together with its expression at very early stages of development suggest that its role might be prominently developmental. In contrast, the point mutant data implicate a role for GRIP while also leaving open the possibility of a role for PICK1. The peptide data implicate a role for PICK1, but do not rule out a role for GRIP. Therefore, the simplest interpretation of our results is that interactions with both GRIP and PICK1 are required to maintain synaptic KAR function.
Differential Regulation of Synaptic AMPARs and KARs
In most experiments, we monitored AMPAR- and KAR-mediated EPSCs simultaneously. The effects of the peptides on EPSCA at mossy fibers were the same as for CA1 synapses (Daw et al., 2000). This suggests that PDZ protein interactions regulate AMPARs in a similar manner at very different types of glutamatergic synapse. Thus, the PICK1-specific inhibitor pep2-EVKI did not affect basal transmission, whereas pep2-SVKI caused a run-up in approximately one-third of synapses. This effect was attributed to insertion of AMPARs into synapses as a result of disruption of subsynaptic GluR2-GRIP interactions. In the present study, infusion of ct-GluR52b, ct-GluR6, and certain mutants that bind GRIP did not cause a sustained run-up in EPSCA. This is consistent with a selective blockade of the KAR-GRIP interaction by these KAR subunit C termini with no significant block of the GluR2-GRIP interaction.
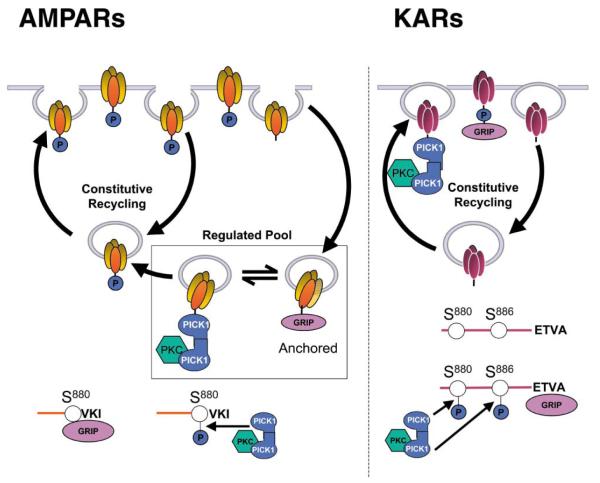
A Model for the Differential Regulation of AMPARs and KARs
The finding that KARs and AMPARs can bind to a common pool of PDZ proteins suggests that these proteins may play important general roles in the regulation of glutamatergic synapses. Based on the present findings and previous work on AMPARs (Chung et al., 2000a; Daw et al., 2000; Kim et al., 2001; Perez et al., 2001), it is possible to speculate on the molecular mechanisms that mediate the differential regulation of AMPARs and KARs by these PDZ proteins (Figure 8). In this scheme, AMPARs are secured in intracellular pools via association of the GluR2 subunit with GRIP and/or ABP. These “gripped” receptors are immobile over the time course of our electrophysiology experiments. PICK1 exchanges for GRIP and targets PKCα, which then phosphorylates S880 of GluR2, thereby preventing the rebinding of GRIP. The S880-phosphorylated AMPARs are mobile and available for surface expression. This model for acute AMPAR regulation is consistent with our current data and in agreement with our previous study of CA1 synapses (Daw et al., 2000; but see also Kim et al., 2001). We propose that KARs are also “gripped” by GRIP, but in this case, PICK1-targetted, PKC-dependent phosphorylation stabilizes the GRIP interaction with GluR5/6 and anchors the receptors at the postsynaptic membrane. These data are entirely consistent with the observations that blockade of either GRIP or PICK1 binding, or inhibition of PKC, results in a rapid decrease in KAR-mediated synaptic currents. We speculate that, whereas phosphorylation of S880 of GluR2 prevents GRIP binding (Chung et al., 2000), phosphorylation of S880 and/or S886 of GluR52b (and/or equivalent residues of GluR6) stabilizes GRIP binding and anchors the receptors at the synapse.
Figure 8.
Model Showing Possible Mechanisms of Differential Regulation of Functional AMPAR and KAR by PDZ Proteins and PKC An explanation of the scheme (adapted from Daw et al., 2000) is given in the text.
