Abstract
CD4+ T-cells from gut-associated lymphoid tissues (GALT) are major targets for HIV-1 infection. Recruitment of excess effector CD8+ T-cells in the proximity of target cells is critical for the control of viral replication. Here, we investigated the colocalization potential of HIV-specific CD8+ and CD4+ T-cells into the GALT and explored the role of retinoic acid (RA) in regulating this process in a cohort of HIV-infected subjects with slow disease progression. The expression of the gut-homing molecules integrin β7, CCR6, and CXCR3 was identified as a “signature” for HIV-specific but not CMV-specific CD4+ T-cells thus providing a new explanation for their enhanced permissiveness to infection in vivo. HIV-specific CD8+ T-cells also expressed high levels of integrin β7 and CXCR3; however CCR6 was detected at superior levels on HIV-specific CD4+ versus CD8+ T-cells. All trans RA (ATRA) upregulated the expression of integrin β7 but not CCR6 on HIV-specific T-cells. Together, these results suggest that HIV-specific CD8+ T-cells may colocalize in excess with CD4+ T-cells into the GALT via integrin β7 and CXCR3, but not via CCR6. Considering our previous findings that CCR6+CD4+ T-cells are major cellular targets for HIV-DNA integration in vivo, a limited ability of CD8+ T-cells to migrate in the vicinity of CCR6+CD4+ T-cells may facilitate HIV replication and dissemination at mucosal sites.
Introduction
The human immunodeficiency virus type 1 (HIV) epidemic remains a major global health problem despite major advances made since the discovery of the virus in 1983 [1]. The HIV infection has been known to be associated to a gastrointestinal pathology since the beginning of the epidemic [2], [3]. Recent studies in HIV-infected individuals and simian models of infection demonstrated that depletion of CD4+ T-cells from gut-associated lymphoid tissues (GALT) occurs very early upon infection [4], [5], [6], [7]. Memory CD4+ T-cells, expressing the HIV coreceptor CCR5, massively infiltrate the GALT and are preferential targets of viral replication and depletion [8], [9]. The alteration of GALT homeostasis in HIV-infected individuals leads to the impairment of mucosal immunity and microbial translocation from the gut, which can drive chronic immune activation [10], [11]. Despite a partial restoration of mucosal immunity in the GALT of individuals receiving long-term antiretroviral therapies (ART) [12], [13], viral reservoirs persist in different cellular and anatomic compartments and represent a major barrier to HIV eradication [14].
A small fraction of HIV-infected individuals control disease progression for a long period of time in the absence of ART and are called long-term nonprogressors (LTNPs) or slow progressors [15]. The LTNPs subjects are generally defined as HIV-infected individuals who remain clinically asymptomatic with non-declining CD4 counts (>500 cells/µl), and undetectable plasma viral loads (<50–75 HIV RNA copies/ml) for >10 years [16]. However, LTNPs are very heterogeneous and include individuals with virological and/or immunological control. Virological controllers (also called “elite controllers”) are characterized by undetectable plasma viral loads regardless of their CD4 counts, while immunological controllers maintain their CD4 counts in the normal range despite detectable plasma viral loads [15], [16]. Mechanisms involved in the control of disease progression in LTNPs have been linked to host genetic factors controlling the quality of innate and adaptive immunity [17], [18]. The ability of CD8+ T-cells to control HIV replication via cytotoxic and non-cytotoxic mechanisms is well documented [19], [20], [21]. However, viral reservoirs persist in LTNPs [22], [23], pointing out the inability of the immune system to achieve HIV eradication. This is consistent with the finding that the GALT remains an important target of HIV replication in LTNPs with functional alterations in this compartment contributing to slow disease progression [24]. Nevertheless, the existence of a group of HIV-exposed uninfected individuals, in which HIV-specific CD8+ T-cell responses were detected in the cervical mucosa [25], provides proof that protective immunity against HIV can be mounted under specific conditions. Thus, the mechanisms of immune protection against HIV require further investigations.
HIV infection is initiated by a small viral founder population that undergoes mutations to escape T-cell responses [26], [27], [28]. Limiting viral dissemination from the portal site of entry very early after infection via robust anti-viral mechanisms is of paramount importance to prevent the establishment of a chronic HIV infection [28]. Recent studies using a model of simian immunodeficiency virus (SIV)-infection and in situ visualization techniques demonstrated that SIV-specific CD8+ T-cells (effectors) are recruited into the vaginal mucosa and lymph nodes in close proximity to SIV-infected CD4+ T-cells (targets) [29]. The spatial proximity of excess effectors versus target cells appears to be critical for the control of SIV replication and dissemination in vivo [29]. By analogy, the colocalization potential of HIV-specific CD8+ and CD4+ T-cells into tissues such as the GALT might determine the extent of viral dissemination and the outcome of disease progression.
Trafficking of peripheral blood T-cells into the GALT is mediated via specific adhesion molecules and chemokine receptors. The integrin α4β7 binds to the mucosal addressin cell adhesion molecule-1 (MadCAM-1) expressed on gut endothelial cells and allows cells to cross the endothelial barrier [30]. The integrin αEβ7 binds to the E-cadherin expressed on the basolateral surface of intestinal epithelial cells and contributes to cell retention in the intraepithelial compartment [31]. The CCR6 is important in the recruitment of T-cells into Peyer's Patches [32], [33], [34], while CCR9 mediates T-cell infiltration into lamina propria [35], [36], [37]. The CCR5 and CXCR3 binding chemokines also regulate infiltration of T-cells into the gut [38], [39]. Previous studies reported the expression of gut-homing molecules on HIV-specific CD8+ or CD4+ T-cells. The HIV-specific CD4+ T-cells express the integrin β7 and CCR5 [40], [41], while HIV-specific CD8+ T-cells from the gut express CCR5 and integrin αEβ7 [42]. In addition, a fraction of HIV-specific CD4+ and CD8+ T-cells express CCR6 [43].
Results from our group and those published by others identified CCR6 as a marker for memory CD4+ T-cells that are highly permissive to HIV infection in vitro [44], [45] and carry superior levels of integrated HIV-DNA in vivo [44], [45]. Also, we demonstrated that treatment with retinoic acid (RA), a metabolite of vitamin A responsible for the imprinting for gut homing [46], [47], significantly increased the permissiveness of CCR6+ but not CCR6− CD4+ T-cells to HIV replication by acting at entry (CCR5 upregulation) and yet unidentified post-entry levels [48]. Thus, CCR6+CD4+ T-cells may represent sites for active HIV replication into the GALT. The ability of HIV-specific CD8+ T-cells to be recruited into the GALT in the vicinity of CCR6+CD4+ T-cells remains unknown and might be predictive of an efficient control of HIV replication in target cells at the portal sites of entry.
In this study, we investigated the potential of total and HIV-specific CD8+ T-cells to colocalize in excess with CCR6+CD4+ T-cells and explored the role of the RA pathway in regulating the gut-homing potential of these cells. We report here a decreased frequency of CD8+ and CD4+ T-cells expressing CCR6 in the peripheral blood of HIV-infected subjects regardless of their clinical characteristics of disease progression. In a cohort of HIV-infected subjects with slow disease progression, HIV-specific versus CMV-specific CD4+ T-cells highly express the gut-homing markers integrin β7, CCR6, and CXCR3, suggesting a link between enhanced permissiveness to infection in HIV-specific CD4+ T-cells [49] and their gut-homing potential. HIV-specific CD8+ T-cells also express the gut-homing molecules integrin β7 and CXCR3 but express low levels of CCR6. Thus, HIV-specific CD8+ T-cells may migrate into the gut via integrin β7 and CXCR3 but exhibit a limited potential to colocalize with CD4+ T-cell in certain GALT sites where recruitment is dependent on CCR6 (e.g., Peyer's Patches) [32], [33], [34]. This is consistent with our previous finding that CCR6+CD4+ T-cells are major sites for HIV-DNA integration in vivo [44]. Together these results suggest that, in addition to other previously described cellular features (e.g., antiviral properties, poly-functionality, and exhaustion), the co-localization potential of HIV-specific CD4+ and CD8+ T-cells might represent a new parameter to consider in order to predict the efficacy of anti-HIV responses. Future therapeutic strategies should aim at increasing the colocalization potential of HIV-specific effector and target cells in mucosal tissues for a better control of HIV dissemination from the portal sites of entry.
Materials and Methods
Study subjects
Included in the study were three groups of HIV-infected subjects: (i) treatment-naïve with slow disease progression (SP, slow progressors; n = 14), (ii) recently infected untreated (RI; n = 18), and chronically infected under viral-suppressive anti-retroviral therapy (CI on ART; n = 20). A cohort of n = 13 HIV-uninfected subjects were included in this study as controls. Tables 1, 2, and 3 contain information on the CD4 and CD8 counts, plasma viral loads, and time since infection of SP, RI, and CI on ART HIV-infected subjects, respectively. At the time of leukapheresis, 9/14 SP subjects satisfied the long-term nonprogressor (LTNP) criteria, which include >7 years time since infection, low to undetectable plasma viral load in the absence of ART, and CD4 counts >500 cells/µl [15], [16], while 5/14 SP subjects had CD4 counts <500 cells/µl (Table 1). Among the later group, three subjects (SP 005, 011, 0108) were infected for >19-years and had detectable plasma viral loads, while two subjects (SP 015, 0102) had undetectable plasma viral loads and time since infection >9-years (Table 1). SP subjects with CD4 counts <500 cells/µl were not treated as they lacked clinical signs of immunological failure and maintained their plasma viral load <104 HIV RNA copies/ml. CI on ART subjects received various antiviral regimens containing a protease inhibitor, a non-nucleoside reverse transcriptase inhibitor, or three nucleoside reverse transcriptase inhibitors, as we previously reported [44]. Plasma viral load was measured using the Amplicor HIV-1 monitor ultrasensitive method (Roche). PBMCs (109–1010 cells) were collected from HIV-infected and uninfected subjects by leukapheresis, as previously reported [50]. HIV-subjects were recruited via the HIV Primo Infection cohort at the McGill University Health Centre, Royal Victoria Hospital, Montreal, or from the Canadian Cohort of HIV+ Slow Progressors.
Table 1. Clinical parameters of HIV-infected subjects with slow disease progression (SP).
| Subjects | CD4 counts# | CD8 counts# | Plasma viral load& | Time since infection* | ART |
| SP 001 | 860 | 1148 | 158 | 15 | No |
| SP 005 | 435 | 694 | 1862 | 19 | No |
| SP 006 | 991 | 921 | <50 | 15 | No |
| SP 007 | 720 | 631 | <50 | 19 | No |
| SP 008 | 670 | 475 | <50 | 15 | No |
| SP 011 | 325 | 941 | 5370 | 19 | No |
| SP 015 | 448 | 403 | <50 | 16 | No |
| SP 0101 | 1080 | 1320 | <50 | 11 | No |
| SP 0102 | 440 | 162 | <50 | 9 | No |
| SP 0105 | 780 | 1020 | 15920 | 12 | No |
| SP 0106 | 520 | 1790 | 86128 | 12 | No |
| SP 0107 | 720 | 2840 | 1769 | 18 | No |
| SP 0108 | 387 | 465 | 2937 | 21 | No |
| SP 0109 | 670 | 740 | 1942 | 19 | No |
| Median | 670 | 831 | 964 | 16 |
, cells/µl;
, HIV RNA copies per ml plasma (log10);
, years; ART, antiretroviral therapy.
Table 2. Clinical parameters of recently HIV-infected (RI) untreated subjects.
| Subjects | CD4 counts# | CD8 counts# | Plasma viral load& | Time since infection* | ART |
| RI 001 | 704 | 1081 | 68412 | 9 | No |
| RI 002 | 310 | 350 | 200363 | 46 | No |
| RI 003 | 522 | 366 | 2021 | 5 | No |
| RI 004 | 691 | 1122 | 8714 | 12 | No |
| RI 005 | 341 | 372 | 16883 | 5 | No |
| RI 006 | 483 | 930 | 366646 | 2 | No |
| RI 007 | 857 | 1499 | 93223 | 3 | No |
| RI 008 | 475 | 640 | 56838 | 2 | No |
| RI 009 | 338 | 1829 | 81984 | 4 | No |
| RI 010 | 378 | 779 | 93706 | 2 | No |
| RI 011 | 443 | 736 | 176557 | 5 | No |
| RI 012 | 442 | 538 | 36349 | 5 | No |
| RI 013 | 571 | 1266 | 5897 | 7 | No |
| RI 014 | 824 | 626 | 1167770 | 6 | No |
| RI 015 | 494 | 1055 | 15703 | 16 | No |
| RI 016 | 730 | 1310 | 97044 | 25 | No |
| RI 017 | 255 | 988 | 52835 | 25 | No |
| RI 018 | 316 | 376 | 57154 | 8 | No |
| Median | 479 | 855 | 62,783 | 5.7 |
, cells/µl;
, HIV RNA copies per ml plasma (log10);
, months; ART, antiretroviral therapy.
