Abstract
Copy-choice RNA recombination occurs during viral RNA synthesis when the viral transcription complex switches templates. We demonstrate that RNA-dependent RNA polymerase from bovine viral diarrhea virus and the replicases from three plant-infecting RNA viruses can produce easily detectable recombination products in vitro by switching templates during elongative RNA synthesis. Template sequence and/or structure, and NTP availability affected the frequency of template switch by the transcription complex. Our results provide biochemical support for copy-choice recombination and establish assays for mechanistic analyses of intermolecular RNA recombination in vitro.
RNA recombination contributes to genetic diversity and pathogenesis of RNA viruses (1–3). Two RNA recombination mechanisms have been proposed: breakage-rejoining and copy-choice (1). The breakage-rejoining mechanism takes place with the splicing of group II introns (4) and can result in the production of some recombinant RNAs by the Qβ replicase (5). Copy-choice recombination occurs when the transcription complex (TC) leaves the first template (the donor) and resumes synthesis on a second template (the acceptor). RNA recombination via template switch is classified as base-pairing-dependent, base-pairing-independent, or base-pairing-assisted (1). A copy-choice mechanism seems to account for the majority of viral RNA recombinants identified thus far (1, 3, 6).
Products of RNA recombination during viral infection have been extensively characterized (7–14). Although informative for the mechanism of RNA recombination, some recombinants will be underrepresented in these analyses because of selection for fitness. Also, characterization of recombinant RNAs from viral infections requires extensive molecular manipulation (14). A biochemical assay would be useful for a mechanistic analysis of RNA recombination. We observed that the recombinant RNA-dependent RNA polymerase (RdRp) (NS5B) from bovine viral diarrhea virus (BVDV) and several viral replicases could efficiently produce RNAs longer than template length in vitro, because of the RdRp switching template either at the 5′ end or within the donor RNA. A similar activity was reported for the recombinant poliovirus RdRp with homopolymeric templates (15). In this work, we examine the features in the donor and acceptor that affect end-to-end and internal template switch during viral RNA synthesis in vitro.
Materials and Methods
Reagents and Molecular Manipulations.
Recombinant BVDV NS5B was prepared from Escherichia coli (16). Enriched viral replicases were prepared from infected plants as described (17–19). RNAs less than 30 nucleotides were chemically synthesized by either Oligos Etc. (Wilsonville, OR) or by Dharmacon (Boulder, CO). Longer RNAs were transcribed by the T7 RNA polymerase (20). All RNAs were purified and checked for quality and quantity as previously described (19). RNA synthesis reactions, electrophoresis, and quantification of the RdRp products were as described in Adkins and Kao (18). RNaseH and RNaseT1 digestions were performed essentially as described in Cheng and Gott (21).
Results
End-to-End Template Switch.
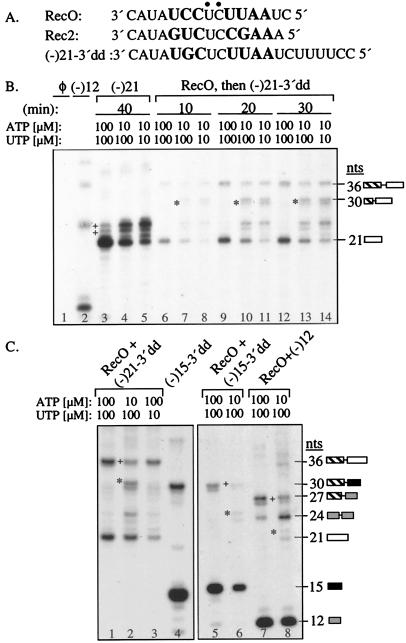
The BVDV RdRp can produce RNA products that are multimeric in length of the input template (22). To distinguish whether the multimeric products are the result of copy-choice template switch by the TC or whether the reactions contained template ligation as well as polymerase activities, we used templates (-)21–3′dd, containing a 3′-dideoxycytidylate and d(-)21, a single-stranded DNA of the same bases as (-)21–3′dd (Fig. 1A). Both templates lack a 5′ phosphate but can direct de novo initiation of RNA synthesis by the BVDV RdRp (22, 23). We found that (-)21–3′dd and d(-)21 produced RNAs that are multiples of 21 nucleotides in length whose abundance increased with template concentration (Fig. 1B). Because a 5′-phosphate and a 3′-ribose hydroxyl are needed for template ligation and primer-dependent synthesis, the multimers are generated by template switch by the TC, not template ligation. Also, it is unlikely that our highly purified recombinant BVDV RdRp is contaminated with both RNA and DNA ligases. Monomers were observed in reactions with high template concentration (Fig. 1B), indicating that the TC will terminate even if acceptors are available. Multimers produced from two or more templates of the same sequence will be called identical recombinants (IR).
