Abstract
Background
Prokaryotic ubiquitin-like protein (Pup), the firstly identified post-translational protein modifier in prokaryotes, is an important signal for the selective degradation of proteins. Recently, large-scale proteomics technology has been applied to identify a large number of pupylated proteins. The development of a database for managing pupylated proteins and pupylation sites is important for further analyses.
Description
A database named PupDB is constructed by collecting experimentally identified pupylated proteins and pupylation sites from published studies and integrating the information of pupylated proteins with corresponding structures and functional annotations. PupDB is a web-based database with tools for browses and searches of pupylated proteins and interactive displays of protein structures and pupylation sites.
Conclusions
The structured and searchable database PupDB is expected to provide a useful resource for further analyzing the substrate specificity, identifying pupylated proteins in other organisms and developing computational tools for predicting pupylation sites. PupDB is freely available at http://cwtung.kmu.edu.tw/pupdb.
Background
Protein-to-protein modifications are essential for regulating protein functions. In eukaryotes, ubiquitylation involved in numerous regulatory functions such as protein degradation, DNA repair, transcription and signal transduction is particular important [1]. Recently, pupylation has been identified as the first post-translational protein-to-protein modification in prokaryotes [2,3]. Similar to ubiquitin, prokaryotic ubiquitin-like protein (Pup) attaches to specific lysine residues of substrate proteins by forming isopeptide bonds to target the proteins for proteasomal degradation [2,3].
Although ubiquitylation and pupylation are functional analogues, the enzymology of ubiquitylation and pupylation is different. In contrast to the three-step reaction of ubiquitylation, pupylation requires only two steps that only two enzymes are involved in pupylation. First, the C-terminal glutamine of Pup is deamidated to glutamine by deamidase of Pup (Dop) [4]. Subsequently, proteasome accessory factor A (PafA) attaches the deamidated Pup to specific lysine residues of substrate proteins [5].
The identification of pupylated proteins and pupylation sites can provide insights into the substrate specificity and functions of pupylation. Recently, large-scale proteomics technology has been applied to identify pupylated proteins and pupylation sites [6-9]. As the number of identified pupylated proteins and sites grows, a structured and searchable database of pupylated proteins and pupylation sites is desirable for further analyzing substrate specificity and functions of pupylated proteins and developing prediction methods for pupylation sites. For this purpose, the freely accessible database named PupDB integrating information of pupylated proteins and pupylation sites, protein structures, functional annotations and tools for browses, searches and interactive displays of protein structures and pupylation sites was constructed.
Construction and content
The PupDB database is implemented using MySQL Server Edition 5.1. The PupDB website is publicly available at http://cwtung.kmu.edu.tw/pupdb. The web interface and all functions are implemented using PHP and Perl languages. The software of Google Chart Tools [10] is utilized to make sortable tables.
Database content
Two kinds of proteins included in PupDB are pupylated proteins and candidate pupylated proteins. All proteins are collected from four large-scale proteomics studies [6-9]. Proteins with experimentally identified pupylation sites are annotated as pupylated proteins. Candidate pupylated proteins are experimentally identified proteins whose pupylation sites are still unknown.
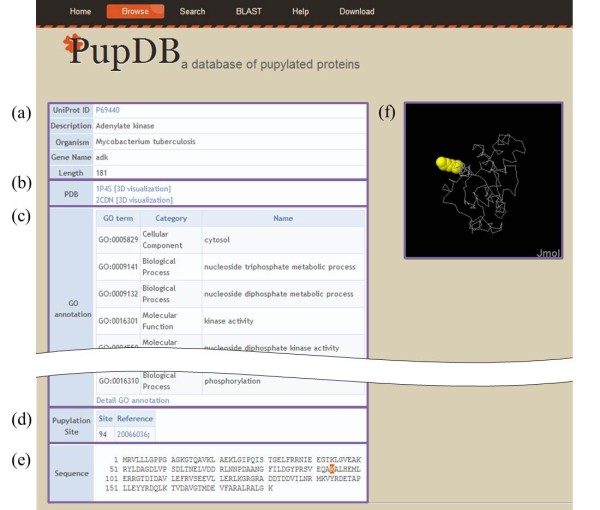
Redundant proteins are removed from PupDB by using CD-HIT [11,12] with a sequence identity threshold of 98%. Currently, PupDB contains 182 pupylated proteins with 215 known pupylation sites and 1,123 candidate pupylated proteins. All proteins belong to three organisms of Mycobacterium smegmatis, Mycobacterium tuberculosis and Escherichia coli. For each protein, the corresponding information consists of six major parts of basic information, PDB ID, gene ontology (GO) annotation, pupylation site, protein sequence and structure as shown in Figure 1. PupDB will be regularly updated with additional data and corrections and analytical tools. Researchers are encouraged to contribute their data and suggestions to PupDB.
