Abstract
CYP24A1 functions in vitamin D target tissues to degrade 1,25-dihydroxyvitamin D3 (1,25(OH)2D3). Thus, the concentration of this enzyme and the regulation of its expression is a primary determinant of the overall biological activity of 1,25(OH)2D3 within cells. The principle regulator of CYP24A1 expression is 1,25(OH)2D3 itself, which functions through the vitamin D receptor to upregulate the transcriptional activity of the Cyp24a1 gene. In this report, we explore the mechanism of this regulation using recently developed ChIP-chip and ChIP-seq techniques that permit an unbiased search for enhancer elements that participate in this transcriptional control. Our studies both confirm a regulatory region defined earlier and located proximal to the transcriptional start site (TSS) of mouse Cyp24a1 (−160 nt and −265 nt) and identify a novel intergenic region located downstream of the transcription unit that contains two enhancers (+35 kb and +37 kb) that facilitate 1,25(OH)2D3-dependent upregulation of Cyp24a1 expression. Interestingly, while C/EBPβ also binds under basal conditions to a site located immediately upstream of the Cyp24a1 promoter (−345 nt), occupancy by this factor is strikingly increased following 1,25(OH)2D3 treatment. The locations and activities of these regulatory regions that mediate 1,25(OH)2D3 actions were confirmed in mice in vivo. We conclude that the mechanism through which 1,25(OH)2D3 induces the CYP24A1 enzyme, thereby autoregulating its own destruction, involves both promoter-proximal as well as downstream-distal enhancers. These findings highlight new concepts regarding the molecular mechanism of action of 1,25(OH)2D3 and other hormonal regulators.
Keywords: Cyp24a1; 1,25(OH)2D3; VDR/RXR heterodimer; C/EBPβ; transcription; distal enhancers; ChIP-seq analysis
Introduction
CYP24A1, otherwise known as the 25-hydroxyvitamin D3-24-hydroxylase, is a mitochondrial P450-containing enzyme that functions exclusively in vitamin D target tissues to degrade 1,25-dihydroxyvitamin D3 (1,25(OH)2D3), the hormonal form of this vitamin (1). Evidence for this activity derives from early studies of CYP24A1 that demonstrated preferential substrate selectivity for both 25-hydroxyvitamin D3 (25OHD3) and 1,25(OH)2D3, yet higher affinity of the enzyme for the latter metabolite (1). This hypothesis has been strengthened more recently through studies which show that genetic removal of the Cyp24a1 gene in mice results in a phenotype characterized by highly elevated circulating levels of 1,25(OH)2D3 that are coincident with exaggerated and prolonged vitamin D target gene activation and which exhibit a reduced ability to metabolize exogenously administered 1,25(OH)2D3 (2, 3). Interestingly, the importance of CYP24A1 in humans has been confirmed most recently in studies which demonstrated that mutations in the coding regions of the CYP24A1 gene that prevent 24-hydroxylase activity are responsible for idiopathic hypercalcemia in infants (4). Given that 1,25(OH)2D3, via its ability to rapidly induce Cyp24a1 expression, is the primary regulator of CYP24A1 abundance (1), these features suggest that the enzyme’s primary physiologic function is not simply to limit the extent and duration of response to 1,25(OH)2D3 when the hormone’s circulating levels are physiologically or pathologically elevated, but is likely also to confine intracellular levels of 1,25(OH)2D3 within a range appropriate for individual tissue response.
Interestingly, basal levels of expression of Cyp24a1 are also determined in a tissue/cell type-specific manner by many additional factors. In the kidney, for example, Cyp24a1 expression is controlled by both PTH (1, 5) and FGF23 (6), two hormones that are intimately involved in coordinating not only the renal production of 1,25(OH)2D3 but also its participation in regulating calcium and phosphorus homeostasis. In other tissues, Cyp24a1 expression can also be regulated by systemic glucocorticoids (7), estrogens (8), and retinoid ligands (9) and a myriad of additional local factors as well (1). Thus, the set point for control of intracellular 1,25(OH)2D3 degradation is influenced by cellular vitamin D hormone levels but can also be modulated in a cell-specific fashion by other factors. The influence of this set point on 1,25(OH)2D3 activity is exemplified most strikingly in disease states such as chronic kidney-mineral bone disease (CK-MBD) (10, 11) or in various cancers (12–19). In these situations, increased basal Cyp24a1 expression is capable of blunting cellular response to 1,25(OH)2D3 (by increasing the rate of cellular degradation) and in some cases by conferring tissue resistance to even high levels of the circulating hormone. Thus, Cyp24a1 in the context of cancer may represent an oncogene, as its protein product is capable of reducing the anti-tumor response to 1,25(OH)2D3 (12). Surprisingly, however, whether in the parathyroid gland or in tumor cells, little is known of either the factors or the mechanisms that function under these or other circumstances to upregulate basal Cyp24a1 expression. Despite this critical lack of understanding, the result is clear; increased Cyp24a1 expression in cells limits the actions of 1,25(OH)2D3 and thus impacts both normal as well as pathological cell function.
The cloning of the CYP24A1 cDNA by Ohyama and colleagues (20) and subsequent identification of the Cyp24a1 promoter (21) set the stage for early molecular studies that focused on understanding how 1,25(OH)2D3 regulated the expression of Cyp24a1. These studies suggested that 1,25(OH)2D3 induced Cyp24a1 expression in trans through the vitamin D receptor (VDR), the transcription factor that mediates virtually all of the transcriptional regulating activity of 1,25(OH)2D3 (22), and in cis through a promoter-proximal regulatory region that contains two closely spaced 15 bp vitamin D responsive elements (VDREs) with which the receptor interacts (23–25). VDR binding to these as well as VDREs for other vitamin D modulated genes is dependent upon heterodimer formation with retinoid X receptor (RXR) which forms an active unit to recruit coregulatory protein complexes that facilitate the entry of RNA polymerase II (RNA pol II) into pre-initiation complexes at the Cyp24a1 promoter (26–28). The promoter-proximal regulatory region also contains DNA binding sites for other transcription factors including CREB, C/EBP, GR, and ETS1 (29–31). Recently, however, we have shown using ChIP-chip analysis that the regulatory activity of 1,25(OH)2D3 and its receptor is not restricted to the promoter-proximal region of Cyp24a1, but rather extends to include clusters of regulatory elements located downstream of the Cyp24a1 genes as well (32). In this report, we characterize these regulatory features of the mouse gene further using ChIP-seq analysis, and demonstrate additional properties both in cells in culture and in mice in vivo. These findings support new concepts of transcriptional regulation by not only 1,25(OH)2D3, but other modulators of gene expression as well.