Figure 1.
End-to-end template switch by a recombinant BVDV RdRp. (A) Sequence of templates used in these reactions. The arrow denotes the initiation cytidylate. (B) End-to-end template switch with (-)21–3′dd as a function of template concentration. φ is a control reaction performed without RNA. (C) Formation of NRs. At the right of the autoradiogram are schematics of the product from the templates: RecO (hatched box), (-)21–3′dd (open box); (-)15, (black box); (-)12, (gray box). Asterisks (*) identify the NRs. (D) (Top) Schematic of the purified dimeric NR generated from reactions containing donor RecO (boxed) and acceptor Rec1 (gray box with positions of radiolabeled CMPs underlined). Arrows indicate potential sites for RNaseT1 cleavage. (Bottom) Autoradiogram of the results of RNaseT1 digestion with the key components in each reaction indicated above each lane. M denotes a reaction by using donor RecO and acceptor Rec1 similar to one that generated the NR that was purified for analysis in lanes 3–5. Lane 5 contains a darker exposure of lane 4 to show the expected RNaseT1 digestion intermediates. (E) (Top) Schematic of the IR generated from template 14–2 emphasizing the sequence near the switch junction. The sequence whose synthesis is directed by donor is boxed, whereas the sequence from acceptor is shaded. The position of the only radiolabeled CMP is underlined. J1 and J2 are DNA oligonucleotides that should and should not, respectively, stably anneal with the sequence near the switch junction in the IR. The smallest radiolabeled RNA fragment that should remain should be 8 nucleotides. (Bottom) Autoradiogram with results from RNaseH digestions. M contains a reaction similar to the one used to generate the IR that was purified and used in the analysis in lanes 2–4.
To confirm end-to-end template switch by the BVDV RdRp, reactions were performed with donor RecO and acceptors of different lengths (Fig. 1). RecO, which lacks any guanylate and does not direct radiolabeled CMP incorporation (Fig. 1 A and C, lane 5), was incubated with the BVDV RdRp for 10 min before addition of acceptors (-)12, (-)15–3′dd, or (-)21–3′dd (Fig. 1C). As expected, the initial incubation with RecO decreased the use of the other potential donor RNAs (compare Fig. 1C lanes 3–4 to 6–8). Reactions with the 15-nt RecO and the acceptors of 12-, 15-, and 21-nt produced nonidentical recombinants (NRs) of ≈27, 30, and 36 nucleotides (Fig. 1C, lanes 6–8). Putative NRs were absent in reactions that contained only (-)12 or (-)21–3′dd (Fig. 1C, lanes 3 and 4).
We used ribonuclease digestion to further characterize the putative recombinant RNAs. Dimer-length NRs of 34 nucleotides were purified from reactions with donor RecO (15 nucleotides) and acceptor Rec1 (19 nucleotides) (Fig. 1A). If this 34-nt RNA is generated by template switch, complete RNaseT1 digestion that selectively cleaves the phosphodiester bond 3′ of guanylates in RNAs should yield an 18-nt RNA, with incomplete digestion products of 33, 28, 27, 25, and 19 nucleotides (Fig. 1D Top). A predominant 18-nt RNA was observed after RNaseT1 digestion (Fig. 1D, lane 4), along with fainter incomplete digestion products, indicating that this NR is the product of an end-to-end switch by the TC (Fig. 1D, lane 5). We used RNaseH digestion to probe the sequence of the recombinant dimeric IRs generated from (-)14–2 as both donor and acceptor (Fig. 1A). The purified dimer was annealed to either the DNA oligonucleotide J1, generating a 13-bp RNA:DNA duplex at the switch junction, or J2, which should not anneal (Fig. 1E Top). RNaseH should then hydrolyze the duplex to generate a labeled fragment of eight nucleotides. Such a band was observed after RNaseH treatment, confirming that template switch by the TC resulted in the production of recombinant products (Fig. 1E, lane 3).
Donor Sequence and Template Switch Efficiency.