Figure 1.
Content of a typical PupDB entry. (a) Basic information. (b) Structure. (c) GO annotation. (d) Pupylation site. (e) Sequence. (f) 3D structure.
Annotations
As shown in Figure 1a, the first part of basic information includes the UniProt AC, description, gene name, organism and sequence length. For further information of protein annotations, PupDB provides links to the corresponding entries of UniProt database [13]. Also, structure information including PDB (Protein Data Bank) ID and hyperlinks to the PDB database [14] is provided in the second part (Figure 1b). The visualization of pupylation sites in a protein structure can provide helpful information for analysis. The protein 3D structure and associated pupylation sites can be viewed in PupDB by clicking the link of '3D visualization'. The java applet-based program Jmol [15] is utilized for interactive displays of protein structure (Figure 1f). The UniProt protein accession numbers and PDB IDs are obtained by using the ID mapping function of UniProt. Currently, there are 766 PDB structures associated with 294 PupDB entries.
The GO annotations [16] can give useful information of molecular function, cellular component and biological process. For a given protein, the corresponding GO annotations can be extracted by using its UniProt accession number. Figure 1c shows the third part of GO annotations for protein P69440. Further GO information can be accessed by clicking the hyperlink of 'Detailed GO annotation' that links to the corresponding entry of QuickGO [17].
The fourth part of pupylation sites includes pupylation sites and corresponding references for pupylated proteins (Figure 1d). References are represented as PubMed IDs with hyperlinks to PubMed database [18]. Instead of showing only references for a candidate pupylated protein whose pupylation sites are still unknown, PupDB highlights pupylation sites in both sequence and structure of a pupylated protein for visualization as shown in Figure 1e and 1f, respectively.
Utility and discussion
PupDB is a database of pupylated proteins and pupylation sites aiming to provide an easily accessible web service for the analysis of pupylated proteins. The analysis of pupylated proteins in PupDB can provide better insights into the specificity of pupylation. For example, Two Sample Logo [19] can be utilized to graphically analyze over- and underrepresented residues surrounding pupylation sites as shown in Figure 2.
Figure 2.
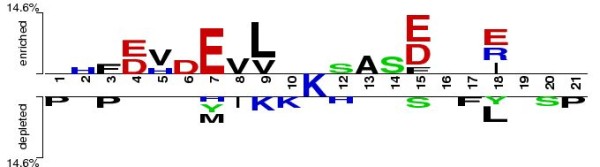
Two-Sample Logo representation of over- (upper half) and underrepresented (lower half) residues surrounding pupylation sites.
Hyperlinks to major protein, structure and annotation databases are provided for accessing related information. Four useful tools are constructed and integrated into PupDB to provide functions of browses, keyword searches, sequence similarity searches and interactive displays of protein structures. The functions of the integrated tools are introduced in the follows.
Browse tool
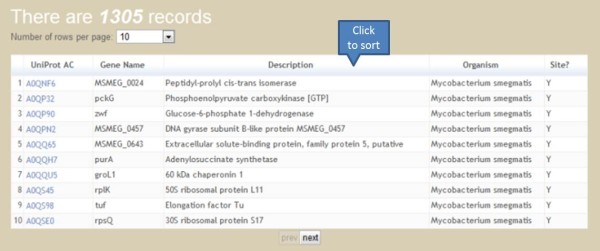
Users can browse PupDB by selecting the 'Browse' option. All proteins will be shown in a sortable table. The entry with 'Y' in the field of 'Site' is a pupylated protein. Otherwise, it is a candidate pupylated protein with 'N' in the field of 'Site'. By clicking the caption of a specific column in a sortable table, the output table will be sorted according to data of the selected column. Furthermore, users can specify the number of rows shown per page (Figure 3).
Figure 3.
Browse tool.