Table 3. Clinical parameters of chronically HIV-infected subjects under long-term viral suppressive ART (CI on ART).
| Subjects | CD4 counts# | CD8 counts# | Plasma viral load& | Time since infection* | ART |
| CI 001 | 890 | 673 | <50 | 57 | Yes |
| CI 002 | 463 | 757 | <50 | 152 | Yes |
| CI 003 | 602 | 767 | <50 | 158 | Yes |
| CI 004 | 563 | 613 | <50 | 86 | Yes |
| CI 005 | 424 | 461 | <50 | 84 | Yes |
| CI 006 | 731 | 413 | <50 | 51 | Yes |
| CI 007 | 834 | 527 | <50 | 38 | Yes |
| CI 008 | 552 | 715 | <50 | 139 | Yes |
| CI 009 | 671 | 1120 | <50 | 242 | Yes |
| CI 010 | 510 | 765 | <50 | 61 | Yes |
| CI 011 | 799 | 1727 | <50 | 62 | Yes |
| CI 012 | 501 | 278 | <50 | 90 | Yes |
| CI 013 | 344 | 642 | <50 | 59 | Yes |
| CI 014 | 604 | 1281 | <50 | 53 | Yes |
| CI 015 | 443 | 322 | <50 | 18 | Yes |
| CI 016 | 599 | 923 | <50 | 86 | Yes |
| CI 017 | 688 | 1273 | <50 | 100 | Yes |
| CI 018 | 434 | 583 | <50 | 165 | Yes |
| CI 019 | 492 | 582 | <50 | 170 | Yes |
| CI 020 | 529 | 690 | <50 | 49 | Yes |
| Median | 558 | 682 | <50 | 85 |
, cells/µl;
, HIV RNA copies per ml plasma (log10);
, months; ART, antiretroviral therapy.
Ethics statement
This study using PBMC samples from HIV-infected and uninfected subjects, was conducted in compliance with the principles included in the Declaration of Helsinki. This study received approval from the Institution Review Board of the McGill University Health Center and CHUM-Research Center, Montreal, Canada. All blood donors provided written informed consent for their participation to the study.
Antibodies and polychromatic flow cytometry analysis
Fluorochrome-conjugated Abs used for polychromatic flow cytometry analysis were CD3-Pacific Blue (UCHT1), CD3-PE/Cy7 (SK7) CD4-Alexa700 (RPA-T4), CD8 APC-Cy7 (SKI), CCR4-PE/Cy7 (1G1), CXCR3-PE/Cy5 (1C6), CD154-PE/Cy5 (89-76), CCR5-PE (2D7) β7-PE/Cy5 (FIB504), CCR6-PE (11A9) and IFN-γ-Alexa700 (B27) (BD Biosciences); CXCR3-FITC (49801) (R&D Systems); β7-PE (FIB504) and IL-17A-PE (ebio64DEC17) and TNF-α-Pacific Blue (MAB11) (eBioscience). The β7 chain may associate with the α4 chain to form the α4β7 integrin or with the αE chain to form the αEβ7 integrin. In previous studies, we demonstrated that the majority of peripheral blood T cells expressed α4 but not the αE chain [48]. Based on this evidence, we can assume the antibody against the β7 chain used in our study identifies the α4β7 dimer.
For extracellular staining, PBMCs were washed with FACS buffer (PBS 1X, 10% FBS (v/v) (Sigma), 0.02% sodium azide (weight/volume)), stained with specific antibodies for 20 minutes at 4°C, washed with FACS buffer, and fixed with a 2% paraformaldehyde buffer. For cell phenotype analysis of antigen-specific T-cells by flow cytometry, 200–5,000 events were acquired using a BD LSRII flow cytometer. A viability stain (Vivid, Invitrogen) was included in the specific staining cocktails to exclude dead cells from our analysis. Results were analyzed using the BD Diva software. Prior to use, all Abs were titrated for an optimal signal to noise ratio. All Abs cocktails were validated by comparing single to multiple staining, and gates were established using fluorescence minus one (FMO), as previously described [44].
HIV-1 peptide pool preparation
Stimulatory peptides were 15-mers with 11 amino acid overlaps corresponding to HIV-1 clade B consensus Gag (n = 123), Nef (n = 49), and Pol (n = 249) (National Institute of Health (NIH) AIDS Research and Reference Reagent Program, Germantown, MD). Each peptide was diluted in DMSO at 12.5–50 mg peptide/ml, depending on the peptide solubility and stored at −80°C. These were used for the preparation of peptide pools (50–100 µg peptide/ml), containing 11 to 28 peptides per pool, as described in Table 4 and Table S1. Pools equivalent to the complete sequence of Nef (100 µg peptide/ml), Gag (100 µg peptide/ml), and Pol (50 µg peptide/ml) proteins were also prepared. Peptide pools were stored at −80°C and used for CD154 assays (10 µg peptide/ml) and proliferation assays (500 ng peptide/ml)
Table 4. Screening for HIV-1 specific CD4+ and CD8+ T-cells responses using the cell proliferation CFSE dilution assay.
| % CFSElow T-cells | ||||||||||
| Subjects | SP 001 | SP 006 | SP 007 | SP 011 | SP 015 | |||||
| T-cells | CD4+ | CD8+ | CD4+ | CD8+ | CD4+ | CD8+ | CD4+ | CD8+ | CD4+ | CD8+ |
| Medium# | 0.06 | 0.12 | 0.12 | 0.21 | 0.13 | 0.1 | 0.14 | 0.13 | 0.11 | 0.15 |
| SEB | 39.8& | 49.9 | 21.5 | 39.01 | 40.76 | 42.07 | 37.36 | 42.57 | 53.01 | 55.27 |
| CMV pp65 | 1.05 | 2.3 | 0.36 | 3.9 | 57.69 | 8.94 | 2.32 | 3.71 | 25.58 | 25.99 |
| HIV p24 | -# | - | - | - | 1.7 | 1.4 | - | - | 0.74 | 1.17 |
| Nef 5139–5187 | 0.23 | 0.57 | - | - | - | 2.87 | - | 0.36 | 0.27 | 0.69 |
| Nef 5139–5163 | 0.2 | 0.32 | - | - | - | 2.1 | - | - | - | 0.44 |
| Nef 5164–5187 | - | 0.34 | - | - | - | 0.52 | - | - | - | - |
| Gag 705–827 | 0.5 | 5.55 | 0.55 | 1.28 | 1.38 | 13.92 | - | 1.26 | 1.1 | 3.03 |
| Gag 705–728 | 0.31 | 1.18 | - | - | 0.29 | 0.5 | - | 1.14 | - | - |
| Gag 729–752 | - | - | - | - | - | 2.91 | - | - | 0.28 | - |
| Gag 753–776 | 0.21 | 2.29 | - | - | 1.73 | 1.48 | - | - | - | - |
| Gag 777–800 | 0.24 | 2.33 | - | - | 0.61 | 19.53 | - | 0.4 | 0.86 | 2.19 |
| Gag 801–827 | - | - | - | - | - | - | - | - | 0.74 | 0.78 |
| Pol 461–709 | 0.54 | 4.13 | 0.25 | 0.89 | 0.38 | 6.38 | - | 1.13 | 0.25 | 2.26 |
| Pol 461–484 | - | - | - | - | - | 0.32 | - | - | - | - |
| Pol 485–508 | 0.19 | - | - | - | - | - | - | - | 0.45 | 0.4 |
| Pol 509–532 | - | - | - | - | - | - | - | - | - | - |
| Pol 533–556 | 0.22 | 0.23 | - | - | - | - | - | - | 0.29 | 4.75 |
| Pol 557–580 | - | - | - | - | - | 3.89 | - | - | - | 0.38 |
| Pol 581–604 | - | - | - | - | - | - | - | - | - | 0.5 |
| Pol 605–628 | 0.39 | 0.39 | 0.3 | - | - | 0.29 | - | - | - | 1.28 |
| Pol 629–652 | 0.45 | 0.47 | - | - | 3.79 | 0.24 | - | - | - | 0.73 |
| Pol 653–674 | 0.54 | 0.47 | - | - | - | 0.21 | - | - | - | 0.31 |
| Pol 675–698 | 0.44 | 2.58 | 0.24 | - | - | 0.22 | - | - | 0.32 | 0.89 |
| Pol 699–709 | 0.42 | 0.37 | - | - | - | - | - | - | - | - |
| Nef Gag Pol | 1.87 | 9.21 | 0.34 | 1.01 | 1.47 | 14.05 | 0.56 | 1.92 | 0.82 | 3.65 |
, background proliferation;
, T-cell proliferation was considered positive when the % of CFSElow T-cells in antigen-stimulated compared to the background was >2-fold higher.
Candida albicans hyphaes culture and lysis
Candida albicans hyphaes was used a s positive control for the induction of IL-17 production by T-cells (Figure S1), as previously described [51]. Candida albicans LAM-1 strain was provided by Dr. Louis de Repentigny (University of Montreal, Montreal, Quebec, Canada), as colonies in a Petri dish. From the isolated colonies, the yeast form was cultured overnight at 37°C in Yeast Peptone Dextrose (YPD) medium (BD Bioscience). To induce the transition from the yeasts form to hyphaes, 0.1–0.5×106 yeasts/ml were cultured in YPD media 20% FBS and incubated for 4 hours at 37°C. The hyphaes generated were washed and resuspended at 2×106 cellules/100 µl in PBS (GIBCO). Micro glass-beads (SIGMA) were added, and cell lysis was performed using the FastPrep FP120 instrument (Thermo Savant, Carlsbad, CA). Cells were lyzed at a speed of 5 meter/second 4 times for 30 seconds and then placed on ice for 2 minutes; this step was repeated 10 times. Lysates were stored frozen at −20°C. Proliferation assays were performed to determine the optimal immunogenic concentrations of Candida albicans hyphae.
CD154/CD40L assay
To identify antigen-specific CD4+ T-cells, the CD154/CD40L assay was performed as previously described (89). Briefly, PBMC from HIV-infected subjects were resuspended in RPMI 1640 (GIBCO), 100 units/ml Penicillin (GIBCO), 100 µg/ml Streptomycin (GIBCO), and 2 mM of L-glutamine (RPMI) with 10% FBS (SIGMA) at 10×106 cells/ml. Cell suspension (200 µl/well) were plated into 96-well plates and stimulated with 1 µg/ml Staphylococcal enterotoxin B (SEB) (Toxin Technology), 5 µg/ml of Cytomegalovirus (CMV) pp65 peptide pool (Miltenyi), 5 µg/ml recombinant HIV-p24 protein (ImmunoDiagnostics, Inc.), or 10 µg/ml of HIV peptide pools (NIAID AIDS Reagent Program) in the presence of 20 µl/well of anti-CD154-PE/Cy5 Abs (BD Biosciences) and 2 µM of monensin (SIGMA) for 16 hrs at 37°C. Cells were then harvested, stained for surface markers with fluorescence-conjugated Abs against CD3, CD4, integrin β7, CCR6, CXCR3, and CCR4, and analyzed by flow cytometry for the expression of homing markers on CD3+CD4+CD154+ T-cells.
CFSE dilution assay
To detect antigen-specific T-cell proliferation, the Carboxy Fluoroscein Succinimidyl Ester (CFSE) dilution assay was performed as previously described [52]. Briefly, PBMC were loaded with 0.5 µM CFSE (Sigma) for 8 minutes at room temperature. The optimal concentration for CFSE was determined by titration for each CFSE lot. Cells were then washed once with PBS and once with RPMI 1640, and then cultured in 5 ml polypropylene tubes (Becton Dickinson) at 2×106 cells/ml in RPMI with 10% human serum (Gemini). Cells were stimulated with HIV peptide pools in which each peptide was at a concentration of 500 ng/ml, 5 µg/ml recombinant HIV-p24 protein (ImmunoDiagnostics), 25 ng/ml SEB (Toxin Technologies) or 1 µg/ml pp65 CMV peptide pool (Miltenyi) for 6 days at 37°C. Cells were harvested, stained with fluorescence-conjugated Abs against CD3, CD4, integrin β7, CCR6, CXCR3, CCR4, and/or CCR5, and analyzed by flow cytometry for the phenotype of CD3+CD4+CFSElow and CD3+CD4−CFSElow cells. In preliminary experiments, we demonstrated that the majority (>95%) of CD3+CD4− cells were CD8+ T-cells. When indicated, a viability stain (Vivid; Invitrogen) was included in staining cocktails to exclude dead cells from analysis.
