Abstract
The desert is an excellent model for studying evolution under extreme environments. We present here the complete genome and ultraviolet (UV) radiation-induced transcriptome of Deinococcus gobiensis I-0, which was isolated from the cold Gobi desert and shows higher tolerance to gamma radiation and UV light than all other known microorganisms. Nearly half of the genes in the genome encode proteins of unknown function, suggesting that the extreme resistance phenotype may be attributed to unknown genes and pathways. D. gobiensis also contains a surprisingly large number of horizontally acquired genes and predicted mobile elements of different classes, which is indicative of adaptation to extreme environments through genomic plasticity. High-resolution RNA-Seq transcriptome analyses indicated that 30 regulatory proteins, including several well-known regulators and uncharacterized protein kinases, and 13 noncoding RNAs were induced immediately after UV irradiation. Particularly interesting is the UV irradiation induction of the phrB and recB genes involved in photoreactivation and recombinational repair, respectively. These proteins likely include key players in the immediate global transcriptional response to UV irradiation. Our results help to explain the exceptional ability of D. gobiensis to withstand environmental extremes of the Gobi desert, and highlight the metabolic features of this organism that have biotechnological potential.
Introduction
The order Deinococcales contains 50 species of extremely ionizing radiation (IR) and UV tolerant bacteria (http://www.bacterio.cict.fr/) [1]. D. radiodurans R1, isolated from canned meat that had spoiled following exposure to X-rays, was sequenced first [2]. D. radiodurans has 200-fold greater resistance to ionizing radiation and 20-fold greater resistance to UV radiation than Escherichia coli [3], but it encodes approximately the same number and types of DNA repair proteins as E. coli, and no unique DNA repair system was found [3], [4]. Recently, the genome sequences of the slightly thermophilic D. geothermalis DSM11300, isolated from a hot spring, D. deserti VCD115, isolated from the Sahara desert, Truepera radiovictrix RQ-24, isolated from hot spring runoff on the Island of Sao Miguel, and D. maricopensis LB-34, isolated from the Sonoran Desert soil, were published [5]–[8]. Besides, the sequence of the complete genome of D. proteolyticus MRP is available under GenBank accession number CP002536. Investigation of the biology and biochemistry of Deinococcus spp. has benefited from the availability of genomic information and the development of genetic tools, but the extreme resistance phenotype of Deinococcus spp. is still not fully understood [9]. Comparative genomics combined with microarray and proteomic analysis suggest that the extreme resistance phenotype results from a combination of different molecular mechanisms [5], [6], [10]–[12].
About 10% of the Earth's terrestrial surface is covered by desert. The Gobi desert of northwestern China is a cold, arid biotope with cycles of extreme temperatures, prolonged dryness, and intense solar radiation [13]. Despite the extreme challenges of the desert, diverse microorganisms have adapted and colonized this harsh environment. Thus, the desert biosphere is an excellent venue for studying evolution under extreme conditions, and it provides a useful gene pool for genetic engineering. One of the major stresses for bacteria inhabiting the surface sands of the desert is intense solar UV radiation-induced damage. In general, the capacity of prokaryotes to withstand significant UV radiation requires a wide array of physiological responses, including transcriptional regulation and cellular repair of irradiation-induced damage [11], [14], [15]. Several studies have focused on cellular recovery following exposure to UV irradiation and have shown that induction and repression of UV radiation-responsive genes occurs in a time-dependent manner [16]–[18]. Currently, very little is known about the immediate transcriptome response to UV irradiation.
We recently characterized a new bacterial species, Deinococcus gobiensis I-0, that was isolated from the upper sand layers of the cold Gobi desert of the Xinjiang region in China [19]. This strain shows higher tolerance for gamma radiation and UV light than all other known Deinococcus strains [19]. To obtain a comprehensive understanding of the molecular mechanisms underlying the resistance phenotype of Deinococcus, the genome of D. gobiensis was sequenced and compared to those of the three most closely related sequenced bacterial strains, D. radiodurans R1, D. geothermalis DSM11300, and D. deserti VCD115, which were isolated from canned meat, a hot spring, and the hot Sahara desert, respectively [3]–[6]. This study also provides the first transcriptome analysis investigating the UV resistance of Deinococcus. In particular, we identified a subset of poorly characterized UV irradiation-induced genes that may provide clues to the adaptation of Deinococcus to extreme environments.
Results
Genome features
The genome of D. gobiensis I-0 is composed of seven replicons: a 3.1 Mb main chromosome and six plasmids from 433 to 53 kb (Figure 1 and Table 1, GenBank accession numbers CP002191–CP002197 for the main Chromosome and Plasmids P1–P6, respectively). The chromosome and the 433 kb plasmid P1 have an average GC content of 71%, higher than that of the six other sequenced Deinococcales species (Table S1), and similar to that of the extreme thermophile Thermus thermophilus [20]. The genome of D. gobiensis contains 4,340 predicted coding sequences (CDSs), 46 tRNA genes, and 15 rRNA genes, and is larger than those of the six other published Deinococcales species (Table S1).
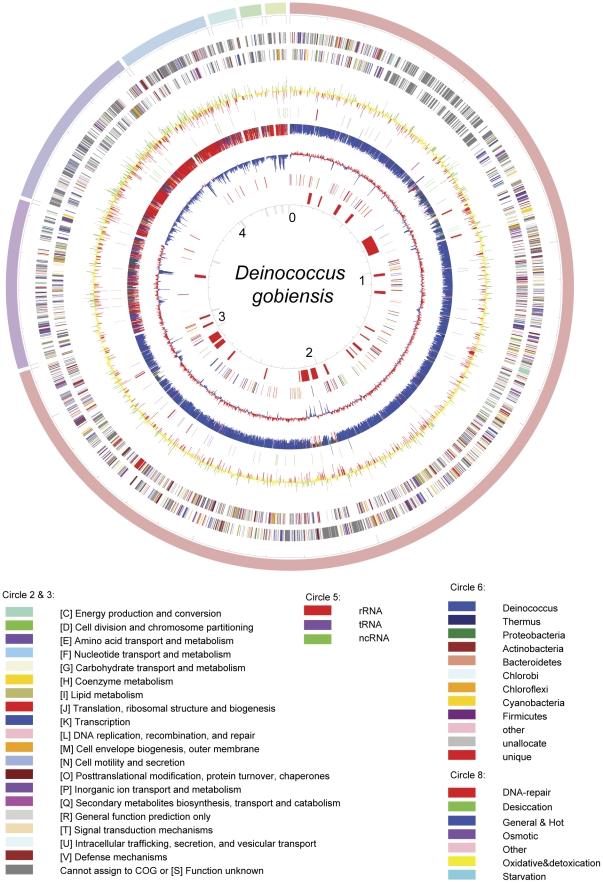
Figure 1. D. gobiensis I-0 genome structure.
The seven replicons were opened at sequence position 1 and concatenated. Circle 1, red, chromosome (3.1 Mb); violet, plasmid 1 (P1, 433 kb); indigo, P2 (425 kb); blue, P3 (232 kb); light blue, P4 (72 kb); dark green, P5 (55 kb); light green, P6 (53 kb). Circles 2 and 3, predicted protein coding sequences (CDSs) clockwise and anticlockwise, respectively. Coloring is according to COG. Circle 4, Fold change in the immediate global transcriptional response to UV irradiation for each gene: green, upgulated; red, down-regulated; yellow, not changed significantly. Circle 5, red, rRNA; purple, tRNA; green, ncRNAs (noncoding). Circle 6, blue, genes with homologues in other Deinococcus genomes; red, genes found only in D. gobiensis I-0; other colors, genes with closest homologues in other phyla. Circle 7, deviation from the average 69.15% total genomic GC content: red, higher; blue, lower. Circle 8, previously reported genes that are involved in DNA repair and stress-responses. Circle 9, location of the 23 genomic islands. Circle 10, Mb scale.