Materials and methods
Reagents
General biochemicals were obtained from Fisher Scientific (Pittsburg, PA) and Sigma Chemical Co (St. Louis, MO). 1,25(OH)2D3 was obtained from SAFC Pharma (Madison, WI). α-MEM was purchased from Invitrogen Corp (Carlsbad, CA). Antibodies to VDR (C-20, SC-1008), RXR (ΔN-197, SC-774), and C/EBPβ (C-19, SC-150) were purchased from Santa Cruz Biotechnology Inc. (Santa Cruz, CA). Antibodies to tetra-acetylated histone H4 (06–866) were obtained from Upstate (Charlottesville, VA). All quantitative real-time PCR (qPCR) reagents (Power SYBR green) were obtained from ABI (Foster City, CA).
Cell culture procedures
Mouse osteoblastic MC3T3-E1 cells were obtained from ATCC (Manassas, VA). (Manassas, VA). Cells were cultured in α-MEM supplemented with 10% fetal bovine serum (FBS) obtained from Hyclone (Logan, UT). Cells were sub-cultured to confluency and then treated with either vehicle (0.1% ETOH) or 10−7 M 1,25(OH)2D3 in 0.1% ETOH for 3 hr prior to harvest. Our earlier studies established the time and concentration of 1,25(OH)2D3 (10−7 M) necessary for maximal VDR/RXR binding to the MC3T3-E1 genome (33). This high concentration, utilized by many other investigators as well, is required to overcome the restrictive ability of vitamin D binding protein to limit entry of the hormone into cells in vitro, as documented recently (34, 35).
Mouse experimentation
C57BL/6 mice were obtained from Harlan (Madison, WI) and maintained in the Biochemistry Animal Care facility at the University of Wisconsin-Madison. Mice were maintained on a standard purified diet (AIN-76A) obtained from Research Diets (New Brunswick, NJ). All animal experiments were conducted according to protocols on file with the University of Wisconsin-Madison Research Animal Resource Center (RARC) and approved by IACUC of this institution. Mice were treated by intraperitoneal injection with either propylene glycol (Gallipot, St. Paul, MN) or 1,25(OH)2D3 (10 ng/g bw) in propylene glycol. Total duodenal tissue (4 cm) from the intestine and both kidneys were harvested after 1 hr for ChIP and 0, 3 or 6 hrs for RNA analysis. The validity of this approach and the time points and concentration of hormone have been established in previous studies (36).
RNA analysis
RNA was isolated from mouse intestine and kidney using TRIZOL reagent. Samples were reverse transcribed using Superscript III Reverse Transcription kit from Invitrogen (Carlsbad, CA) and analyzed using primer sets to mouse Cyp24a1 and β-actin.
Chromatin immunoprecipitation (ChIP) coupled to ChIP-chip and ChIP-seq analysis
ChIP and ChIP-chip analyses were performed as described in depth previously (32, 37, 38). ChIP-seq analysis was also performed as described in earlier studies (39). Briefly, ChIP-DNA was prepared and amplified using the Illumina ChIP-seq DNA preparation kit (1003473, #11257047 RevA), clusters formed and sequenced on the Illumina GAIIx sequencers by the University of Wisconsin-Madison DNA Sequencing Facility in the University of Wisconsin-Madison Biotechnology Center. DNA clusters were generated using either a Standard Cluster Generation kit (ver. 4) on an Illumina cluster station (Illumina, San Diego, CA), [for all samples sequenced before April, 2010] or using a cBot Single Read Cluster Generation kit on an Illumina cBot (Illumina) [after April, 2010] according to the manufacturer’s instructions, to obtain an average of 2.0×107 clusters for each lane on a flowcell. All sequencing runs for 36mers were performed on an Illumina Genome Analyzer IIx or Illumina HiSeq 2000 using the Illumina Sequencing kit (ver. 4). Fluorescent images were analyzed using the Illumina base-calling pipeline 1.6.0 to obtain FASTQ formatted sequence data. Sequences were mapped to the mouse genome (NCBI37/mm9) using BOWTIE (Bowtie 0.12.5 –-q – best – m 1) (40). Further data analysis was performed using QuEST and HOMER packages (41, 42). Genome-wide data will be published elsewhere.
Bioinformatic and statistical analysis
Positive peak regions were interrogated further using the Genomatix Software Suite (43, 44) as well as HOMER (42). Motif analysis (de novo and known), was performed using the HOMER software.
Results
1,25(OH)2D3 induces VDR/RXR binding at promoter-proximal and downstream-distal sites within the Cyp24a1 gene locus
Previous studies using ChIP-chip analysis revealed that in addition to 1,25(OH)2D3-induced VDR binding near the promoter for both mouse and human Cyp24a1, DNA-bound VDR could also be identified at multiple intergenic sites located downstream of the Cyp24a1 transcription units as well (32). Importantly, while neither the locations of these clusters nor their number were conserved across the two species, their contributions to the regulation of CYP24a1 expression by 1,25(OH)2D3 were verified using large, stably integrated DNA segments containing mouse and human CYP24A1 gene loci in context with their individual regulatory regions. To confirm and extend these findings in mouse bone cells, we treated MC3T3-E1 cells with either vehicle or 1,25(OH)2D3 for 3 hr, subjected the cells to chromatin immunoprecipitation (ChIP) using antibodies to either VDR or RXR and then evaluated the co-precipitated DNA using deep sequencing techniques (ChIP-seq). This concentration of 1,25(OH)2D3 as well as the time point selected was selected for maximal response, as indicated earlier (33). Sequenced segments were mapped to the mouse genome (NCBI37/mm9) using Bowtie and further processed with QuEST and HOMER. The results in Figure 1 document the abundance of these sequenced segments (normalized to 107 reads) for each condition across a segment of chromosome 2 containing the Cyp24a1 gene locus, as illustrated in the figure. Importantly, they demonstrate that 1,25(OH)2D3 induces VDR and RXR co-occupancy at both a region near the transcriptional start site of the Cyp24a1 gene and at two regions located intergenically +35 kb and +37 kb downstream of the TSS as well. This finding confirms VDR binding at both proximal and downstream sites across the mouse Cyp24a1 locus but also extends the observation to include co-occupancy with RXR as well. The results are therefore consistent with the emerging theme of gene regulation which suggests that many, if not most, genes are regulated by multiple enhancers located at intronic or intergenic sites which are frequently 10’s if not 100’s of kilobases from the promoters they regulate (32, 39, 45).
Figure 1.