Intracellular staining for cytokines
CFSE loaded PBMC were stimulated with antigen for 5 days and then restimulated with 50 ng/ml PMA (SIGMA) and 1 µg/ml Ionomycin (SIGMA) in the presence of 10 µg/ml Brefeldin A (SIGMA) for 18 hours. The production of IFN-γ, TNF-α, and IL-17A was measured by intracellular staining with appropriate Abs using the BD Cytofix/Cytoperm kit (BD Biosciences) according to the manufacturer's protocol.
Statistics
The significance of differences observed between-groups was assessed using Mann-Whitney tests (for unpaired samples) and Paired t-test (for paired samples) as specified in figure legends. The correlation between study variables was assessed using a Spearman correlation test and linear regression models. All statistical analyses were performed using the GraphPad Prism 5 software. P-values <0.05 were considered significant.
Results
Decreased frequency of CCR6-expressing CD8+ and CD4+ T-cells in the peripheral blood of HIV-infected subjects with slow and rapid disease progression: We previously identified CCR6 as a marker for memory CD4+ T-cells being highly permissive to HIV infection in vitro and major sites for HIV-DNA integration in infected subjects [44], [48]. As a consequence, the frequency of circulating CCR6+CD4+ T-cells is dramatically reduced from the early stages of HIV infection and the normalization of this frequency is not observed under viral suppressive ART [44]. Considering the antiviral properties of CD8+ T-cells [19], [53], we hypothesized that a robust control of HIV disease progression is dependent on the ability of CD8+ T-cells to co-localize with CCR6-expressing CD4+ T-cells. To test this hypothesis, the expression of CCR6 was first quantified on peripheral blood CD8+ and CD4+ T-cells from HIV-infected subjects with slow and rapid disease progression. The cohort of slow progressors (SP; n = 14) included HIV-infected subjects with a median time since infection of 16 years, median CD4 counts of 670 cells/µl, median CD4 counts of 831 cells/µl, and undetectable or low plasma viral loads (median: 964 HIV RNA copies/ml) in the absence of antiretroviral therapy (ART) (Table 1). The cohort of HIV-infected progressors included recently infected untreated (RI; n = 18; median CD4 counts: 479 cells/µl; median CD8 counts: 855 cells/µl; median plasma viral load: 62,783 HIV RNA copies/ml; median time since infection: 5.7 months) (Table 2) and chronically infected under long-term (>1-year) viral-suppressive ART (CI on ART; n = 20; median CD4 counts: 558 cells/µl; median CD8 counts: 682 cells/µl; median plasma viral load: <50 HIV RNA copies/ml; median time since infection: 85 months) subjects (Table 3). The frequency of CCR6-expressing CD8+ and CD4+ T-cells and the CD8/CD4 ratios within total and CCR6+ T-cell fractions were compared between HIV-uninfected and the three groups of HIV-infected subjects.
The frequency of CCR6-expressing CD8+ and CD4+ T-cells was significantly decreased in RI and CI on ART subjects compared to uninfected controls; unexpectedly, this frequency was also significantly decreased in SP compared to uninfected controls and CI on ART subjects (Figure 1A–B). The CD8/CD4 ratios within the total T-cell population were significantly higher in RI (median: 1.4), CI on ART (median: 1.4), and SP subjects (median: 2.3) compared to uninfected controls (median: 0.9) and also in SP compared to CI on ART subjects (Figure 1C). This suggests the potential recruitment of excess CD8+ T-cells in the vicinity of CD4+ T-cells. In contrast, the median CD8/CD4 ratios within the CCR6+ fraction were <1 in HIV-infected and uninfected subjects, with no significant differences between RI, CI on ART, and SP subjects (Figure 1D). No significant correlations were found between CD4 counts or plasma viral loads and all four parameters investigated in Figure 1 within the three HIV-infected groups (data not shown). These results demonstrate an alteration in the frequency of CCR6-expressing CD8+ and CD4+ T-cells in HIV-infected subjects regardless of their clinical characteristics of disease progression. These results suggest the inability of CD8+ T-cells to be recruited in excess in the proximity of CCR6+CD4+ T-cells, and this even in subjects with slow disease progression.
Figure 1. The frequency of CD8+ and CD4+ T-cells expressing CCR6 is decreased in HIV infected subjects with slow and rapid disease progression.
Peripheral blood mononuclear cells (PBMC) from HIV-uninfected (HIV-; n = 13) and HIV+ subjects recently infected untreated (RI, n = 18), chronically infected under long-term viral suppressive antiretroviral therapy (CI on ART; n = 20), and slow progressors (SP, n = 14) were stained with a cocktail of fluorescence-conjugated CD3, CD4, and CCR6 Abs. The frequency of (A) CD3+CD4− T-cells (referred to as CD8+ T-cells) and (B) CD3+CD4+ T-cells (referred to as CD4+ T-cells) expressing CCR6 was quantified by polychromatic flow cytometry and compared between HIV-uninfected controls and RI, CI on ART, and SP HIV-infected subjects. The CD8/CD4 T-cell ratios were calculated within the total (C) and CCR6+ T-cell fractions (D) in the group of HIV-uninfected controls and the three groups of HIV-infected subjects. Horizontal bars indicate median values. Mann-Whitney p-values are indicated in the figures.
CD4+ T-cells specific for HIV versus CMV preferentially express gut-homing markers: HIV preferentially infects HIV-specific CD4+ T-cells, while CMV-specific CD4+ T-cells are relatively resistant to infection in vivo [41], [49], [54]. This coincides with the fact that HIV-specific CD4+ T-cells express higher levels of the HIV CCR5 coreceptor and produce lower levels of CCR5 binding chemokines than do CMV-specific CD4+ T-cells [41], [49], [54]. We hypothesized that differences in viral permissiveness between HIV-specific versus CMV-specific CD4+ T-cells are also related to their distinct ability to home into anatomic sites of active viral replication, such as the GALT. To test this hypothesis, we investigated the gut-homing potential on CD4+ T-cells specific for HIV versus CMV. The SEB superantigen is known to induce polyclonal T-cell activation [52] and was used as a positive control. The tissue-specific homing molecules studied were integrin β7 for the migration across the GALT endothelium [30], [39], [46], CCR6 for the migration into the GALT Peyer's Patches [33], [55], CXCR3 for the migration into inflammatory sites, including the GALT [30], [39], [46], [56], and CCR4 for the migration into the skin [57]. Experiments were performed with PBMC from seven HIV-infected treatment-naïve subjects with slow disease progression (Table 1), because they exhibited relatively high frequencies of HIV-specific CD4+ and CD8+ T-cells (Table 4 and Table S1). This choice is also justified by the fact that the frequency of CD4+ and CD8+ T-cells expressing CCR6 is also altered in the peripheral blood of SP subjects compared to uninfected controls (Figure 1).
In a first experimental approach, antigen-specific CD4+ T-cells were identified based on their expression of CD154 (CD40 ligand, CD40L) using flow cytometry analysis (Figure S1A), as previously described by others [58]. The PBMCs from HIV-infected individuals were screened for the ability to respond to an antigen panel that included HIV Nef (n = 3), Gag (n = 6) and Pol (n = 11) overlapping peptide pools, HIV-p24 recombinant protein, SEB, and CMV-pp65 recombinant protein (Table S1). The PBMCs were then stimulated with the most immunogenic antigenic panel and the expression of integrin β7, CCR6, CXCR3, and CCR4 was analyzed on antigen-specific CD154+CD4+ T-cells by polychromatic flow cytometry (Figure S1A–B). The phenotype of CD154+CD4+ T-cells specific for different HIV peptide pools was highly heterogeneous within the same donor. Also, inter-donor variations were observed in the expression of homing molecules on CD154+CD4+ T-cells, even those specific for the same HIV peptide pool (Figure S1C). Regardless of this heterogeneity, statistical analysis of homing molecule expression demonstrated that HIV-specific compared to CMV-specific CD154+CD4+ T-cells expressed significantly higher levels of the integrin β7 (Figure S1D). In addition, HIV-specific compared to SEB-specific T-cells displayed increased expression of the integrin β7, CCR6, CXCR3, and CCR4 (Figure S1D). These results demonstrate that HIV-specific CD154+CD4+ T-cells distinguish from cells of other antigenic specificities (CMV, SEB) by their high expression of both gut-homing markers integrin β7 and CCR6.
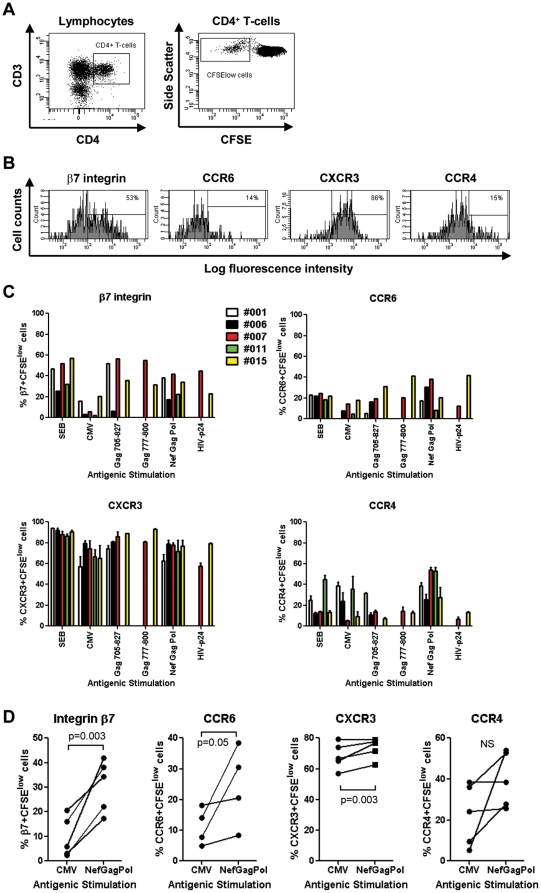
In a second experimental approach, antigen-specific CD4+ T-cells were identified based on their proliferation potential (CFSElow phenotype) using the CFSE dilution assay (Figure 2A), as previously described [52]. The PBMCs from HIV-infected individuals were screened for the ability to respond to an antigen panel that included HIV Nef (n = 3), Gag (n = 6) and Pol (n = 11) overlapping peptide pools, HIV-p24 recombinant protein, SEB, and CMV-pp65 recombinant protein (Table 4). The PBMCs were then stimulated with the most immunogenic antigenic panel and the expression of integrin β7, CCR6, CXCR3, and CCR4 was quantified on CFSElow CD4+ T-cells by polychromatic flow cytometry (Figure 2B). Similar to data obtained on HIV-specific CD154+CD4+ T-cells (Figure S1C), inter- and intra-donor variations were observed in the phenotype of CFSElow CD4+ T-cells specific for different HIV peptide pools (Figure 2C). CXCR3 was expressed by the majority of antigen-specific cells, while the expression of integrin β7, CCR6 and CCR4 was limited to a fraction of cells (Figure 2C). The CD4+ T-cells proliferating in response to the HIVNefGagPol peptide pool versus CMV from matched donors expressed significantly higher levels of integrin β7, CCR6, and CXCR3, with not significant differences regarding CCR4 expression (Figure 2D). The same trend was observed when CD4+ T-cells specific for all HIV peptide pools were compared with those specific for CMV from five different donors (Figure S2A). The HIV-specific CD4+ T-cells identified as CD154+ versus CFSElow cells differed in the expression of homing receptors; these differences were observed when cells were stimulated with distinct (Nef versus Gag, respectively) or the same HIV antigenic pools (HIV-p24) (Figure S1C and Figure 2C). However, the expression at high levels of both integrin β7 and CCR6 remained a unique feature of HIV-specific CD4+ T-cells when compared to CMV-specific cells (Figures S1D and S2A, and Figure 2D). This unique particularity of HIV-specific CD4+ T-cells may confer them the ability to migrate into the GALT, a major site of HIV replication in vivo [8], [59].