Table 1. General features of the D. gobiensis genome.
| Molecule | Chromosome | Plasmids | All | |||||
| P1 | P2 | P3 | P4 | P5 | P6 | |||
| Size(bp) | 3,137,147 | 432,699 | 424,524 | 231,600 | 72,036 | 54,602 | 53,428 | 4,406,036 |
| GC content (%) | 70.8 | 69.81 | 63.89 | 62.97 | 60.44 | 61.87 | 54.72 | 69.15 |
| Coding density (%) | 84.8 | 86 | 83.8 | 83.7 | 80.2 | 74.9 | 74.1 | 85.4 |
| Protein-coding genes | 2959 | 383 | 523 | 282 | 70 | 79 | 44 | 4340 |
| (Average length, nt) | (899) | (971) | (679) | (687) | (825) | (511) | (900) | (863) |
| Pseudo genes | 19 | 0 | 0 | 0 | 0 | 0 | 0 | 19 |
| tRNAs | 46 | - | - | - | - | - | - | 46 |
| 5 S rRNA | 5 | - | - | - | - | - | - | 5 |
| 16 S rRNA | 5 | - | - | - | - | - | - | 5 |
| 23 S rRNA | 5 | - | - | - | - | - | - | 5 |
| ncRNA | 19 | 7 | 7 | 5 | 1 | 0 | 6 | 46 |
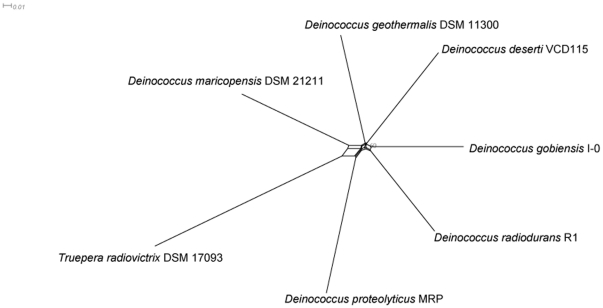
Phylogenetic analyses using only the orthologous proteins that occur in 14 sequenced strains from the phylum Deinococcus-Thermus showed that Deinococcus strains belong to the same branch (Figure S1). Further phylogenetic analyses showed that D. gobiensis, D. radiodurans, D. geothermalis and D. deserti belong to the same deeper branch and D. gobiensis was more closely related to D. radiodurans than to D. deserti and D. geothermalis (Figure 2). Pairwise comparisons of the Deinococcus genomes revealed limited synteny (Figure S2). The most striking feature was the cross patterns that indicate frequent symmetrical exchanges of genes, possibly by recombination between the bidirectional replication forks [21], [22]. There were no long stretches of synteny, indicating that the Deinococcus genomes exhibit remarkable plasticity involving gene rearrangements, acquisition, and loss (Figure S2). A 354-kb stretch of the main chromosome that is populated by 316 genes lacks a single functional COG prediction (Figure 1, circles 2 and 3; grey DGo_CA0296–DGo_CA0612, 306 kb–660 kb). BLAST searches revealed that 262 (83%) of the 316 genes have counterparts in other Deinococcus species, although they were dispersed throughout the genomes. We suggest the name “Grey Heaven of D. gobiensis” for this intriguing gene cluster which appears to underpin the unprecedented resistance of deinococci.
Figure 2. Unrooted Deinococcales neighbor-joining phylogenetic tree deduced from the nucleotide acid sequences of the orthologous proteins that occur in all 14 sequenced strains from the phylum Deinococcus-Thermus.
D. gobiensis and D. radiodurans are most closely related. Numbers indicate bootstrap values below 100.
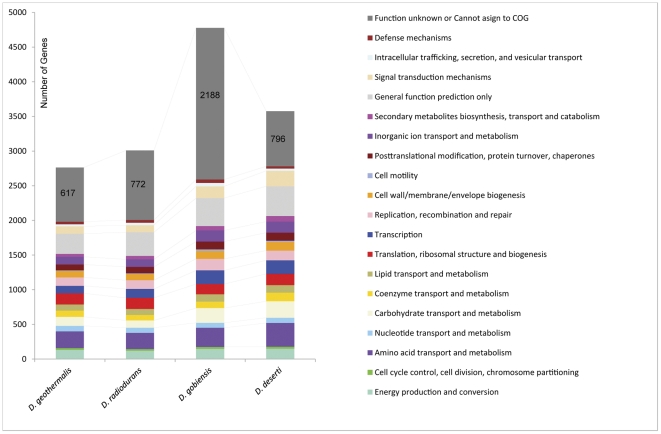
Of the 4,340 CDSs, 55% (2,376/4,340) were classified by Clusters of Orthologous Groups (COGs), a lower percentage than for the other deinococci (75% in D. radiodurans, 85% in D. deserti and 79% in D. geothermalis). 1,534 (35%) of the D. gobiensis CDSs were hypothetical proteins, and 834 (19%) were orphans without precedent. To identify the functional gene categories that characterize the cold desert adaptation in deinococci, we compared the genomes of D. gobiensis and other three related Deinococcus species (Figure 3). D. gobiensis and D. deserti are similar in each COG category. Notably, the two desert species have more genes than the other two published species in most categories, especially in energy production and conversion, carbohydrate transport and metabolism, transcription, cell wall/membrane/envelope biogenesis, inorganic ion transport and metabolism, signal transduction mechanisms. D. gobiensis also has more genes for replication, recombination and repair, cell motility, intracellular trafficking, secretion, and vesicular transport, and defense mechanisms, but fewer genes for translation and nucleotide metabolism genes. This COG characteristic is similar to functional gene categories that are associated with cold adaptation of the psychrophilic Methanococcoides burtonii [23].
Figure 3. COG functional categories in the four Deinococcus species.
Standard colors were used to indicate the COG functional categories. The numbers of unassigned and function unknown genes are indicated in the grey areas.
Transcriptome profiling
The transcriptome was analyzed using high throughput cDNA sequencing (RNA-Seq), that uses deep sequencing to assess the transcriptional activity of annotated genes, reveal previously unannotated genes and identify non-coding RNAs (ncRNAs) [24], [25]. We found that 4,325 of the 4,340 CDSs and 45 non-coding RNAs were detected under normal growth conditions (The RNA-Seq raw data of are available from the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE29088). The accuracy of the RNA-seq data was verified by quantitative real-time RT-PCR analysis (using 16 S rRNA as a control) of eight selected genes qRT-PCR (Table S2). In total, 638 plausible transcriptional start sites (TSS) were identified on the chromosome, and 362 on the six plasmids (Table S3). Furthermore, cDNAs for almost all of the CDSs of unknown function (2,029/2,041) were detected.