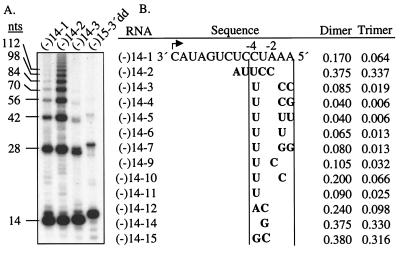
We seek to identify features in the donor that affect the frequency of end-to-end template switch. To examine the role of the sequence near the 5′-terminus of the donor, a series of 14-nt RNAs that varied in the 5′-terminal 7 nucleotides were used as template for the BVDV RdRp as tested. The amount of dimeric and trimeric RNAs was normalized to that of monomer, taking into account the number of radiolabeled CMP that could be incorporated (Fig. 2). The 5′-most nucleotide will be designated position 0 whereas preceding nucleotides will be referred to as negative integers. The 13 templates produced a wide range of multimeric RNAs (Fig. 2A), indicating that the sequence at the switch site in the donor can dramatically impact the ability of the TC to switch templates. With templates (-)14–3 through (-)14–7 that are different only at positions 0 and -1, nucleotide substitutions resulted in only 2-fold relative effect on dimeric RNA production. Two uridylates at these positions [(-)14–5] resulted in a similar amount of dimeric and trimeric RNAs as did (-)14–14, which has a cytidylate and a guanylate at position 0 and −1. Therefore, nucleotides at the very 5′-terminus did not dramatically affect template switch. However, the three nucleotides from positions -2 and -4 affected the formation of recombinants in a more obvious way: one or more guanylate or cytidylate increased the frequency of template switch. For example (-)14–1, with a -4 cytidylate produced twice as much recombinants as did (-)14–11, which has a -4 uridylate. In general, templates 14–3, -4, -5,-6,-7 and -11 that lacked a guanylate or cytidylate within positions -2 to -4 produced dimers from 4 to 9% whereas those template that had one or more guanylate or cytidylate produced dimers from 10.5 to 38% (Fig. 2B). An exception to this simple rule is (-)14–10, which directed dimer production at 20% while lacking a guanylate or cytidylate from -2 to -4. (-)14–10 does, however, contain a cytidylate at the -1 position.
Figure 2.
Frequency of end-to end switch by the TC on different templates. (A) Autoradiogram of RdRp products from selected templates. (B) Templates used to quantify template switch during RNA synthesis. In RNAs (-)14–2 through (-)14–15, only the nucleotides that are different from (-)14–1 are shown. The percentage of dimer and trimer produced by each template RNA is adjusted to be relative to the amount of monomer directed by the same RNA and normalized for CMP incorporation.
Changes near the 3′-terminus of the acceptor RNA were examined for effects on frequency of template switch. Only the NR produced by different donor and acceptors were used for these analyses to rule out effects on the initiation of RNA synthesis. The amounts of dimeric NR were normalized to that from donor RecO and acceptor (-)21–3′dd. Nucleotide substitution of 3′ +1C in the acceptor reduced end-to-end switch by more than 10-fold (Table 1). Changes of the +1C also severely affected de novo initiation of RNA synthesis (22), suggesting that an initiation cytidylate in the acceptor confers an advantage for template switch. Also, different frequencies of NR formation were observed when the order of addition of donor and acceptor templates was reversed (Table 1). Template switch is thus affected by both the donor and acceptor sequences.
Table 1.
Effects of different donor and acceptors on the relative frequency of NR dimers produced by the BVDV RdRp
| Donor | Acceptor | % dimeric NR |
|---|---|---|
| RecO | (−)21−3′dd | 100 |
| (−)21+1C/U | 7 | |
| (−)21+1C/G | 5 | |
| (−)21, 3′AA | 7 | |
| (−)12 | 53 | |
| (−)12 | RecO | 140 |
| (−)21dd | RecO | 18 |
Internal Template Switch.
The frequency of template switch is likely coupled to the rate of synthesis by the TC in a manner dependent on substrate availability and the information within the template (24). Lowering the concentration of one or more NTPs may affect internal template switch by causing polymerase pausing (24, 25). We manipulated ATP and UTP concentrations and examined internal template switch by the BVDV RdRp from donor RecO to acceptor (-)21–3′dd (Fig. 3 A and B). RecO contains sequence UUAA at positions from +10 to +13 from the initiation (+1) cytidylate, which may cause the RdRp to pause before it reaches position +10 when ATP and UTP concentration are decreased (Fig. 3A). If a TC that produced an 8- to 9-nt nascent RNA switches template and completes synthesis on (-)21–3′dd, then internal recombinants (IntR) of 29–30 nucleotides would be expected.