Figure 2. Preferential gut-homing potential of HIV-specific versus CMV-specific CD4+ T-cells.
PBMC from SP subjects were loaded in CFSE (0.5 µM) and stimulated with different HIV Nef, Gag, Pol peptide pools (500 ng/ml), recombinant HIV-p24 (5 µg/ml), SEB (25 ng/ml), or the recombinant CMV-pp65 peptide pool (1 µg/ml) for 6 days at 37°C. Antigen-specific T-cells were identified as CFSElow cells, as previously described [52]. Cells were stained with a cocktail of fluorescence-conjugated CD3, CD4, integrin β7, CCR6, CXCR3, and CCR4 Abs and analyzed by polychromatic flow cytometry for (A) the frequency of CFSElowCD3+CD4+ T-cells (referred to as CD4+ T-cells) and (B–D) the expression of integrin β7, CCR6, CXCR3, and CCR4 on antigen-specific CFSElowCD4+ T-cells. (A–B) Shown are results from one donor (i.e., SP 007) generated upon stimulation of PBMC with HIV Gag705–827 peptide pool, representative of results generated with cells from five different donors. (C) Shown is the expression of the homing receptors on CFSElowCD4+ T-cells specific for SEB, CMV and different HIV peptide pools in five different SP subjects. (D) Shown is the homing molecule expression on matched CFSElowCD4+ T-cells specific for CMV versus HIVNefGagPol peptide pool in four-five different SP subjects. Paired T-test p-values are indicated in the figures.
CCR6 is a well established marker for Th17 and Th1Th17 cells with a CCR4+CCR6+ and CXCR3+CCR6+ phenotype [60]. Our results demonstrated that only a minority of CFSElow HIV-specific CD4+ T-cells produced IL-17, while a larger fraction produced IFN-γ and TNF-α (Figure S3). These results indicate that HIV-specific CD4+ T-cells exhibit a Th1Th17 polarization profile. We previously demonstrated that a Th1Th17 profile is favorable to active HIV replication in vitro [44]. Therefore, HIV-specific cells from SP subjects may be highly permissive to HIV, as previously demonstrated in HIV progressors [41], [49], [54].
The colocalization potential of HIV-specific CD4+ and CD8+ T-cells is mediated by the integrin β7 but not CCR6: The antiviral properties of CD8+ T-cells are well characterized [19], [20], [21] and depend on their ability to colocalize in excess with target cells, such as CD4+ T-cells [28], [29]. We next investigated the expression of gut-homing molecules on HIV-specific CD8+ T-cells in order to evaluate their ability to colocalize with HIV-specific CD4+ T-cells for an efficient control of HIV replication in vivo. In preliminary experiments, we demonstrated that the majority of CD8+ T-cells exhibited a CD3+CD4− phenotype (data not shown). Antigen-specific CD8+ T-cells were identified as CFSElow cells (Figure 3A) and tested for their expression of the integrin β7, CCR6, CXCR3, and CCR4 (Figure 3B). Similar to antigen-specific CD4+ T-cells (Figure 2C), the expression of the homing receptors on CFSElow CD8+ T-cells was subject to inter-donor variations (Figure 3C). Despite this heterogeneity, HIVNefGagPol-specific versus CMV-specific CFSElow CD8+ T-cells from matched donors expressed significantly higher levels of integrin β7, while no significant differences were observed in the levels of CCR6, CXCR3, and CCR4 expression (Figure 3D). The same results were observed when CD8+ T-cells specific for all HIV peptide pools were compared with those specific for CMV (Figure S2B). These results suggest an increased ability of HIV-specific versus CMV-specific CD8+ T-cells to migrate via integrin β7 into the GALT, which is likely a site for the initial priming of HIV-specific T-cells.
Figure 3. Homing potential of CD8+ T-cells proliferating in response to HIV peptides.
PBMC from five SP subjects were stimulated and stained with Abs as in Figure 2. At day 6 of stimulation, cells were analyzed by polychromatic flow cytometry for (A) the frequency of CFSElowCD3+CD4− T-cells (referred to as CD8+ T-cells) and (B–D) the expression of β7 integrin, CCR6, CXCR3, and CCR4 on CFSElowCD8+ T-cells. (A–B) Shown are results from one donor (i.e., SP 007) generated upon stimulation with HIV Gag705–827 peptide pool. (C) Shown is the expression of the homing receptors on CFSElowCD8+ T-cells specific for SEB, CMV, and different HIV peptide pools in five different SP subjects. (D) Shown is the homing molecule expression on matched CFSElowCD8+ T-cells specific for CMV versus HIVNefGagPol peptide pool in five different SP subjects. Paired T-test p-values are indicated in the figures.
To further investigate the colocalization potential of HIV-specific CD4+ and CD8+ T-cells, the frequency of cells expressing gut-homing molecules was compared between matched CD4+ and CD8+ T-cells proliferating in response to specific HIV peptide pools. Results depicted in Figure 4A show that HIV-specific CD8+ compared to CD4+ T-cells express significantly higher levels of integrin β7, lower levels of CCR6 and CCR4, and similarly high levels of CXCR3. Spearman correlation and linear regression models were applied and demonstrated a positive correlation between the frequency of HIV-specific CD4+ and CD8+ T-cells expressing the integrin β7 and CXCR3; this correlation was not observed for CCR6 and CCR4 (Figure 4B). Moreover, there was a positive correlation between the frequency of HIV-specific CD4+ and CD8+ T-cells co-expressing the integrin β7 and CXCR3 (Figure 4C). Furthermore, the β7+CXCR3+ phenotype appears to be a unique feature of HIV-specific T-cells since the frequency of β7+CXCR3+ cells was significantly higher in HIVNefGagPol-specific versus CMV-specific CD4+ and CD8+ T-cells from four matched SP subjects (Figure S2C). The proliferation of CD8+ compared to CD4+ T-cells in response to a specific HIV peptide pool was significantly higher in all five HIV-infected subjects (Figure 4D), with median CD8/CD4 ratios of 3.2, 2.3, 4.1, 5.6, and 2.8 in SP subjects 001, 006, 007, 011, and 015, respectively (data not shown). These results suggest that a significant fraction of HIV-specific CD8+ T-cells may colocalize with CD4+ T-cells (at high CD8/CD4 ratios) in certain anatomic sites of the GALT where homing depends on integrin β7 and CXCR3. In contrast, HIV-specific CD8+ T-cells express low levels of CCR6 and CCR4 and thus may be impaired in their ability to colocalize and control viral replication in CCR6+CCR4+ CD4+ T-cells, such as Th17 cells [51] which are highly permissive to infection [44], [45]. Thus, the low expression of CCR6 on CD8+ T-cells may reflect their limited ability to control HIV replication in CD4+ T-cells from certain GALT sites such as the Peyer's Patches, where homing depends on CCR6 [32], [33], [34].
Figure 4. Potential colocalisation of HIV-specific CD8+ and CD4+ T-cells via integrin β7 but not CCR6.
(A–C) PBMC from five SP subjects were stimulated with different HIV peptide pools or recombinant HIV-p24 and analyzed by polychromatic flow cytometry for the expression of homing molecules, as described in Figures 2 and 3. HIV-specific CD4+ and CD8+ T-cells were analyzed for (A–C) the expression of the homing molecules β7 integrin, CCR6, CXCR3, and CCR4 (D) the proliferation potential. (A) Shown is the expression of homing molecules on matched CD4+ versus CD8+ T-cells specific for different HIV peptide pools in five different SP subjects. Shown are correlations between (B) the frequency of matched HIV-specific CD4+ and CD8+ T-cells expressing β7 integrin, CCR6, CXCR3, or CCR4 alone, and (C) co-expressing the β7 integrin and CXCR3 molecules (β7+CXCR3+ phenotype). (D) Shown is the percentage of matched CD4+ versus CD8+ T-cells proliferating (CFSElow) in response to different HIV peptide pools. Paired T-test p-values are indicated in the Figures A and D. Spearman correlation r and p-values and linear regression r2 values are indicated in the Figures B and C.
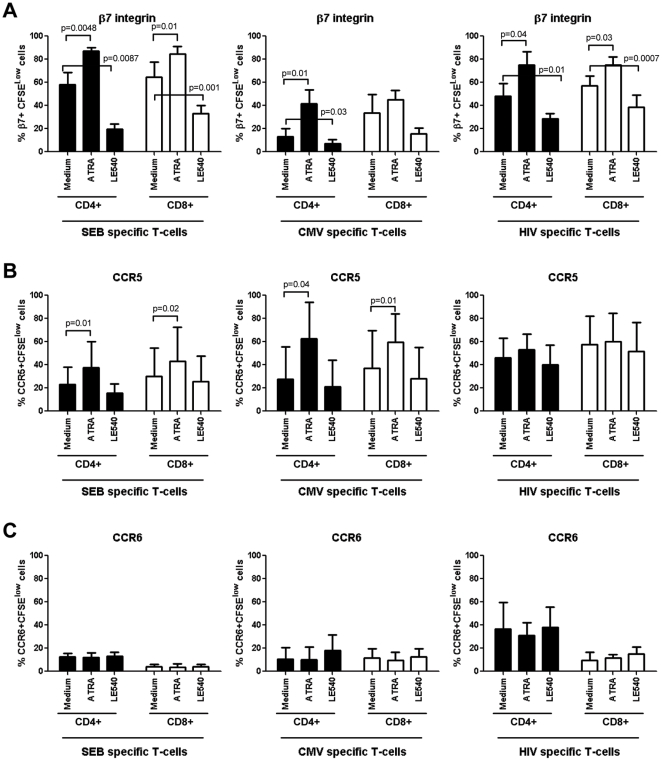
The retinoic acid pathway regulates expression of integrin β7 but not CCR6 and CCR5 on HIV-specific T-cells: The imprinting for gut-homing is regulated at least in part by RA, a derivate of vitamin A metabolism produced by the intestinal dendritic cells [46], [47]. Of note, exposure of CD4+ T-cells to RA upregulates integrin β7 and CCR5 expression and renders them highly permissive to HIV replication [61], [62]. Here, we investigated whether RA can be used to manipulate the colocalization potential of HIV-specific CD4+ and CD8+ T-cells via integrin β7 and CCR6. With this in mind, PBMC from SP subjects were exposed to the HIVNefGagPol peptide pool, SEB, or CMV in the presence or absence of all-trans RA (ATRA) or the RA antagonist LE540. In preliminary experiments, we demonstrated that at physiological dose (10 nM [61], [63]), ATRA did not have any significant effect on cell proliferation, while LE540 at 1 µg/ml [64] decreased integrin β7 expression on SEB-specific T-cells without interfering with cell viability (data not shown). The expression of integrin β7, CCR5, and CCR6 was quantified on CFSElow T-cells by multicolor flow cytometry. Exposure to ATRA and LE540 led to a significant increase and decrease, respectively, in the integrin β7 expression on CD4+ T-cells specific for HIV-, SEB, and CMV and also on CD8+ T-cells specific for HIV and SEB (Figure 5A). The ATRA also increased expression of CCR5 on SEB-specific and CMV-specific but not on HIV-specific T-cells (Figure 5B), where levels of CCR5 expression were higher, although not statistically significant due to donor-to-donor variability, compared to those on SEB-specific and CMV-specific T-cells (Paired t-Test p = 0.04 and p = 0.1, respectively). In contrast, ATRA and LE540 treatment did not interfere with the expression of CCR6 on antigen-specific CD4+ or CD8+ T-cells (Figure 5C). These results demonstrate that the RA pathway regulates the expression of integrin β7 but does not interfere with CCR5 and CCR6 expression on HIV-specific CD4+ and CD8+ T-cells.
Figure 5. Retinoic acid activation pathway regulates integrin β7 expression on HIV-specific T-cells.
PBMC from SP subjects were loaded in CFSE (0.5 µM) and stimulated with HIVNef-Gag-Pol peptide pool (500 ng/ml), SEB (25 ng/ml), or the recombinant CMV-pp65 peptide pool (1 µg/ml) for 6 days at 37°C in the presence or absence of all-trans-retinoic acid (ATRA; 10 nM) or the RA antagonist LE540 (1 µg/ml). Cells were stained with a cocktail of fluorochrome-conjugated CD3, CD4, integrin β7, CCR5 or CCR6 Abs and analyzed for the expression of (A) integrin β7, (B) CCR5, and (C) CCR6 on CFSElow CD4+ and CD8+ T-cells specific for SEB, CMV, and HIV antigens. Experiments were performed with cells from four SP subjects (mean±SD). Paired t-Test p-values are indicated in the figures.
