Abstract
Introduction
Preoperative chemoradiotherapy represents the standard treatment for patients with locally advanced rectal cancer. Unfortunately, the response of individual tumors to multimodal treatment is not uniform and ranges from complete response to complete resistance. This poses a particular problem for patients with a priori resistant tumors because they may be exposed to irradiation and chemotherapy, treatment regimens that are both expensive and at times toxic, without benefit. Accordingly, there is a strong need to establish molecular biomarkers that predict the response of an individual patient’s tumor to multimodal treatment and that indicate treatment-associated toxicities prior to therapy. Such biomarkers may guide clinicians in choosing the best possible treatment for each individual patient. In addition, these biomarkers could be used to identify novel molecular targets and thereby assist in implementing novel strategies to sensitize a priori resistant tumors to multimodal treatment regimens.
Objective
The aim of this review is to summarize recent findings about the molecular basis of treatment resistance and treatment toxicity in patients with rectal cancer. Whole-genome, as well as single-biomarker or multibiomarker, analyses and their potential implications will be highlighted. At the end, we will outline a future vision of rectal cancer treatment in the era of personalized medicine.
Keywords: Rectal cancer, Preoperative chemoradiotherapy, Chemoradiosensitivity and chemoradioresistance, Response prediction, Molecular biomarker
Introduction
For rectal cancers, both surgical and nonsurgical treatment concepts have been improved considerably over the last decades. In this respect, the surgical concept of total mesorectal excision (TME) [1] and the implementation of preoperative treatment regimens can be considered as cornerstones of modern and optimized treatment for locally advanced rectal cancer. Randomized multicenter trials independently demonstrated that preoperative application of radiotherapy, either as short-term radiotherapy or as long-term irradiation accompanied by infusional 5-fluorouracil (5-FU), significantly decreases the rate of local recurrence [2, 3]. Consequently, preoperative (chemo)radiotherapy now represents the standard treatment for patients with locally advanced rectal cancer in Europe and the USA [4].
With respect to long-term preoperative chemoradiotherapy, it was soon realized that the histopathological response to chemoradiotherapy varied tremendously from one patient to another, ranging from complete regression to complete resistance, and that, for many patients, the extent of regression was correlated to clinical outcome [5]. In order to increase the response rates to preoperative chemoradiotherapy, agents such as oxaliplatin and antibody-based regimens have been incorporated into multimodal treatment concepts, and these strategies are currently under extensive evaluation [4]. However, at the same time, the inclusion of more toxic agents increased the acute toxicity and long-term side effects of these treatments [4]. In some cases, acute organ toxicity necessitates a dose reduction or even termination of therapy. Furthermore, any higher grade of acute toxicity impairs the quality of life of the individual patient.
While the variability of both treatment-associated tumor regression and toxicity from one patient to another remains a major clinical problem, it obviously draws attention to the important possibility to individualize rectal cancer treatment. In this respect, some patients may require an intensified regimen to increase tumor response, whereas standard 5-FU-based chemoradiotherapy may be sufficient for others. However, advancing accurate stratification is strongly dependent on the identification of reliable clinical parameters and molecular biomarkers that allow pretherapeutic stratification with high certainty. The aim of this review is to briefly summarize the general principles of chemoradiosensitivity and to highlight recent findings about the molecular basis of treatment response and toxicity in patients with rectal cancer. At the end, we will discuss potential implications of these findings for an individualized treatment of rectal cancer patients.
General principles of chemoradiosensitivity
The predominant local effects of chemoradiotherapy, which is designed to achieve tumor cell damage, are primarily elicited by irradiation, whereas concomitant chemotherapy may serve as a radiosensitizer, most often without or with only small direct effects on tumor cell killing. The effects of chemoradiotherapy are largely the result of DNA damage, which either occurs directly through ionization within the DNA molecule or indirectly from the action of chemical radicals, which are also formed during irradiation [6, 7]. Through these mechanisms, several alterations, like base damage, DNA-protein cross-links, and single-strand or double-strand breaks, are generated and contribute to the antitumor effects and side effects [8].
However, as already discussed above, response to chemoradiotherapy differs widely. In general, four major explanations for these different sensitivities of solid tumors have been invoked, which could be causative alone or in combination:
Tumors with poor response to chemoradiotherapy often show a high content of hypoxic cells and/or fail to reoxygenate during fractionated multimodal treatments. For this reason, the necessary DNA damage caused by chemical radicals formed as a result of local ionization is not sufficient to kill all tumor cells. In general, this fact prevails with increasing tumor sizes including larger hypoxic and necrotic areas with lower oxygen pressure [9, 10].
It is well known that cells located in proliferating cell cycle segments are more sensitive to chemoradiotherapy compared to cells resting or degenerating. Therefore, the extent of tumor regression during treatment may be explained in part by the proportion of proliferating cells. A higher growth fraction in solid tumors might lead to a larger turnover rate with a higher proportion of cell loss in comparison to normal tissues [11].
Varying levels of repair capacity of DNA damage caused by chemoradiotherapy have been specified in almost all tumor cell lines and found to correlate with the clinical radiocurability. In other words, less curable tumors often show a strong potential for DNA damage recovery. A confirmation of this fact in larger clinical trials, which could justify its use as predictive marker for tumor response and outcome, has not been achieved so far. This might in part be due to the fact that any given tumor always harbors a mixed cell population, with mitotic or resting cells and stem cells with differing sensitivity to antineoplastic agents and irradiation-induced damage [12].
There seems to be an inherent chemoradiosensitivity, which is associated with the individual genetic sensitivity of every single patient. Assuming that all malignant cells arise from formerly normal tissue cells, it should thus be possible to determine the individual response to multimodal therapies via the patient’s individual genetic profile [13]. This fact is underlined by recent literature clearly showing a correlation between acute organ toxicity, which is a measure of inherent chemoradiosensitivity, and outcome for different tumor entities. For example, overall survival in patients with inoperable head and neck cancer was significantly associated with treatment-related high-grade acute organ toxicity [14].
Molecular basis of chemoradiosensitivity in rectal cancer
Despite the clinical importance of preoperative chemoradiotherapy in multimodal treatment concepts for patients with rectal cancer, our understanding of both the genetic basis of chemoradiosensitivity and the molecular events leading to chemoradioresistance remains relatively sparse. Relevant investigations that have been performed to identify molecular biomarkers differentiating responsive and resistant tumors will be discussed below. From a systematic point of view, high-throughput analyses (aim: whole-genome analysis) can be distinguished from low-throughput analyses (aim: single-biomarker or multibiomarker analysis).
Since complex phenotypes, such as tumor responsiveness to chemoradiotherapy, likely do not depend on the alteration or deregulated expression of single genes, high-throughput technologies have emerged as a central tool in deciphering the molecular basis of this clinically important phenotype because they offer the possibility to identify genomic differences between two groups of patients. However, due to the high number of observed genomic features, it represents a nontrivial task to determine which of these features are actually relevant, and this kind of analysis generally requires a high number of patients. Therefore, in situations where there is already prior biological knowledge pointing to a certain biomarker of interest or when preliminary evidence suggests the involvement of certain pathways, it can be advantageous to focus on a single or few selected biomarkers. Such studies, with a limited number of biomarkers of interest, require lower case numbers in order to reach statistical significance since correction for multiple testing, which is mandatory for high-throughput analyses, can be omitted [15].
Whole-genome analyses
Gene expression profiling
Expression microarrays are commonly used to comprehensively interrogate complex genetic pathways and networks [16, 17]. Consequently, several investigators have used gene expression profiling to analyze the genetics of (colo)rectal cancer response to chemoradiotherapy (recently reviewed by Kuremsky et al. [18], Nannini et al. [19], and Brettingham-Moore et al. [20]). From a systematic point of view, in vivo studies (profiling of primary rectal cancers) need to be examined separately from in vitro studies (profiling of cancer cell lines).
In vivo studies
In a first published report, pretherapeutic biopsies from 30 patients with locally advanced rectal carcinomas were profiled using a cDNA microarray [21]. All patients participated in the CAO/ARO/AIO-94 trial of the German Rectal Cancer Study Group (GRCSG) [3]. Tumor response was defined as T-level downsizing, and a set of 54 genes was found to be differentially expressed between responsive and resistant tumors. Using a leave-one-out cross-validation strategy, response was correctly predicted in 83% of patients. A follow-up study indicated that these genes could also assist in predicting local recurrence and disease-free survival [22]. These data are currently being validated in a much larger set of patients (n > 200) who participated in the CAO/ARO/AIO-04 trial of the GRCSG, treated within different institutions. Preliminary analyses of this cohort confirm the effectiveness of expression profiling for predicting outcome (Gaedcke and Ghadimi, unpublished data).