Search and BLAST tools
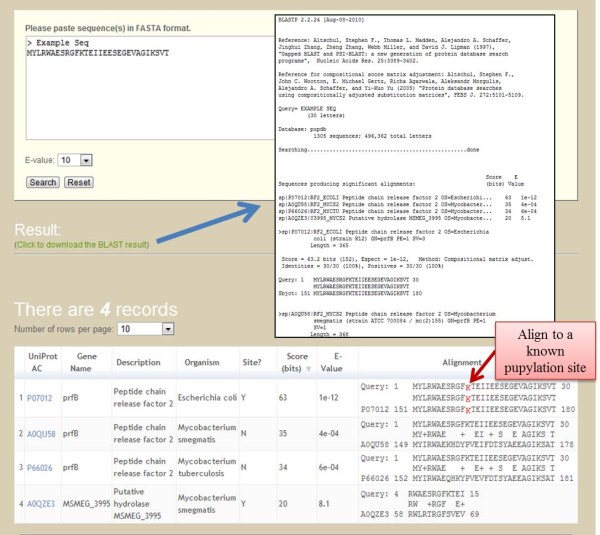
For retrieving entries of interest, PupDB provides two search tools of keyword and similarity searches. 1) The tool of keyword search can be accessed by selecting the 'Search' option. There are six fields for searching PupDB including description, UniProt AC, gene name, organism, protein type and protein with structure. By entering keywords for any one or combination of the fields, PupDB will return search results as a sortable table according to the user input keywords. 2) Users can enter a protein sequence of interest in FASTA format to perform a BLAST [20] search against PupDB to fetch entries with a user-defined threshold of E-value. The BLAST tool can serve as a potentially useful tool for predicting promising pupylation sites by sequence similarity. In addition to the protein information, three additional columns of scores, E-values and alignments obtained from the BLAST search are included in the output sortable table. The detailed information of BLAST sequence alignment can be downloaded by clicking the download link. Figure 4 shows an example of BLAST search. In the query sequence, lysines aligned to known pupylation sites will be marked in red color. Users can submit proteins in other organisms to predict pupylation sites.
Figure 4.
BLAST tool.
Interactive tool for protein structure
PupDB incorporates the Jmol applet of latest version 12.2 for interactive displays of protein structures. By default, PupDB represents protein structures and pupylation sites in grey and yellow colors, respectively. Users can either use the user interface or scripting console to manipulate protein structures.
Conclusions
The PupDB database is a comprehensive repository of pupylated proteins and pupylation sites with a web-based user interface. The built-in tools for browses, searches and interactive displays of protein structures and pupylation sites make PupDB a useful resource for further analyzing the substrate specificity, identifying pupylated proteins in other organisms and developing computational tools for predicting pupylation sites. In addition to the graphical analysis using two-sample logos, advanced machine learning methods such as string kernels [21] can also be utilized to further analyze the specificity of pupylation. The exported dataset of pupylated proteins is downloadable at PupDB.
Post-translational modification databases serve as good data source for developing prediction tools. For example, the construction of UbiPred [22] for predicting ubiquitylation sites is based on dataset of UbiProt [23]. Although a predictor GPS-PUP [24] is available for predicting pupylation sites, PupDB with 215 pupylation sites can be utilized to further improve GPS-PUP trained on only 127 pupylation sites. Future works are two-fold. First, the development and integration of prediction tools based on the dataset of PupDB would be useful for analyzing and predicting pupylation sites. Second, the incorporation of orthology relationships and locations of functional domains can largely improve PupDB.
Availability and requirements
The PupDB is freely available at http://cwtung.kmu.edu.tw/pupdb. The website has been tested with browsers of Safari, Opera, Internet Explorer 7 or later, Firefox and Google Chrome. The Java Runtime Environment (JRE) is required for interactive displays of protein 3D structures by Jmol.
Competing interests
The author declares that they have no competing interests.
Authors' contributions
CWT designed and implemented the database, performed the analysis and wrote the manuscript.
Acknowledgements
CWT would like to thank the National Science Council (NSC 101-2311-B-037-001-MY2) of Taiwan and Kaohsiung Medical University Research Foundation (KMU-Q110015 and KMU-ER013) for financially supporting this research. CWT thanks the anonymous reviewers for their valuable comments and suggestions to improve this work.