To gain more insights into the regulation of trafficking potential of HIV-specific T-cells, we studied the effects of RA and LE540 on the frequency of HIV-specific T-cells with a β7+CCR5+ and β7+CCR6+ phenotype. The CD4+ and CD8+ T-cells proliferating in response to HIV differed from CMV- or SEB-specific T-cells by an increased frequency of β7+CCR5+ T-cells (Figure 6A and C). The pool of HIV-specific CD4+ T-cells included higher frequencies of cells with a β7+CCR6+ phenotype compared to cells specific for CMV or SEB, while CD8+ T-cells specific for HIV, CMV, or SEB included very low frequencies of β7+CCR6+ T-cells (Figure 6B). Exposure to ATRA and LE540 led to a significant increase and decrease, respectively, in the frequency of HIV-specific CD4+ and CD8+ T-cells with a β7+CCR5+ phenotype (Figure 6D). Since ATRA does not modulate CCR5 expression on HIV-specific cells (Figure 5C), the increase in the frequency of β7+CCR5+ cells (Figure 6D) is likely due to the upregulation of integrin β7 expression on existing CCR5+ T-cells. Finally, ATRA and LE540 treatment had no significant effects on the frequencies of HIV-specific CD4+ or CD8+ T-cells with a β7+CCR6+ phenotype (Figure 6E). These results provide evidence that ATRA may be used to enhance recruitment of HIV-specific CD4+ and CD8+ T-cells across the intestinal endothelium via the integrin β7. In contrast, ATRA does not interfere with CCR6 expression and cannot facilitate the in situ colocalization of CD8+ T-cells with CCR6+CD4+ T-cells.
Figure 6. Retinoic acid upregulates the frequency of HIV-specific T-cells with a β7+CCR5+ but not β7+CCR6+ phenotype.
PBMC from SP subjects were loaded in CFSE (0.5 µM) and stimulated as in Figure 5. Cells were stained with a cocktail of fluorochrome-conjugated CD3, CD4, and integrin β7, and CCR5 or CCR6 Abs. CFSElow CD4+ and CD8+ T-cells specific for SEB, CMV, and HIVNef-Gag-Pol peptide pool were analyzed for the co-expression of (A) integrin β7 and CCR5 and (B) integrin β7 and CCR6. The effects of RA and LE540 on the frequency of Ag-specific CD4+ and CD8+ T-cells exhibiting a β7+CCR5+ or β7+CCR6+ phenotype were then analyzed. (A–B) Shown are results from one representative SP subject and (C–E) statistical analysis of results from experiments performed with cells from five SP subjects (mean±SD). Paired t-Test p-values are indicated in the figures.
Discussion
The GALT is a major site for HIV replication in vivo [8], [59], with HIV-specific CD4+ T-cells being highly permissive to infection [49]. The recruitment of effector CD8+ T-cells in the proximity of target CD4+ T-cells is a prerequisite for an efficient control of viral replication in vivo [29], [65]. In this study, we investigated the potential of HIV-specific CD8+ T-cells to colocalize in excess with CD4+ T-cells in the GALT and explored the role of the retinoic acid (RA) activation pathway in regulating this process. We demonstrated that a large fraction of HIV-specific CD4+ and CD8+ T-cells express the gut-homing molecules integrin β7 and CXCR3 while CCR6, a marker of HIV permissiveness in CD4+ T-cells [44], [48], was expressed at high and low levels on HIV-specific CD4+ and CD8+ T-cells, respectively. We also demonstrated that RA upregulated integrin β7 expression but did not affect CCR6 expression. Our data support a model in which HIV-specific CCR6+CD4+ T-cells escape the antiviral control of CD8+ T-cells in certain GALT sites (e.g., Peyer's Patches) where migration is dependent on CCR6 [32], [33], [34] (Figure 7). Considering the critical role played by CCR6+ Th17 cells in mucosal immunity [66], uncontrolled HIV replication in these cells likely leads to dramatic alterations of mucosal immunity and microbial translocation [13], [59], [67], [68], [69]. These observations were made in a cohort of SP subjects with a median time since infection 16 years. Whether CD8+ T-cells colocalize in excess with CCR6+CD4+ T-cells for an efficient control of HIV replication in SP subjects during the first years of infection or in HIV-exposed uninfected individuals remains to be determined.
Figure 7. Proposed model for the differential colocalization of HIV-specific CD4+ and CD8+ T-cells into the GALT.
The control of viral replication is dependent on the in situ colocalization of excess effector versus target cells [28]. Given the results included in Figures 1– 6 of the present manuscript, we propose a model where (A) HIV replication in CD4+ T-cells may be controlled by CD8+ T-cells in certain GALT sites (e.g., lamina propria), where recruitment is dependent on integrin β7, CXCR3 and CCR5 because of an increased ratio between HIV-specific CD8+ and CD4+ T-cells. In contrast, (B) HIV-specific CD4+ T-cells recruited into other GALT sites via CCR6 (e.g., Peyers's Patches) may escape the CD8+ T-cell-mediated antiviral control due to a limited CCR6-dependent colocalization potential of CD4+ and CD8+ T-cells. This model is in line with our previous findings that CCR6+CD4+ T-cells harbor the highest levels of integrated HIV-DNA in vivo [44] and suggests that novel therapeutic strategies aimed at increasing CCR6 expression on CD8+ T-cells may lead to a better control of HIV replication in CCR6+CD4+ T-cells.
Our initial hypothesis was that the colocalization potential of CD8+ T-cells with CCR6+CD4+ T-cells was altered in HIV-infected subjects with disease progression but not in slow progressors (SP). To test this hypothesis, the expression of CCR6 was quantified on peripheral blood CD8+ and CD4+ T-cells from SP subjects and two other cohorts of HIV progressors, recently infected untreated (RI) and chronically infected under viral suppressive ART subjects (CI on ART). Unexpectedly, we found an alteration in the frequency of CCR6-expressing CD8+ and CD4+ T-cells in HIV-infected subjects regardless of their clinical characteristics of disease progression. This suggested the inability of CD8+ T-cells to be recruited in excess in the proximity of CCR6+CD4+ T-cells, and this even in subjects with slow disease progression. This is consistent with studies reporting immunological alterations related to HIV persistence in CD4+ T-cells and chronic immune activation in SP subjects, especially after many years of infection in the absence of ART [23], [24], [70], [71].
According to the paradigm of tissues-specific homing, effector memory T-cells are imprinted with the ability to recirculate into peripheral tissues where the initial antigen encounter took place [37], [46], [56], [72]. A specific combination of adhesion molecules and chemokine receptors regulate the multi-step process of tissue-specific homing of T-cells [73], [74]. Also, local presentation of antigens by endothelial cells contribute to the recruitment of T-cells into specific sites [75]. In this study, we demonstrated that HIV-specific CD154+CD4+ T-cells express molecules regulating migration into the GALT (integrin β7, CCR6, CXCR3) and skin (CCR4). Considering the fact that the GALT [59], [69] but not the skin are sites of HIV replication in vivo, this broad homing potential of HIV-specific CD154+CD4+ T-cells was unexpected and inconsistent with the paradigm of tissue-specific homing of pathogen-specific T-cells. This exception from the rule is not unique. In fact, T-cells induced upon subcutaneous yellow fever immunization have a dynamic migration program and home into multiple distal tissues, including the GALT [76].
CD154 was proposed as a surrogate marker for the identification of cytokine-producing T-cells in response to an antigenic stimulation [58]. However, we found a differential expression of homing molecules on recently activated CD154+ compared to proliferating CD4+ T-cells (CFSElow) in response to HIV, SEB, or CMV. The skin-homing receptor CCR4 was highly expressed on CD154+ compared to CFSElow CD4+ T-cells specific for HIV and CMV. This is consistent with the paradigm that T-cells are originally imprinted with a skin-homing potential that is lost during the process of differentiation into specialized Ag-specific cells [46]. Also, the expression of CCR6 was higher on CMV-specific CD154+ compared to CFSElow CD4+ T-cells, while the integrin β7 expression was lower on SEB-specific CD154+ compared to CFSElow CD4+ T-cells. The finding that some antigens induced either CD154+ or CFSElow CD4+ T-cells but not both suggests that CD154 expression does not predict the ability of a cell to proliferate. Accordingly, CD154+CD4+ T-cells were mainly triggered by HIVNef peptide pools, while CFSElowCD4+ T-cells were selectively induced by HIVGag peptide pools (Table 4 and Table S1). Thus, HIV-specific CD154+ and CFSElow CD4+ T-cells exhibit distinct homing potential and antigenic specificity and therefore may represent different stages of CD4+ T-cell differentiation with distinct roles in antiviral immunity. The molecular determinism underlying these differences remains unclear but might be related to the anatomic site of original antigenic priming.
The HIV establishes a persistent infection by mechanisms that are not clearly understood, and viral eradication is not achieved under current antiretroviral therapies [14], [77]. The CD4+ T-cells play a critical role in HIV pathogenesis [77]. The HIV-specific compared to CMV-specific CD4+ T-cells are preferentially infected with HIV in vivo [49]. This is because HIV-specific CD4+ T-cells express high levels of the HIV coreceptor CCR5 [41] and produce low levels of CCR5 binding chemokines and therefore fail to protect themselves from infection in an autocrine manner [54]. Consistent with the evidence that the GALT is a major site of HIV replication [59], [69], we observed that HIV-specific compared to CMV-specific CD154+CD4+ T-cells expressed at high levels both gut-homing molecules integrin β7 and CCR6. We also observed increased expression of integrin β7 and CCR6 on CD4+ T-cells proliferating in response to HIV compared to CMV antigens. In addition to their role in gut-homing, the integrin β7 was identified as a new HIV-gp120 binding receptor [61], [62], and its expression on HIV-specific CD4+ T-cells might favor HIV binding on these cells and viral dissemination from the portal sites of entry. The CCR6 is a marker for memory CD4+ T-cells with a Th17 and Th1Th17 lineage polarization profile [51]. We found that a small fraction of HIV-specific cells produced IL-17, with the majority of cells producing IFN-γ and TNF-α. Thus, HIV-specific CD4+ T-cells exhibit a Th1Th17 polarization profile. Indeed other studies demonstrated that very few HIV-specific CD4+ T-cells produce IL-17 [78]. Considering our previous findings that HIV replicates actively in T-cells with a Th1Th17 polarization profile [44], these results suggest that HIV-specific CD4+ T-cells in SP subjects are also permissive to infection. Consistent with this prediction, the expression of the HIV co-receptor CCR5 was relatively high on HIV-specific CD4+ T-cells from SP subjects. This may render them extremely permissive to infection and may explain why some of the SP subjects begin loosing their CD4 counts, especially after many years of infection in the absence of ART [16]. Hence, the expression of integrin β7, CCR6, and CCR5 represents a unique “signature” for HIV-specific T-cells. The relationship between imprinting for gut-homing and viral permissiveness was recently demonstrated for adenovirus serotype 5 (AD5)-specific CD4+ T-cells generated upon AD5-HIV vaccination (STEP trial), as cells exhibited an integrin α4β7+CCR5+ phenotype and high susceptibility to HIV infection [79]. The molecular mechanisms that control homing potential of HIV-specific T-cells are likely related to the cellular/tissue environment in which these cells initially encountered antigen. Indeed, the GALT dendritic cells produce RA, which is known to trigger integrin α4β7 expression and upregulate CCR5 expression on T-cells [47], [48], [61]. Similarly, the GALT environment is rich in Th17 polarizing cytokines (TGF-β, IL-1, IL-6) [51], [80], [81] that may trigger CCR6 expression on HIV-specific CD4+ T-cells.