Shortly thereafter, a Japanese group published a gene expression analysis of 52 rectal cancer patients treated with preoperative radiotherapy [23]. They reported that 33 genes were differentially expressed between responders and nonresponders (based on histopathological regression grading of surgically resected specimens), with a class prediction accuracy of 88.6%.
Kim and colleagues described the identification of 261 genes that were differentially expressed between 20 partial responders to preoperative chemoradiotherapy, based on tumor regression grading, and 11 complete responders (defined as the training set). This set was validated in a test set of 15 patients. The authors reported class prediction accuracies of 84% (training set) and 87% (test set), respectively [24].
Similarly, Rimkus and colleagues profiled pretherapeutic biopsies from 43 patients with locally advanced rectal cancers. Using histopathologic response as endpoint for comparison with gene expression, they identified a 42-gene signature [25]. In the most recent study, Brettingham-Moore and colleagues analyzed pretherapeutic biopsies from 51 locally advanced rectal cancers and generated gene expression classifiers based on tumor regression grade, metabolic response, and UICC downstaging [26]. The sensitivity and specificity of these classifiers to predict outcome after preoperative chemoradiotherapy centered around 82% and 89%, respectively. Interestingly, the authors also tested the effectiveness of previously published gene expression signatures to predict outcome in their data set, but the results were rather unfruitful.
It is important to note that there was only a very limited overlap of genes from within these signatures. This could be due to several reasons: First, there are many different ways of defining “response” and “resistance.” Second, case numbers were different. Third, there are differences in the number and identity of spotted genes on the respective microarrays. Fourth, there is the problem of high dimensionality of the data. In most settings, the number of patients is naturally limited (i.e., in the order of tens or hundreds), while the number of measured features, i.e., gene transcripts, is usually very high (in the order of thousands or tens of thousands). It is, therefore, very likely that several gene expression signatures exist that are able to accurately predict the clinical outcome. This may be particularly true since genes are often highly interrelated, and homologs or isotypes may serve similar functions in different pathways. In the process of selecting a gene expression signature, it is, therefore, sometimes a matter of random choice which genes will end up in the classification signature and which genes will not. In the past, this has lead to enormous confusion and debate in the field [27–29]. Some more recent methods actually try to avoid this problem by guiding the gene selection process using prior knowledge, based, for example, on pathway and functional network databases [30, 31].
Despite these limitations, gene expression analyses hold considerable promise to unveil the underlying complex genetics of chemoradioresistance and to play a future role in stratifying rectal cancer patients. In this respect, breast cancer constitutes a prominent biological precedent to demonstrate the feasibility of using expression signatures in clinical decision-making (proof-of-principle). For this disease entity, a prognostic signature, consisting of 70 genes, has been established [32, 33] and subsequently extensively validated [34, 35], resulting in the initiation of a multicenter trial confirming the clinical effectiveness of this gene set (“Microarray in Node-Negative Disease May Avoid Chemotherapy” [MINDACT] trial).
In vitro studies
Cancer cell lines are widely used as model systems for target screening, drug discovery, and functional analyses because many features of primary tumors are recapitulated in derived cell lines [36–39]. Eschrich and colleagues were the first to report a gene expression-based model for in vitro sensitivity of colorectal cancer cell lines to irradiation, although their analysis only included seven colorectal cancer cell lines [40, 41]. The remaining 41 cell lines were derived from different entities, including breast, ovarian, renal, prostate, and non-small cell lung cancers, leukemia, melanoma, and tumors of the central nervous system. Building on their own earlier work [42], the authors integrated the respective surviving fractions of these 48 cell lines after irradiation at 2 Gy by pretreatment gene expression profiles, KRAS and TP53 mutation status, and tissue of origin and extracted a network of 10 signature genes. This expression model of intrinsic radiosensitivity, which included prominent target genes such as JUN, STAT1, and CDK1, was subsequently validated in three cohorts of patients with rectal, esophageal, and head and neck cancer [41].
In a similar study, Spitzner and colleagues reported the identification of a gene expression signature for sensitivity of colorectal cancer cell lines to chemoradiotherapy [43]. A panel of 12 cell lines was exposed to doses of both 5-FU and radiation that were similar to the ones used in the clinic, i.e., 3 μM of 5-FU and 2 Gy of radiation, and the respective surviving fractions were correlated with pretreatment gene expression profiles. Analysis of this chemoradiosensitivity signature revealed many genes involved in mitogen-activated protein kinase (MAPK), insulin, and Wnt signaling, cell cycle genes, and novel potential target genes such as STAT3 or ERBB2 [43].
As already pointed out for primary tumors (see the “In vivo studies” section), there is only a very limited overlap between the respective in vitro sensitivity signatures. In addition to the reasons discussed above, there are other potential explanations. First, Spitzner and colleagues correlated gene expression and sensitivity to both chemotherapy and irradiation, while Eschrich and colleagues established a signature of radiosensitivity. Second, Eschrich and colleagues included cell lines that were mismatch repair (MMR) proficient as well as cell lines that were MMR deficient, although these pathways are genetically different [44, 45]. Third, Eschrich and colleagues analyzed a panel of cell lines from various tumor entities.
Chromosomal aberrations
Chromosomal aneuploidy is a defining feature of colorectal carcinomas [46, 47]. This is reflected by tumor- and stage-specific genomic copy number aberrations [48], which are virtually identical in colon and rectal cancers [49, 50]. Accordingly, it may be speculated that differences in treatment responses can be correlated with differences on the DNA level.
In one of the first studies to address this question, pretherapeutic biopsies from 42 patients with locally advanced rectal cancers were analyzed using metaphase comparative genomic hybridization (CGH). Based on downsizing of the T-category, chromosomal gains of 7q32–q36 and 7q11–q31 as well as amplifications of 20q11–q13 were associated with responsiveness to preoperative chemoradiotherapy [51]. However, the authors reported a high probability that these genomic copy number changes were detected by chance, therefore requiring independent validation in a larger patient population and with a higher resolution. In a more recent study, Chen and colleagues used oligonucleotide array-based CGH to screen for chromosomal copy number alterations correlated with pathologic complete response (pCR). Analyzing DNA from 95 rectal cancers, the authors observed that chromosomal loss of 15q11.1–q26.3 was associated with non-pCR, while loss of 12p13.31 was associated with pCR [52].
Single-biomarker and multibiomarker analyses
DNA mutations in the RAS–MAPK pathway
The v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog (KRAS), a member of a large family of GTP-binding proteins involved in signal transduction [53, 54], plays an important role in colorectal carcinogenesis because a high percentage of colorectal carcinomas are characterized by activating mutations of this oncogene [45]. These mutations result in the constitutive activation of the MAPK pathway. Because preliminary evidence indicated an ability of the RAS oncogene to enhance radioresistance in vitro [55–58], several studies investigated the potential relevance of the RAS–MAPK pathway for the response of primary rectal cancers to irradiation.
Luna-Perez and colleagues correlated the response of 37 rectal cancers to preoperative chemoradiotherapy with KRAS mutation status [59]. Analyzing codons 12, 13, and 61, they could show that tumors with wild-type KRAS were more likely to be responsive than tumors with mutant KRAS. It should be noted that the authors used irradiated tumor tissue for their analysis, although recent evidence suggests that preoperative multimodal treatment does not alter KRAS mutation status [60]. In contrast, Zauber and colleagues screened pretherapeutic biopsies from 53 patients with stage I–III rectal cancers and detected KRAS mutations in 18 patients (34%). The presence of a KRAS mutation, however, was not indicative of tumor regression after preoperative chemoradiotherapy [61].
Shortly thereafter, Gaedcke and colleagues observed a KRAS mutation frequency of 48% (n = 45) in pretherapeutic biopsies of 94 patients with locally advanced rectal cancers [62]. In contrast to Zauber and colleagues, these authors specifically reported the affected codons: Twenty-nine mutations (64%) were located in codon 12, 10 mutations (22%) in codon 13, and 3 mutations (7%) each in codons 61 and 146. The presence of none of these mutations was correlated with response to preoperative chemoradiotherapy. However, Gaedcke et al. detected differential sensitivities when the mutations were grouped based on the respective amino acid exchange; G12V mutations appeared to be associated with higher rates of tumor regression than G13D mutations (p = 0.012). Most recently, Garcia-Aguilar and colleagues published their analysis of pretherapeutic biopsies from 132 patients with locally advanced rectal cancers and reported that KRAS mutations were more likely in tumors from patients without pCR, i.e., resistant tumors [63].