ChIP-seq analysis of VDR and RXR binding at the mouse Cyp24a1 gene locus. MC3T3-E1 cells were treated with either vehicle or 1,25(OH)2D3 (10−7 M) for 3 hr and then subjected to ChIP-seq analysis using antibodies to either VDR or RXR. The genomic interval on mouse chromosome 2 and the location of the Cyp24a1 transcription unit (12 exons) including the direction of transcription (arrow) is shown at the top. An infrequently utilized promoter for the gene Pfdn4 and the direction of its transcription (not regulated by 1,25(OH)2D3) is also shown. Tracks indicate tag densities (normalized to 107 reads) for vehicle-or 1,25(OH)2D3-treated VDR or RXR binding. Note the scale for peak height is different for each track (shown in red) to highlight VDR and RXR peak activities. DNA sequences for the putative VDREs located promoter distal (two validated VDREs) and single VDREs at +35 and +37 kb found at the peak height are indicated below the tracks. TSS, transcriptional start site.
DNA sequences within the VDR/RXR binding regions confirm the presence of motifs representative of VDREs
ChIP-seq analysis provides DNA sequence resolution that is far superior to that of ChIP-chip analysis. Thus, we searched the sites of VDR/RXR binding at both the TSS and in the two sites downstream of the gene for VDREs. As can be seen in Figure 1, VDR/RXR binding at the TSS coincides with two VDREs, one positioned at −161 nt (AGGTGA GTG AGGGCG) and the other located slightly upstream at −280 nt (GGTTCA GCG GGTGCG). These sequences represent highly conserved mouse versions of those identified functionally in the rat gene by Ohyama and colleagues (46) and the human CYP24a1 gene by Zierold and colleagues (25) and likely represent the binding sites responsible for VDR and RXR co-localization in these ChIP-seq analyses. Investigation of the two regions located downstream of the Cyp24a1 gene also revealed several VDRE-like motifs likely responsible for VDR/RXR binding at these sites. They include a potential VDRE at −35 kb (TCAACC CAG TGACTC) read on the opposite strand, and a motif at −37 kb (AGGGCA CTG AGTTCT) read on the forward strand. Interestingly, all four motifs retain a highly conserved G residue at position 9 within the VDRE sequence itself. These results are consistent with the concept that has emerged using genome-wide approaches that the predominant, although not exclusive, structural VDRE motif is represented by two hexanucleotide repeats separated by three base pairs (39, 47). DNA motifs capable of binding other transcription factors were also evident in these regions (data not shown).
Residual C/EBPβ binding near the Cyp24a1 promoter region is enhanced by 1,25(OH)2D3
Previous studies by Christakos and colleagues identified a DNA sequence immediately upstream of the mouse Cyp24a1 TSS that is activated by C/EBPβ (7, 29). Interestingly, co-localization of C/EBPβ at sites of VDR/RXR activity represents a more general phenomenon, since recent genome-wide studies suggest that a very high percentage of enhancers that binds VDR/RXR contains C/EBPβ binding motifs and interacts directly with C/EBPβ (39, 48, 49). To explore this observation further, we conducted a ChIP-seq analysis of C/EBPβ in MC3T3-E1 cells examined the binding of C/EBPβ at the Cyp24a1 locus in the presence and absence of 1,25(OH)2D3. As can be seen in Figure 2, C/EBPβ binds in untreated bone cells near the Cyp24a1 TSS immediately upstream of the more distal VDRE. This site (located at −353 nt) contains a binding motif (TTGCACAATCG) that is similar to that of a C/EBPβ consensus element (Figure 2). Interestingly, 1,25(OH)2D3 treatment not only induces VDR/RXR binding, but strongly upregulates C/EBPβ occupancy at its respective binding site at the Cyp24a1 gene as well. As the upregulation of C/EBPβ binding in response to 1,25(OH)2D3 also occurs frequently at other vitamin D target genes, for example at the Tnfsf11 (Rankl) gene (49), we speculate that both VDR/RXR and C/EBPβ may comprise a functional complex that together orchestrates the recruitment of additional protein components that are necessary for upregulation of genes such as Cyp24a1. The specific role of C/EBPβ in this putative complex, however, remains to be defined. Despite this uncertainty, C/EBPβ does not represent a residual or inducible component of all Cyp24a1 enhancers to which the VDR binds (39). Thus, as seen in Figure 2a, C/EBPβ does not appear to bind to the two downstream enhancers at −35 kb and −37 kb. This observation suggests the possibility that these downstream regulatory elements may function differently from those located near the Cyp24a1 promoter. It is also consistent with the concept that regulatory regions of genes are capable of binding multiple transcription factors, and that the interactions between these factors are critical to the modulation of gene expression.
Figure 2.
Chip-seq analysis of C/EBPβ binding at the mouse Cyp24a1 gene locus. MC3T3-E1 cells were treated as in Figure 1 and then subjected to ChIP-seq analysis using antibody to C/EBPβ. Normalized tag densities for VDR and RXR are shown for reference together with tag densities for C/EBPβ across the indicated interval. A C/EBPβ response element located immediately upstream of the Cyp24a1 promoter is indicated below the tracks. Additional illustrative details are identical to those in Figure 1.