The CD8+ T-cells control HIV replication in target cells via cytotoxic and non-cytotoxic mechanisms [19], [20], [21]. Recent studies using visualization techniques demonstrated that recruitment of excess viral-specific effectors in the vicinity of target cells is critical for the control of viral replication and disease progression in an SIV model of infection [29]. Our results reveal that matched HIV-specific CD8+ and CD4+ T-cells may colocalize to anatomic sites where recruitment is mediated by integrin β7, CXCR3 and/or CCR5 (Figure 7A). The expression of integrin β7 on both HIV-specific CD8+ and CD4+ T-cells supports the idea that these cells are primed with the antigen within the GALT, where they are likely exposed to factors that imprint cells with a gut-homing potential [46], [47]. The CXCR3 is responsible for leukocyte migration into the inflammatory sites, including the gut [56]. Of note, a decreased frequency of CXCR3+CD8+ T-cells was reported in advanced HIV-1 infection that might contribute to cytotoxic T-lymphocyte dysfunction [82]. In our SP cohort, HIV-specific T-cells expressed maximal levels of CXCR3, and this suggests their functional competence in vivo. In contrast, we observed that CCR6 was expressed at high levels on HIV-specific CD4+ but not CD8+ T-cells. The low expression of CCR6 on HIV-specific CD8+ T-cells was consistent with the decreased frequency of memory CCR6+CD8+ T-cells in the peripheral blood of HIV-infected subjects with slow and rapid disease progression compared to uninfected individuals. This points to the fact that regardless of the clinical characteristics of HIV disease progression, CD8+ T-cells have a limited ability to colocalize with CCR6+CD4+ T-cells and therefore to control HIV replication in these cells. This may explain in part why CCR6+CD4+ T-cells are highly permissive to HIV-DNA integration in vivo [44]. The escape of CCR6+CD4+ T-cells from the non-cytotoxic antiviral control by CD8+ T-cells may also explain the preferential depletion of these cells during disease progression [44], likely via a virus-induced toxicity mechanism [83].
Several previous studies demonstrated that HIV-specific CD8+ T cells from SP subjects are efficient in controlling viral replication ex vivo [15], [16], [19]. This is consistent with our observations in vitro that HIV replication in antigen-stimulated PBMC from the seven SP subjects was controlled (undetectable HIV-p24 levels, as quantified by ELISA), likely by CD8+ T-cells (Ancuta, unpublished observations). However, the situation in vivo may be different. Our results support a model in which the low expression of CCR6 on HIV-specific CD8+ T-cells is exploited by HIV-1 for its efficient replication in CCR6+CD4+ T-cells in some GALT sites such as the Peyer's Patches (Figure 7B). In this context, it is of interest to identify ways to increase the ability of HIV-specific CD8+ T-cell to colocalize with HIV-specific CD4+ T-cells. We found that RA increased the expression of integrin β7 on T-cells specific for HIV, CMV and SEB and the frequency of T-cells specific for CMV and SEB with a β7+CCR5+ phenotype. The HIV-specific CD4+ and CD8+ T-cells included a relatively high fractions of β7+CCR5+, and their frequency remained high upon RA treatment. In contrast, HIV-specific CD4+ compared to CD8+ T-cells included an increased frequency of cells with a β7+CCR6+ phenotype and the physiological dose of RA used (10 nM, [61], [63]) did not upregulate CCR6 expression on CD8+ nor CD4+ T-cells. Other factors responsible for Th17 differentiation, such as TGF-β, IL-1, IL-6 [51], [80], [81], might be involved in regulating CCR6 expression on HIV-specific CD8+ T-cells and remain to be identified.
Together, these results suggest the ability of HIV-specific CD8+ and CD4+ T-cells to colocalize into the GALT (e.g., lamina propria) via integrin β7, CCR5, and CXCR3 and reveal a limited potential of HIV-specific CD8+ T-cells to migrate into other GALT sites (e.g., Peyer's Patches), where recruitment is dependent on CCR6 [32], [33], [34]. Studies on gut biopsies are required to validate our proposed model (Figure 7), which is consistent with the preferential permissiveness of CCR6+CD4+ T-cells to HIV infection in vivo [44]. In this context, understanding molecular mechanisms regulating CCR6 expression on HIV-specific CD8+ T-cells, together with the expression of the CCR6 ligands into the GALT, is of paramount importance for the design of new therapeutic strategies aimed at HIV eradication. However, caution must be taken when designing such strategies to avoid an increased recruitment of HIV targets at the portal sites of mucosal entry. Finally, studies evaluating the functionality of the immune system in response to ART and vaccination may gain in physiological relevance if they consider monitoring the in situ colocalization potential of HIV-specific CD8+ and CD4+ T-cells as a new correlate of protection.
Supporting Information
The HIV-specific versus CMV-specific CD154+CD4+ T-cells preferentially express a gut-homing potential. PBMC from SP subjects were stimulated with different HIV Nef, Gag, Pol peptide pools (10 µg/ml), recombinant HIV-p24 protein (5 µg/ml), SEB (1 µg/ml), or CMV-pp65 peptide pool (5 µg/ml) for 18 hours at 37°C in the presence of fluorescence conjugated anti-CD154-PE/Cy5 Abs (20 µl/2×106 cells/0.2 ml/well). Antigen-specific T-cells were identified as CD154+ cells, as previously described [58]. Cells were harvested, stained with a cocktail of fluorochrome-conjugated CD3, CD4, and β7 integrin, CCR6, CXCR3, or CCR4 Abs and analyzed by polychromatic flow cytometry for (A) the expression of CD154 on CD3+CD4+ T-cells and (B–D) the expression of homing molecules on CD3+CD4+CD154+ T-cells. (A–B) Shown are results from one SP subjects (SP 015 stimulated with the HIV Nef5164-5187 peptide pool), representative of results generated with cells from five different donors. (C) The expression (%) of homing receptors was analyzed on CD154+ T-cells specific for SEB, CMV, and different HIV peptide pools in five different SP subjects. (D) Shown are statistical analyses of the homing molecule expression on CD154+CD4+ T-cells specific for SEB, CMV, and HIV (all peptide pools) in five different SP subjects (box & whisker graph: range and median). Mann-Whitney p-values are indicated in the figures.
(TIF)
Homing potential of CD4+ and CD8+ T-cells proliferating in response to HIV peptides. PBMC from SP subjects were stimulated with different antigens and analyzed by polychromatic flow cytometry for the expression of homing molecules as in Figures 2 and 3. Shown are statistical analyses of the homing molecule expression on (A) CFSElowCD4+ and (B) CFSElowCD8+ T-cells specific for SEB, CMV, and HIV (all peptide pools) in five different SP subjects (box & whisker graph: range and median). Mann-Whitney p-values are indicated in the figures. (C) Shown are statistical analyses of the integrin β7 and CXCR3 co-expression on matched CD4+ and CD8+ T-cells proliferating (CFSElow) in response to CMV versus HIVNefGagPol peptide pool in four different SP subjects. Paired T-test p-values are indicated in the figures.
(TIF)
The HIV-specific CD4+ T-cells exhibit a Th1Th17 polarization profile. PBMCs from SP subjects were loaded in CFSE (0.5 µM) and stimulated with different HIV Nef, Gag, Pol peptide pools (500 ng/ml), recombinant HIV-p24 (5 µg/ml), SEB (25 ng/ml), a peptide pool spanning the CMV pp65 protein (1 µg/ml), or C. albicans hyphae (25 µl of protein lysate) for 5 days at 37°C and further stimulated with PMA (50 ng/ml) and Ionomycin (1 µg/ml) in the presence of Brefeldin A (10 µg/ml) for 18 hours at 37°C. Cells were stained on the surface with CD3 and CD8 Abs as well as intracellularly with IFN-γ, TNF-α, and IL-17 Abs and then analyzed by polychromatic flow cytometry for the expression of cytokines in CD3+CD8− (referred as CD4+ T-cells) cells. Shown is (A) the gating strategy for CD4+ T-cells identification and (B) representative dot plots of IFN-γ, TNF-α, and IL-17 production by HIV-specific and C. albicans-specific CD4+ T-cells. (C) Shown is the intracellular expression of cytokines by CFSElowCD4+ T-cells specific for SEB, CMV, C. albicans, and different HIV peptide pools in five different SP subjects.
(TIF)
Screening for HIV-1 specific CD4+ T-cells responses using the CD154 co-culture assay. To identify antigen-specific CD4+ T-cells, the CD154/CD40L assay was performed as previously described (89). To this aim, PBMC from five HIV-infected SP subjects were stimulated with SEB (1 µg/ml), CMV-pp65 peptide pool (5 µg/ml), recombinant HIV-p24 protein (5 µg/ml), or HIV peptide pools (10 µg/ml) in the presence of anti-CD154-PE/Cy5 Abs (20 µl/well) and monensin (2 µM) for 16 hrs at 37°C. Cells were then harvested, stained for surface markers with fluorescence-conjugated Abs against CD3, CD4, integrin β7, CCR6, CXCR3, and CCR4, and analyzed by flow cytometry for the expression of homing markers on CD3+CD4+CD154+ T-cells. Results are expressed as % of CD4+CD154+ T-cells. Values included in the table were >1.5-fold higher than the background CD154 expression observed for cells cultured in the absence of antigen.