Figure 3.
Internal template switch by the BVDV RdRp. (A) RNAs used in the experiments described below. Positions in the RNAs that may be affected by varying NTP concentrations are shown in bold. The dots identify positions at which the TC switches to an acceptor template. (B) Autoradiogram showing the internal template switch recombinants at different times. RNAs and the concentrations of ATP and UTP used in each reaction are above the autoradiogram. Size markers were generated from templates (-)12 and from (-)21-3′dd (see Fig. 1A for sequences). Asterisks identify putative IntR of 29 and 30 nucleotides. The RdRp products are to the right of the autoradiogram. The schematics from the template and their symbols in parentheses are as follows: RecO (hatched box); (-)21-3′dd (open box). (C) Internal template switch from donor RecO to acceptors of (-)21-3′dd, (-)15-3′dd, and (-)12 reactions with varying ATP and/or UTP concentrations. Sizes of RNAs are listed on the right with diagrams. The products from the template and their symbols in parentheses are as follows: RecO (hatched box), (-)21-3′dd (open box), (-)15-3′dd, (black box), and (-)12 (gray box). Asterisks (*) denote intR and NR, respectively.
RNA syntheses were examined over time in sets of three reactions that varied by only ATP and UTP concentrations: (i) 100 μM of both; (ii) 10 μM of ATP and 100 μM of UTP; and (iii) 10 μM of both (Fig. 3B). RNAs of 29 and 30 nucleotides were detected only in reactions containing 10 μM ATP (Fig. 3B, lanes 6–14). Reactions with 10 μM UTP and 100 μM ATP did not produce increased amounts of IntR (Fig. 3C, lane 3). Therefore, both NTP concentration and the position of the TC along the template could affect IntR formation. The IntR from RecO and (-)21–3′dd appear as two bands differing by one nucleotide, which would be expected from template switch being induced by ATP limitations when the RdRp encounters the two uridylates at positions +10 and +11 of RecO.
Other products expected of this complex reaction are present and accounted for. The monomeric 21-nt RNAs resulting from de novo initiation and correct termination from (-)21–3′dd are observed, as are the ≈36-nt end-to-end switch products by using donor RecO and acceptor (-)21–3′dd. Products likely generated by polymerase stuttering or nontemplated addition are also observed when ATP and UTP concentrations are decreased (Fig. 3B, lanes 3–5) but cannot account for the 29- and 30-nt RNAs. Premature termination on acceptor (-)21–3′dd after end-to-end template switch from donor RecO also could not account for IntR production, because the expected products of 24 and 25 nucleotides were not observed. Also, if premature termination in acceptor (-)21–3′dd occurred because of the decreased NTP concentration, then reactions with 10 μM of UTP and 100 μM of ATP should produce RNAs 1–2 nucleotides longer than those produced from the reaction with 10 μM of ATP and 10 μM of UTP. This was also not observed (Fig. 3C, lane 3). Thus, internal template switch from RecO followed by complete synthesis on acceptor (-)21–3′dd is the most likely explanation for the 29- and 30-nt RdRp products.
These results beg the question of why the two uridylates at positions +10 and +11 of RecO are better at inducing polymerase switch than the two uridylates present at the identical positions in (-)21–3′dd. We speculate that the additional 5′ sequence in (-)21–3′dd may have an effect. Alternatively, substrate requirements may differ with the position of the TC along the template. After the TC switches from RecO to (-)21–3′dd, ≈18 phosphodiester bonds are made by the time the uridylates in (-)21–3′dd are encountered, in contrast to the 9 bonds when the TC encounters the two uridylates in RecO.
The position promoting the internal template switch on donor RecO was further confirmed. The IntR should vary with the lengths of the acceptors. Putative IntRs of 24 and 25 nucleotides and 22 and 21 nucleotides were observed in reactions containing donor RecO and acceptors of 15 and 12 nucleotides (Fig. 3C, lanes 6 and 8). Again, these IntRs are present only in reactions with 10 μM ATP (Fig. 3C, lanes 5–8), confirming that the internal template switch preferentially takes place at positions +8 and +9 of donor RecO. The junction for the internal template switch was verified by using Rec2, which contains the sequence CGAA immediately 5′ of the switch junction in RecO (Fig. 3A). When CTP and UTP concentrations are decreased, IntRs of 22 to 24 nucleotides were formed (data not shown). This indicates that the internal template switch occurred before positions +10 and +11 of the donor, even with nucleotide changes at these template positions.