Bacteria generally have a global transcriptional response after UV irradiation that initiates subsequent recovery from the radiation damage. Immediately following UV irradiation, a total of 390 genes (9% of the genome) were induced, and 754 genes (17%) were repressed (Table S4). Approximately 8% of all D. gobiensis genes were predicted to be involved in regulation, which is the same as for D. radiodurans and also for E. coli. The regulators include six sigma factors (rpoD, sigE, sigK, sig3, sig4 and sig5), 54 sensor kinases and 129 transcriptional regulators. Among those, 30 were induced, and 60 were repressed. In bacteria, noncoding RNAs often coordinate the adaptation to environmental changes, integrate environmental signals, and control target-gene expression [26]. We observed that 13 ncRNAs were upregulated, and 22 were downregulated. Moreover, a total of 61 predicted proteins were constitutively expressed at high levels (≥1% of total mRNA) before and after UV irradiation, including a heat shock protein (IbpA), a phage shock protein (PspA), cold shock proteins (PprM1 and PprM2), DNA damage response proteins (DdrE and DdrO), chaperones (DnaK, DnaJ, GrpE and GroEL) and CDSs of unknown function, some of which may be responsible for cold desert adaptation.
Deinococcus-specific genes and horizontal gene transfer
The six sequenced Deinococcus strains share 1,474 genes. Among them, 39 genes of unknown function had no close orthologs in other species that don't belong to Deinococcales (Table S5). Furthermore, 10 previously predicted DNA damage-responsive genes (ddr) induced in response to IR and desiccation in D. radiodurans [14] were also found in D. gobiensis. Recently, ddrB was identified as a member of a new family of bacterial single-stranded DNA binding proteins which are induced by ionizing radiation [27], [28]. We observed that the expression of some ddr genes was induced significantly by UV, including ddrA (a double-strand break repair protein), ddrB (a radiation-induced single-stranded DNA binding protein), ddrC, ddrE, ddrI and ddrP. Although the precise functions of these genes are not clear, they may contribute to a novel mechanism for adaptation to the extreme desert environment.
Horizontal gene transfer has played a major role in the evolution of bacteria and archaea. The total genome of D. gobiensis is about 1 Mb larger than the other Deinococcus genomes, probably due to gene acquisition by horizontal gene transfer. D. gobiensis contains more DNA sequences that are indicative of mobile elements than the other deinococci, including 77 putative transposases, 17 integrases or recombinases, and 2 prophages. In addition, we found that the D. gobiensis plasmid P1 was similar to the D. deserti plasmid P2, the D. geothermalis plasmid pDSM11300 (DG574) and the D. radiodurans chromosome 2, suggesting that these similar sequences may have come from a common ancestor (Figure 1 and Figure S2). The D. gobiensis plasmids P2–P6 contain a high proportion of genes that were exclusive to D. gobiensis (Figure 1). Furthermore, most of the genes on D. gobiensis P3–P6 had no homologs in the NCBI database. This indicated that the D. gobiensis plasmids have different evolutionary histories. Furthermore, the D. gobiensis genome contained 23 sequences, which were classified as genetic islands (GI) and may have been acquired by horizontal gene transfer (Table S6). For example, genes for tRNAs flank GI 19, while the others contain integrase genes (GI 7–10 and 15). In addition, GIs 9 and 10 resemble prophages, and most of their genes have their closest homologs outside the genus Deinococcus.
The closest ortholog of the 2,728 CDSs in the D. gobiensis genome was in the phylum Deinococcus-Thermus. However, 1,612 CDSs were indicative of horizontal gene transfer, 691 of which seemed to originate from Proteobacteria, Actinobacteria and Firmicutes. These horizontally acquired genes are mostly involved in sugar transport, signal transduction, transcriptional regulation and transposition. Notably, the two desert species D. gobiensis and D. deserti contain more insertion sequence (IS) associated orphan genes without Deinococcus orthologs than D. radiodurans. This difference likely reflects the evolutionary development of the efficient adaptive strategy derived by IS-associated horizontal acquisition of additional genes. Further transcriptome analysis indicated that the expression of more than 35% of the horizontally acquired genes were changed by UV light, strongly suggesting the involvement of these genes in resistance to UV irradiation.
Metabolic pathways and transport
As expected, the D. gobiensis genome possesses most of the genes for the central metabolic pathways, including glycolysis, the pentose phosphate (PP) pathway, the tricarboxylic acid (TCA) cycle, biosynthesis of amino acids, and de novo purine and pyrimidine biosynthesis. This set of predicted pathways is similar to that found in the other Deinococcus strains. Transport systems are key components for the communication of bacteria with their environments. For example, D. gobiensis has approximately 300 genes coding for transporters. We found that the rbs genes encoding ribose ABC transporters are present in two desert species but absent from D. radiodurans and D. geothermalis. In addition to having more Mn ABC transporter proteins that are similar to those found in D. deserti, D. gobiensis is also rich in ABC transporters for oligopeptides, amino acids and sugars relative to D. radiodurans and D. geothermalis. Eighteen of the 80 amino acid or peptide transporter proteins and 16 of the 54 proteins for sugar transport are specific to D. gobiensis.
Significant induction of most of the genes encoding metabolic pathways was not detected immediately after UV irradiation. Specifically, among the 286 transporters, 22, including two for K+ (TrkG and KdpC) and PO4 3− (PstA and PstC), were induced and 45, including two glycerol transporters and three ribose transporters, were repressed. In addition, we observed strong repression of the zwf gene (glucose 6-phosphate dehydrogenase), which is the rate-limiting enzyme of the PP pathway required for removal of reactive oxygen species (ROS). We also found that the two adjacent operons nuoAB and nuoC-N encoding NADH-quinone oxidoreductase were significantly downregulated, and NADH:quinone oxidoreductase plays a pivotal role in cellular energy production. Also among repressed genes were the aceB gene that encodes a malate synthase for the glyoxylic acid shunt, the genes responsible for the synthesis of amino acids (including arginine, ornithine, tryptophan, aspartic acid and lysine), and the genes fabZ and fabG that encode NADPH-dependent enzymes involved in the synthesis of fatty acids and biotin. This repression can be explained by the induction of relA, an inducer of the stringent response involved in ppGpp synthesis. We hypothesize that desert-dwelling bacteria could have specialized mechanisms to reduce their metabolism. This metabolic switch may be a general response for coping with solar UV radiation in a desert environment and could be the subject of further experimental work.
DNA repair and associated systems
The extremely radiation-resistant bacteria must have highly efficient and specialized DNA repair systems [3]–[6]. D. gobiensis contains sets of genes encoding proteins for various DNA repair pathways (Table S7 A) and, like D. radiodurans, D. gobiensis has only 15 orthologs of the 31 genes that comprise the LexA-RecA-mediated SOS response to UV in E. coli [29]; D. gobiensis lacks homologs of the error-prone DNA polymerase V genes umuC and umuD, which are important for the SOS response in many bacteria [30]. We found, however, many Deinococcus-specific genes that are probably involved in DNA repair and extreme resistance. Unexpectedly, most of these genes were not changed in their expression immediately after UV irradiation.
UV radiation generates two major DNA damage products, the cyclobutane pyrimidine dimer and the pyrimidine pyrimidinone dimer. Photoreactivation mediated by photolyase is one of the simplest and oldest repair systems for UV-induced cyclobutane-pyrimidine dimers. The two desert species, D. gobiensis and D. deserti, contain a homolog (DGo_PA0134 and Deide_3p02150) of the splB gene encoding an active spore photoproduct lyase belonging to the radical S-adenosylmethionine superfamily [31], which is absent from both D. radiodurans and D. geothermalis. Additionally, D. gobiensis encodes another photolyase (PhrB, DGo_CA0607) whose closest homolog is from the Archaea. In contrast to photoreactivation, excision repair pathways are much more complex and can be separated into base excision repair (BER) and nucleotide excision repair (NER). The D. gobiensis genome contains sets of the excision repair genes, including nine genes for BER and six for NER (Table S7 A), which may play in important role in UV-induced DNA repair.