These conflicting data indicate that it may not be sufficient to solely determine the mutation status of KRAS, but rather to group patients according to the respective nucleotide-specific amino acid exchange. This interpretation is supported by previous investigations suggesting that the level of aggressiveness depends on the mutation form [64, 65] and that these specific mutation forms may activate distinct downstream targets and different oncogenic pathways [66]. Furthermore, De Roock and colleagues recently reported that chemotherapy-refractory metastatic colorectal cancers harboring a G13D KRAS mutation were more sensitive to treatment with the epidermal growth factor receptor (EGFR) inhibitor cetuximab compared to tumors with other KRAS mutations [67]. This was confirmed in vitro, and G12V-mutated cancer cells were resistant to cetuximab, whereas G13D-mutated and KRAS wild-type cancer cells were sensitive. Whether the KRAS mutation status also influences the response of rectal cancers to multimodal treatment concepts that include EGFR inhibitors remains elusive, particularly as the clinical relevance of these combinations remains to be determined [68, 69].
Single-nucleotide polymorphisms
Single-nucleotide polymorphisms (SNPs) are sites in the genome sequence where individuals differ by a single base [70]. The total number of these sites in the human genome is estimated to be roughly 10 million, and these SNPs are distributed at an overall frequency of 1 in every 300 to 1,000 base pairs [71]. Importantly, it has been demonstrated that specific haplotypes and genetic polymorphisms are associated with clinical phenotypes. For instance, the presence of a G allele within the SNP rs6983267, located on chromosome 8q24, confers an increased risk for the development of colorectal cancer [72–74]. Due to the growing body of evidence suggesting that genetic variation between individuals can account for differences in drug response [75, 76], it has been speculated that genetic polymorphisms in genes encoding drug- or radiation-related responses may influence the individual’s response to chemoradiotherapy [77].
Most prominently, thymidylate synthase (TS) has been analyzed in this respect, but the results are conflicting. Villafranca and colleagues were the first to correlate polymorphisms in the TS promoter and tumor response to preoperative chemoradiotherapy. Analyzing tumor DNA from pretreatment biopsies of 65 patients, the authors observed that the TS genotype was predictive for tumor downstaging following preoperative chemoradiotherapy [78].
Terrazzino and colleagues, as well as the other investigators mentioned within this section, analyzed germline (blood) DNA from rectal cancer patients. However, there was no correlation with histopathological tumor regression [79]. This result has recently been confirmed by Conradi and colleagues who also failed to demonstrate any association between TS genotype and relevant clinical parameters such as local response, tumor regression grading, or disease-free and overall survival [80].
In contrast, Spindler and colleagues demonstrated that the TS genotype had a significant impact on the rate of complete pathological response following preoperative chemoradiotherapy [81]. Similar results were subsequently published by Stoehlmacher and colleagues who also reported a correlation between TS genotype and histopathological tumor regression [82]. Data from Hur and colleagues confirm the interpretation that SNPs within the TS enhancer region affect the response of rectal cancers to preoperative chemoradiotherapy [83]. Very recently, Tan and colleagues reported the prospective use of TS genotyping to direct preoperative chemoradiotherapy in a single-institution phase II study [84].
As a second example, recent data indicated that germline polymorphisms in the TGFB1 gene are associated with quality of life-impairing acute organ toxicity in patients with locally advanced rectal cancer. Analyzing DNA from two independent cohorts of patients participating in the CAO/ARO/AIO-94 and -04 trials (n = 88 and n = 75), Schirmer and colleagues demonstrated that all patients carrying the TGFB1 Pro25 variant developed high-grade acute organ toxicity during preoperative 5-FU-based chemoradiotherapy [85]. The positive predictive value for acute toxicity in the presence of this SNP is 100%, which highlights the potential clinical importance of this observation.
Immunohistochemistry
A plethora of studies has been published which focused on a single immunohistochemical marker or a combination of a few. A comprehensive summary of these studies would extend beyond the scope of this review, and we refer the reader to recent comprehensive reviews [18, 86, 87]. Very briefly, primary focus was the analysis of proteins involved in DNA damage repair, proliferation, angiogenesis, and apoptosis, including Ki-67, cyclin E, p21, p53, survivin, Bcl-2, BAX, EGFR, VEGF, PCNA, XIAP, PTGS2 (COX-2), HIF-1α, TS, and PROM1 (CD133). However, for most marker studies, the results are conflicting and still remain inconclusive.
Novel molecular targets for chemoradiosensitization
As discussed above, there is a clinical need to establish molecular biomarkers that differentiate responsive and resistant tumors because such biomarkers could be used pretherapeutically to predict the response of an individual patient’s tumor to multimodal treatment (diagnostic approach). In addition, genes that are differentially expressed between resistant and responsive tumors could be used to identify novel therapeutic targets and thereby assist in implementing novel therapeutic strategies (therapeutic approach). For instance, genes that are overexpressed in resistant tumors could be repressed via RNA interference (RNAi)-based approaches [88, 89] or using chemical/small molecule inhibitors, potentially leading to sensitization to chemoradiotherapy. In this context, both survivin and T cell-specific factor 4 (TCF4) represent two prominent and promising examples.
Survivin
The baculoviral inhibitor of apoptosis repeat-containing 5 (BIRC5), more commonly referred to as survivin, encodes for the smallest and structurally unique member of the inhibitors of apoptosis family of proteins [90]. Survivin is overexpressed in a variety of human tumors, and it plays a prominent role in regulating apoptosis, during cell division, and during adaptation to stress.
Following up on the observation that the expression of survivin was inversely correlated with spontaneous and radiation-induced apoptosis [91], Rödel and colleagues used siRNA-mediated gene silencing to demonstrate that inhibition of survivin sensitizes colorectal cancer cells to radiation therapy, accompanied by increased levels of G2/M phase arrest and increased levels of DNA double-strand breaks after irradiation [92]. Very recently, the same authors could show that survivin rapidly accumulates in the nucleus following irradiation where it subsequently interacts with members of the DNA double-strand break repair machinery in order to regulate the activity of DNA-dependent protein kinase [93]. Because survivin inhibitors are currently being investigated in clinical trials, future studies will ultimately demonstrate whether its inhibition represents an effective strategy for (chemo)radiosensitization [94]. In this respect, the potential relevance of survivin for monitoring response to preoperative chemoradiotherapy has recently been confirmed by Sprenger and colleagues who could show that high survivin expression in pretreatment biopsies correlated with advanced posttherapeutical tumor and UICC stage and decreased disease-free survival [95].
TCF4
As discussed above, Ghadimi and colleagues reported the identification of a 54-gene signature that differentiated resistant and responsive rectal cancers from patients who had been treated with preoperative chemoradiotherapy [21]. Interestingly, within this signature, the transcription factor TCF4 was found to be significantly overexpressed in resistant tumors. TCF4, also known as TCF7L2, represents a key downstream effector that mediates canonical Wnt signaling, a pathway that plays a central role in colorectal tumorigenesis and tumor progression [96, 97].
In order to explore the functional relevance of this overexpression for mediating treatment resistance, Kendziorra and colleagues recently silenced TCF4 in resistant colorectal cancer cell lines and could show that RNAi-mediated inhibition of TCF4 caused a significant radiosensitization of colorectal cancer cells with high TCF reporter activity (Fig. 1). Follow-up experiments revealed that the effect of radiosensitization was associated with a G2/M phase arrest, an impaired ability to adequately halt cell cycle progression after irradiation, and a compromised DNA double-strand break repair [98]. These data indicate a novel role of the Wnt transcription factor TCF4 in mediating radioresistance and, if further validated, suggest that TCF4 is a promising therapeutic target.
Fig. 1.
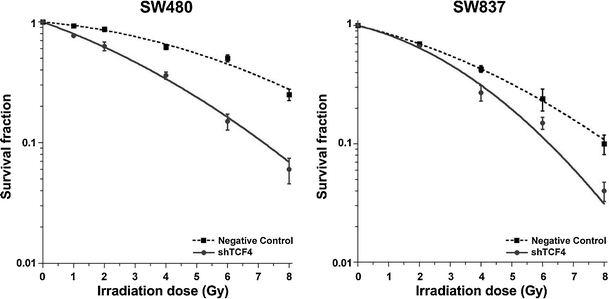
RNAi-mediated silencing of TCF4 results in radiosensitization. TCF4 was silenced in SW837 and SW480 cells using shRNA constructs, and stable single-cell clones were subsequently established. A standard colony-forming assay demonstrated that silencing of TCF4 significantly increased the sensitivity of SW480 and SW837 cells to clinically relevant doses of X-rays
Conclusions and future perspective
The genetic diversity of rectal cancer is associated with varying responses to chemoradiotherapy, and varying toxicity rates. This offers a wide range of options to pretherapeutically assess both response and toxicity for the individual patient. Consequently, a plethora of potential biomarkers has already been evaluated using whole-genome and single-marker or multimarker analyses, some of which have great potential to stratify rectal cancer patients for multimodal treatment regimens and to implement targeted therapeutics (Fig. 2).