These differences in the molecular consequences of PKC-mediated phosphorylation of AMPARs and KARs can explain the differential regulation in opposite directions of the functional synaptic responses. Our results showing that, at the same population of synapses, disruption of PDZ protein interactions results in an increase in EPSCA, and a simultaneous decrease in EPSCK suggests that these proteins may act to regulate the relative proportions of AMPARs and KARs at synapses. Physiologically, given the distinct biophysical and functional profiles of AMPARs and KARs, the dynamic regulation of these interactions will play important roles in the modulation of basal glutamatergic synaptic transmission. Furthermore, it has been reported previously that some forms of developmental and activity-dependent synaptic plasticity involve a switch from functionally expressed KARs to AMPARs (Kidd and Isaac, 1999). The differential effects of PDZ-interacting proteins demonstrated here on these two receptor types provide an attractive molecular mechanism to account for these developmental and activity-dependent changes in the AMPAR and KAR complement at synapses.
Experimental Procedures
cDNA Constructs
cDNAs encoding the C terminus cytoplasmic domain of the rat KAR subunit GluR52b (residues 841–905), both wild-type and mutants, as well as of the C termini of GluR52a (residues 841–856), GluR52c (residues 841–934), GluR6a (residues 841–908), and GluR7a (residues 811–888) were cloned by PCR using specific primers and inserted in-frame into the pBTM116 vector (bait vector).
The rat syntenin and PICK1 cDNAs were isolated by the yeast two-hybrid screen of an adult rat brain cDNA library cloned into the GAL4 activation domain vector pGAD10 (fish vector; Clontech) with pBTM116-ct-GluR52b (Nishimune et al., 1998). GRIP[4-7] (residues 430-1112 in pGAD10) was isolated from a yeast two-hybrid screen performed using ct-GluR2 as a bait. To map the interaction sites, truncation mutants of either PICK1 or syntenin were prepared by PCR using specific primers and subcloned in-frame into the pGAD10 vector. All constructs were verified by sequencing.
The FLAG tag was introduced at the N terminus of PICK1 by PCR and subcloned into the mammalian expressing vector pCIneo (Dev et al., 1999). Myc and HA epitope tags were introduced to the N terminus of GRIP[4-7] and full-length syntenin by subcloning them from pGAD10 into pCMV-myc or pCMV-HA (Clontech) respectively. Myc-tagged full-length PSD95 was from S. Grant (University of Edinburgh).
Yeast Two-Hybrid System
Screens were performed as described previously (Nishimune et al., 1998). Positive GluR52b interactors were selected by growth on triple dropout media (–Trp/–Leu/–His) and by β-galactosidase filter assays.
Membrane Preparations
Brains of adult Wistar rats were dissected on ice and homogenized in 320 mM sucrose containing protease inhibitors (Complete, Boehringer-Mannheim). Subcellular fractionations were obtained by differential centrifugation (Gray and Whittaker, 1962). The PSD fractions were prepared as described previously (Carlin et al., 1980), with the following modifications. The synaptosome fraction was solubilized in ice-cold 0.5% Triton X-100 for 15 min and centrifuged at 32,000 × g for 20 min to obtain the PSD I pellet. PSD II and PSD III pellets were obtained by resuspending the PSD I pellet in 0.5% Triton X-100 and ice-cold 3% Sarcosyl, respectively. After 10 min incubation on ice, the insoluble fractions were separated by 1 hr centrifugation at 201,800 × g. All pellets were resuspended in either PBS or 40 mM Tris-HCl (pH 8.0). Protein concentrations were determined using the BCA protein assay (Pierce).
Preparation of Solubilized Proteins
FLAG-PICK1, myc-GRIP[4-7], myc-PSD95, and HA-Syntenin soluble recombinant proteins were obtained from cell lysates of COS7 cells transiently transfected using FuGene6 (Roche). Cells pellets were resuspended in PTxE (PBS containing 0.1 mM EDTA, 1% Triton X-100 [2% TritonX-100 for the extraction of myc-GRIP[4-7]], pH 7.4), sonicated 6 × 10 s on ice, solubilized by rotation for 1hr at 4°C and centrifuged at 15,000 rpm for 20 min at 4°C. The supernatant was used in pull-down assays.