(DOC)
Acknowledgments
The authors thank Laurence Lejeune (Flow Cytometry Core Facility, CHUM-Research Center, Saint-Luc Hospital, Montréal, QC, Canada) for expert technical support with polychromatic flow cytometry analysis and sorting, Anne Vassal for help with ethics approvals and informed consents, and Mario Legault, Maryse Lainesse, Véronique Lafontaine for the management of HIV-infected patient cohorts, and Dr. Louis de Repentigny for the gift of Candida albicans LAM-1 strain. The authors also thank Heather Yampolsky for English corrections and Dr Aurélie Cleret for critical reading of the manuscript and valuable discussions. Finally, the authors acknowledge HIV-infected and uninfected donors for their gift of leukapheresis.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the Cooley's Anemia Foundation (CAF), the Associazione Veneta Lotta alla Talassemia (AVLT; Veneta Association for the Fight Against Thalassemia — Italy) (to L. Breda). This work is supported by grants from the Carlo and Micol Schejola Foundation, the Children's Cancer and Blood Foundation and United States National Institutes of Health grants 5K01DK063992 and NHLBI-5R01HL102449 (to S. Rivella). Additional support was provided by the Clinical and Translational Science Center (CTSC; NIH grant UL1RR024992) and the Carlo and Micol Schejola Foundation (to S. Rivella). RG was funded by Telethon (contract #GGP10124). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Barre-Sinoussi F. HIV: a discovery opening the road to novel scientific knowledge and global health improvement. Virology. 2010;397:255–259. doi: 10.1016/j.virol.2009.08.033. [DOI] [PubMed] [Google Scholar]
- 2.Brenchley JM, Price DA, Douek DC. HIV disease: fallout from a mucosal catastrophe? Nat Immunol. 2006;7:235–239. doi: 10.1038/ni1316. [DOI] [PubMed] [Google Scholar]
- 3.Lackner AA, Mohan M, Veazey RS. The gastrointestinal tract and AIDS pathogenesis. Gastroenterology. 2009;136:1965–1978. doi: 10.1053/j.gastro.2008.12.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brenchley JM, Schacker TW, Ruff LE, Price DA, Taylor JH, et al. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J Exp Med. 2004;200:749–759. doi: 10.1084/jem.20040874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mehandru S, Poles MA, Tenner-Racz K, Horowitz A, Hurley A, et al. Primary HIV-1 infection is associated with preferential depletion of CD4+ T lymphocytes from effector sites in the gastrointestinal tract. J Exp Med. 2004;200:761–770. doi: 10.1084/jem.20041196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Q, Duan L, Estes JD, Ma ZM, Rourke T, et al. Peak SIV replication in resting memory CD4+ T cells depletes gut lamina propria CD4+ T cells. Nature. 2005;434:1148–1152. doi: 10.1038/nature03513. [DOI] [PubMed] [Google Scholar]
- 7.Mattapallil JJ, Douek DC, Hill B, Nishimura Y, Martin M, et al. Massive infection and loss of memory CD4+ T cells in multiple tissues during acute SIV infection. Nature. 2005;434:1093–1097. doi: 10.1038/nature03501. [DOI] [PubMed] [Google Scholar]
- 8.Veazey RS, Lackner AA. Getting to the guts of HIV pathogenesis. J Exp Med. 2004;200:697–700. doi: 10.1084/jem.20041464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lackner AA, Veazey RS. Current concepts in AIDS pathogenesis: insights from the SIV/macaque model. Annu Rev Med. 2007;58:461–476. doi: 10.1146/annurev.med.58.082405.094316. [DOI] [PubMed] [Google Scholar]
- 10.Brenchley JM, Price DA, Schacker TW, Asher TE, Silvestri G, et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med. 2006;12:1365–1371. doi: 10.1038/nm1511. [DOI] [PubMed] [Google Scholar]
- 11.Ancuta P, Kamat A, Kunstman KJ, Kim EY, Autissier P, et al. Microbial translocation is associated with increased monocyte activation and dementia in AIDS patients. PLoS ONE. 2008;3:e2516. doi: 10.1371/journal.pone.0002516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sheth PM, Chege D, Shin LY, Huibner S, Yue FY, et al. Immune reconstitution in the sigmoid colon after long-term HIV therapy. Mucosal Immunol. 2008;1:382–388. doi: 10.1038/mi.2008.23. [DOI] [PubMed] [Google Scholar]
- 13.Macal M, Sankaran S, Chun TW, Reay E, Flamm J, et al. Effective CD4+ T-cell restoration in gut-associated lymphoid tissue of HIV-infected patients is associated with enhanced Th17 cells and polyfunctional HIV-specific T-cell responses. Mucosal Immunol. 2008;1:475–488. doi: 10.1038/mi.2008.35. [DOI] [PubMed] [Google Scholar]
- 14.Trono D, Van Lint C, Rouzioux C, Verdin E, Barre-Sinoussi F, et al. HIV persistence and the prospect of long-term drug-free remissions for HIV-infected individuals. Science. 2010;329:174–180. doi: 10.1126/science.1191047. [DOI] [PubMed] [Google Scholar]
- 15.Deeks SG, Walker BD. Human immunodeficiency virus controllers: mechanisms of durable virus control in the absence of antiretroviral therapy. Immunity. 2007;27:406–416. doi: 10.1016/j.immuni.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 16.Migueles SA, Connors M. Long-term nonprogressive disease among untreated HIV-infected individuals: clinical implications of understanding immune control of HIV. JAMA. 2010;304:194–201. doi: 10.1001/jama.2010.925. [DOI] [PubMed] [Google Scholar]
- 17.Pereyra F, Jia X, McLaren PJ, Telenti A, de Bakker PI, et al. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science. 2010;330:1551–1557. doi: 10.1126/science.1195271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bashirova AA, Thomas R, Carrington M. HLA/KIR restraint of HIV: surviving the fittest. Annu Rev Immunol. 2011;29:295–317. doi: 10.1146/annurev-immunol-031210-101332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saez-Cirion A, Pancino G, Sinet M, Venet A, Lambotte O. HIV controllers: how do they tame the virus? Trends Immunol. 2007;28:532–540. doi: 10.1016/j.it.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 20.Migueles SA, Osborne CM, Royce C, Compton AA, Joshi RP, et al. Lytic granule loading of CD8+ T cells is required for HIV-infected cell elimination associated with immune control. Immunity. 2008;29:1009–1021. doi: 10.1016/j.immuni.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Killian MS, Johnson C, Teque F, Fujimura S, Levy JA. Natural suppression of human immunodeficiency virus type 1 replication is mediated by transitional memory CD8+ T cells. J Virol. 2011;85:1696–1705. doi: 10.1128/JVI.01120-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hatano H, Delwart EL, Norris PJ, Lee TH, Dunn-Williams J, et al. Evidence for persistent low-level viremia in individuals who control human immunodeficiency virus in the absence of antiretroviral therapy. J Virol. 2009;83:329–335. doi: 10.1128/JVI.01763-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hunt PW, Hatano H, Sinclair E, Lee TH, Busch MP, et al. HIV-specific CD4+ T cells may contribute to viral persistence in HIV controllers. Clin Infect Dis. 2011;52:681–687. doi: 10.1093/cid/ciq202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sankaran S, Guadalupe M, Reay E, George MD, Flamm J, et al. Gut mucosal T cell responses and gene expression correlate with protection against disease in long-term HIV-1-infected nonprogressors. Proc Natl Acad Sci U S A. 2005;102:9860–9865. doi: 10.1073/pnas.0503463102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaul R, Plummer FA, Kimani J, Dong T, Kiama P, et al. HIV-1-specific mucosal CD8+ lymphocyte responses in the cervix of HIV-1-resistant prostitutes in Nairobi. J Immunol. 2000;164:1602–1611. doi: 10.4049/jimmunol.164.3.1602. [DOI] [PubMed] [Google Scholar]
- 26.Goonetilleke N, Liu MK, Salazar-Gonzalez JF, Ferrari G, Giorgi E, et al. The first T cell response to transmitted/founder virus contributes to the control of acute viremia in HIV-1 infection. J Exp Med. 2009;206:1253–1272. doi: 10.1084/jem.20090365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salazar-Gonzalez JF, Salazar MG, Keele BF, Learn GH, Giorgi EE, et al. Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. J Exp Med. 2009;206:1273–1289. doi: 10.1084/jem.20090378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haase AT. Early events in sexual transmission of HIV and SIV and opportunities for interventions. Annu Rev Med. 2011;62:127–139. doi: 10.1146/annurev-med-080709-124959. [DOI] [PubMed] [Google Scholar]
- 29.Li Q, Skinner PJ, Ha SJ, Duan L, Mattila TL, et al. Visualizing antigen-specific and infected cells in situ predicts outcomes in early viral infection. Science. 2009;323:1726–1729. doi: 10.1126/science.1168676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berlin C, Berg EL, Briskin MJ, Andrew DP, Kilshaw PJ, et al. Alpha 4 beta 7 integrin mediates lymphocyte binding to the mucosal vascular addressin MAdCAM-1. Cell. 1993;74:185–185. doi: 10.1016/0092-8674(93)90305-a. [DOI] [PubMed] [Google Scholar]
- 31.Agace WW. T-cell recruitment to the intestinal mucosa. Trends Immunol. 2008;29:514–522. doi: 10.1016/j.it.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 32.Kunkel EJ, Campbell DJ, Butcher EC. Chemokines in lymphocyte trafficking and intestinal immunity. Microcirculation. 2003;10:313–323. doi: 10.1038/sj.mn.7800196. [DOI] [PubMed] [Google Scholar]
- 33.Westphal S, Lugering A, von Wedel J, von Eiff C, Maaser C, et al. Resistance of chemokine receptor 6-deficient mice to yersinia enterocolitica infection: evidence of defective m-cell formation in vivo. Am J Pathol. 2008;172:671–680. doi: 10.2353/ajpath.2008.070393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang C, Kang SG, Lee J, Sun Z, Kim CH. The roles of CCR6 in migration of Th17 cells and regulation of effector T-cell balance in the gut. Mucosal Immunol. 2009;2:173–183. doi: 10.1038/mi.2008.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iwata M, Hirakiyama A, Eshima Y, Kagechika H, Kato C, et al. Retinoic acid imprints gut-homing specificity on T cells. Immunity. 2004;21:527–538. doi: 10.1016/j.immuni.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 36.Stenstad H, Ericsson A, Johansson-Lindbom B, Svensson M, Marsal J, et al. Gut-associated lymphoid tissue-primed CD4+ T cells display CCR9-dependent and -independent homing to the small intestine. Blood. 2006;107:3447–3454. doi: 10.1182/blood-2005-07-2860. [DOI] [PubMed] [Google Scholar]
- 37.Sigmundsdottir H, Butcher EC. Environmental cues, dendritic cells and the programming of tissue-selective lymphocyte trafficking. Nat Immunol. 2008;9:981–987. doi: 10.1038/ni.f.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Olsson J, Poles M, Spetz AL, Elliott J, Hultin L, et al. Human immunodeficiency virus type 1 infection is associated with significant mucosal inflammation characterized by increased expression of CCR5, CXCR4, and beta-chemokines. J Infect Dis. 2000;182:1625–1635. doi: 10.1086/317625. [DOI] [PubMed] [Google Scholar]
- 39.Dwinell MB, Lugering N, Eckmann L, Kagnoff MF. Regulated production of interferon-inducible T-cell chemoattractants by human intestinal epithelial cells. Gastroenterology. 2001;120:49–59. doi: 10.1053/gast.2001.20914. [DOI] [PubMed] [Google Scholar]
- 40.Zaunders JJ, Ip S, Munier ML, Kaufmann DE, Suzuki K, et al. Infection of CD127+ (interleukin-7 receptor+) CD4+ cells and overexpression of CTLA-4 are linked to loss of antigen-specific CD4 T cells during primary human immunodeficiency virus type 1 infection. J Virol. 2006;80:10162–10172. doi: 10.1128/JVI.00249-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zaunders JJ, Munier ML, Kaufmann DE, Ip S, Grey P, et al. Early proliferation of CCR5(+) CD38(+++) antigen-specific CD4(+) Th1 effector cells during primary HIV-1 infection. Blood. 2005;106:1660–1667. doi: 10.1182/blood-2005-01-0206. [DOI] [PubMed] [Google Scholar]
- 42.Shacklett BL, Cox CA, Sandberg JK, Stollman NH, Jacobson MA, et al. Trafficking of human immunodeficiency virus type 1-specific CD8+ T cells to gut-associated lymphoid tissue during chronic infection. J Virol. 2003;77:5621–5631. doi: 10.1128/JVI.77.10.5621-5631.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lecureuil C, Combadiere B, Mazoyer E, Bonduelle O, Samri A, et al. Trapping and apoptosis of novel subsets of memory T lymphocytes expressing CCR6 in the spleen of HIV-infected patients. Blood. 2007;109:3649–3657. doi: 10.1182/blood-2006-01-035717. [DOI] [PubMed] [Google Scholar]
- 44.Gosselin A, Monteiro P, Chomont N, Diaz-Griffero F, Said EA, et al. Peripheral blood CCR4+ CCR6+ and CXCR3+ CCR6+ CD4+ T cells are highly permissive to HIV-1 infection. J Immunol. 2010;184:1604–1616. doi: 10.4049/jimmunol.0903058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.El Hed A, Khaitan A, Kozhaya L, Manel N, Daskalakis D, et al. Susceptibility of human Th17 cells to human immunodeficiency virus and their perturbation during infection. J Infect Dis. 2010;201:843–854. doi: 10.1086/651021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mora JR, von Andrian UH. Retinoic acid: an educational “vitamin elixir” for gut-seeking T cells. Immunity. 2004;21:458–460. doi: 10.1016/j.immuni.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 47.Manicassamy S, Pulendran B. Retinoic acid-dependent regulation of immune responses by dendritic cells and macrophages. Semin Immunol. 2009;21:22–27. doi: 10.1016/j.smim.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Monteiro P, Gosselin A, Wacleche VS, El-Far M, Said EA, et al. Memory CCR6+CD4+ T cells are preferential targets for productive HIV type 1 infection regardless of their expression of integrin beta7. J Immunol. 2011;186:4618–4630. doi: 10.4049/jimmunol.1004151. [DOI] [PubMed] [Google Scholar]