Fig. 2.
Potential pathways and proteins regulating and mediating resistance of rectal cancer cells to chemoradiotherapy
However, there are several drawbacks of these findings that still impede transition to routine clinical practice: First, conflicting results were obtained by different investigators. Second, virtually all biomarkers described to date have been identified in retrospective studies and lack independent validation in a prospective setting using standardized analytical procedures. This represents the most challenging hurdle to the implementation of these biomarkers, once further validated, into a clinical setting.
In this respect, the TransValid-KFO179/GRCSG-Trials (TransValid A, TransValid B) are the first biomarker-driven clinical trials for patients with rectal cancer (Fig. 3). Funded by the Deutsche Forschungsgemeinschaft and initiated and promoted by the institutions of the GRCSG as well as the Clinical Research Unit 179 (Klinische Forschergruppe, KFO179), the aim of these multicenter studies is to prospectively implement the validation of previously identified molecular and clinical biomarkers into a highly standardized clinical setting. To achieve this goal, patients with locally advanced (cUICC II/III) cancers of the lower two thirds of the rectum as well as patients with resectable synchronous liver metastases (UICC IV) can be enrolled into one of these trials based on the responsible clinicians’ discretion. In TransValid A (“validation study”), patients are treated with standard 5-FU-based chemoradiotherapy followed by standardized TME surgery according to the German S3 Guideline (Fig. 3). Adjuvant chemotherapy consists of either 5-FU monotherapy or, in selected cases based on the clinicians’ assessment, a shortened FOLFOX regimen. The aim of TransValid B (“feasibility study,” phase I/II) is to establish the feasibility of an intensified preoperative chemoradiotherapy regimen (radiation, 5-FU, and oxaliplatin) combined with a shortened FOLFOX regimen prior to standardized TME surgery (Fig. 3). One rationale for this approach is the fact that today many patients do not receive, or receive limited doses of, adjuvant chemotherapy. Ascertainment of biomaterial at various time points is a major inclusion criterion for the TransValid trials (Fig. 3). This biomaterial will be processed according to previously established strict standard operating procedures, which includes performing these experiments on a week-by-week basis (prospective data generation).
Fig. 3.
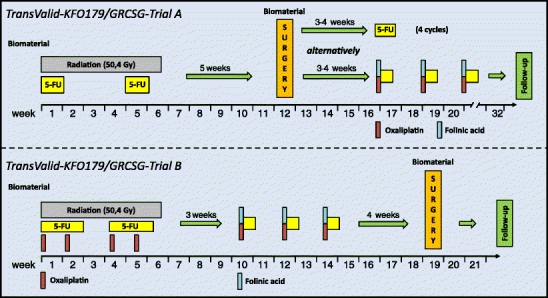
Outline of the TransValid-KFO179/GRCSG-Trials (TransValid A, TransValid B). TransValid A (validation study): 200 patients will be treated with 5-FU-based (1,000 mg/m2, 120 h continuous i.v. on days 1–5 and 29–33) chemoradiotherapy (radiation, 28 × 1.8 Gy) followed by radical surgery. Adjuvant therapy consists of either four cycles of 5-FU (500 mg/m2, bolus i.v. on days 1–5, repeat on day 29) or, in selected cases based on the clinicians’ discretion, six applications of a shortened FOLFOX regimen (folinic acid 400 mg/m2, 2 h continuous i.v.; oxaliplatin 100 mg/m2, 2 h continuous i.v.; 5-FU 2,400 mg/m2, 46 h continuous i.v.; on days 1, 15, 30, 45, 60, and 75). TransValid B (feasibility study, phase I/II): 50 patients will be treated with chemoradiotherapy (radiation, 28 × 1.8 Gy; 5-FU 250 mg/m2, continuous i.v. on days 1–14 and 22–35; oxaliplatin, 50 mg/m2, 2 h continuous i.v. on days 1, 8, 22, and 29), followed by three applications of a shortened FOLFOX regimen on days 1, 15, and 30 and radical surgery
From a personal perspective, we strongly believe that molecular biomarkers will be implemented into clinical decision-making in the near future. In a potential scenario, pretherapeutic patient material from both tumor and normal tissue will be ascertained at the initial diagnosis and subjected to multilayer genomic analyses (Fig. 4). Based on the results of these analyses (aim: prediction of both response and toxicity), the individual patient will be stratified into different alternative treatment concepts (personalized medicine). In this setting, patients with a biomarker profile indicating “responder to standard treatment” are subjected to a low-toxicity preoperative regimen. In contrast, for patients with a biomarker profile indicating “nonresponder to standard treatment,” a more aggressive approach is needed. For instance, an intensified regimen could be pursued, including the application of more effective systemic agents such as oxaliplatin. An induction combination chemotherapy (preoperative chemoradiotherapy followed by chemotherapy with sufficient dose and intensity prior to surgery) would be another interesting option because many patients do not receive adjuvant chemotherapy after preoperative chemoradiotherapy and surgical resection (either due to surgical complications, patients’ refusal, or investigators’ discretion). For patients predicted to be “nonresponder to standard treatment” and to develop high acute organ toxicity, primary surgery may be an option. With respect to novel therapeutic target genes, there are examples of molecular targets such as survivin and TCF4 that have the potential to be incorporated into treatment concepts, although this requires extensive validation and testing.
Fig. 4.
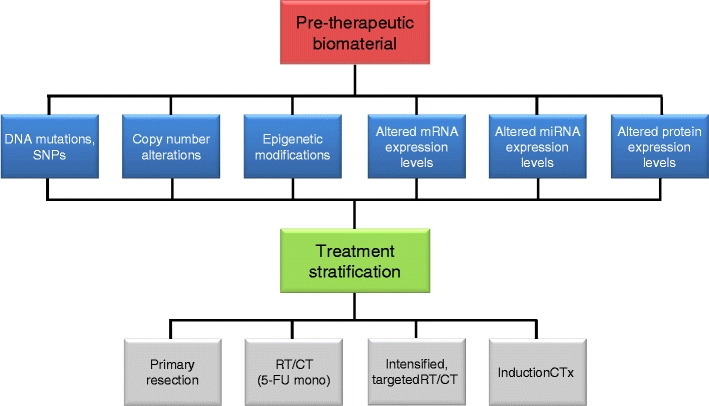
Future vision for the treatment of patients with locally advanced rectal cancers. Pretherapeutic patient material (tumor and normal tissue) will be subjected to multilayer genomic analyses. Based on the results of these analyses, patients will be stratified into different (preoperative) treatment concepts (personalized medicine)
In any case, rectal cancer represents a prominent example on how to individualize multimodal treatment regimens. Once demonstrated that predictive biomarkers can be examined in a week-by-week setting with high quality and reproducibility and in a cost-effective manner, and once these biomarkers have been prospectively validated utilizing sufficient patient numbers, a personalized medicine is within reach. This holds considerable promise to improve the outcome of patients with this disease.
Acknowledgements
The authors thank Dr. Melanie Spitzner, Birte Roesler, and Emil Kendziorra for the editorial assistance, Dr. Tim Beissbarth for the helpful discussions, and Dr. Reinhard Ebner for critically reading the manuscript. The work of the authors is supported by the Deutsche Forschungsgemeinschaft (KFO179).
Conflicts of interest
None.