For pull-downs using native proteins, adult rat brain homogenates were prepared in 320 mM sucrose, 4 mM HEPES, 1 mM EDTA, 1 mM EGTA, pH 7.4, using a glass/Teflon homogenizer (ten passes). Homogenates were centrifuged at 4°C, 1,000 × g, for 10 min, and the supernatant (S1) was centrifuged at 4°C, 48,000 × g, for 30 min. The resulting pellet (P2) was resuspended in PTxE (PBS containing 0.1 mM EDTA, 1% TritonX-100, pH 7.4), sonicated (6 × 10 s on ice), and solubilized by rotation for 1 hr at 4°C. After centrifugation (100,000 × g for 1 hr at 4°C), the supernatant (S3) was used for pull-downs assays.
For in vivo coimmunoprecipitation, the P2 fractions from 14-day-old rats were resuspended in 50 mM Tris, 150 mM NaCl, 0.5 mM EDTA, 1.5% CHAPS, and 0.75% n-octyl-β-D-glucopyranoside, pH 8.0, with protease inhibitors. The resuspended membranes were solubilized using a glass/Teflon homogenizer (two series of ten passes, interrupted with 10 min incubation on ice), followed by 1 hr gentle rotation at 4°C. In the experiments performed using either adult or 3-day-old myc-GluR6 transgenic mice (Coussen et al., 2002), the membrane fractions were extracted and solubilized using 1% Triton X-100 as detergent in the presence or absence of 0.5 mM Ca2+. After the rotation, the suspension was centrifuged at 13,000 rpm for 45 min at 4°C, and the resulting supernatant (S3) was used for immunoprecipitation.
Affinity Chromatography (GST Pull-Downs)
The ct-GluR52a, GluR52b, GluR52c, GluR6, and GluR7a were subcloned into the pGEX-4T-1 plasmid (Pharmacia), expressed in E. coli strain BL21, and extracted using BugBuster (Novagen). GST alone and GST fusion proteins were bound to 25 μl (50 μl in pull-downs from brain) glutathione Sepharose 4B (Pharmacia) in the presence of 1 mg/ml BSA for 30 min at 4°C. The coupled Sepharose was then washed twice with 1 ml PTx[0.1] (PBS containing 0.1% Triton X-100, pH 7.4). Supernatants containing the tagged or native proteins of interest were then incubated with the coupled beads in the presence of 2 mg/ml BSA. After 1 hr (overnight in pull-downs from brain) rotation at 4°C, the suspensions were washed five times with 1 ml PTx[0.1] buffer before being processed for Western blotting.
Coimmunoprecipitation
4-10 μl of the anti-GluR5/6/7 (Chemicon, CA) was incubated with the rat brain S3 fraction for 90 min at 4°C. 100 μl of 50% slurry anti-Mouse IgM agarose (Sigma, St Louis, MO) was then added and the incubation continued for overnight at 4°C. The suspensions were washed six times with 20 mM Tris, 1 mM EDTA, 100 mM NaCl, 0.5% Nonidet P-40, pH 7.4. For immunoprecipitation from myc-GluR6 transgenic mice, 8 μg of anti-myc antibody (clone 9E10; Roche) was incubated with precleared S3 fractions for 60 min at 4°C. 50 μl of 50% slurry protein-G agarose (Sigma) was added and left overnight at 4°C. Beads were washed with 20 mM HEPES, 1% TritonX-100, 150 mM NaCl, and 0.15 mM EDTA, pH 7.5, containing protease inhibitor cocktail.
Western Blotting and Antibodies
Proteins were separated on 10% SDS-polyacrylamide gels, transferred onto PVDF membrane (Millipore), and blocked for 1 hr at room temperature with 5% nonfat dry milk in TBS-T (200 mM Tris, 137 mM NaCl, 0.1% Tween20, pH 7.4) prior to incubation overnight at 4°C with the primary antibody. The primary antibodies were: (i) affinity-purified anti-PICK1 rabbit Ab (Dev et al., 1999) (1:200 dilution); (ii) anti-HA Ab (Santa-Cruz, CA); (iii) anti-GRIP (Upstate Biotechnologies); (iv) anti-syntenin serum (1:200 dilution); (v) anti-GluR6/7 Ab (Upstate Biotechnologies); (vi) anti-GluR6/7(C18) Ab (Santa-Cruz, CA); (vii) anti-bassoon mAb (Stressgen); (viii) anti-PSD95 mAb (Chemicon), (ix) anti-FLAG M2 mAb (Sigma); and (x) anti-myc mAb (Oncogene). Appropriate HRP-conjugated secondary antibodies were used for Western blotting (Sigma) and detected using ECL. The intensity of bands on immunoblots was quantified by densitometry using the Gel-Doc system (Biorad).