A recombinational repair pathway is also operative in various organisms. As a part of the recombinational DNA repair of UV-lesions, E. coli RecA protein has a regulatory role in lesion bypass through coprotease activity which includes stimulation of self-cleavage of the repressor LexA. In D. gobiensis we observed a two-fold induction of lexA by UV irradiation, while recA was unchanged. This observation was very different from the response of E. coli in which both genes were induced by UV irradiation [16]. RecB is critical for the enzyme activity of the multifunctional exonuclease V (RecBCD) involved in DNA degradation [32]. D. gobiensis contains two plasmid-encoded homologs of recB (DGo_PB0022 and DGo_PC0098), and the expression of DGo_PC0098 was three-fold induced by UV irradiation. In a previous study, expressing the E. coli recB in D. radiodurans increased UV resistance [33]. Because the other Deinococcus strains lack recB, UV irradiation-induced expression of recB may contribute to the extreme UV resistance, and it is worth noticing that recB is absent in the other three published Deinococcus genomes. The two desert species, D. gobiensis and D. deserti, have a homolog of polB encoding DNA polymerase II (DGo_PC0151 and Deide_1p00180), which is essential for resumption of DNA replication after UV exposure [34] and may also be involved in DNA repair. However, D. radiodurans and D. geothermalis lack polB. Further analysis showed that expression of polB increased significantly following UV irradiation, whereas dnaE encoding the alpha subunit of DNA polymerase III, which is required for misincorporation and bypass during UV mutagenesis, was slightly repressed.
Reactive oxygen species detoxification
The ability to survive acute or chronic exposure to ionizing and UV irradiation and desiccation can be attributed to prevention, tolerance, and repair mechanisms. Scavenging oxygen radicals is an important component of prevention mechanism because reactive oxygen species (ROS) are key intermediates in the damage to cells caused by ionizing and UV radiation and desiccation. Several such prevention gene products are present in the D. gobiensis genome, including five catalases, four superoxide dismutases, and several regulatory genes, for example OxyR of the LysR family of proteins that activates the transcription of genes involved in peroxide metabolism and protection (katG, ahpC, ahpF, and dps), redox balance (grxA, and trxC). Genes involved in carotenoid biogenesis have been shown to confer a modest level of radiation resistance by scavenging electrons from ROSs [35]. D. gobiensis produces carotenoids, and the carotenoid biosynthetic pathway is similar to that found in D. radiodurans [36]. In addition, we observed that genes for secondary metabolite biosynthesis of carotenoids, vitamin B1, vitamin B12, NAD+ and cytochrome P450 were strongly downregulated after UV irradiation.
D. gobiensis possesses two of the three types of known Mn2+ transporters: one from the natural resistance-associated macrophage family and one from the ATP dependent ABC-type transporter family. Interestingly, we observed that D. gobiensis (the Mn/Fe ratio of 1.60±0.02) accumulated 1.1 times more Mn and 2.4 times less Fe than D. radiodurans R1 (Mn/Fe ratio of 0.60±0.04). Previous studies of stress response systems in D. radiodurans demonstrated that the dose-response relationship for desiccation killing in bacteria isolated from desert environments parallels the levels of protein oxidation and the Mn/Fe ratios [37]. A first line of defence against ionizing radiation might be the accumulation of manganese complexes, which can prevent the production of iron-dependent reactive oxygen species [38]. More recently, the quantitative measurement of proteome oxidation (i.e., protein carbonylation) in D. radiodurans exposed to ionizing radiation or UVC light has revealed a consistent correlation with cell killing [39]. A comprehensive outlook on D. radiodurans strategies of combating oxidative stress suggests that the level of protein damage together with the cellular ROS-scavenging capacity determine the radiation survival of bacteria [12]. Thus, it is likely that in D. gobiensis, accumulation of high level of Mn may contribute to the enhanced tolerance to ionizing radiation or UV light.
Discussion
The surface sands of the desert are exposed to intense solar radiation, cycles of extreme temperatures, and desiccation. Such extreme conditions cause stress-induced damage to DNA and proteins, which is lethal to most organisms. Therefore, desert-dwelling bacteria protect DNA and proteins from damage and/or repair them efficiently. Two striking results of this work came from comparison of D. gobiensis with three other sequenced Deinococcus species isolated from canned meat, hot springs and the Sahara desert, respectively. Despite their phylogenetic differences, the two desert strains, D. gobiensis I-0 and D. deserti VCD115, have a large repertoire of similar genes. The two desert strains contain surprisingly large numbers of probably horizontally acquired genes and diverse mobile elements. Many genes shared by the two desert strains are associated with putative mobile elements that aided the parallel evolution of the two desert species. However, D. gobiensis and D. deserti were isolated from very different deserts: the cold Gobi, and the hot Sahara. Comparative analyses of the two desert strains revealed two distinct gene sets: a core of shared orthologous genes and, species-specific genes. Interestingly, D. gobiensis contains 1,541 genes that are missing from D. deserti, including genes for a glucose-6-phosphate dehydrogenase, two transketolases, four catalases, two superoxide dismutases, an alkyl hydroperoxide reductase, and a putative glutathione-S-transferase, which are probably involved in adaptation to a cold desert environment. Notably, in D. gobiensis, phrB and recB were induced immediately after UV irradiation, whereas the other Deinococcus strains lacked the phrB and recB genes. This seemed advantageous for D. gobiensis to make use of the repair systems associated with phrB and recB, since phrB encodes a photolyase that breaks pyrimidine dimers typically caused by UV exposure [40] and recBCD encodes a multifunctional enzyme involved in DNA degradation [32].
Depletion of the stratospheric ozone layer causes increases in UV radiation at the Earth's surface, and the molecular basis of extremely UV radiation-resistant phenotypes is one the intriguing problems of modern biology. The success of D. gobiensis in the cold Gobi desert is probably due to its specialized metabolism, complex regulatory mechanisms, and robust repair systems. Indeed, D. gobiensis was isolated from the upper sand layers of a cold desert where bacteria are frequently exposed to long-lasting solar UV irradiation, necessitating a specific regulatory response that precedes the cellular recovery after UV irradiation damage. Several studies on UV radiation resistance have been conducted on exponential phase cells recovering from ionizing and UV radiation. In one such study, transcriptome dynamics of D. radiodurans recovering from ionizing radiation indicated that the maximum response for most functional gene groups occurred concurrently at approximately three hours post-exposure [11]. UV radiation induces both upregulation of the nos gene and cellular nitric oxide (NO) production in D. radiodurans, and subsequently NO upregulates obgE, a gene for an essential GTPase involved in the regulation of many growth-related processes [18]. These studies have provided important information about cellular recovery after UV irradiation. However, more studies of the transcriptome response immediately after UV irradiation are needed to establish a detailed understanding of the regulatory networks underlying the extreme resistance of deinococci. In the present study, we showed that the expression of most of the previously characterized genes, including nos and obgE, was not induced immediately after UV irradiation. Notably, the 30 regulatory genes induced immediately after UV irradiation included well-known regulators, such as two transcriptional activators (CarD and PhoR), a glucose-inhibited division protein A (GidA), a cognate response regulator (CitB) and a tryptophan repressor (TrpR), and also two uncharacterized protein kinases and 12 ncRNAs. The carD gene encodes an essential regulator of rRNA transcription necessary for the mycobacterial stringent response to oxidative stress, DNA damage, and nutrient limitation [41]. PhoR activates genes of the E. coli phosphate regulon in response to phosphate deprivation [42]. The gidA gene encodes the glucose-inhibited cell division protein A that controls the posttranscriptional regulation of quorum-sensing genes via RhlR-dependent and RhlR-independent pathways in Pseudomonas aeruginosa [43]. These proteins are likely key players in the global transcriptional response immediately following UV irradiation, preceding the cellular recovery of UV irradiation damage (Table 2).