1,25(OH)2D3 induces VDR binding at similar sites across the Cyp24a1 locus in mice in vivo and promotes both RNA pol II recruitment and histone H4 acetylation
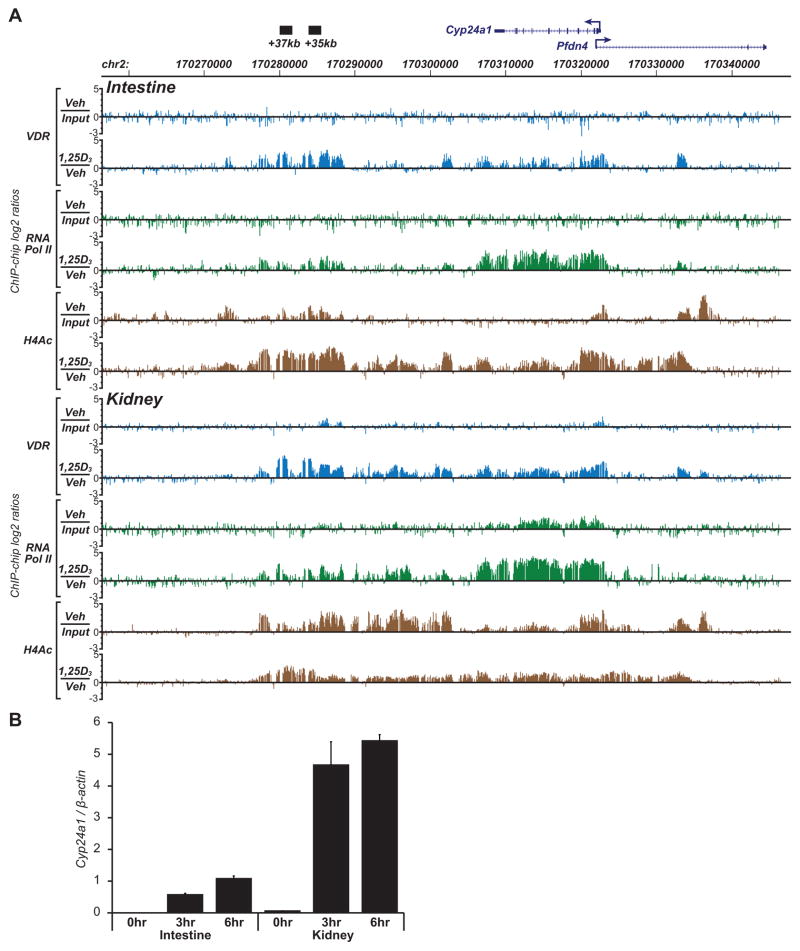
Based upon the above characterization of elements that participate in 1,25(OH)2D3-mediated upregulation of Cyp24a1 expression in bone cells, we examined in a final set of experiments the ability of 1,25(OH)2D3 to induced VDR binding to those same sites on the mouse genome in vivo. Accordingly, mice were subjected to a single IP injection of either vehicle (polypropylene glycol) or 1,25(OH)2D3 (10 ng/g bw in polypropylene glycol). Both duodenal and kidney tissues were extracted 1 hr later and subjected to chromatin immunoprecipitation using antibodies to VDR, RNA pol II or tetra-acetylated histone H4. The immunoprecipitated DNA was then amplified and samples co-hybridized to a set of microarrays containing tiled 70-mer oligonucleotides that spanned the mouse Cyp24a1 gene locus at 100 bp resolution from +100 kb to −100 kb relative to the Cyp24a1 TSS (ChIP-chip analysis). Vehicle samples were co-hybridized to the arrays with un-precipitated input samples to establish baseline activity whereas 1,25(OH)2D3-treated samples were co-hybridized to the arrays with vehicle treated samples to establish increased levels of VDR, RNA pol II or histone H4 acetylation. As can be seen in Figure 3a, while baseline VDR binding is not evident across the Cyp24a1 gene in ChIP samples derived from intestinal and only modestly evident in kidney tissue of vehicle-treated mice, 1,25(OH)2D3 strongly upregulated VDR binding when compared to baseline activity at sites near the promoter as well as at sites located +35 kb and +37 kb downstream. Typical of ChIP-chip analysis, however, overall basal activity is much noisier and VDR binding at these sites is much broader than that identified by ChIP-seq analysis (37, 48, 49). It is clear, nevertheless, that 1,25(OH)2D3 induces VDR binding in all the regions identified as a result of the cell culture studies, thereby confirming the binding capabilities of the downstream enhancers in vivo. To assess whether this binding activity leads to a functional outcome, we also examined the effects of VDR binding on histone H4 acetylation and on the recruitment of RNA pol II, activities that generally correlate with increased transcriptional output. As can also be seen in Figure 3a, 1,25(OH)2D3 strongly induces a rapid increase in both RNA pol II recruitment, a requirement for the synthesis of transcripts and in histone H4 acetylation, indicative of chromatin decondensation necessary for transcriptional upregulation. Interestingly, RNA pol II recruitment was not limited to the Cyp24a1 promoter, but can be seen dispersed across the Cyp24a1 transcription unit, as expected, and also broadly at the downstream enhancers. The recruitment of RNA pol II to enhancers may represent a mechanism to facilitate an increase in the local concentrations of the enzyme (50), although alternatively it may serve to induce the transcription of non coding RNAs that may play a role in gene regulation (51). Histone H4 acetylation manifests an extended pattern as well. We assume that all of these events precede the upregulation of Cyp24a1 mRNA that can be measured in intestine and kidney as early as 3 hr following 1,25(OH)2D3 treatment, as documented in Figure 3b. Collectively, these data support the idea that as in bone cells in culture, the upregulation of Cyp24a1 expression by 1,25(OH)2D3 in both the intestine and kidney is initiated through VDR occupancy at both promoter-proximal and downstream-distal sites across the Cyp24a1 gene itself which leads to chromatin consequences that are associated with transcriptional upregulation.
Figure 3.
ChIP-chip analysis of VDR binding, RNA pol II recruitment, and histone H4 tetra-acetylation levels at the Cyp24a1 gene locus in the intestine and kidney of mice in vivo. A) C57BL/6 mice were administered a single dose (IP) of either vehicle or 1,25(OH)2D3 (see Materials and Methods), sacrifice 1 hr later and utilized to obtain total duodenum and total kidney. Tissues were subjected to ChIP-chip analysis using antibodies to VDR, RNA pol II, or tetra-acetylated histone H4 (H4Ac). A schematic diagram and additional details of the mouse Cyp24a1 gene locus are as shown in Figure 1. Individual VDR, RNA pol II and tetra-acetylated H4 hybridization tracks for both intestine and kidney from vehicle and 1,25(OH)2D3 treated single mice are indicated. Data represent the log2 ratio of fluorescence obtained following co-hybridization of vehicle-treated vs input samples (basal activity) or 1,25(OH)2D3-treated vs vehicle-treated samples (1,25(OH)2D3 induced activity). Tracks are representative of several separate experiments. B) Total RNA obtained from duodenal or kidney tissues of mice treated as above for 0, 3 or 6 hr were evaluated for Cyp24a1 mRNA activity by RT-PCR analysis and normalize to β-actin. n= 5, ± se.
Discussion
The results described in this report document a mechanism whereby 1,25(OH)2D3 induces Cyp24a1 upregulation in mouse cells. Using ChIP-chip and ChIP-seq techniques, we show that the VDR/RXR heterodimer binds both to promoter proximal elements defined in earlier studies (23) as well as to distal sites located intergenically +35 and +37 kb downstream of the Cyp24a1 transcription unit (32). Each of these sites contained DNA sequence motifs that are similar to VDRE consensus elements and likely represented the sites to which the VDR binds in these studies. We also discovered that 1,25(OH)2D3 strongly induced C/EBPβ binding at an element near the Cyp24a1 promoter that is similar to that identified previously in the human CYP24A1 gene (29). Collectively, these data establish an increased complexity with regard to how 1,25(OH)2D3 regulates Cyp24a1 expression, and identify additional enhancers that could participate, based upon the characteristic of modularity, in the gene’s basal expression as well as its upregulation by factors other than 1,25(OH)2D3.