Template Switch by Viral Replicases.
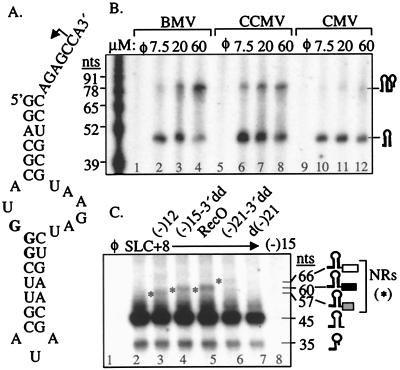
Although the recombinant BVDV RdRp provides a well-defined system to examine the basic requirements for template switch, viral RNA replication takes place with multisubunit replicases. Intermolecular template switch with viral replicases was examined in vitro by using RNA SLC+8, an RNA of defined structure that can direct synthesis by using replicases from brome mosaic virus (BMV), cucumber mosaic virus (CMV), and cowpea chlorotic mottle virus (CCMV) (Fig. 4A; refs. 19 and 26). Along with the correctly initiated and terminated RNAs of 45 nucleotides, longer replicase products were observed in a manner dependent on SLC+8 concentration (Fig. 4B), suggesting that the replicases switched templates by a copy-choice mechanism. Fewer recombinants were observed with the CMV replicase in comparison to the BMV and CCMV replicases (compare lanes 10–12 to 2–8). A complete end-to-end template switch should result in IRs of 90 nucleotides, but IRs of ≈80 nucleotides were detected (Fig. 4B). This is likely because of premature termination following the end-to-end switch because of low CTP concentration in the reaction. SLC+8 contains 3 consecutive guanylates at positions +35 to +37 that may cause premature termination (Fig. 4A). Consistent with this claim, RNAs of ≈35 nucleotides were observed (Fig. 4C). We cannot rule out more complex mechanisms responsible for the 80-nt RNAs.
Figure 4.
End-to-end template switch by three viral replicases. (A) Sequence and secondary structure of SLC+8. The initiation cytidylate is denoted with an arrow, and three guanylates at positions +35 to +37 are in bold. (B) An autoradiogram showing IRs from SLC+8 by three viral replicases. Concentrations of SLC+8 (in μM) used in each reaction are at the top of the autoradiogram. Size markers and schematics of monomeric and IR replicase products are shown (Left and Right) of the autoradiogram, respectively. Each hairpin represents one product from SLC+8. (C) An autoradiogram showing monomer synthesized from SLC+8 and NRs from donor SLC+8 and several acceptors. Schematics of the NRs generated in reactions with SLC+8 and acceptors (-)12 (gray box), (-)15–3′dd (black box), (-)21 (open box) are to the left of the autoradiogram. Asterisks (*) identify NRs.
Next, we examined whether replicases can switch from donor SLC+8 to nonidentical acceptors. SLC+8 was incubated with BMV replicases 10 min before the addition of one of five acceptors that lack the replicase recognition elements within SLC+8 and cannot direct RNA synthesis by the three viral replicases (Fig. 4C, data not shown). Reactions with acceptors (-)12, (-)15–3′dd, and RecO produced RNAs of 57, 60, and 66 nucleotides, respectively (Fig. 4C, lanes 3–6), lengths that correspond to expected end-to-end template switch products. Therefore, single-stranded RNA lacking sequence homology and replicase recognition elements can be accepted for end-to-end template switch by the BMV replicase. However, in comparison to SLC+8 as an acceptor, the non-BMV RNA acceptors formed recombinants at levels severalfold less (Fig. 4 B and C). Single-stranded DNA d(-)21 did not produce detectable levels of IR (Fig. 4C, lane 7).