In Vitro Phosphorylation Assays
In vitro PKC-mediated phosphorylation assays were performed on 5 μg purified GST fusions of GluR52a, GluR52b, and GluR6 using a previously published protocol (Nakajima et al., 1999). Briefly, reactions were performed at 30°C for 30 min in phosphorylation buffer (40 mM Tris-HCl, pH 7.5, 20 mM MgCl2, 200 μg·ml−1 L-α-phosphatidyl-L-serine, 40 μg·ml−1 1,2-dioleoyl-sn-glycerol), purified PKCα (1.5 μg·ml−1; Calbiochem), and 100 μM ATP. All reactions included 5–10 μCi of [γ-32P]ATP and were stopped with SDS-PAGE sample buffer. The phosphorylated fusion proteins were resolved by SDS-PAGE and visualized by autoradiography.
Hippocampal Slice Electrophysiology
Hippocampal slices were prepared from 13- to 15-day-old Wistar rats. Rats were anaesthetised, decapitated, and the brain placed in ice-cold extracellular solution. Transverse hippocampal slices (400 μm thick) were prepared on a vibratome and placed in a recovery chamber, submerged in extracellular solution, for 1–3 hr. Slices were then placed in the recording chamber where they were continuously superfused with extracellular solution at room temperature (23°C–25°C). The extracellular solution was as follows: 124.0 mM NaCl, 3.0 mM KCl, 1.0 mM MgSO4, 1.25 mM NaH2PO4, 26.0 mM NaHCO3, 2.0 mM CaCl2, 15 mM D-glucose, 2.0 mM ascorbate, 100 μM picrotoxin, and 50 μM D-AP5, saturated with 95% O2:5% CO2, 295 mOsm, pH 7.4. Whole-cell voltage-clamp recordings were made from CA3 pyramidal cells using electrodes with a resistance of 2–4 MΩ filled with the intracellular solution: 130.0 mM CsMethanesulphonate, 10.0 mM HEPES, 0.5 mM EGTA, 8.0 mM NaCl, 5.0 mM QX-314Cl, 4.0 mM MgATP, and 0.3 mM NaGTP, 285 mOsm, pH 7.2. The intracellular solution also contained the protease inhibitors bestatin (100 μM), leupeptin (100 μM), pepstatin-A (100 μM), and the relevant purified GST-ct-fusion proteins (100 nM) or the synthetic peptides (100 μM).
Dual AMPA and kainate receptor-mediated synaptic currents were evoked by afferent stimulation of the dentate granule cell layer (mossy fiber pathway) with five shocks at 100 Hz, at an interval of 40 s. Data were collected and analyzed on-line using the LTP program (www.ltp-program.com) and series resistance was estimated on-line (Anderson and Collingridge, 2001). Afferent stimulation commenced immediately following the obtainment of whole-cell recording. Baseline values were obtained by averaging the first five to six responses. The amplitudes of the KAR-mediated component of the EPSC (EPSCK) were measured 110–120 ms after the last stimulus in the train. The pharmacologically isolated EPSCK was measured at the peak. In some experiments, AMPAR-mediated EPSCs (EPSCA) were measured as the peak of the EPSC evoked by single shock stimuli. These were collected (10–20 at 0.1 Hz) at the start of recordings (after the initial baseline of five to six responses was collected using the five shock stimulus protocol) and also at the end of the recordings. In other experiments, EPSCA was measured throughout the experiment by monitoring the peak amplitude of the fifth EPSC in the train. Data are expressed as mean ± SEM. For statistical tests, paired, two-tailed Student’s t tests were used.
Peptides were custom synthesized (Sigma) with purity higher than 98%. GST and GST fusion proteins were prepared described above dialyzed at 4°C for 20 hr (2 × 2 liters) against 5 mM Na2HPO4, 0.9 mM KH2PO4, and 0.5 mM EGTA (pH 7.2) and then concentrated using the Centiprep system (Millipore). Purified proteins were then immediately aliquoted and stored at −20°C until use. All peptide/GST fusion protein-containing intracellular solutions were made up in parallel and stored in exactly the same way, and the pH and osmolarity of the solutions were checked.