Direct evidence that activity at these sites contributed to mouse Cyp24a1 upregulation was established previously using large recombinant BAC clones containing the entire Cyp24a1 gene locus together with a luciferase reporter inserted into the final 3′ non coding exon (32). In these studies, a natural locus as well as several additional versions containing mutations or deletions in both the promoter-proximal as well as downstream-distal enhancers were stably integrated into the MC3T3-E1 cell line and their basal and 1,25(OH)2D3-inducible activities assessed. The results suggest that mutation of the promoter proximal VDREs and deletion of the downstream enhancers both reduced but did not eliminate the ability of 1,25(OH)2D3 to upregulate the reporter activity associated with the clonal segments. However, coupling both sets of mutations resulted in complete loss of regulatory capacity by 1,25(OH)2D3. Future studies are focused upon evaluating the putative VDREs located in the downstream enhancers. Final confirmation, however, will necessitate creating enhancer-null mutations in the mouse genome in vivo and exploring the consequence of these mutations on the biologic phenotype of the resulting mice (52, 53).
The location of distal enhancers raises the question as to how they function within the cell. Recent studies using multiple versions of chromosome confirmation capture (3C) analysis suggests that distal enhancers are likely to be located in the nucleus in situ near the promoters they regulate, and that the large intervening segments of the DNA are looped out (54, 55). Indeed, studies using 3C analysis suggested that the downstream cluster of enhancer in the human CYP24A1 gene were located structurally in close proximity to the gene’s promoter (32). Thus, distal enhancers might be capable of directly modulating the promoters they regulate. Studies have also suggested that many enhancers produce both short and long RNA transcripts (51, 56, 57). While some of these RNA transcripts may provide regulatory function, either in cis or in trans, alternative explanations for this transcription phenomenon is that this transcriptional activity is essential for modulating chromatin structure. In the human CYP24A1 gene, we have noted both the recruitment of RNA pol II to these downstream regions as well as the synthesis of long RNA transcripts (data not shown), although their function remains to be determined. Finally, we note an abundance of histone H4 acetylation across the Cyp24a1 locus. These covalent epigenetic modifications to histones likely function to facilitate an open chromatin structure, providing enhanced access of transcription factors to the gene’s regulatory machinery (58). Although the co-regulatory proteins that mediate this activity remain unknown for the mouse gene, histone acetyltransferases such as SRC1 and CBP were shown to be recruited to all the regulatory regions of the human CYP24A1 gene (32).
ChIP-seq analysis revealed residual C/EBPβ occupancy near the Cyp24a1 promoter that was strikingly increased in response to 1,25(OH)2D3. While the interaction of C/EBPβ at this site in the human gene was identified earlier (7, 29), the inducibility of this binding by 1,25(OH)2D3 suggests that this unique transcription factor may play a key role in Cyp24a1 expression. Interestingly, recent genome-wide studies of 1,25(OH)2D3 induced VDR/RXR binding suggest that C/EBPβ is a frequent participant in the regulation of many of the genes that are induced by 1,25(OH)2D3 (39). One possible role is that C/EBPβ may facilitate chromatin modifications that are imposed by nucleosome remodeling complexes, as suggested by Christakos and colleagues (59). Regardless of function, however, the question arises as to the mechanism by which 1,25(OH)2D3 induces C/EBPβ binding to specific target genes such as Cyp24a1. This phenomenon is not likely to be due to upregulation of C/EBPβ protein levels, which are delayed relative to the upregulation of DNA binding. It is possible that C/EBPβ may represent a component of a preformed complex that contains the VDR/RXR heterodimer. Alternatively, the DNA binding and functional capabilities of C/EBPβ might be directly induced by 1,25(OH)2D3 via the latter’s ability to activate directly one or more of the protein kinases that are known to regulate C/EBPβ activation and/or phosphorylation (60). Perhaps a careful time course of both C/EBPβ and VDR binding following 1,25(OH)2D3 treatment might resolve whether one or the other of these possibilities is correct, although this would not be a trivial undertaking.
Recent studies have suggested that the human CYP24a1 gene contains SNPs that affect vitamin D metabolism and are associated with increased cancer risk (61–63). As none of these SNPs are located in the 12 exons that encode CYP24A1, it seems likely that these correlations, if correct, may simply influence the expression of the gene. One SNP in particular has been identified in the promoter-proximal region of the human gene (64). Molecular biological studies suggest that this SNP influences the basal activity of CYP24A1 expression, although these studies involve artificial DNA constructs and are very difficult to interpret. This SNP particular was not located within either the two VDREs or in the C/EBPβ binding site, however, and therefore has not been informative with respect to functional mechanism. The discovery of additional regulatory elements downstream of the CYP24A1 gene defines new regions that will have to be explored and imposes additional complexity in linking single SNPs in this gene locus mechanistically to the expression of CYP24A1.
In summary, we have shown using ChIP-seq analysis that 1,25(OH)2D3 induces Cyp24a1 expression via promotion of VDR/RXR at promoter-proximal and downstream-distal enhancers as well as upregulation of C/EBPβ binding at a site near the promoter and immediately upstream of the VDREs. Both local as well as distal sites were confirmed using ChIP-chip analysis in mouse intestine and kidney in vivo following treatment with 1,25(OH)2D3. These studies expand our understanding of how 1,25(OH)2D3 induces Cyp24a1, which plays a key role at the local level in the regulation of the biological activity of 1,25(OH)2D3. On a more fundamental level, however, they support emerging genome-wide studies of 1,25(OH)2D3 action which suggest that vitamin D target genes are regulated through the VDR/RXR heterodimer by multiple enhancers that are frequently located at remote sites, but which converge on the promoters they regulate via DNA looping.
Highlights.
1,25(OH)2D3 regulates the expression of Cyp24a1 through multiple proximal and distal enhancers.
Activation induces VDR and RXR binding to VDREs located in each of the enhancers.
1,25(OH)2D3 co-induces binding of C/EBPβ to the enhancer located near the promoter.
Similar vitamin D regulated enhancers are utilized in vivo in mouse intestine and kidney.
Acknowledgments
We thank members of the Pike laboratory for their helpful contributions to the studies herein. This work was supported National Institutes of Health grants DK-072281 and DK-073995 to J.W.P.