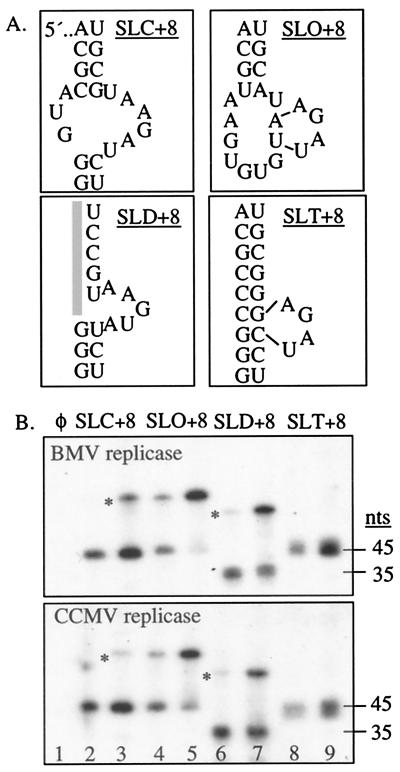
To identify why SLC+8 is better at allowing template switch, four SLC+8 variants were examined for RNA synthesis by the BMV and CCMV replicases (Fig. 5A). SLO+8 and SLD+8 have weakened or nonexistent base pairing in the stems flanking the rightward-facing bulge in SLC+8. The substitutions in SLT+8 resulted in a more stable stem (26). Consistent with previous results, the BMV and CCMV replicases produced significant amounts of IR in reactions with higher template concentration (Fig. 5B, lanes 2 and 3 of Top and Bottom). However, the amount of IR formation differed with the four RNAs; SLO+8 produced the most and SLT+8 the least (Fig. 5B, lanes 2–9 of Top and Bottom). These results demonstrate that changes in RNA structure and/or the sequence affect the efficiency of template switch by viral replicases.
Figure 5.
Efficiency of template switch depends on RNA structure and/or sequence. (A) Structures and sequences examined in this experiment. Only the sequences that are different in the four RNAs are shown. (B) Autoradiograms showing monomers and IRs from SLC+8, SLO+8, SLD+8, and SLT+8. Reactions in even- and odd-numbered lanes contained at 5 and 60 μM template, except lane 1, which lacked template. Asterisks (*) identify IRs.
Discussion
We have demonstrated that a recombinant RdRp and three viral replicases can synthesize RNAs that are longer than the length of a template, likely by the TC not terminating synthesis at the 5′-terminus of the donor, and then resuming synthesis at some frequency on an acceptor. This activity mimics observations of RNA recombination in vivo, including the preference for A-U-rich sequences and the production of multimeric satellite RNAs (6, 9). The RdRp can also switch templates internal to a donor RNA then continue synthesis on an acceptor. Our results are consistent with base-pairing-independent copy-choice RNA recombination (1). The ability to switch templates in vitro has been reported for several polymerases (14, 15, 27–29) and thus is a universal activity of template-dependent polymerases. Being able to observe the products of template switch in a simple viral RNA synthesis assay will help elucidate the mechanism for RNA recombination in vitro. Furthermore, we can now observe the products of template switch without the bias generated by the selection of the reproductively competent RNAs.
Sequence within 7 nucleotides from the 5′-terminus of the donor RNA affects the frequency of template switch. The strength of base pairing between the nascent and template RNA may partially account for the propensity for template switch. Weaker A-U-rich base pairing at positions -2 to -4 generally produced less IRs in comparison to templates that allow some G-C-rich base pairing at these positions. These results suggest that lower stability of the RNA duplex within specific region in the TC favors the termination of synthesis. No simple correlation, however, existed between the template switch frequency and the number of base-pairing hydrogen bonds between the template and nascent RNA at positions -2 to -4 of the donor, suggesting that the TC is stabilized by other factors in addition to nascent RNA–template interaction.
Features in RNA that are required for efficient RNA synthesis appear to facilitate efficient template switch by the TC. Initiation-debilitated templates and RNAs that lack the motifs needed for recognition by viral replicases were poor acceptors for BVDV RdRp and viral replicases, respectively (Table 1, Fig. 5). This agrees with the observations of Nagy et al. (30), who found that an enhancer of RNA synthesis increased the frequency of RNA recombination. We want to emphasize, however, replicase structure/function and/or subunit composition also affects the frequency of template switch, because the cucumber mosaic virus replicase produced 8- to 10-fold less recombinant RNAs than did the BMV and CCMV replicases. This conclusion is in agreement with the observations of Figlerowicz et al., who demonstrated that mutations in viral RNA replication proteins could significantly affect both the types and the frequencies of RNA recombination (31, 32).