Acknowledgments
We thank Michael Hollman and Steve Heinemann for the generous gift of KAR clones and Seth Grant for providing the myc-PSD95 clone. We are grateful to Atsushi Nishimune for helpful discussion. The contribution of each author was as follows: S.B., original Y2H screening showing PICK1 and syntenin interactions; K.K.D., construction of some of the plasmids and mutants; V.C., assistance in the early stages of the project; G.M., GRIP constructs; J.T.R.I., direction of electrophysiological experiments; G.L.C., funding and direction of electrophysiological experiments; and J.M.H., conception, funding, and direction of project. We acknowledge the Wellcome Trust, the MRC, and the EU (KARTRAP) for financial support. H.H. was supported by the AFRT, France Alzheimer, and Wellcome Trust Fellowships. S.E.L. and K.K.D. were supported by Wellcome Trust Fellowships. J.C.F. and V.C. were supported by BBSRC studentships.
References
- Anderson WW, Collingridge GL. The LTP Program: a data acquisition program for on-line analysis of long-term potentiation and other synaptic events. J. Neurosci. Methods. 2001;108:71–83. doi: 10.1016/s0165-0270(01)00374-0. [DOI] [PubMed] [Google Scholar]
- Bahn S, Volk B, Wisden W. Kainate receptor gene expression in the developing rat brain. J. Neurosci. 1994;14:5525–5547. doi: 10.1523/JNEUROSCI.14-09-05525.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bliss T, Collingridge G. A synaptic model of memory: long-term potentiation in the hippocampus. Nature. 1993;361:31–39. doi: 10.1038/361031a0. [DOI] [PubMed] [Google Scholar]
- Bortolotto ZA, Clarke VR, Delany CM, Parry MC, Smolders I, Vignes M, Ho KH, Miu P, Brinton BT, Fantaske R, et al. Kainate receptors are involved in synaptic plasticity. Nature. 1999;402:297–301. doi: 10.1038/46290. [DOI] [PubMed] [Google Scholar]
- Braithwaite SP, Meyer G, Henley JM. Interactions between AMPA receptors and intracellular proteins. Neuropharmacology. 2000;39:919–930. doi: 10.1016/s0028-3908(99)00171-9. [DOI] [PubMed] [Google Scholar]
- Brenman JE, Chao DS, Gee SH, McGee AW, Craven SE, Santillano DR, Wu Z, Huang F, Xia H, Peters MF, et al. Interaction of nitric oxide synthase with the postsynaptic density protein PSD-95 and alpha1-syntrophin mediated by PDZ domains. Cell. 1996;84:757–767. doi: 10.1016/s0092-8674(00)81053-3. [DOI] [PubMed] [Google Scholar]
- Carlin RK, Grab DJ, Cohen RS, Siekevitz P. Isolation and characterization of postsynaptic densities from various brain regions: enrichment of different types of postsynaptic densities. J. Cell Biol. 1980;86:831–845. doi: 10.1083/jcb.86.3.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo PE, Malenka RC, Nicoll RA. Kainate receptors mediate a slow postsynaptic current in hippocampal CA3 neurons. Nature. 1997;388:182–188. doi: 10.1038/40645. [DOI] [PubMed] [Google Scholar]
- Chen L, Chetkovich DM, Petralia RS, Sweeney NT, Kawasaki Y, Wenthold RJ, Bredt DS, Nicoll RA. Stargazing regulates synaptic targeting of AMPA receptors by two distinct mechanisms. Nature. 2000;408:936–943. doi: 10.1038/35050030. [DOI] [PubMed] [Google Scholar]
- Chetkovich DM, Chen L, Stocker TJ, Nicoll RA, Bredt DS. Phosphorylation of the postsynaptic density-95 (PSD-95)/discs large/zona occludens-1 binding site of stargazin regulates binding to PSD-95 and synaptic targeting of AMPA receptors. J. Neurosci. 2002;22:5791–5796. doi: 10.1523/JNEUROSCI.22-14-05791.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chittajallu R, Vignes M, Dev KK, Barnes JM, Collingridge GL, Henley JM. Regulation of glutamate release by presynaptic kainate receptors in the hippocampus. Nature. 1996;379:78–81. doi: 10.1038/379078a0. [DOI] [PubMed] [Google Scholar]