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
References
- 1.Heald RJ, Husband EM, Ryall RD. The mesorectum in rectal cancer surgery—the clue to pelvic recurrence? Br J Surg. 1982;69(10):613–616. doi: 10.1002/bjs.1800691019. [DOI] [PubMed] [Google Scholar]
- 2.Kapiteijn E, Marijnen CA, Nagtegaal ID, Putter H, Steup WH, Wiggers T, Rutten HJ, Pahlman L, Glimelius B, van Krieken JH, Leer JW, van de Velde CJ. Preoperative radiotherapy combined with total mesorectal excision for resectable rectal cancer. N Engl J Med. 2001;345(9):638–646. doi: 10.1056/NEJMoa010580. [DOI] [PubMed] [Google Scholar]
- 3.Sauer R, Becker H, Hohenberger W, Rodel C, Wittekind C, Fietkau R, Martus P, Tschmelitsch J, Hager E, Hess CF, Karstens JH, Liersch T, Schmidberger H, Raab R. Preoperative versus postoperative chemoradiotherapy for rectal cancer. N Engl J Med. 2004;351(17):1731–1740. doi: 10.1056/NEJMoa040694. [DOI] [PubMed] [Google Scholar]
- 4.Cunningham D, Atkin W, Lenz HJ, Lynch HT, Minsky B, Nordlinger B, Starling N. Colorectal cancer. Lancet. 2010;375(9719):1030–1047. doi: 10.1016/S0140-6736(10)60353-4. [DOI] [PubMed] [Google Scholar]
- 5.Rodel C, Martus P, Papadoupolos T, Fuzesi L, Klimpfinger M, Fietkau R, Liersch T, Hohenberger W, Raab R, Sauer R, Wittekind C. Prognostic significance of tumor regression after preoperative chemoradiotherapy for rectal cancer. J Clin Oncol. 2005;23(34):8688–8696. doi: 10.1200/JCO.2005.02.1329. [DOI] [PubMed] [Google Scholar]
- 6.Seiwert TY, Salama JK, Vokes EE. The concurrent chemoradiation paradigm—general principles. Nat Clin Pract Oncol. 2007;4(2):86–100. doi: 10.1038/ncponc0714. [DOI] [PubMed] [Google Scholar]
- 7.Katz D, Ito E, Liu FF. On the path to seeking novel radiosensitizers. Int J Radiat Oncol Biol Phys. 2009;73(4):988–996. doi: 10.1016/j.ijrobp.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 8.Begg AC, Stewart FA, Vens C. Strategies to improve radiotherapy with targeted drugs. Nat Rev Cancer. 2011;11(4):239–253. doi: 10.1038/nrc3007. [DOI] [PubMed] [Google Scholar]
- 9.Vaupel P, Schlenger K, Knoop C, Hockel M. Oxygenation of human tumors: evaluation of tissue oxygen distribution in breast cancers by computerized O2 tension measurements. Cancer Res. 1991;51(12):3316–3322. [PubMed] [Google Scholar]
- 10.Steel GG, Peacock JH. Why are some human tumours more radiosensitive than others? Radiother Oncol. 1989;15(1):63–72. doi: 10.1016/0167-8140(89)90119-9. [DOI] [PubMed] [Google Scholar]
- 11.Zaider M, Hanin L. Tumor control probability in radiation treatment. Med Phys. 2011;38(2):574–583. doi: 10.1118/1.3521406. [DOI] [PubMed] [Google Scholar]
- 12.Weichselbaum RR, Little JB. Radioresistance in some human tumor cells conferred in vitro by repair of potentially lethal X-ray damage. Radiology. 1982;145(2):511–513. doi: 10.1148/radiology.145.2.7134460. [DOI] [PubMed] [Google Scholar]
- 13.Steel GG, Peckham MJ. Exploitable mechanisms in combined radiotherapy-chemotherapy: the concept of additivity. Int J Radiat Oncol Biol Phys. 1979;5(1):85–91. doi: 10.1016/0360-3016(79)90044-0. [DOI] [PubMed] [Google Scholar]
- 14.Wolff HA, Daldrup B, Jung K, Overbeck T, Hennies S, Matthias C, Hess CF, Roedel RM, Christiansen H. High-grade acute organ toxicity as positive prognostic factor in adjuvant radiation and chemotherapy for locally advanced head and neck cancer. Radiology. 2011;258(3):864–871. doi: 10.1148/radiol.10100705. [DOI] [PubMed] [Google Scholar]
- 15.Cheng C, Pounds S. False discovery rate paradigms for statistical analyses of microarray gene expression data. Bioinformation. 2007;1(10):436–446. doi: 10.6026/97320630001436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Quackenbush J. Microarray analysis and tumor classification. N Engl J Med. 2006;354(23):2463–2472. doi: 10.1056/NEJMra042342. [DOI] [PubMed] [Google Scholar]
- 17.van’t Veer LJ, Bernards R. Enabling personalized cancer medicine through analysis of gene-expression patterns. Nature. 2008;452(7187):564–570. doi: 10.1038/nature06915. [DOI] [PubMed] [Google Scholar]
- 18.Kuremsky JG, Tepper JE, McLeod HL. Biomarkers for response to neoadjuvant chemoradiation for rectal cancer. Int J Radiat Oncol Biol Phys. 2009;74(3):673–688. doi: 10.1016/j.ijrobp.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 19.Nannini M, Pantaleo MA, Maleddu A, Astolfi A, Formica S, Biasco G. Gene expression profiling in colorectal cancer using microarray technologies: results and perspectives. Cancer Treat Rev. 2009;35(3):201–209. doi: 10.1016/j.ctrv.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 20.Brettingham-Moore KH, Duong CP, Heriot AG, Thomas RJ, Phillips WA. Using gene expression profiling to predict response and prognosis in gastrointestinal cancers—the promise and the perils. Ann Surg Oncol. 2011;18(5):1484–1491. doi: 10.1245/s10434-010-1433-1. [DOI] [PubMed] [Google Scholar]
- 21.Ghadimi BM, Grade M, Difilippantonio MJ, Varma S, Simon R, Montagna C, Fuzesi L, Langer C, Becker H, Liersch T, Ried T. Effectiveness of gene expression profiling for response prediction of rectal adenocarcinomas to preoperative chemoradiotherapy. J Clin Oncol. 2005;23(9):1826–1838. doi: 10.1200/JCO.2005.00.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liersch T, Grade M, Gaedcke J, Varma S, Difilippantonio MJ, Langer C, Hess CF, Becker H, Ried T, Ghadimi BM. Preoperative chemoradiotherapy in locally advanced rectal cancer: correlation of a gene expression-based response signature with recurrence. Cancer Genet Cytogenet. 2009;190(2):57–65. doi: 10.1016/j.cancergencyto.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Watanabe T, Komuro Y, Kiyomatsu T, Kanazawa T, Kazama Y, Tanaka J, Tanaka T, Yamamoto Y, Shirane M, Muto T, Nagawa H. Prediction of sensitivity of rectal cancer cells in response to preoperative radiotherapy by DNA microarray analysis of gene expression profiles. Cancer Res. 2006;66(7):3370–3374. doi: 10.1158/0008-5472.CAN-05-3834. [DOI] [PubMed] [Google Scholar]
- 24.Kim IJ, Lim SB, Kang HC, Chang HJ, Ahn SA, Park HW, Jang SG, Park JH, Kim DY, Jung KH, Choi HS, Jeong SY, Sohn DK, Kim DW, Park JG. Microarray gene expression profiling for predicting complete response to preoperative chemoradiotherapy in patients with advanced rectal cancer. Dis Colon Rectum. 2007;50(9):1342–1353. doi: 10.1007/s10350-007-277-7. [DOI] [PubMed] [Google Scholar]
- 25.Rimkus C, Friederichs J, Boulesteix AL, Theisen J, Mages J, Becker K, Nekarda H, Rosenberg R, Janssen KP, Siewert JR. Microarray-based prediction of tumor response to neoadjuvant radiochemotherapy of patients with locally advanced rectal cancer. Clin Gastroenterol Hepatol. 2008;6(1):53–61. doi: 10.1016/j.cgh.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 26.Brettingham-Moore KH, Duong CP, Greenawalt DM, Heriot AG, Ellul J, Dow CA, Murray WK, Hicks RJ, Tjandra J, Chao M, Bui A, Joon DL, Thomas RJ, Phillips WA. Pretreatment transcriptional profiling for predicting response to neoadjuvant chemoradiotherapy in rectal adenocarcinoma. Clin Cancer Res. 2011;17(9):3039–3047. doi: 10.1158/1078-0432.CCR-10-2915. [DOI] [PubMed] [Google Scholar]
- 27.Borst P, Wessels L. Do predictive signatures really predict response to cancer chemotherapy? Cell Cycle. 2010;9(24):4836–4840. doi: 10.4161/cc.9.24.14326. [DOI] [PubMed] [Google Scholar]