Internal template switch by the BVDV RdRp occurred in a manner determined by NTP concentrations (Fig. 3). The observation that preferential internal switch by the BVDV RdRp occurs after the synthesis of 8- to 9-phosphodiester bonds suggests that the distance of the switch junction from the initiation site may affect template switch. RdRps will transit from initiation to processive elongative synthesis in a manner similar to RNA polymerases (reviewed in ref. 32). Preliminary evidence suggests that the BVDV RdRp transits to processive synthesis between the synthesis of 10- to 12-phosphodiester bonds (ref. 23 and M.-J.K., unpublished results), a value similar to that reported for the BMV replicase (33, 34). The internal template switch observed with RecO may be caused by the TC not being committed to elongation by position +10. However, because the nascent RNA is used to prime synthesis at the 3′-end of the acceptor, the complex of nascent RNA and RdRp may translocate to the acceptor. This mechanism could explain the repair of deleted 3′-termini of turnip crinkle virus satellite RNAs in vivo (35).
That RdRp and RNA replicases can switch template efficiently and synthesize products longer than the template may explain some previously published results. Products longer than template length were observed with the replicases from BMV (26), dengue virus (36), turnip crinkle virus (37), and the RdRps from hepatitis C virus (38, 39) and poliovirus (40). If all of the unexplained dimeric products are caused by template switch, then RNA recombination in vitro can be directly studied with RNA viruses for which RNA synthesis assays are available.
Acknowledgments
We thank P. Nagy for helpful comments and encouragement. The National Science Foundation and U.S. Department of Agriculture provided funding for this work.
Abbreviations
- TC
transcription complex
- RdRp
RNA-dependent RNA polymerase
- BVDV
bovine viral diarrhea virus
- BMV
brome mosaic virus
- CCMV
cowpea chlorotic mottle virus
- IR
identical recombinants
- IntR
internal recombinants, NR, nonidentical recombinants
References
- 1.Nagy P D, Simon A E. Virology. 1997;235:1–9. doi: 10.1006/viro.1997.8681. [DOI] [PubMed] [Google Scholar]
- 2.Jarvis T C, Kirkegaard K. Trends Genet. 1991;7:181–191. doi: 10.1016/0168-9525(91)90434-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lai M M C. Microbiol Rev. 1992;56:61–79. doi: 10.1128/mr.56.1.61-79.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morl M, Schmelzer C. Nucleic Acids Res. 1990;18:6545–6551. doi: 10.1093/nar/18.22.6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chetverina A, Chetverina H, Demidenko A A, Ugarov V. Cell. 1997;88:503–513. doi: 10.1016/S0092-8674(00)81890-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuroda T, Natsuaki T, Wang W-Q, Okuda S. J Gen Virol. 1997;78:941–946. doi: 10.1099/0022-1317-78-4-941. [DOI] [PubMed] [Google Scholar]
- 7.Bujarski J J. In: Molecular Biology of Plant Viruses. Mandahar C L, editor. Dordrecht, The Netherlands: Kluwer; 1999. [Google Scholar]
- 8.Simon A E, Nagy P D. Semin Virol. 1997;7:373–379. [Google Scholar]
- 9.Nagy P, Bujarski J J. J Virol. 1997;71:3799–3810. doi: 10.1128/jvi.71.5.3799-3810.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nagy P D, Bujarski J J. J Virol. 1998;72:1122–1130. doi: 10.1128/jvi.72.2.1122-1130.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cascone P J, Carpenter C D, Li X-H, Simon A E. EMBO J. 1990;9:1709–1715. doi: 10.1002/j.1460-2075.1990.tb08294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carpenter C D, Oh J-W, Zhang C, Simon A E. J Mol Biol. 1995;245:608–622. doi: 10.1006/jmbi.1994.0050. [DOI] [PubMed] [Google Scholar]
- 13.Nagy P D, Ogiela C, Bujarski J J. Virology. 1999;254:92–104. doi: 10.1006/viro.1998.9545. [DOI] [PubMed] [Google Scholar]