- Chittajallu R, Braithwaite SP, Clarke VRJ, Henley JM. Kainate receptors: subunits, synaptic localisation and function. Trends Pharmacol. Sci. 1999;20:544–553. doi: 10.1016/s0165-6147(98)01286-3. [DOI] [PubMed] [Google Scholar]
- Chung HJ, Xia J, Scannevin RH, Zhang X, Huganir RL. Phosphorylation of the AMPA receptor subunit GluR2 differentially regulates its interaction with PDZ domain-containing proteins. J. Neurosci. 2000;20:7258–7267. doi: 10.1523/JNEUROSCI.20-19-07258.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke V, Ballyk B, Hoo K, Mandelzys A, Pellizzari A, Bath C, Thomas J, Sharpe E, Davies C, Ornstein P, et al. A hippocampal GluR5 kainate receptor regulating inhibitory synaptic transmission. Nature. 1997:599–603. doi: 10.1038/39315. [DOI] [PubMed] [Google Scholar]
- Contractor A, Swanson GT, Sailer A, O’Gorman S, Heinemann SF. Identification of the kainate receptor subunits underlying modulation of excitatory synaptic transmission in the CA3 region of the hippocampus. J. Neurosci. 2000;20:8269–8278. doi: 10.1523/JNEUROSCI.20-22-08269.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contractor A, Swanson G, Heinemann SF. Kainate receptors are involved in short- and long-term plasticity at mossy fiber synapses in the hippocampus. Neuron. 2001;29:209–216. doi: 10.1016/s0896-6273(01)00191-x. [DOI] [PubMed] [Google Scholar]
- Coussen F, Normand E, Marchal C, Costet P, Choquet D, Lambert M, Mege RM, Mulle C. Recruitment of the kainate receptor subunit glutamate receptor 6 by cadherin/catenin complexes. J. Neurosci. 2002;22:6426–6436. doi: 10.1523/JNEUROSCI.22-15-06426.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daw MI, Chittajallu R, Bortolotto ZA, Dev KK, Duprat F, Henley JM, Collingridge GL, Isaac JT. PDZ proteins interacting with C-terminal GluR2/3 are involved in a PKC- dependent regulation of AMPA receptors at hippocampal synapses. Neuron. 2000;28:873–886. doi: 10.1016/s0896-6273(00)00160-4. [DOI] [PubMed] [Google Scholar]
- Dev KK, Nishimune A, Henley JM, Nakanishi S. The protein kinase Cα binding protein PICK1 interacts with short but not long form alternative splice varients of AMPA receptor subunits. Neuropharmacology. 1999;38:635–644. doi: 10.1016/s0028-3908(98)00230-5. [DOI] [PubMed] [Google Scholar]
- Dev KK, Nakajima Y, Kitano J, Braithwaite SP, Henley JM, Nakanishi S. PICK1 interacts with and regulates PKC phosphorylation of mGLUR7. J. Neurosci. 2000;20:7252–7257. doi: 10.1523/JNEUROSCI.20-19-07252.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Husseini AE, Schnell E, Chetkovich DM, Nicoll RA, Bredt DS. PSD-95 involvement in maturation of excitatory synapses. Science. 2000;290:1364–1368. [PubMed] [Google Scholar]
- Garcia EP, Mehta S, Blair LA, Wells DG, Shang J, Fukushima T, Fallon JR, Garner CC, Marshall J. SAP90 binds and clusters kainate receptors causing incomplete desensitization. Neuron. 1998;21:727–739. doi: 10.1016/s0896-6273(00)80590-5. [DOI] [PubMed] [Google Scholar]
- Garner CC, Nash J, Huganir RL. PDZ domains in synapse assembly and signalling. Trends Cell Biol. 2000;10:274–280. doi: 10.1016/s0962-8924(00)01783-9. [DOI] [PubMed] [Google Scholar]
- Gray EG, Whittaker VP. The isolation of nerve endings from brain: an electron-microscopic study of cell fragments derived by homogenisation and centrifugation. J. Anat. 1962;96:79–88. [PMC free article] [PubMed] [Google Scholar]
- Grootjans JJ, Zimmermann P, Reekmans G, Smets A, Degeest G, Durr J, David G. Syntenin, a PDZ protein that binds syndecan cytoplasmic domains. Proc. Natl. Acad. Sci. USA. 1997;94:13683–13688. doi: 10.1073/pnas.94.25.13683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanley JG, Khatri L, Hanson PI, Ziff EB. NSF AT-Pase and alpha-/beta-SNAPs disassemble the AMPA receptor-PICK1 complex. Neuron. 2002;34:53–67. doi: 10.1016/s0896-6273(02)00638-4. [DOI] [PubMed] [Google Scholar]