- 28.Koscielny S. Why most gene expression signatures of tumors have not been useful in the clinic. Sci Transl Med. 2010;2(14):14ps12. doi: 10.1126/scitranslmed.3000313. [DOI] [PubMed] [Google Scholar]
- 29.Gonen M. Statistical aspects of gene signatures and molecular targets. Gastrointest Cancer Res. 2009;3(2 Suppl):S19–S21. [PMC free article] [PubMed] [Google Scholar]
- 30.Johannes M, Brase JC, Frohlich H, Gade S, Gehrmann M, Falth M, Sultmann H, Beissbarth T. Integration of pathway knowledge into a reweighted recursive feature elimination approach for risk stratification of cancer patients. Bioinformatics. 2010;26(17):2136–2144. doi: 10.1093/bioinformatics/btq345. [DOI] [PubMed] [Google Scholar]
- 31.Porzelius C, Johannes M, Binder H, Beissbarth T. Leveraging external knowledge on molecular interactions in classification methods for risk prediction of patients. Biom J. 2011;53(2):190–201. doi: 10.1002/bimj.201000155. [DOI] [PubMed] [Google Scholar]
- 32.van de Vijver MJ, He YD, van’t Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, Parrish M, Atsma D, Witteveen A, Glas A, Delahaye L, van der Velde T, Bartelink H, Rodenhuis S, Rutgers ET, Friend SH, Bernards R. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347(25):1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 33.van’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, Schreiber GJ, Kerkhoven RM, Roberts C, Linsley PS, Bernards R, Friend SH. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415(6871):530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 34.Buyse M, Loi S, Veer L, Viale G, Delorenzi M, Glas AM, d’Assignies MS, Bergh J, Lidereau R, Ellis P, Harris A, Bogaerts J, Therasse P, Floore A, Amakrane M, Piette F, Rutgers E, Sotiriou C, Cardoso F, Piccart MJ. Validation and clinical utility of a 70-gene prognostic signature for women with node-negative breast cancer. J Natl Cancer Inst. 2006;98(17):1183–1192. doi: 10.1093/jnci/djj329. [DOI] [PubMed] [Google Scholar]
- 35.Bueno-de-Mesquita JM, van Harten WH, Retel VP, van’t Veer LJ, van Dam FS, Karsenberg K, Douma KF, van Tinteren H, Peterse JL, Wesseling J, Wu TS, Atsma D, Rutgers EJ, Brink G, Floore AN, Glas AM, Roumen RM, Bellot FE, van Krimpen C, Rodenhuis S, van de Vijver MJ, Linn SC. Use of 70-gene signature to predict prognosis of patients with node-negative breast cancer: a prospective community-based feasibility study (RASTER) Lancet Oncol. 2007;8(12):1079–1087. doi: 10.1016/S1470-2045(07)70346-7. [DOI] [PubMed] [Google Scholar]
- 36.Shoemaker RH. The NCI60 human tumour cell line anticancer drug screen. Nat Rev Cancer. 2006;6(10):813–823. doi: 10.1038/nrc1951. [DOI] [PubMed] [Google Scholar]
- 37.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, Speed T, Spellman PT, DeVries S, Lapuk A, Wang NJ, Kuo WL, Stilwell JL, Pinkel D, Albertson DG, Waldman FM, McCormick F, Dickson RB, Johnson MD, Lippman M, Ethier S, Gazdar A, Gray JW. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10(6):515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Benson JD, Chen YN, Cornell-Kennon SA, Dorsch M, Kim S, Leszczyniecka M, Sellers WR, Lengauer C. Validating cancer drug targets. Nature. 2006;441(7092):451–456. doi: 10.1038/nature04873. [DOI] [PubMed] [Google Scholar]
- 39.Solomon DA, Kim JS, Ressom HW, Sibenaller Z, Ryken T, Jean W, Bigner D, Yan H, Waldman T. Sample type bias in the analysis of cancer genomes. Cancer Res. 2009;69(14):5630–5633. doi: 10.1158/0008-5472.CAN-09-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eschrich S, Zhang H, Zhao H, Boulware D, Lee JH, Bloom G, Torres-Roca JF. Systems biology modeling of the radiation sensitivity network: a biomarker discovery platform. Int J Radiat Oncol Biol Phys. 2009;75(2):497–505. doi: 10.1016/j.ijrobp.2009.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eschrich SA, Pramana J, Zhang H, Zhao H, Boulware D, Lee JH, Bloom G, Rocha-Lima C, Kelley S, Calvin DP, Yeatman TJ, Begg AC, Torres-Roca JF. A gene expression model of intrinsic tumor radiosensitivity: prediction of response and prognosis after chemoradiation. Int J Radiat Oncol Biol Phys. 2009;75(2):489–496. doi: 10.1016/j.ijrobp.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Torres-Roca JF, Eschrich S, Zhao H, Bloom G, Sung J, McCarthy S, Cantor AB, Scuto A, Li C, Zhang S, Jove R, Yeatman T. Prediction of radiation sensitivity using a gene expression classifier. Cancer Res. 2005;65(16):7169–7176. doi: 10.1158/0008-5472.CAN-05-0656. [DOI] [PubMed] [Google Scholar]
- 43.Spitzner M, Emons G, Kramer F, Gaedcke J, Rave-Frank M, Scharf JG, Burfeind P, Becker H, Beissbarth T, Ghadimi BM, Ried T, Grade M. A gene expression signature for chemoradiosensitivity of colorectal cancer cells. Int J Radiat Oncol Biol Phys. 2010;78(4):1184–1192. doi: 10.1016/j.ijrobp.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walther A, Johnstone E, Swanton C, Midgley R, Tomlinson I, Kerr D. Genetic prognostic and predictive markers in colorectal cancer. Nat Rev Cancer. 2009;9(7):489–499. doi: 10.1038/nrc2645. [DOI] [PubMed] [Google Scholar]
- 45.Markowitz SD, Bertagnolli MM. Molecular origins of cancer: molecular basis of colorectal cancer. N Engl J Med. 2009;361(25):2449–2460. doi: 10.1056/NEJMra0804588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Albertson DG, Collins C, McCormick F, Gray JW. Chromosome aberrations in solid tumors. Nat Genet. 2003;34(4):369–376. doi: 10.1038/ng1215. [DOI] [PubMed] [Google Scholar]
- 47.Grade M, Becker H, Liersch T, Ried T, Ghadimi BM. Molecular cytogenetics: genomic imbalances in colorectal cancer and their clinical impact. Cell Oncol. 2006;28(3):71–84. doi: 10.1155/2006/173815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ried T, Knutzen R, Steinbeck R, Blegen H, Schrock E, Heselmeyer K, du Manoir S, Auer G. Comparative genomic hybridization reveals a specific pattern of chromosomal gains and losses during the genesis of colorectal tumors. Genes Chromosomes Cancer. 1996;15(4):234–245. doi: 10.1002/(SICI)1098-2264(199604)15:4<234::AID-GCC5>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 49.Grade M, Ghadimi BM, Varma S, Simon R, Wangsa D, Barenboim-Stapleton L, Liersch T, Becker H, Ried T, Difilippantonio MJ. Aneuploidy-dependent massive deregulation of the cellular transcriptome and apparent divergence of the Wnt/beta-catenin signaling pathway in human rectal carcinomas. Cancer Res. 2006;66(1):267–282. doi: 10.1158/0008-5472.CAN-05-2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grade M, Hormann P, Becker S, Hummon AB, Wangsa D, Varma S, Simon R, Liersch T, Becker H, Difilippantonio MJ, Ghadimi BM, Ried T. Gene expression profiling reveals a massive, aneuploidy-dependent transcriptional deregulation and distinct differences between lymph node-negative and lymph node-positive colon carcinomas. Cancer Res. 2007;67(1):41–56. doi: 10.1158/0008-5472.CAN-06-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grade M, Gaedcke J, Wangsa D, Varma S, Beckmann J, Liersch T, Hess C, Becker H, Difilippantonio MJ, Ried T, Ghadimi BM. Chromosomal copy number changes of locally advanced rectal cancers treated with preoperative chemoradiotherapy. Cancer Genet Cytogenet. 2009;193(1):19–28. doi: 10.1016/j.cancergencyto.2009.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen Z, Liu Z, Li W, Qu K, Deng X, Varma MG, Fichera A, Pigazzi A, Garcia-Aguilar J. Chromosomal copy number alterations are associated with tumor response to chemoradiation in locally advanced rectal cancer. Genes Chromosomes Cancer. 2011;50(9):689–699. doi: 10.1002/gcc.20891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Malumbres M, Barbacid M. RAS oncogenes: the first 30 years. Nat Rev Cancer. 2003;3(6):459–465. doi: 10.1038/nrc1097. [DOI] [PubMed] [Google Scholar]