- 14.Tang R S, Barton D J, Flanegan J B, Kirkegaard K. RNA. 1997;3:624–633. [PMC free article] [PubMed] [Google Scholar]
- 15.Arnold J J, Cameron C E. J Biol Chem. 1999;274:2706–2716. doi: 10.1074/jbc.274.5.2706. [DOI] [PubMed] [Google Scholar]
- 16.Zhong W, Gutshall L L, DelVecchio A M. J Virol. 1999;72:9365–9369. doi: 10.1128/jvi.72.11.9365-9369.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun J, Adkins S, Faurote G, Kao C. Virology. 1996;226:1–12. doi: 10.1006/viro.1996.0622. [DOI] [PubMed] [Google Scholar]
- 18.Adkins S, Kao C. Virology. 1998;252:1–8. doi: 10.1006/viro.1998.9449. [DOI] [PubMed] [Google Scholar]
- 19.Sivakumaran K, Bao Y, Roossinck M, Kao C. J Virol. 2000;74:10323–10331. doi: 10.1128/jvi.74.22.10323-10331.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Milligan J F, Groebe D R, Witherell G W, Uhlenbeck O C. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng Y-W, Gott J M. Nucleic Acids Res. 2000;28:3695–3701. doi: 10.1093/nar/28.19.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kao C, Del Vecchio A M, Zhong W. Virology. 1999;253:1–7. doi: 10.1006/viro.1998.9517. [DOI] [PubMed] [Google Scholar]
- 23.Kim M-J, Zhong W, Hong Z, Kao C. J Virol. 2000;74:10312–10322. doi: 10.1128/jvi.74.22.10312-10322.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mooney R, Artsimovitch A, Landick R. J Bacteriol. 1998;180:3265–3272. doi: 10.1128/jb.180.13.3265-3275.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levin J R, Chamberlin M J. J Mol Biol. 1987;196:61–84. doi: 10.1016/0022-2836(87)90511-0. [DOI] [PubMed] [Google Scholar]
- 26.Kim C-H, Kao C C, Tinoco I. Nat Struct Biol. 2000;7:415–423. doi: 10.1038/75202. [DOI] [PubMed] [Google Scholar]
- 27.Nudler E, Avetissova E, Markovtsov V, Goldfarb A. Science. 1996;273:211–217. doi: 10.1126/science.273.5272.211. [DOI] [PubMed] [Google Scholar]
- 28.Anderson J A, Teufel R J, Yin P D, Hu W S. J Virol. 1998;72:1186–1194. doi: 10.1128/jvi.72.2.1186-1194.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murthy V, Meijer W J J, Blanco L, Salas M. Mol Microbiol. 1998;29:787–798. doi: 10.1046/j.1365-2958.1998.00972.x. [DOI] [PubMed] [Google Scholar]
- 30.Nagy P D, Pogany J, Simon A E. EMBO J. 1999;20:5653–5665. doi: 10.1093/emboj/18.20.5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Figlerowicz M, Nagy P D, Bujarski J J. Proc Natl Acad Sci USA. 1997;94:2073–2078. doi: 10.1073/pnas.94.5.2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Figlerowicz M, Nagy P D, Tang N, Kao C, Bujarski J. J Virol. 1998;72:9192–9200. doi: 10.1128/jvi.72.11.9192-9200.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adkins S, Stawicki S, Faurote G, Siegel R, Kao C. RNA. 1998;4:455–470. [PMC free article] [PubMed] [Google Scholar]
- 34.Sun J S, Kao C. Virology. 1997;233:63–73. doi: 10.1006/viro.1997.8583. [DOI] [PubMed] [Google Scholar]
- 35.Nagy P D, Carpenter C, Simon A E. Proc Natl Acad Sci USA. 1997;94:1113–1118. doi: 10.1073/pnas.94.4.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.You S, Padmanabhan R. J Biol Chem. 1999;274:33714–33722. doi: 10.1074/jbc.274.47.33714. [DOI] [PubMed] [Google Scholar]
- 37.Song C, Simon A E. Proc Natl Acad Sci USA. 1994;91:8792–8796. doi: 10.1073/pnas.91.19.8792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lohmann V, Overton H, Bartenschlager R. J Biol Chem. 1999;274:10807–10815. doi: 10.1074/jbc.274.16.10807. [DOI] [PubMed] [Google Scholar]
- 39.Kao C, Yang X, Kline A, Wang M, Barket D, Heinz B. J Virol. 2000;74:11121–11128. doi: 10.1128/jvi.74.23.11121-11128.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Young D C, Tuschall D M, Flanagan J B. J Virol. 1985;54:256–264. doi: 10.1128/jvi.54.2.256-264.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]