References
- Herrmann J, Lerman LO, Lerman A. Ubiquitin and ubiquitin-like proteins in protein regulation. Circ Res. 2007;100(9):1276–1291. doi: 10.1161/01.RES.0000264500.11888.f0. [DOI] [PubMed] [Google Scholar]
- Pearce MJ, Mintseris J, Ferreyra J, Gygi SP, Darwin KH. Ubiquitin-like protein involved in the proteasome pathway of Mycobacterium tuberculosis. Science. 2008;322(5904):1104–1107. doi: 10.1126/science.1163885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns KE, Liu WT, Boshoff HI, Dorrestein PC, Barry CE. Proteasomal protein degradation in Mycobacteria is dependent upon a prokaryotic ubiquitin-like protein. J Biol Chem. 2009;284(5):3069–3075. doi: 10.1074/jbc.M808032200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Striebel F, Imkamp F, Sutter M, Steiner M, Mamedov A, Weber-Ban E. Bacterial ubiquitin-like modifier Pup is deamidated and conjugated to substrates by distinct but homologous enzymes. Nat Struct Mol Biol. 2009;16(6):647–651. doi: 10.1038/nsmb.1597. [DOI] [PubMed] [Google Scholar]
- Guth E, Thommen M, Weber-Ban E. Mycobacterial ubiquitin-like protein ligase PafA follows a two-step reaction pathway with a phosphorylated pup intermediate. J Biol Chem. 2011;286(6):4412–4419. doi: 10.1074/jbc.M110.189282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerda-Maira FA, McAllister F, Bode NJ, Burns KE, Gygi SP, Darwin KH. Reconstitution of the Mycobacterium tuberculosis pupylation pathway in Escherichia coli. EMBO Rep. 2011;12(8):863–870. doi: 10.1038/embor.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Festa RA, McAllister F, Pearce MJ, Mintseris J, Burns KE, Gygi SP, Darwin KH. Prokaryotic ubiquitin-like protein (Pup) proteome of Mycobacterium tuberculosis. PLoS One. 2010;5(1):e8589. doi: 10.1371/journal.pone.0008589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen C, Akhter Y, Jeon AH, Schmitt-Ulms G, Meyer HE, Stefanski A, Stuhler K, Wilmanns M, Song YH. Proteome-wide identification of mycobacterial pupylation targets. Mol Syst Biol. 2010;6:386. doi: 10.1038/msb.2010.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watrous J, Burns K, Liu WT, Patel A, Hook V, Bafna V, Barry CE, Bark S, Dorrestein PC. Expansion of the mycobacterial "PUPylome". Mol Biosyst. 2010;6(2):376–385. doi: 10.1039/b916104j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Google Chart Tools. http://code.google.com/intl/zh-TW/apis/chart/index.html
- Huang Y, Niu B, Gao Y, Fu L, Li W. CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics. 2010;26(5):680–682. doi: 10.1093/bioinformatics/btq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22(13):1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- Magrane M, Consortium U. UniProt Knowledgebase: a hub of integrated protein data. Database Oxford. 2011;2011:bar009. doi: 10.1093/database/bar009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The protein data bank. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jmol: an open-source Java viewer for chemical structures in 3D. http://www.jmol.org/
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT. et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binns D, Dimmer E, Huntley R, Barrell D, O'Donovan C, Apweiler R. QuickGO: a web-based tool for Gene Ontology searching. Bioinformatics. 2009;25(22):3045–3046. doi: 10.1093/bioinformatics/btp536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PubMed. http://www.ncbi.nlm.nih.gov/pubmed/
- Vacic V, Iakoucheva LM, Radivojac P. Two Sample Logo: a graphical representation of the differences between two sets of sequence alignments. Bioinformatics. 2006;22(12):1536–1537. doi: 10.1093/bioinformatics/btl151. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung CW, Ziehm M, Kamper A, Kohlbacher O, Ho SY. POPISK: T-cell reactivity prediction using support vector machines and string kernels. BMC Bioinforma. 2011;12:446. doi: 10.1186/1471-2105-12-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung CW, Ho SY. Computational identification of ubiquitylation sites from protein sequences. BMC Bioinforma. 2008;9:310. doi: 10.1186/1471-2105-9-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernorudskiy AL, Garcia A, Eremin EV, Shorina AS, Kondratieva EV, Gainullin MR. UbiProt: a database of ubiquitylated proteins. BMC Bioinforma. 2007;8:126. doi: 10.1186/1471-2105-8-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Ma Q, Cao J, Gao X, Ren J, Xue Y. GPS-PUP: computational prediction of pupylation sites in prokaryotic proteins. Mol Biosyst. 2011;7(10):2737–2740. doi: 10.1039/c1mb05217a. [DOI] [PubMed] [Google Scholar]