Table 2. The D. gobiensis genes implicated in the immediate global transcriptional response to UV irradiation.
| Locus_tag | Gene | Product description | Fold Change** |
| Regulation | |||
| DGo_CA0550 | carD | Transcriptional regulator, CarD family | 2.2 |
| DGo_CA0552 | ccpA | Catabolite control protein A | 3.4 |
| DGo_CA0357 | citB | Cognate response regulator | 2.0 |
| DGo_CA1040 | ddrI | Transcriptional regulator, Crp/Fnr family | 3.5 |
| DGo_CA0719 | deoR * | DeoR-family transcriptional regulator | 2.1 |
| DGo_CA2738 | fliY | ABC-type amino acid transport/signal transduction system, periplasmic component | 2.2 |
| DGo_CA2290 | gidA | Glucose-inhibited division protein A | 2.4 |
| DGo_CA2808 | lspA | Lipoprotein signal peptidase | 4.9 |
| DGo_PC0175 | phoR | Sensor protein, transcriptional activators | 2.7 |
| DGo_CA0725 | sig4 | RNA polymerase sigma factor | 2.0 |
| DGo_CA0977 | str | Streptomycin 3′-kinase | 2.1 |
| DGo_PB0002 | trpR * | Tryptophan repressor, LysR family transcriptional regulator | 3.4 |
| DNA repair | |||
| DGo_CA2046 | ddrA | Double-strand break repair protein | 1.9 |
| DGo_CA0350 | ddrB | Single-stranded DNA binding protein | 2.2 |
| DGo_CA0002 | dnaN | DNA polymerase III, beta subunit | 2.4 |
| DGo_CA1041 | gyrA | putative DNA topoisomerase subunit A | 2.0 |
| DGo_CA0873 | gyrB | putative DNA topoisomerase subunit B | 2.0 |
| DGo_PC0001 | lexA | Repressor LexA | 1.9 |
| DGo_CA0607 | phrB * | Deoxyribodipyrimidine photo-lyase type II | 3.2 |
| DGo_PC0151 | polB | DNA polymerase II | 2.0 |
| DGo_PC0098 | recB * | Predicted nuclease, RecB family | 3.0 |
| DGo_CA0376 | yqgF | Putative Holliday junction resolvase | 2.7 |
| Other functions | |||
| DGo_CA0071 | ddrC | Uncharacterized DNA damage response protein | 2.1 |
| DGo_CA0988 | ddrE | Putative zinc metal lopeptidase | 1.9 |
| DGo_CA2239 | ddrP | Uncharacterized DNA damage response protein | 1.9 |
Genes that are identified in D. gobiensis but not in other published Deinococcus species.
Change values are means of values obtained from two independent experiments.
Various DNA repair and stress response-related genes previously identified and many new gene products of potential interest for biotechnological applications were also found in D. gobiensis (Table S7). For example, the D. gobiensis genome encodes a Deinococcus-specific global regulator (DGo_CA2805) that is similar to IrrE, a global regulator from the extremely radiation-resistant D. radiodurans that confers enhanced salt tolerance in both E. coli and in the plant Brassica napus [44]. D. gobiensis contains two similar cold shock proteins (DGo_CA1136 and DGo_PA0041), with 65–70% identity to CspA from E. coli and CspB from B. subtilis that confer abiotic stress tolerance in transgenic plants and improved grain yield in maize under water-limited conditions [45]. Furthermore, approximately 47% of the D. gobiensis genes encode proteins of unknown function. We identified a subset of previously uncharacterized genes induced immediately following UV irradiation, suggesting that the organism's extreme resistance phenotype may be attributable to still unknown genes and pathways. It would be intriguing to investigate which of their products are required for the extreme resistance phenotype. To our knowledge, this is the first report of a transcriptome analysis immediately following UV irradiation. Taken together, our results highlight the exceptional ability of D. gobiensis to withstand environmental extremes. These findings may have significant potential for biotechnological and agricultural applications. Further investigations will reveal commonalities in the genetic basis of the UV response and provide insight into the molecular mechanisms underlying the extreme resistance phenotype of the genus Deinococcus.
Materials and Methods
Bacteria and growth conditions
Cells of Deinococcus gobiensis I-0 ( = DSM 21396) were grown in TGY broth (1.0% peptone, 0.5% yeast extract, 0.1% glucose) at 30°C. For genome sequencing, cells were harvested at the early stationary phase (5–10×108 colony-forming units (CFUs) ml−1). For irradiation, cells were harvested at the late-log phase (≈2×108 CFUs ml−1) and then washed twice with equal volumes of potassium phosphate buffer (100 mM, pH 7.0) as described previously [19]. UV light (254 nm, 200 µW cm−2) was used to irradiate a 20 ml suspension for 5 min in a 9 cm plate with stirring. As a control, an additional non-irradiated suspension was incubated for the same duration. After treatment, cells were harvested by centrifugation at 8,000 rpm for 3 min.
To test the concentrations of Mn and Fe, cells of strains I-0 and R1 were collected from late-log phase TGY cultures and dried by low-temperature vacuum drying. Measurement was performed at the Micro Structure Analytical Lab (Beijing) using Wavelength Dispersive X-Ray Fluorescence (WDXRF).
To test the utilization of aromatic substrates, cells from the TGY 48 h culture were washed twice and subsequently inoculated into Mineral Salt medium (benzoic acid 2 mmol/L, NaNO3 0.5 g/L, K2HPO4 0.65 g/L, KH2PO4 0.17 g/L, MgSO4 0.10 g/L) at a dilution of 1∶100.
Genomic DNA extraction and whole-genome shotgun sequencing
Total DNA was isolated from D. gobiensis I-0 ( = DSM 21396) according to a published method described for bacteria [46]. Genome sequencing was performed at Tianjin Research Center for Functional Genomics and Biochip (Tianjin, China) using the Sanger/pyrosequencing strategy described previously [47]. The Roche 454 FLX gene sequencer (454 Life Sciences, Branford, CT) was used to generate 329,480 reads that were assembled into 287 contigs using the Newbler assembler. Artificial 1 kb reads representing the Roche/454 assembly were generated using mktrace in the Consed package (www.phrap.org/) and assembled with 11,444 ABI3730 reads (3.2 kb library) using PhredPhrap (www.phrap.org/). Possible misassemblies were corrected according to the mate-pair relationships of ABI3730 reads; gaps between contigs were closed by editing in Consed, custom primer walks, or PCR amplification.