- Hirbec H, Perestenko O, Nishimune A, Meyer G, Nakanishi S, Henley JM, Dev KK. The PDZ proteins PICK1, GRIP, and syntenin bind multiple glutamate receptor subtypes. Analysis of PDZ binding motifs. J Biol Chem. 2002;277:15221–15224. doi: 10.1074/jbc.C200112200. [DOI] [PubMed] [Google Scholar]
- Kidd FL, Isaac JTR. Developmental and activity-dependent regulation of kainate receptors at thalamocortical synapses. Nature. 1999;400:569–573. doi: 10.1038/23040. [DOI] [PubMed] [Google Scholar]
- Kidd FL, Coumis U, Collingridge GL, Crabtree JW, Isaac JT. A presynaptic kainate receptor is involved in regulating the dynamic properties of thalamocortical synapses during development. Neuron. 2002;34:635–646. doi: 10.1016/s0896-6273(02)00699-2. [DOI] [PubMed] [Google Scholar]
- Kim CH, Chung HJ, Lee HK, Huganir RL. Interaction of the AMPA receptor subunit GluR2/3 with PDZ domains regulates hippocampal long-term depression. Proc. Natl. Acad. Sci. USA. 2001;98:11725–11730. doi: 10.1073/pnas.211132798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Sheng M. Differential K+ channel clustering activity of PSD-95 and SAP97, two related membrane-associated putative guanylate kinases. Neuropharmacology. 1996;35:993–1000. doi: 10.1016/0028-3908(96)00093-7. [DOI] [PubMed] [Google Scholar]
- Koroll M, Rathjen FG, Volkmer H. The neural cell recognition molecule neurofascin interacts with syntenin-1 but not with syntenin-2, both of which reveal self- associating activity. J. Biol. Chem. 2001;276:10646–10654. doi: 10.1074/jbc.M010647200. [DOI] [PubMed] [Google Scholar]
- Lauri SE, Bortolotto ZA, Bleakman D, Ornstein PL, Lodge D, Isaac JT, Collingridge GL. A critical role of a facilitatory presynaptic kainate receptor in mossy fiber LTP. Neuron. 2001;32:697–709. doi: 10.1016/s0896-6273(01)00511-6. [DOI] [PubMed] [Google Scholar]
- Lerma J, Paternain AV, Rodriguez-Moreno A, Lopez-Garcia JC. Molecular physiology of kainate receptors. Physiol. Rev. 2001;81:971–998. doi: 10.1152/physrev.2001.81.3.971. [DOI] [PubMed] [Google Scholar]
- Li P, Kerchner GA, Sala C, Wei F, Huettner JE, Sheng M, Zhuo M. AMPA receptor-PDZ interactions in facilitation of spinal sensory synapses. Nat. Neurosci. 1999;2:972–977. doi: 10.1038/14771. [DOI] [PubMed] [Google Scholar]
- Malinow R, Malenka RC. AMPA receptor trafficking and synaptic plasticity. Annu. Rev. Neurosci. 2002;25:103–126. doi: 10.1146/annurev.neuro.25.112701.142758. [DOI] [PubMed] [Google Scholar]
- Mulle C, Sailer A, Perez-Otano I, Dickinson-Anson H, Castillo PE, Bureau I, Maron C, Gage FH, Mann JR, Bettler B, et al. Altered synaptic physiology and reduced susceptibility tokainate-induced seizures in GluR6-deficient mice. Nature. 1998;392:601–605. doi: 10.1038/33408. [DOI] [PubMed] [Google Scholar]
- Nakajima Y, Yamamoto T, Nakayama T, Nakanishi S. A relationship between protein kinase C phosphorylation and calmodulin binding to the metabotropic glutamate receptor subtype 7. J. Biol. Chem. 1999;274:27573–27577. doi: 10.1074/jbc.274.39.27573. [DOI] [PubMed] [Google Scholar]
- Nishimune A, Isaac JTR, Molnar E, Noel J, Nash SR, Tagaya M, Collingridge GL, Nakanishi S, Henley JM. NSF binding to GluR2 regulates synaptic transmission. Neuron. 1998;21:87–97. doi: 10.1016/s0896-6273(00)80517-6. [DOI] [PubMed] [Google Scholar]