- 54.Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat Rev Cancer. 2011;11(11):761–774. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McKenna WG, Weiss MC, Bakanauskas VJ, Sandler H, Kelsten ML, Biaglow J, Tuttle SW, Endlich B, Ling CC, Muschel RJ. The role of the H-ras oncogene in radiation resistance and metastasis. Int J Radiat Oncol Biol Phys. 1990;18(4):849–859. doi: 10.1016/0360-3016(90)90407-B. [DOI] [PubMed] [Google Scholar]
- 56.Miller AC, Kariko K, Myers CE, Clark EP, Samid D. Increased radioresistance of EJras-transformed human osteosarcoma cells and its modulation by lovastatin, an inhibitor of p21ras isoprenylation. Int J Cancer. 1993;53(2):302–307. doi: 10.1002/ijc.2910530222. [DOI] [PubMed] [Google Scholar]
- 57.Bernhard EJ, McKenna WG, Hamilton AD, Sebti SM, Qian Y, Wu JM, Muschel RJ. Inhibiting Ras prenylation increases the radiosensitivity of human tumor cell lines with activating mutations of ras oncogenes. Cancer Res. 1998;58(8):1754–1761. [PubMed] [Google Scholar]
- 58.Gupta AK, Bakanauskas VJ, Cerniglia GJ, Cheng Y, Bernhard EJ, Muschel RJ, McKenna WG. The Ras radiation resistance pathway. Cancer Res. 2001;61(10):4278–4282. [PubMed] [Google Scholar]
- 59.Luna-Perez P, Segura J, Alvarado I, Labastida S, Santiago-Payan H, Quintero A. Specific c-K-ras gene mutations as a tumor-response marker in locally advanced rectal cancer treated with preoperative chemoradiotherapy. Ann Surg Oncol. 2000;7(10):727–731. doi: 10.1007/s10434-000-0727-0. [DOI] [PubMed] [Google Scholar]
- 60.Ondrejka SL, Schaeffer DF, Jakubowski MA, Owen DA, Bronner MP. Does neoadjuvant therapy alter KRAS and/or MSI results in rectal adenocarcinoma testing? Am J Surg Pathol. 2011;35(9):1327–1330. doi: 10.1097/PAS.0b013e3182253800. [DOI] [PubMed] [Google Scholar]
- 61.Zauber NP, Marotta SP, Berman E, Grann A, Rao M, Komati N, Ribiero K, Bishop DT (2009) Molecular genetic changes associated with colorectal carcinogenesis are not prognostic for tumor regression following preoperative chemoradiation of rectal carcinoma. Int J Radiat Oncol Biol Phys. doi:10.1016/j.ijrobp.2008.08.020 [DOI] [PubMed]
- 62.Gaedcke J, Grade M, Jung K, Schirmer M, Jo P, Obermeyer C, Wolff HA, Herrmann MK, Beissbarth T, Becker H, Ried T, Ghadimi M. KRAS and BRAF mutations in patients with rectal cancer treated with preoperative chemoradiotherapy. Radiother Oncol. 2010;94(1):76–81. doi: 10.1016/j.radonc.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Garcia-Aguilar J, Chen Z, Smith DD, Li W, Madoff RD, Cataldo P, Marcet J, Pastor C. Identification of a biomarker profile associated with resistance to neoadjuvant chemoradiation therapy in rectal cancer. Ann Surg. 2011;254(3):486–492. doi: 10.1097/SLA.0b013e31822b8cfa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guerrero S, Casanova I, Farre L, Mazo A, Capella G, Mangues R. K-ras codon 12 mutation induces higher level of resistance to apoptosis and predisposition to anchorage-independent growth than codon 13 mutation or proto-oncogene overexpression. Cancer Res. 2000;60(23):6750–6756. [PubMed] [Google Scholar]
- 65.Bazan V, Migliavacca M, Zanna I, Tubiolo C, Grassi N, Latteri MA, La Farina M, Albanese I, Dardanoni G, Salerno S, Tomasino RM, Labianca R, Gebbia N, Russo A. Specific codon 13 K-ras mutations are predictive of clinical outcome in colorectal cancer patients, whereas codon 12 K-ras mutations are associated with mucinous histotype. Ann Oncol. 2002;13(9):1438–1446. doi: 10.1093/annonc/mdf226. [DOI] [PubMed] [Google Scholar]
- 66.Horsch M, Recktenwald CV, Schadler S, Hrabe de Angelis M, Seliger B, Beckers J. Overexpressed vs mutated Kras in murine fibroblasts: a molecular phenotyping study. Br J Cancer. 2009;100(4):656–662. doi: 10.1038/sj.bjc.6604882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.De Roock W, Jonker DJ, Di Nicolantonio F, Sartore-Bianchi A, Tu D, Siena S, Lamba S, Arena S, Frattini M, Piessevaux H, Van Cutsem E, O’Callaghan CJ, Khambata-Ford S, Zalcberg JR, Simes J, Karapetis CS, Bardelli A, Tejpar S. Association of KRAS p.G13D mutation with outcome in patients with chemotherapy-refractory metastatic colorectal cancer treated with cetuximab. JAMA. 2010;304(16):1812–1820. doi: 10.1001/jama.2010.1535. [DOI] [PubMed] [Google Scholar]
- 68.Weiss C, Arnold D, Dellas K, Liersch T, Hipp M, Fietkau R, Sauer R, Hinke A, Rodel C. Preoperative radiotherapy of advanced rectal cancer with capecitabine and oxaliplatin with or without cetuximab: a pooled analysis of three prospective phase I–II trials. Int J Radiat Oncol Biol Phys. 2010;78(2):472–478. doi: 10.1016/j.ijrobp.2009.07.1718. [DOI] [PubMed] [Google Scholar]
- 69.Debucquoy A, Machiels JP, McBride WH, Haustermans K. Integration of epidermal growth factor receptor inhibitors with preoperative chemoradiation. Clin Cancer Res. 2010;16(10):2709–2714. doi: 10.1158/1078-0432.CCR-09-1622. [DOI] [PubMed] [Google Scholar]
- 70.Manolio TA, Brooks LD, Collins FS. A HapMap harvest of insights into the genetics of common disease. J Clin Invest. 2008;118(5):1590–1605. doi: 10.1172/JCI34772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Botstein D, Risch N. Discovering genotypes underlying human phenotypes: past successes for mendelian disease, future approaches for complex disease. Nat Genet. 2003;33(Suppl):228–237. doi: 10.1038/ng1090. [DOI] [PubMed] [Google Scholar]
- 72.Tomlinson I, Webb E, Carvajal-Carmona L, Broderick P, Kemp Z, Spain S, Penegar S, Chandler I, Gorman M, Wood W, Barclay E, Lubbe S, Martin L, Sellick G, Jaeger E, Hubner R, Wild R, Rowan A, Fielding S, Howarth K, Silver A, Atkin W, Muir K, Logan R, Kerr D, Johnstone E, Sieber O, Gray R, Thomas H, Peto J, Cazier JB, Houlston R. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat Genet. 2007;39(8):984–988. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- 73.Zanke BW, Greenwood CM, Rangrej J, Kustra R, Tenesa A, Farrington SM, Prendergast J, Olschwang S, Chiang T, Crowdy E, Ferretti V, Laflamme P, Sundararajan S, Roumy S, Olivier JF, Robidoux F, Sladek R, Montpetit A, Campbell P, Bezieau S, O’Shea AM, Zogopoulos G, Cotterchio M, Newcomb P, McLaughlin J, Younghusband B, Green R, Green J, Porteous ME, Campbell H, Blanche H, Sahbatou M, Tubacher E, Bonaiti-Pellie C, Buecher B, Riboli E, Kury S, Chanock SJ, Potter J, Thomas G, Gallinger S, Hudson TJ, Dunlop MG. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat Genet. 2007;39(8):989–994. doi: 10.1038/ng2089. [DOI] [PubMed] [Google Scholar]