Genome annotation and analysis
Glimmer3 was initially used to identify putative CoDing Sequences (CDS), and tRNAs were predicted using tRNAscan-SE, and Artemis [48] was used to collate data and facilitate annotation. Function predictions were based on BLASTp similarity searches (E-value <10−5) in the non-redundant GenBank protein database (www.ncbi.nlm.nih.gov/protein), the SwissProt protein database (http://www.ebi.ac.uk/swissprot/), the clusters of orthologous groups (COG) database (www.ncbi.nlm.nih.gov/COG) and KEGG database (www.genome.ad.jp/kegg).
Pairwise genome comparisons of D. gobiensis with three other Deinococcus species were made using nucmer in Mummer 3 [49]. The minimum length of a cluster of matches, break length and maximum gap distance were set to 30 bp, 3 kb and 3 kb, respectively.
To analyze the taxonomic affiliations of D. gobiensis proteins, the BLAST hits with at least 95% of the highest score (E-value <10−4) to the RefSeq database (www.ncbi.nlm.nih.gov/RefSeq) were collected for each of the D. gobiensis proteins. For each query, if the taxonomic affiliations of all hits at the phylum level were the same, the query was considered affiliated with this taxon; otherwise, the taxon affiliation of the query was considered unresolved. For each of the genes belonging to the phylum Deinococcus-Thermus, the query was further assigned to the order Deinococcales or Thermales according to the taxonomic affiliation of the best hit.
D. gobiensis and another eight publicly available Deinococcus-Thermus genomes from NCBI databases were used in the comparisons. To ensure consistency, the annotations of all genomes were verified based on the similarity with proteins in D. gobiensis using tBLASTn [50], [51]. The sets of orthologous protein-coding genes were defined as mutual fully transitive reciprocal BLASTp [52] hits (E-value <10−4) [53]. The amino acid (for the whole phylum) and nucleic acid sequence (within the order Deinococcales)of each orthologous group was aligned using the CLUSTALW program version 2.0 [54]. For each data set, the phylogenetic relationship was estimated and tested in one thousand bootstrap samples using TREE-PUZZLE version 5.3 (general time reversible (GTR) +Γ4+I model of evolution with a BIONJ starting tree) [55]. The bi-partitions with at least 70% supports in the bootstrap test for each data set were recorded as “0/1” status and used to reconstruct the consensus sequence. The phylogenetic relationship of the consensus sequence was built using SplitsTREE 4 with the BioNJ model [56].
Isolation and enrichment of mRNA, RNA processing and transcriptome sequencing
Bacterial cells were collected and ground into a fine powder in liquid nitrogen. Total RNA was isolated using Trizol reagent (Invitrogen), subsequently purified using RNeasy MinElute Cleanup Kit (Qiagen) and eluted in RNase-free water. Bacterial ribosomal RNAs were removed via a mixed treatment using the MICROB Express kit (Ambion) and the mRNA-ONLY Prokaryotic mRNA isolation Kit (Epeicentre® Biotech.) according to the manufacturer's instructions. Heating at 94°C fragmented the mRNA. First strand cDNA was synthesized with random hexamer primers, and second strand cDNA was synthesized with DNA polymerase. Double strand cDNA was end-repaired, a single adenosine was added, and the Illumina adapters were ligated. Gel-electrophoresis was used to select DNA fragments between 200–250 bp. Libraries were amplified by PCR using Phusion polymerase. Sequencing libraries were denatured with sodium hydroxide and diluted in hybridization buffer for loading onto a single lane of an Illumina GA flow cell. Cluster formation, primer hybridization and pair-end, 101×2 cycle, sequencing were performed using proprietary reagents according to the manufacturers' recommended protocols (https://icom.illumina.com/).
Transcriptomics analysis and qRT-PCR verification
High-throughput cDNA sequencing was performed using the Firecrest, Bustard and GERALD programs [24]. The low quality bases (Q<5) at the ends of the reads were trimmed. The reads that were longer than 20 bps were kept and aligned to the D. gobiensis genome using Burrows-Wheeler Aligner (BWA) [57]. The reads that were mapped into the rRNA regions were not included in further analysis. A transcript coverage map was calculated based on the alignment of whole transcript reads. For each of the genes, the 5′-end of the translation regions were defined as positions supported by at least 5 reads summarized in both of the samples.
To identify the non-coding RNA, the continuous regions (≥30 bp) with an average sequencing depth of ≥15 times/bp in the intergenic regions were extracted and compared against GenBank using blastx and blastn. Those regions that lacked similarities to known protein coding genes or were similar to known ncRNAs were classified as possible non-coding RNAs.
To compare the different samples, the fragments per kb of CDS per million mapped reads (FPKM) value were used to normalize the data and represent the overall gene expression. The differently expressed genes between the two samples were selected according to their significance in Chi-square tests (p = 0.05, with Bonferroni correction) and at least 2-fold differences.
One hundred micrograms of total RNA was then synthesized into cDNA using ProtoScript® M-MuLV First Strand cDNA Synthesis Kit (NEB). The primers were designed by Perlprimer v1.1.19 [58]. The expression levels of the selected genes were determined using the 7500 Real-Time PCR System (Applied Biosystems) according to SYBR® Premix ExTaq™ Kit's manual (Takara) using 20 microliter system.
Supporting Information
Unrooted neighbor-joining phylogenetic tree deduced from the orthologous proteins that occur in all 14 sequenced strains from the phylum Deinococcus-Thermus. D. gobiensis and D. radiodurans are most closely related. Numbers indicate bootstrap values below 100.
(TIF)
Synteny plots comparing D. gobiensis I-0 and the other three Deinococcus genomes. The dot plots represent nucmer alignments generated by MUMMER 3 of D. gobiensis on the x-axis and three other Deinococcus species on the y-axis. Forward matches are shown in red, and reverse matches are shown in blue. dnaA isat the bottom left of each plot. The red marks on the horizontal line representing the D. gobiensis genome indicate genomic islands.
(TIF)
General features of the genomes of Deinococcales species.
(DOC)
The qRT-PCR verification.
(XLS)
Transcriptional start sites (TSS). First column presents the TSS location; the TSS and its strand are listed in the next two columns.
(DOC)
Functional description of 1144 genes induced or repressed after UV-irradiation.
(DOC)
Deinococcales-specific genes.
(XLS)
Genomic islands in D. gobiensis.
(DOC)
DNA repair genes (A), stress response-related genes (B) and additional enzymes of possible biotechnological interest genes (C) identified in sequenced deinococci.