Footnotes
Disclosure: The authors state that they have nothing to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Prosser DE, Jones G. Enzymes involved in the activation and inactivation of vitamin D. Trends Biochem Sci. 2004;29:664–673. doi: 10.1016/j.tibs.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 2.Dardenne O, Prud’homme J, Arabian A, Glorieux F, St-Arnaud R. Targeted inactivation of the 25-hydroxyvitamin D3-1α-hydroxylase gene (CYP27B1) creates an animal model of pseudovitamin D-deficiency rickets. Endocrinology. 2001;142:3135–3141. doi: 10.1210/endo.142.7.8281. [DOI] [PubMed] [Google Scholar]
- 3.Masuda S, Byford V, Arabian A, Sakai Y, Demay MB, St-Arnaud R, Jones G. Altered pharmacokinetics of 1alpha,25-dihydroxyvitamin D3 and 25-hydroxyvitamin D3 in the blood and tissues of the 25-hydroxyvitamin D-24-hydroxylase (Cyp24a1) null mouse. Endocrinology. 2005;146:825–834. doi: 10.1210/en.2004-1116. [DOI] [PubMed] [Google Scholar]
- 4.Schlingmann KP, Kaufmann M, Weber S, Irwin A, Goos C, John U, Misselwitz J, Klaus G, Kuwertz-Bröking E, Fehrenbach H, Wingen AM, Güran T, Hoenderop JG, Bindels RJ, Prosser DE, Jones G, Konrad M. Mutations in CYP24A1 and idiopathic infantile hypercalcemia. N Engl J Med. 2011;365:410–421. doi: 10.1056/NEJMoa1103864. [DOI] [PubMed] [Google Scholar]
- 5.Zierold C, Mings JA, DeLuca HF. Regulation of 25-hydroxyvitamin D3-24-hydroxylase mRNA by 1,25-dihydroxyvitamin D3 and parathyroid hormone. J Cell Biochem. 2003;88:234–237. doi: 10.1002/jcb.10341. [DOI] [PubMed] [Google Scholar]
- 6.Haussler MR, Haussler CA, Whitfield GK, Hsieh JC, Thompson PD, Barthel TK, Bartik L, Egan JB, Wu Y, Kubicek JL, Lowmiller CL, Moffet EW, Forster RE, Jurutka PW. The nuclear vitamin D receptor controls the expression of genes encoding factors which feed the “Fountain of Youth” to mediate healthful aging. J Steroid Biochem Mol Biol. 2010;121:88–97. doi: 10.1016/j.jsbmb.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dhawan P, Christakos S. Novel regulation of 25-hydroxyvitamin D3 24-hydroxylase (24(OH)ase) transcription by glucocorticoids: cooperative effects of the glucocorticoid receptor, C/EBPβ, and the Vitamin D receptor in 24(OH)ase transcription. J Cell Biochem. 2010;110:1314–1323. doi: 10.1002/jcb.22645. [DOI] [PubMed] [Google Scholar]
- 8.Nashold FE, Spach KM, Spanier JA, Hayes CE. Estrogen controls vitamin D3-mediated resistance to experimental autoimmune encephalomyelitis by controlling vitamin D3 metabolism and receptor expression. J Immunol. 2009;183:3672–3681. doi: 10.4049/jimmunol.0901351. [DOI] [PubMed] [Google Scholar]
- 9.Allegretto E, Shevde N, Zou A, Howell S, Boehm M, Hollis B, Pike J. Retinoid X receptor acts as a hormone receptor in vivo to induce a key metabolic enzyme for 1,25-dihydroxyvitamin D3. J Biol Chem. 1995;270:23906–23909. doi: 10.1074/jbc.270.41.23906. [DOI] [PubMed] [Google Scholar]
- 10.Helvig CF, Cuerrier D, Hosfield CM, Ireland B, Kharebov AZ, Kim JW, Ramjit NJ, Ryder K, Tabash SP, Herzenberg AM, Epps TM, Petkovich M. Dysregulation of renal vitamin D metabolism in the uremic rat. Kidney Int. 2010;78:463–472. doi: 10.1038/ki.2010.168. [DOI] [PubMed] [Google Scholar]
- 11.Posner GH, Helvig C, Cuerrier D, Collop D, Kharebov A, Ryder K, Epps T, Petkovich M. Vitamin D analogues targeting CYP24 in chronic kidney disease. J Steroid Biochem Mol Biol. 2010;121:13–19. doi: 10.1016/j.jsbmb.2010.03.065. [DOI] [PubMed] [Google Scholar]
- 12.Albertson DG, Ylstra B, Segraves R, Collins C, Dairkee SH, Kowbel D, Kuo WL, Gray JW, Pinkel D. Quantitative mapping of amplicon structure by array CGH identifies CYP24 as a candidate oncogene. Nat Genet. 2000;25:144–146. doi: 10.1038/75985. [DOI] [PubMed] [Google Scholar]
- 13.Mimori K, Tanaka Y, Yoshinaga K, Masuda T, Yamashita K, Okamoto M, Inoue H, Mori M. Clinical significance of the overexpression of the candidate oncogene CYP24 in esophageal cancer. Ann Oncol. 2004;15:236–241. doi: 10.1093/annonc/mdh056. [DOI] [PubMed] [Google Scholar]
- 14.Anderson MG, Nakane M, Ruan X, Kroeger PE, Wu-Wong JR. Expression of VDR and CYP24A1 mRNA in human tumors. Cancer Chemother Pharmacol. 2006;57:234–240. doi: 10.1007/s00280-005-0059-7. [DOI] [PubMed] [Google Scholar]
- 15.Cross HS, Pavelka M, Slavik J, Peterlik M. Growth control of human colon cancer cells by vitamin D and calcium in vitro. J Natl Cancer Inst. 1992;84:1355–1357. doi: 10.1093/jnci/84.17.1355. [DOI] [PubMed] [Google Scholar]
- 16.Parise RA, Egorin MJ, Kanterewicz B, Taimi M, Petkovich M, Lew AM, Chuang SS, Nichols M, El-Hefnawy T, Hershberger PA. CYP24, the enzyme that catabolizes the antiproliferative agent vitamin D, is increased in lung cancer. Int J Cancer. 2006;119:1819–1828. doi: 10.1002/ijc.22058. [DOI] [PubMed] [Google Scholar]
- 17.Lou YR, Qiao S, Talonpoika R, Syvälä H, Tuohimaa P. The role of Vitamin D3 metabolism in prostate cancer. J Steroid Biochem Mol Biol. 2004;92:317–325. doi: 10.1016/j.jsbmb.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 18.Yee SW, Campbell MJ, Simons C. Inhibition of Vitamin D3 metabolism enhances VDR signalling in androgen-independent prostate cancer cells. J Steroid Biochem Mol Biol. 2006;98:228–235. doi: 10.1016/j.jsbmb.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 19.Chung I, Karpf AR, Muindi JR, Conroy JM, Nowak NJ, Johnson CS, Trump DL. Epigenetic silencing of CYP24 in tumor-derived endothelial cells contributes to selective growth inhibition by calcitriol. J Biol Chem. 2007;282:8704–8714. doi: 10.1074/jbc.M608894200. [DOI] [PubMed] [Google Scholar]