- Perez JL, Khatri L, Chang C, Srivastava S, Osten P, Ziff EB. PICK1 targets activated protein kinase Calpha to AMPA receptor clusters in spines of hippocampal neurons and reduces surface levels of the AMPA-type glutamate receptor subunit 2. J. Neurosci. 2001;21:5417–5428. doi: 10.1523/JNEUROSCI.21-15-05417.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP, Phillips C, Davies KE, Blake DJ. PDZ domains: targeting signalling molecules to sub-membranous sites. Bioessays. 1997;19:469–479. doi: 10.1002/bies.950190606. [DOI] [PubMed] [Google Scholar]
- Raymond LA, Blackstone CD, Huganir RL. Phosphorylation and modulation of recombinant GluR6 glutamate receptors by cAMP-dependent protein kinase. Nature. 1993;361:637–641. doi: 10.1038/361637a0. [DOI] [PubMed] [Google Scholar]
- Scannevin RH, Huganir RL. Postsynaptic organization and regulation of excitatory synapses. Nat. Rev. Neurosci. 2000;1:133–141. doi: 10.1038/35039075. [DOI] [PubMed] [Google Scholar]
- Schmitz D, Frerking M, Nicoll RA. Synaptic activation of presynaptic kainate receptors on hippocampal mossy fiber synapses. Neuron. 2000;27:327–338. doi: 10.1016/s0896-6273(00)00040-4. [DOI] [PubMed] [Google Scholar]
- Schmitz D, Mellor J, Nicoll RA. Presynaptic kainate receptor mediation of frequency facilitation at hippocampal mossy fiber synapses. Science. 2001;291:1972–1976. doi: 10.1126/science.1057105. [DOI] [PubMed] [Google Scholar]
- Setou M, Seog D, Tanaka Y, Kanai Y, Takei Y, Kawagishi M, Hirokawa N. Glutamate-receptor-interacting protein GRIP1 directly steers kinesin to dendrites. Nature. 2002;417:83–87. doi: 10.1038/nature743. [DOI] [PubMed] [Google Scholar]
- Sheng M. Molecular organization of the postsynaptic specialization. Proc. Natl. Acad. Sci. USA. 2001;98:7058–7061. doi: 10.1073/pnas.111146298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudinger J, Zhou J, Burgess R, Elledge SJ, Olson EN. PICK1: a perinuclear binding protein and substrate for protein kinase C isolated by the yeast two-hybrid system. J. Cell Biol. 1995;128:263–271. doi: 10.1083/jcb.128.3.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudinger J, Lu J, Olson EN. Specific interaction of the PDZ domain protein PICK1 with the COOH terminus of protein kinase C. J. Biol. Chem. 1997;272:32019–32024. doi: 10.1074/jbc.272.51.32019. [DOI] [PubMed] [Google Scholar]
- Tezuka T, Umemori H, Akiyama T, Nakanishi S, Yamamoto T. PSD-95 promotes Fyn-mediated tyrosine phosphorylation of the N-methyl-D-aspartate receptor subunit NR2A. Proc. Natl. Acad. Sci. USA. 1999;96:435–440. doi: 10.1073/pnas.96.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignes M, Collingridge GL. The synaptic activation of kainate receptors. Nature. 1997;388:179–182. doi: 10.1038/40639. [DOI] [PubMed] [Google Scholar]
- Vignes M, Bleakman D, Lodge D, Collingridge GL. The synaptic activation of the GluR5 subtype of kainate receptor in area CA3 of the rat hippocampus. Neuropharmacology. 1997;36:1477–1481. doi: 10.1016/s0028-3908(97)00158-5. [DOI] [PubMed] [Google Scholar]
- Wang LY, Taverna FA, Huang XP, Macdonald JF, Hampson DR. Phosphorylation and modulation of a kainate receptor (GluR6) by cAMP-dependent protein kinase. Science. 1993;259:1173–1175. doi: 10.1126/science.8382377. [DOI] [PubMed] [Google Scholar]
- Xia J, Zhang X, Staudinger J, Huganir RL. Clustering of AMPA receptors by the synaptic PDZ domain-containing protein PICK1. Neuron. 1999;22:179–187. doi: 10.1016/s0896-6273(00)80689-3. [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Tomatis D, Rosas M, Grootjans J, Leenaerts I, Degeest G, Reekmans G, Coomans C, David G. Characterization of syntenin, a syndecan-binding PDZ protein, as a component of cell adhesion sites and microfilaments. Mol. Biol. Cell. 2001;12:339–350. doi: 10.1091/mbc.12.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]