(DOC)
Acknowledgments
We would like to thank Prof. Lei Wang for helpful discussions and Dr. Tobias Kieser for critical reading of the manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by grants from the National Basic Research (973) Program of China (No. 2010CB126504 and 2007CB109203), the National Natural Science Foundation of China (No. 30925002 and 30970069) and the National Research and Development Project of Transgenic Crops of China (2011ZX08009-003 and 2009ZX08009-075B). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Battista JR. Against all odds: The survival strategies of Deinococcus radiodurans. Annual Review of Microbiology. 1997;51:203–224. doi: 10.1146/annurev.micro.51.1.203. [DOI] [PubMed] [Google Scholar]
- 2.Anderson A, Nordan H, Cain R, Parrish G, Duggan D. Studies on a radio-resistant Micrococcus. I. Isolation, morphology, cultural characteristics, and resistance to gamma radiation. Food Technol. 1956;10:575–578. [Google Scholar]
- 3.White O, Eisen JA, Heidelberg JF, Hickey EK, Peterson JD, et al. Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science. 1999;286:1571–1577. doi: 10.1126/science.286.5444.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Makarova KS, Aravind L, Wolf YI, Tatusov RL, Minton KW, et al. Genome of the extremely radiation-resistant bacterium Deinococcus radiodurans viewed from the perspective of comparative genomics. Microbiology and Molecular Biology Reviews. 2001;65:44–79. doi: 10.1128/MMBR.65.1.44-79.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Makarova KS, Omelchenko MV, Gaidamakova EK, Matrosova VY, Vasilenko A, et al. Deinococcus geothermalis: the pool of extreme radiation resistance genes shrinks. PLos One. 2007;2 doi: 10.1371/journal.pone.0000955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Groot A, Dulermo R, Ortet P, Blanchard L, Guerin P, et al. Alliance of proteomics and genomics to unravel the specificities of Sahara bacterium Deinococcus deserti. PLoS Genetics. 2009;5 doi: 10.1371/journal.pgen.1000434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ivanova N, Rohde C, Munk C, Nolan M, Lucas S, et al. Complete genome sequence of Truepera radiovictrix type strain (RQ-24T). Standards in Genomic Sciences. 2011;4:91. doi: 10.4056/sigs.1563919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pukall R, Zeytun A, Lucas S, Lapidus A, Hammon N, et al. Complete genome sequence of Deinococcus maricopensis type strain (LB-34T). Standards in Genomic Sciences. 2011;4:163. doi: 10.4056/sigs.1633949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cox MM, Battista JR. Deinococcus radiodurans: the consummate survivor. Nature Reviews Microbiology. 2005;3:882–892. doi: 10.1038/nrmicro1264. [DOI] [PubMed] [Google Scholar]
- 10.Lipton MS, Pasa-Tolic L, Anderson GA, Anderson DJ, Auberry DL, et al. Global analysis of the Deinococcus radiodurans proteome by using accurate mass tags. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:11049–11054. doi: 10.1073/pnas.172170199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu YQ, Zhou JZ, Omelchenko MV, Beliaev AS, Venkateswaran A, et al. Transcriptome dynamics of Deinococcus radiodurans recovering from ionizing radiation. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:4191–4196. doi: 10.1073/pnas.0630387100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Slade D, Radman M. Oxidative Stress Resistance in Deinococcus radiodurans. Microbiol Mol Biol Rev. 2011;75:133–191. doi: 10.1128/MMBR.00015-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Warren Rhodes K, Rhodes K, Boyle L, Pointing S, Chen Y, et al. Cyanobacterial ecology across environmental gradients and spatial scales in China's hot and cold deserts. FEMS microbiology ecology. 2007;61:470–482. doi: 10.1111/j.1574-6941.2007.00351.x. [DOI] [PubMed] [Google Scholar]
- 14.Tanaka M, Earl AM, Howell HA, Park MJ, Eisen JA, et al. Analysis of Deinococcus radiodurans's transcriptional response to ionizing radiation and desiccation reveals novel proteins that contribute to extreme radioresistance. Genetics. 2004;168:21–33. doi: 10.1534/genetics.104.029249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sinha RP, Häder D-P. UV-induced DNA damage and repair: a review. Photochemical & Photobiological Sciences. 2002;1:225–236. doi: 10.1039/b201230h. [DOI] [PubMed] [Google Scholar]
- 16.Courcelle J, Khodursky A, Peter B, Brown PO, Hanawalt PC. Comparative gene expression profiles following UV exposure in wild-type and SOS-deficient Escherichia coli. Genetics. 2001;158:41–64. doi: 10.1093/genetics/158.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qiu XY, Sundin GW, Wu LY, Zhou JZ, Tiedje JM. Comparative analysis of differentially expressed genes in Shewanella oneidensis MR-1 following exposure to UVC, UVB, and UVA radiation. Journal of Bacteriology. 2005;187:3556–3564. doi: 10.1128/JB.187.10.3556-3564.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patel BA, Moreau M, Widom J, Chen H, Yin L, et al. Endogenous nitric oxide regulates the recovery of the radiation-resistant bacterium Deinococcus radiodurans from exposure to UV light. Proceedings of the National Academy of Sciences. 2009;106:18183. doi: 10.1073/pnas.0907262106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuan M, Zhang W, Dai S, Wu J, Wang Y, et al. Deinococcus gobiensis sp. nov., an extremely radiation-resistant bacterium. International Journal of Systematic and Evolutionary Microbiology. 2009;59:1513. doi: 10.1099/ijs.0.004523-0. [DOI] [PubMed] [Google Scholar]
- 20.Henne A, Bruggemann H, Raasch C, Wiezer A, Hartsch T, et al. The genome sequence of the extreme thermophile Thermus thermophilus. Nature biotechnology. 2004;22:547–553. doi: 10.1038/nbt956. [DOI] [PubMed] [Google Scholar]
- 21.Tillier ERM, Collins RA. Genome rearrangement by replication-directed translocation. Nature genetics. 2000;26:195–197. doi: 10.1038/79918. [DOI] [PubMed] [Google Scholar]
- 22.Mackiewicz P, Mackiewicz D, Kowalczuk M, Cebrat S. Flip-flop around the origin and terminus of replication in prokaryotic genomes. Genome Biology. 2001;2 doi: 10.1186/gb-2001-2-12-interactions1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Allen MA, Lauro FM, Williams TJ, Burg D, Siddiqui KS, et al. The genome sequence of the psychrophilic archaeon, Methanococcoides burtonii: the role of genome evolution in cold adaptation. The ISME Journal. 2009;3:1012–1035. doi: 10.1038/ismej.2009.45. [DOI] [PubMed] [Google Scholar]
- 24.Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature. 2010;464:250–255. doi: 10.1038/nature08756. [DOI] [PubMed] [Google Scholar]
- 25.Filiatrault MJ, Stodghill PV, Bronstein PA, Moll S, Lindeberg M, et al. Transcriptome analysis of Pseudomonas syringae identifies new genes, ncRNAs, and antisense activity. Journal of Bacteriology. 2010 doi: 10.1128/JB.01445-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Norais CA, Chitteni-Pattu S, Wood EA, Inman RB, Cox MM. DdrB protein, an alternative Deinococcus radiodurans SSB induced by ionizing radiation. Journal of Biological Chemistry. 2009;284:21402. doi: 10.1074/jbc.M109.010454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sugiman-Marangos S, Junop MS. The structure of DdrB from Deinococcus: a new fold for single-stranded DNA binding proteins. Nucleic Acids Research. 2010;38:3432–3440. doi: 10.1093/nar/gkq036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fernández de Henestrosa A, Ogi T, Aoyagi S, Chafin D, Hayes J, et al. Identification of additional genes belonging to the LexA regulon in Escherichia coli. Molecular Microbiology. 2000;35:1560–1572. doi: 10.1046/j.1365-2958.2000.01826.x. [DOI] [PubMed] [Google Scholar]