- 20.Ohyama Y, Noshiro M, Okuda K. Cloning and expression of cDNA encoding 25-hydroxyvitamin D3 24-hydroxylase. FEBS Lett. 1991;278:195–198. doi: 10.1016/0014-5793(91)80115-j. [DOI] [PubMed] [Google Scholar]
- 21.Ohyama Y, Noshiro M, Eggertsen G, Gotoh O, Kato Y, Björkhem I, Okuda K. Structural characterization of the gene encoding rat 25-hydroxyvitamin D3 24-hydroxylase. Biochemistry. 1993;32:76–82. doi: 10.1021/bi00052a011. [DOI] [PubMed] [Google Scholar]
- 22.Pike J, Meyer M, Martowicz M, Bishop K, Lee S, Nerenz R, Goetsch P. Emerging regulatory paradigms for control of gene expression by 1,25-dihydroxyvitamin D3. J Steroid Biochem Mol Biol. 2010;121:130–135. doi: 10.1016/j.jsbmb.2010.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohyama Y, Ozono K, Uchida M, Yoshimura M, Shinki T, Suda T, Yamamoto O. Functional Assessment of Two Vitamin D-responsive Elements in the Rat 25-Hydroxyvitamin D3 24-Hydroxylase Gene. J Biol Chem. 1996;271:30381–30385. doi: 10.1074/jbc.271.48.30381. [DOI] [PubMed] [Google Scholar]
- 24.Ozono K, Yamagata M, Ohyama Y, Nakajima S. Direct repeat 3-type element lacking the ability to bind to the vitamin D receptor enhances the function of a vitamin D-responsive element. J Steroid Biochem Mol Biol. 1998;66:263–269. doi: 10.1016/s0960-0760(98)00055-7. [DOI] [PubMed] [Google Scholar]
- 25.Zierold C, Darwish HM, DeLuca HF. Two vitamin D response elements function in the rat 1,25-dihydroxyvitamin D 24-hydroxylase promoter. J Biol Chem. 1995;270:1675–1678. doi: 10.1074/jbc.270.4.1675. [DOI] [PubMed] [Google Scholar]
- 26.Pike JW. Genome-scale techniques highlight the epigenome and redefine fundamental principles of gene regulation. J Bone Miner Res. 2011;26:1155–1162. doi: 10.1002/jbmr.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pike JW, Meyer MB. The vitamin D receptor: new paradigms for the regulation of gene expression by 1,25-dihydroxyvitamin D3. Endocrinol Metab Clin North Am. 2010;39:255–269. doi: 10.1016/j.ecl.2010.02.007. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pike JW, Meyer MB, Bishop KA. Regulation of target gene expression by the vitamin D receptor-an update on mechanisms. Rev Endocr Metab Disord. 2011 doi: 10.1007/s11154-011-9198-9. [DOI] [PubMed] [Google Scholar]
- 29.Dhawan P, Peng X, Sutton AL, MacDonald PN, Croniger CM, Trautwein C, Centrella M, McCarthy TL, Christakos S. Functional cooperation between CCAAT/enhancer-binding proteins and the vitamin D receptor in regulation of 25-hydroxyvitamin D3 24-hydroxylase. Mol Cell Biol. 2005;25:472–487. doi: 10.1128/MCB.25.1.472-487.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dwivedi P, Omdahl J, Kola I, Hume D, May B. Regulation of rat cytochrome P450C24 (CYP24) gene expression. Evidence for functional cooperation of Ras-activated Ets transcription factors with the vitamin D receptor in 1,25-dihydroxyvitamin D3-mediated induction. J Biol Chem. 2000;275:47–55. doi: 10.1074/jbc.275.1.47. [DOI] [PubMed] [Google Scholar]
- 31.Dwivedi PP, Hii CS, Ferrante A, Tan J, Der CJ, Omdahl JL, Morris HA, May BK. Role of MAP kinases in the 1,25-dihydroxyvitamin D3-induced transactivation of the rat cytochrome P450C24 (CYP24) promoter. Specific functions for ERK1/ERK2 and ERK5. J Biol Chem. 2002;277:29643–29653. doi: 10.1074/jbc.M204561200. [DOI] [PubMed] [Google Scholar]
- 32.Meyer MB, Goetsch PD, Pike JW. A downstream intergenic cluster of regulatory enhancers contributes to the induction of CYP24A1 expression by 1α,25-dihydroxyvitamin D3. J Biol Chem. 2010;285:15599–15610. doi: 10.1074/jbc.M110.119958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim S, Shevde NK, Pike JW. 1,25-Dihydroxyvitamin D3 stimulates cyclic vitamin D receptor/retinoid X receptor DNA-binding, co-activator recruitment, and histone acetylation in intact osteoblasts. J Bone Miner Res. 2005;20:305–317. doi: 10.1359/JBMR.041112. [DOI] [PubMed] [Google Scholar]
- 34.Zella LA, Shevde NK, Hollis BW, Cooke NE, Pike JW. Vitamin D-binding protein influences total circulating levels of 1,25-dihydroxyvitamin D3 but does not directly modulate the bioactive levels of the hormone in vivo. Endocrinology. 2008;149:3656–3667. doi: 10.1210/en.2008-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zella LA, Meyer MB, Nerenz RD, Pike JW. The enhanced hypercalcemic response to 20-epi-1,25-dihydroxyvitamin D3 results from a selective and prolonged induction of intestinal calcium-regulating genes. Endocrinology. 2009;150:3448–3456. doi: 10.1210/en.2009-0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meyer MB, Zella LA, Nerenz RD, Pike JW. Characterizing early events associated with the activation of target genes by 1,25-dihydroxyvitamin D3 in mouse kidney and intestine in vivo. J Biol Chem. 2007;282:22344–22352. doi: 10.1074/jbc.M703475200. [DOI] [PubMed] [Google Scholar]
- 37.Kim S, Yamazaki M, Zella LA, Meyer MB, Fretz JA, Shevde NK, Pike JW. Multiple enhancer regions located at significant distances upstream of the transcriptional start site mediate RANKL gene expression in response to 1,25-dihydroxyvitamin D3. J Steroid Biochem Mol Biol. 2006;103:430–434. doi: 10.1016/j.jsbmb.2006.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meyer MB, Watanuki M, Kim S, Shevde NK, Pike JW. The human transient receptor potential vanilloid type 6 distal promoter contains multiple vitamin D receptor binding sites that mediate activation by 1,25-dihydroxyvitamin D3 in intestinal cells. Mol Endocrinol. 2006;20:1447–1461. doi: 10.1210/me.2006-0031. [DOI] [PubMed] [Google Scholar]