- 74.Haiman CA, Le Marchand L, Yamamato J, Stram DO, Sheng X, Kolonel LN, Wu AH, Reich D, Henderson BE. A common genetic risk factor for colorectal and prostate cancer. Nat Genet. 2007;39(8):954–956. doi: 10.1038/ng2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Weinshilboum R. Inheritance and drug response. N Engl J Med. 2003;348(6):529–537. doi: 10.1056/NEJMra020021. [DOI] [PubMed] [Google Scholar]
- 76.Evans WE, McLeod HL. Pharmacogenomics—drug disposition, drug targets, and side effects. N Engl J Med. 2003;348(6):538–549. doi: 10.1056/NEJMra020526. [DOI] [PubMed] [Google Scholar]
- 77.Barnett GC, Coles CE, Elliott RM, Baynes C, Luccarini C, Conroy D, Wilkinson JS, Tyrer J, Misra V, Platte R, Gulliford SL, Sydes MR, Hall E, Bentzen SM, Dearnaley DP, Burnet NG, Pharoah PD, Dunning AM, West CM. Independent validation of genes and polymorphisms reported to be associated with radiation toxicity: a prospective analysis study. Lancet Oncol. 2012;13(1):65–77. doi: 10.1016/S1470-2045(11)70302-3. [DOI] [PubMed] [Google Scholar]
- 78.Villafranca E, Okruzhnov Y, Dominguez MA, Garcia-Foncillas J, Azinovic I, Martinez E, Illarramendi JJ, Arias F, Martinez Monge R, Salgado E, Angeletti S, Brugarolas A. Polymorphisms of the repeated sequences in the enhancer region of the thymidylate synthase gene promoter may predict downstaging after preoperative chemoradiation in rectal cancer. J Clin Oncol. 2001;19(6):1779–1786. doi: 10.1200/JCO.2001.19.6.1779. [DOI] [PubMed] [Google Scholar]
- 79.Terrazzino S, Agostini M, Pucciarelli S, Pasetto LM, Friso ML, Ambrosi A, Lisi V, Leon A, Lise M, Nitti D. A haplotype of the methylenetetrahydrofolate reductase gene predicts poor tumor response in rectal cancer patients receiving preoperative chemoradiation. Pharmacogenet Genomics. 2006;16(11):817–824. doi: 10.1097/01.fpc.0000230412.89973.c0. [DOI] [PubMed] [Google Scholar]
- 80.Conradi LC, Bleckmann A, Schirmer M, Sprenger T, Jo P, Homayounfar K, Wolff HA, Rothe H, Middel P, Becker H, Ghadimi MB, Beissbarth T, Liersch T. Thymidylate synthase as a prognostic biomarker for locally advanced rectal cancer after multimodal treatment. Ann Surg Oncol. 2011;18(9):2442–2452. doi: 10.1245/s10434-011-1608-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Spindler KL, Nielsen JN, Lindebjerg J, Jakobsen A. Germline polymorphisms may act as predictors of response to preoperative chemoradiation in locally advanced T3 rectal tumors. Dis Colon Rectum. 2007;50(9):1363–1369. doi: 10.1007/s10350-007-0264-z. [DOI] [PubMed] [Google Scholar]
- 82.Stoehlmacher J, Goekkurt E, Mogck U, Aust DE, Kramer M, Baretton GB, Liersch T, Ehninger G, Jakob C. Thymidylate synthase genotypes and tumour regression in stage II/III rectal cancer patients after neoadjuvant fluorouracil-based chemoradiation. Cancer Lett. 2008;272(2):221–225. doi: 10.1016/j.canlet.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 83.Hur H, Kang J, Kim NK, Min BS, Lee KY, Shin SJ, Keum KC, Choi J, Kim H, Choi SH, Lee MY. Thymidylate synthase gene polymorphism affects the response to preoperative 5-fluorouracil chemoradiation therapy in patients with rectal cancer. Int J Radiat Oncol Biol Phys. 2011;81(3):669–676. doi: 10.1016/j.ijrobp.2010.06.049. [DOI] [PubMed] [Google Scholar]
- 84.Tan BR, Thomas F, Myerson RJ, Zehnbauer B, Trinkaus K, Malyapa RS, Mutch MG, Abbey EE, Alyasiry A, Fleshman JW, McLeod HL. Thymidylate synthase genotype-directed neoadjuvant chemoradiation for patients with rectal adenocarcinoma. J Clin Oncol. 2011;29(7):875–883. doi: 10.1200/JCO.2010.32.3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schirmer MA, Nadine Mergler CP, Rave-Frank M, Herrmann MK, Hennies S, Gaedcke J, Conradi LC, Jo P, Beissbarth T, Hess CF, Becker H, Ghadimi M, Brockmoller J, Christiansen H, Wolff HA (2011) Acute toxicity of radiochemotherapy in rectal cancer patients: a risk particularly for carriers of the TGFB1 Pro25 variant. Int J Radiat Oncol Biol Phys. doi:10.1016/j.ijrobp.2011.05.063 [DOI] [PubMed]
- 86.Berardi R, Maccaroni E, Onofri A, Giampieri R, Pistelli M, Bittoni A, Scartozzi M, Pierantoni C, Mandolesi A, Bearzi I, Cascinu S. Locally advanced rectal cancer: from molecular profiling to clinical practice. A literature review: part 2. Expert Opin Pharmacother. 2009;10(15):2467–2478. doi: 10.1517/14656560903143784. [DOI] [PubMed] [Google Scholar]
- 87.Kapur P. Tailoring treatment of rectal adenocarcinoma: immunohistochemistry for predictive biomarkers. Anticancer Drugs. 2011;22(4):362–370. doi: 10.1097/CAD.0b013e3283433764. [DOI] [PubMed] [Google Scholar]
- 88.Petrocca F, Lieberman J. Promise and challenge of RNA interference-based therapy for cancer. J Clin Oncol. 2011;29(6):747–754. doi: 10.1200/JCO.2009.27.6287. [DOI] [PubMed] [Google Scholar]
- 89.Pecot CV, Calin GA, Coleman RL, Lopez-Berestein G, Sood AK. RNA interference in the clinic: challenges and future directions. Nat Rev Cancer. 2011;11(1):59–67. doi: 10.1038/nrc2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Altieri DC. Survivin, cancer networks and pathway-directed drug discovery. Nat Rev Cancer. 2008;8(1):61–70. doi: 10.1038/nrc2293. [DOI] [PubMed] [Google Scholar]
- 91.Rodel C, Haas J, Groth A, Grabenbauer GG, Sauer R, Rodel F. Spontaneous and radiation-induced apoptosis in colorectal carcinoma cells with different intrinsic radiosensitivities: survivin as a radioresistance factor. Int J Radiat Oncol Biol Phys. 2003;55(5):1341–1347. doi: 10.1016/S0360-3016(02)04618-7. [DOI] [PubMed] [Google Scholar]
- 92.Rodel F, Hoffmann J, Distel L, Herrmann M, Noisternig T, Papadopoulos T, Sauer R, Rodel C. Survivin as a radioresistance factor, and prognostic and therapeutic target for radiotherapy in rectal cancer. Cancer Res. 2005;65(11):4881–4887. doi: 10.1158/0008-5472.CAN-04-3028. [DOI] [PubMed] [Google Scholar]
- 93.Capalbo G, Dittmann K, Weiss C, Reichert S, Hausmann E, Rodel C, Rodel F. Radiation-induced survivin nuclear accumulation is linked to DNA damage repair. Int J Radiat Oncol Biol Phys. 2010;77(1):226–234. doi: 10.1016/j.ijrobp.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 94.Rodel F, Reichert S, Sprenger T, Gaipl US, Mirsch J, Liersch T, Fulda S, Rodel C. The role of survivin for radiation oncology: moving beyond apoptosis inhibition. Curr Med Chem. 2011;18(2):191–199. doi: 10.2174/092986711794088362. [DOI] [PubMed] [Google Scholar]
- 95.Sprenger T, Rodel F, Beissbarth T, Conradi LC, Rothe H, Homayounfar K, Wolff HA, Ghadimi BM, Yildrim M, Becker H, Rodel C, Liersch T (2011) Failure of down-regulation of survivin following neoadjuvant radiochemotherapy in rectal cancer is associated with distant metastases and shortened survival. Clin Cancer Res. doi:10.1158/1078-0432.CCR-10-2592 [DOI] [PubMed]
- 96.Moon RT, Kohn AD, De Ferrari GV, Kaykas A. WNT and beta-catenin signalling: diseases and therapies. Nat Rev Genet. 2004;5(9):691–701. doi: 10.1038/nrg1427. [DOI] [PubMed] [Google Scholar]
- 97.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127(3):469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 98.Kendziorra E, Ahlborn K, Spitzner M, Rave-Frank M, Emons G, Gaedcke J, Kramer F, Wolff HA, Becker H, Beissbarth T, Ebner R, Ghadimi BM, Pukrop T, Ried T, Grade M. Silencing of the Wnt transcription factor TCF4 sensitizes colorectal cancer cells to (chemo-) radiotherapy. Carcinogenesis. 2011;32(12):1824–1831. doi: 10.1093/carcin/bgr222. [DOI] [PMC free article] [PubMed] [Google Scholar]