- 30.Jiang QF, Karata K, Woodgate R, Cox MM, Goodman MF. The active form of DNA polymerase V is UmuD ' C-2-RecA-ATP. Nature. 2009;460:359–363. doi: 10.1038/nature08178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buis JM, Cheek J, Kalliri E, Broderick JB. Characterization of an active spore photoproduct lyase, a DNA repair enzyme in the radical S-adenosylmethionine superfamily. Journal of Biological Chemistry. 2006;281:25994. doi: 10.1074/jbc.M603931200. [DOI] [PubMed] [Google Scholar]
- 32.Dillingham MS, Kowalczykowski SC. RecBCD Enzyme and the Repair of Double-Stranded DNA Breaks. Microbiology and Molecular Biology Reviews. 2008;72:642–671. doi: 10.1128/MMBR.00020-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khairnar NP, Kamble VA, Misra HS. RecBC enzyme overproduction affects UV and gamma radiation survival of Deinococcus radiodurans. DNA Repair. 2008;7:40–47. doi: 10.1016/j.dnarep.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 34.Rangarajan S, Woodgate R, Goodman M. A phenotype for enigmatic DNA polymerase II: A pivotal role for pol II in replication restart in UV-irradiated Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:9224. doi: 10.1073/pnas.96.16.9224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bagwell CE, Bhat S, Hawkins GM, Smith BW, Biswas T, et al. Survival in nuclear waste, extreme resistance, and potential applications gleaned from the genome sequence of Kineococcus radiotolerans SRS30216. PLos One. 2008;3 doi: 10.1371/journal.pone.0003878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun ZT, Shen SC, Wang C, Wang H, Hu YP, et al. A novel carotenoid 1,2-hydratase (CruF) from two species of the non-photosynthetic bacterium Deinococcus. Microbiology-Sgm. 2009;155:2775–2783. doi: 10.1099/mic.0.027623-0. [DOI] [PubMed] [Google Scholar]
- 37.Fredrickson JK, Li SMW, Gaidamakova EK, Matrosova VY, Zhai M, et al. Protein oxidation: key to bacterial desiccation resistance? ISME Journal. 2008;2:393–403. doi: 10.1038/ismej.2007.116. [DOI] [PubMed] [Google Scholar]
- 38.Daly MJ. A new perspective on radiation resistance based on Deinococcus radiodurans. Nature Reviews Microbiology. 2009;7:237–245. doi: 10.1038/nrmicro2073. [DOI] [PubMed] [Google Scholar]
- 39.Krisko A, Radman M. Protein damage and death by radiation in Escherichia coli and Deinococcus radiodurans. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:14373–14377. doi: 10.1073/pnas.1009312107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Osburne MS, Holmbeck BM, Frias-Lopez J, Steen R, Huang K, et al. UV hyper-resistance in Prochlorococcus MED4 results from a single base pair deletion just upstream of an operon encoding nudix hydrolase and photolyase. Environmental microbiology. 2010;12:1978–1988. doi: 10.1111/j.1462-2920.2010.02203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stallings CL, Stephanou NC, Chu L, Hochschild A, Nickels BE, et al. CarD Is an Essential Regulator of rRNA Transcription Required for Mycobacterium tuberculosis Persistence. Cell. 2009;138:146–159. doi: 10.1016/j.cell.2009.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Makino K, Shinagawa H, Amemura M, Kawamoto T, Yamada M, et al. Signal transduction in the phosphate regulon of Escherichia coli involves phosphotransfer between PhoR and PhoB proteins* 1. Journal of Molecular Biology. 1989;210:551–559. doi: 10.1016/0022-2836(89)90131-9. [DOI] [PubMed] [Google Scholar]
- 43.Gupta R, Gobble T, Schuster M. GidA posttranscriptionally regulates rhl quorum sensing in Pseudomonas aeruginosa. Journal of Bacteriology. 2009;191:5785–5792. doi: 10.1128/JB.00335-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pan J, Wang J, Zhou Z, Yan Y, Zhang W, et al. IrrE, a global regulator of extreme radiation resistance in Deinococcus radiodurans, enhances salt tolerance in Escherichia coli and Brassica napus. PLos One. 2009;4:e4422. doi: 10.1371/journal.pone.0004422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Castiglioni P, Warner D, Bensen RJ, Anstrom DC, Harrison J, et al. Bacterial RNA chaperones confer abiotic stress tolerance in plants and improved grain yield in maize under water-limited conditions. Plant Physiology. 2008;147:446–455. doi: 10.1104/pp.108.118828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ferenci T, Zhou Z, Betteridge T, Ren Y, Liu Y, et al. Genomic Sequencing Reveals Regulatory Mutations and Recombinational Events in the Widely Used MC4100 Lineage of Escherichia coli K-12. Journal of Bacteriology. 2009;191:4025–4029. doi: 10.1128/JB.00118-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goldberg SMD. A Sanger/pyrosequencing hybrid approach for the generation of high-quality draft assemblies of marine microbial genomes. Proceedings of the National Academy of Sciences. 2006;103:11240–11245. doi: 10.1073/pnas.0604351103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carver T, Berriman M, Tivey A, Patel C, Bohme U, et al. Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics. 2008;24:2672–2676. doi: 10.1093/bioinformatics/btn529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kurtz S, Phillippy A, Delcher A, Smoot M, Shumway M, et al. Versatile and open software for comparing large genomes. Genome Biology. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iguchi A, Thomson N, Ogura Y, Saunders D, Ooka T, et al. Complete genome sequence and comparative genome analysis of enteropathogenic Escherichia coli O127: H6 strain E2348/69. Journal of Bacteriology. 2009;191:347. doi: 10.1128/JB.01238-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou Z, Li X, Liu B, Beutin L, Xu J, et al. Derivation of Escherichia coli O157:H7 from its O55:H7 precursor. PLos One. 2010;5:e8700. doi: 10.1371/journal.pone.0008700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhaxybayeva O, Gogarten J. Bootstrap, Bayesian probability and maximum likelihood mapping: exploring new tools for comparative genome analyses. BMC Genomics. 2002;3:4. doi: 10.1186/1471-2164-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schmidt H, Strimmer K, Vingron M, Von Haeseler A. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics. 2002;18:502. doi: 10.1093/bioinformatics/18.3.502. [DOI] [PubMed] [Google Scholar]
- 56.Huson D, Bryant D. Application of phylogenetic networks in evolutionary studies. Molecular Biology and Evolution. 2006;23:254. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 57.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marshall OJ. PerlPrimer: cross-platform, graphical primer design for standard, bisulphite and real-time PCR. Bioinformatics. 2004;20:2471. doi: 10.1093/bioinformatics/bth254. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Unrooted neighbor-joining phylogenetic tree deduced from the orthologous proteins that occur in all 14 sequenced strains from the phylum Deinococcus-Thermus. D. gobiensis and D. radiodurans are most closely related. Numbers indicate bootstrap values below 100.
(TIF)
Synteny plots comparing D. gobiensis I-0 and the other three Deinococcus genomes. The dot plots represent nucmer alignments generated by MUMMER 3 of D. gobiensis on the x-axis and three other Deinococcus species on the y-axis. Forward matches are shown in red, and reverse matches are shown in blue. dnaA isat the bottom left of each plot. The red marks on the horizontal line representing the D. gobiensis genome indicate genomic islands.
(TIF)
General features of the genomes of Deinococcales species.
(DOC)
The qRT-PCR verification.
(XLS)
Transcriptional start sites (TSS). First column presents the TSS location; the TSS and its strand are listed in the next two columns.
(DOC)
Functional description of 1144 genes induced or repressed after UV-irradiation.
(DOC)
Deinococcales-specific genes.
(XLS)
Genomic islands in D. gobiensis.
(DOC)
DNA repair genes (A), stress response-related genes (B) and additional enzymes of possible biotechnological interest genes (C) identified in sequenced deinococci.
(DOC)