- 39.Meyer MB, Goetsch P, Pike J. Overlapping VDR/RXR and TCF4/β-catenin cistromes in colonic cells of colorectal tumor origin: Impact on c-FOS and c-MYC gene regulation. Mol Endocrinol. 2011 doi: 10.1210/me.2011-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Valouev A, Johnson D, Sundquist A, Medina C, Anton E, Batzoglou S, Myers R, Sidow A. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat Methods. 2008;5:829–834. doi: 10.1038/nmeth.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heinz S, Benner C, Spann N, Bertolino E, Lin Y, Laslo P, Cheng J, Murre C, Singh H, Glass C. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- 44.Quandt K, Frech K, Karas H, Wingender E, Werner T. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim S, Yamazaki M, Zella LA, Shevde NK, Pike JW. Activation of receptor activator of NF-kappaB ligand gene expression by 1,25-dihydroxyvitamin D3 is mediated through multiple long-range enhancers. Mol Cell Biol. 2006;26:6469–6486. doi: 10.1128/MCB.00353-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ohyama Y, Ozono K, Uchida M, Yoshimura M, Shinki T, Suda T, Yamamoto O. Functional assessment of two vitamin D-responsive elements in the rat 25-hydroxyvitamin D3 24-hydroxylase gene. J Biol Chem. 1996;271:30381–30385. doi: 10.1074/jbc.271.48.30381. [DOI] [PubMed] [Google Scholar]
- 47.Heikkinen S, Väisänen S, Pehkonen P, Seuter S, Benes V, Carlberg C. Nuclear hormone 1α,25-dihydroxyvitamin D3 elicits a genome-wide shift in the locations of VDR chromatin occupancy. Nucleic Acids Res. 2011;39:9181–9193. doi: 10.1093/nar/gkr654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zella LA, Meyer MB, Nerenz RD, Lee SM, Martowicz ML, Pike JW. Multifunctional enhancers regulate mouse and human vitamin D receptor gene transcription. Mol Endocrinol. 24:128–147. doi: 10.1210/me.2009-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martowicz ML, Meyer MB, Pike JW. The mouse RANKL gene locus is defined by a broad pattern of histone H4 acetylation and regulated through distinct distal enhancers. J Cell Biochem. 2011;112:2030–2045. doi: 10.1002/jcb.23123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Szutorisz H, Dillon N, Tora L. The role of enhancers as centres for general transcription factor recruitment. Trends Biochem Sci. 2005;30:593–599. doi: 10.1016/j.tibs.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 51.Wang X, Song X, Glass CK, Rosenfeld MG. The Long Arm of Long Noncoding RNAs: Roles as Sensors Regulating Gene Transcriptional Programs. Cold Spring Harb Perspect Biol. 2010;3:a003756. doi: 10.1101/cshperspect.a003756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fu Q, Manolagas SC, O’Brien CA. Parathyroid hormone controls receptor activator of NF-kappaB ligand gene expression via a distant transcriptional enhancer. Mol Cell Biol. 2006;26:6453–6468. doi: 10.1128/MCB.00356-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Galli C, Zella LA, Fretz JA, Fu Q, Pike JW, Weinstein RS, Manolagas SC, O’Brien CA. Targeted deletion of a distant transcriptional enhancer of the receptor activator of nuclear factor-κB ligand gene reduces bone remodeling and increases bone mass. Endocrinology. 2008;149:146–153. doi: 10.1210/en.2007-0734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miele A, Dekker J. Mapping Cis-and Trans-Chromatin Interaction Networks Using Chromosome Conformation Capture (3C) Methods Mol Biol. 2009;464:105–121. doi: 10.1007/978-1-60327-461-6_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lieberman-Aiden E, van Berkum N, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie B, Sabo P, Dorschner M, Sandstrom R, Bernstein B, Bender M, Groudine M, Gnirke A, Stamatoyannopoulos J, Mirny L, Lander E, Dekker J. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ong CT, Corces VG. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat Rev Genet. 2011;12:283–293. doi: 10.1038/nrg2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, Ching KA, Antosiewicz-Bourget JE, Liu H, Zhang X, Green RD, Lobanenkov VV, Stewart R, Thomson JA, Crawford GE, Kellis M, Ren B. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dhawan P, Wieder R, Weider R, Christakos S. CCAAT enhancer-binding protein alpha is a molecular target of 1,25-dihydroxyvitamin D3 in MCF-7 breast cancer cells. J Biol Chem. 2009;284:3086–3095. doi: 10.1074/jbc.M803602200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zanello L, Norman A. 1alpha,25(OH)2 Vitamin D3 actions on ion channels in osteoblasts. Steroids. 2006;71:291–297. doi: 10.1016/j.steroids.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 61.Dong LM, Ulrich CM, Hsu L, Duggan DJ, Benitez DS, White E, Slattery ML, Farin FM, Makar KW, Carlson CS, Caan BJ, Potter JD, Peters U. Vitamin D related genes, CYP24A1 and CYP27B1, and colon cancer risk. Cancer Epidemiol Biomarkers Prev. 2009;18:2540–2548. doi: 10.1158/1055-9965.EPI-09-0228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McCullough ML, Bostick RM, Mayo TL. Vitamin D gene pathway polymorphisms and risk of colorectal, breast, and prostate cancer. Annu Rev Nutr. 2009;29:111–132. doi: 10.1146/annurev-nutr-080508-141248. [DOI] [PubMed] [Google Scholar]
- 63.Holick CN, Stanford JL, Kwon EM, Ostrander EA, Nejentsev S, Peters U. Comprehensive association analysis of the vitamin D pathway genes, VDR, CYP27B1, and CYP24A1, in prostate cancer. Cancer Epidemiol Biomarkers Prev. 2007;16:1990–1999. doi: 10.1158/1055-9965.EPI-07-0487. [DOI] [PubMed] [Google Scholar]
- 64.Roff A, Wilson RT. A novel SNP in a vitamin D response element of the CYP24A1 promoter reduces protein binding, transactivation, and gene expression. J Steroid Biochem Mol Biol. 2008;112:47–54. doi: 10.1016/j.jsbmb.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]