- 49.Douek DC, Brenchley JM, Betts MR, Ambrozak DR, Hill BJ, et al. HIV preferentially infects HIV-specific CD4+ T cells. Nature. 2002;417:95–98. doi: 10.1038/417095a. [DOI] [PubMed] [Google Scholar]
- 50.Boulassel MR, Spurll G, Rouleau D, Tremblay C, Edwardes M, et al. Changes in immunological and virological parameters in HIV-1 infected subjects following leukapheresis. J Clin Apher. 2003;18:55–60. doi: 10.1002/jca.10051. [DOI] [PubMed] [Google Scholar]
- 51.Acosta-Rodriguez EV, Rivino L, Geginat J, Jarrossay D, Gattorno M, et al. Surface phenotype and antigenic specificity of human interleukin 17-producing T helper memory cells. Nat Immunol. 2007;8:639–646. doi: 10.1038/ni1467. [DOI] [PubMed] [Google Scholar]
- 52.Younes SA, Yassine-Diab B, Dumont AR, Boulassel MR, Grossman Z, et al. HIV-1 viremia prevents the establishment of interleukin 2-producing HIV-specific memory CD4+ T cells endowed with proliferative capacity. J Exp Med. 2003;198:1909–1922. doi: 10.1084/jem.20031598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Levy JA. The search for the CD8+ cell anti-HIV factor (CAF). Trends Immunol. 2003;24:628–632. doi: 10.1016/j.it.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 54.Casazza JP, Brenchley JM, Hill BJ, Ayana R, Ambrozak D, et al. Autocrine production of beta-chemokines protects CMV-Specific CD4 T cells from HIV infection. PLoS Pathog. 2009;5:e1000646. doi: 10.1371/journal.ppat.1000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Williams IR. CCR6 and CCL20: partners in intestinal immunity and lymphorganogenesis. Ann N Y Acad Sci. 2006;1072:52–61. doi: 10.1196/annals.1326.036. [DOI] [PubMed] [Google Scholar]
- 56.Woodland DL, Kohlmeier JE. Migration, maintenance and recall of memory T cells in peripheral tissues. Nat Rev Immunol. 2009;9:153–161. doi: 10.1038/nri2496. [DOI] [PubMed] [Google Scholar]
- 57.Reiss Y, Proudfoot AE, Power CA, Campbell JJ, Butcher EC. CC chemokine receptor (CCR)4 and the CCR10 ligand cutaneous T cell-attracting chemokine (CTACK) in lymphocyte trafficking to inflamed skin. J Exp Med. 2001;194:1541–1547. doi: 10.1084/jem.194.10.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chattopadhyay PK, Yu J, Roederer M. A live-cell assay to detect antigen-specific CD4+ T cells with diverse cytokine profiles. Nat Med. 2005;11:1113–1117. doi: 10.1038/nm1293. [DOI] [PubMed] [Google Scholar]
- 59.Brenchley JM, Douek DC. HIV infection and the gastrointestinal immune system. Mucosal Immunol. 2008;1:23–30. doi: 10.1038/mi.2007.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Annunziato F, Cosmi L, Santarlasci V, Maggi L, Liotta F, et al. Phenotypic and functional features of human Th17 cells. J Exp Med. 2007;204:1849–1861. doi: 10.1084/jem.20070663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Arthos J, Cicala C, Martinelli E, Macleod K, Van Ryk D, et al. HIV-1 envelope protein binds to and signals through integrin alpha4beta7, the gut mucosal homing receptor for peripheral T cells. Nat Immunol. 2008;9:301–309. doi: 10.1038/ni1566. [DOI] [PubMed] [Google Scholar]
- 62.Cicala C, Martinelli E, McNally JP, Goode DJ, Gopaul R, et al. The integrin alpha4beta7 forms a complex with cell-surface CD4 and defines a T-cell subset that is highly susceptible to infection by HIV-1. Proc Natl Acad Sci U S A. 2009;106:20877–20882. doi: 10.1073/pnas.0911796106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Napoli JL. Quantification of physiological levels of retinoic acid. Methods Enzymol. 1986;123:112–124. doi: 10.1016/s0076-6879(86)23015-3. [DOI] [PubMed] [Google Scholar]
- 64.Manicassamy S, Ravindran R, Deng J, Oluoch H, Denning TL, et al. Toll-like receptor 2-dependent induction of vitamin A-metabolizing enzymes in dendritic cells promotes T regulatory responses and inhibits autoimmunity. Nat Med. 2009;15:401–409. doi: 10.1038/nm.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Haase AT. Targeting early infection to prevent HIV-1 mucosal transmission. Nature. 2010;464:217–223. doi: 10.1038/nature08757. [DOI] [PubMed] [Google Scholar]
- 66.Ancuta P, Monteiro P, Sekaly RP. Th17 lineage commitment and HIV-1 pathogenesis. Curr Opin HIV AIDS. 2010;5:158–165. doi: 10.1097/COH.0b013e3283364733. [DOI] [PubMed] [Google Scholar]
- 67.Brenchley JM, Paiardini M, Knox KS, Asher AI, Cervasi B, et al. Differential Th17 CD4 T-cell depletion in pathogenic and nonpathogenic lentiviral infections. Blood. 2008;112:2826–2835. doi: 10.1182/blood-2008-05-159301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Raffatellu M, Santos RL, Verhoeven DE, George MD, Wilson RP, et al. Simian immunodeficiency virus-induced mucosal interleukin-17 deficiency promotes Salmonella dissemination from the gut. Nat Med. 2008;14:421–428. doi: 10.1038/nm1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Douek DC, Roederer M, Koup RA. Emerging concepts in the immunopathogenesis of AIDS. Annu Rev Med. 2009;60:471–484. doi: 10.1146/annurev.med.60.041807.123549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hunt PW, Brenchley J, Sinclair E, McCune JM, Roland M, et al. Relationship between T cell activation and CD4+ T cell count in HIV-seropositive individuals with undetectable plasma HIV RNA levels in the absence of therapy. J Infect Dis. 2008;197:126–133. doi: 10.1086/524143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sauce D, Larsen M, Fastenackels S, Pauchard M, Ait-Mohand H, et al. HIV disease progression despite suppression of viral replication is associated with exhaustion of lymphopoiesis. Blood. 2011;117:5142–5151. doi: 10.1182/blood-2011-01-331306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sallusto F, Mackay CR. Chemoattractants and their receptors in homeostasis and inflammation. Curr Opin Immunol. 2004;16:724–731. doi: 10.1016/j.coi.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 73.Butcher EC, Picker LJ. Lymphocyte homing and homeostasis. Science. 1996;272:60–66. doi: 10.1126/science.272.5258.60. [DOI] [PubMed] [Google Scholar]
- 74.Luscinskas FW, Lim YC, Lichtman AH. Wall shear stress: the missing step for T cell transmigration? Nat Immunol. 2001;2:478–480. doi: 10.1038/88663. [DOI] [PubMed] [Google Scholar]
- 75.Marelli-Berg FM, Cannella L, Dazzi F, Mirenda V. The highway code of T cell trafficking. J Pathol. 2008;214:179–189. doi: 10.1002/path.2269. [DOI] [PubMed] [Google Scholar]
- 76.Masopust D, Choo D, Vezys V, Wherry EJ, Duraiswamy J, et al. Dynamic T cell migration program provides resident memory within intestinal epithelium. J Exp Med. 2010;207:553–564. doi: 10.1084/jem.20090858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chomont N, Dafonseca S, Vandergeeten C, Ancuta P, Sekaly RP. Maintenance of CD4+ T-cell memory and HIV persistence: keeping memory, keeping HIV. Curr Opin HIV AIDS. 2011;6:30–36. doi: 10.1097/COH.0b013e3283413775. [DOI] [PubMed] [Google Scholar]
- 78.Yue FY, Merchant A, Kovacs CM, Loutfy M, Persad D, et al. Virus-specific interleukin-17-producing CD4+ T cells are detectable in early human immunodeficiency virus type 1 infection. J Virol. 2008;82:6767–6771. doi: 10.1128/JVI.02550-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Benlahrech A, Harris J, Meiser A, Papagatsias T, Hornig J, et al. Adenovirus vector vaccination induces expansion of memory CD4 T cells with a mucosal homing phenotype that are readily susceptible to HIV-1. Proc Natl Acad Sci U S A. 2009;106:19940–19945. doi: 10.1073/pnas.0907898106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Manel N, Unutmaz D, Littman DR. The differentiation of human T(H)-17 cells requires transforming growth factor-beta and induction of the nuclear receptor RORgammat. Nat Immunol. 2008;9:641–649. doi: 10.1038/ni.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wilson NJ, Boniface K, Chan JR, McKenzie BS, Blumenschein WM, et al. Development, cytokine profile and function of human interleukin 17-producing helper T cells. Nat Immunol. 2007;8:950–957. doi: 10.1038/ni1497. [DOI] [PubMed] [Google Scholar]
- 82.Brainard DM, Tager AM, Misdraji J, Frahm N, Lichterfeld M, et al. Decreased CXCR3+ CD8 T cells in advanced human immunodeficiency virus infection suggest that a homing defect contributes to cytotoxic T-lymphocyte dysfunction. J Virol. 2007;81:8439–8450. doi: 10.1128/JVI.00199-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Doitsh G, Cavrois M, Lassen KG, Zepeda O, Yang Z, et al. Abortive HIV infection mediates CD4 T cell depletion and inflammation in human lymphoid tissue. Cell. 2010;143:789–801. doi: 10.1016/j.cell.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The HIV-specific versus CMV-specific CD154+CD4+ T-cells preferentially express a gut-homing potential. PBMC from SP subjects were stimulated with different HIV Nef, Gag, Pol peptide pools (10 µg/ml), recombinant HIV-p24 protein (5 µg/ml), SEB (1 µg/ml), or CMV-pp65 peptide pool (5 µg/ml) for 18 hours at 37°C in the presence of fluorescence conjugated anti-CD154-PE/Cy5 Abs (20 µl/2×106 cells/0.2 ml/well). Antigen-specific T-cells were identified as CD154+ cells, as previously described [58]. Cells were harvested, stained with a cocktail of fluorochrome-conjugated CD3, CD4, and β7 integrin, CCR6, CXCR3, or CCR4 Abs and analyzed by polychromatic flow cytometry for (A) the expression of CD154 on CD3+CD4+ T-cells and (B–D) the expression of homing molecules on CD3+CD4+CD154+ T-cells. (A–B) Shown are results from one SP subjects (SP 015 stimulated with the HIV Nef5164-5187 peptide pool), representative of results generated with cells from five different donors. (C) The expression (%) of homing receptors was analyzed on CD154+ T-cells specific for SEB, CMV, and different HIV peptide pools in five different SP subjects. (D) Shown are statistical analyses of the homing molecule expression on CD154+CD4+ T-cells specific for SEB, CMV, and HIV (all peptide pools) in five different SP subjects (box & whisker graph: range and median). Mann-Whitney p-values are indicated in the figures.
(TIF)
Homing potential of CD4+ and CD8+ T-cells proliferating in response to HIV peptides. PBMC from SP subjects were stimulated with different antigens and analyzed by polychromatic flow cytometry for the expression of homing molecules as in Figures 2 and 3. Shown are statistical analyses of the homing molecule expression on (A) CFSElowCD4+ and (B) CFSElowCD8+ T-cells specific for SEB, CMV, and HIV (all peptide pools) in five different SP subjects (box & whisker graph: range and median). Mann-Whitney p-values are indicated in the figures. (C) Shown are statistical analyses of the integrin β7 and CXCR3 co-expression on matched CD4+ and CD8+ T-cells proliferating (CFSElow) in response to CMV versus HIVNefGagPol peptide pool in four different SP subjects. Paired T-test p-values are indicated in the figures.
(TIF)
The HIV-specific CD4+ T-cells exhibit a Th1Th17 polarization profile. PBMCs from SP subjects were loaded in CFSE (0.5 µM) and stimulated with different HIV Nef, Gag, Pol peptide pools (500 ng/ml), recombinant HIV-p24 (5 µg/ml), SEB (25 ng/ml), a peptide pool spanning the CMV pp65 protein (1 µg/ml), or C. albicans hyphae (25 µl of protein lysate) for 5 days at 37°C and further stimulated with PMA (50 ng/ml) and Ionomycin (1 µg/ml) in the presence of Brefeldin A (10 µg/ml) for 18 hours at 37°C. Cells were stained on the surface with CD3 and CD8 Abs as well as intracellularly with IFN-γ, TNF-α, and IL-17 Abs and then analyzed by polychromatic flow cytometry for the expression of cytokines in CD3+CD8− (referred as CD4+ T-cells) cells. Shown is (A) the gating strategy for CD4+ T-cells identification and (B) representative dot plots of IFN-γ, TNF-α, and IL-17 production by HIV-specific and C. albicans-specific CD4+ T-cells. (C) Shown is the intracellular expression of cytokines by CFSElowCD4+ T-cells specific for SEB, CMV, C. albicans, and different HIV peptide pools in five different SP subjects.
(TIF)
Screening for HIV-1 specific CD4+ T-cells responses using the CD154 co-culture assay. To identify antigen-specific CD4+ T-cells, the CD154/CD40L assay was performed as previously described (89). To this aim, PBMC from five HIV-infected SP subjects were stimulated with SEB (1 µg/ml), CMV-pp65 peptide pool (5 µg/ml), recombinant HIV-p24 protein (5 µg/ml), or HIV peptide pools (10 µg/ml) in the presence of anti-CD154-PE/Cy5 Abs (20 µl/well) and monensin (2 µM) for 16 hrs at 37°C. Cells were then harvested, stained for surface markers with fluorescence-conjugated Abs against CD3, CD4, integrin β7, CCR6, CXCR3, and CCR4, and analyzed by flow cytometry for the expression of homing markers on CD3+CD4+CD154+ T-cells. Results are expressed as % of CD4+CD154+ T-cells. Values included in the table were >1.5-fold higher than the background CD154 expression observed for cells cultured in the absence of antigen.
(DOC)