Abstract
Diabetes mellitus is a metabolic disorder caused due to insulin deficiency. Banana flower is a rich source of flavonoids that exhibit anti diabetic activity. Insulin receptor is a tetramer that belongs to a family of receptor tyrosine kinases. It contains two alpha subunits that form the extracellular domain and two beta subunits that constitute the intracellular tyrosine kinase domain. Insulin binds to the extracellular region of the receptor and causes conformational changes that lead to the activation of the tyrosine kinase. This leads to autophosphorylation, a step that is crucial in insulin signaling pathway. Hence, compounds that augment insulin receptor tyrosine kinase activity would be useful in the treatment of diabetes mellitus. The 3D structure of IR tyrosine kinase was obtained from PDB database. The list of flavonoids found in banana flower was obtained from USDA database. The structures of the flavonoids were obtained from NCBI Pubchem. Docking analysis of the flavonoids was performed using Autodock 4.0 and Autodock Vina. The results indicate that few of the flavonoids may be potential activators of IR tyrosine kinase.
Keywords: Diabetes mellitus, Banana flower, flavonoids, Insulin Receptor, Tyrosine kinase
Background
Diabetes mellitus is a chronic metabolic disorder caused due to insulin deficiency or insulin resistance. In Type 1 diabetes autoimmune destruction of the beta cells of pancreas leads to insulin deficiency. Type 2 diabetes involves insulin resistance or decreased insulin secretion. Insulin is essential for maintaining blood glucose and regulating carbohydrate metabolism. Insulin receptor (IR) is a tetrameric protein consisting of two extracellular alpha subunits and two transmembrane beta subunits [1].The binding of insulin to alpha subunit of IR causes conformational changes in the receptor leading to the activation of tyrosine kinase beta subunit. The activated IR has the ability to autophosphorylate and phosphorylate intracellular substrates that are essential for initiating other cellular responses of insulin [2–4].These events lead to the activation of downstream signaling molecules that participate in the insulin signaling pathway [5]. Insulin signaling, including activation of IR tyrosine kinase activity, is impaired in most patients with diabetes mellitus. This resistance to insulin then leads to hyperglycemia and other metabolic abnormalities of the disease [6, 7]. Hence, compounds that augment insulin receptor tyrosine kinase activity would be useful in the treatment of diabetes mellitus.
Bio-flavonoids are well-known for their multi-directional biological activities including anti-diabetic efficacy. The flavonoids, ubiquitous in plants, are the largest class of polyphenols, with a common structure of diphenylpropanes (C6-C3-C6), consisting of two aromatic rings linked through three carbons. The six major subclasses of flavonoids include the flavones (e.g. apigenin, luteolin), flavonols (e.g. quercetin, myricetin), flavanones (e.g. naringenin and hesperitin), flavanols (or catechins) (e.g. epicatechin and gallocatechin), anthocyanidins (e.g. cyanidin and pelargonidin) and isoflavones (e.g. genistein and daidezin) [8]. It has been demonstrated that flavonoids can act per se as insulin secretagogues or insulin mimetics, by influencing the pleiotropic mechanisms [9].
Musa sapientum commonly known as ‘banana’ is widely used in Indian folk medicine for the treatment of diabetes mellitus [10]. The available literature confirms that flavonoids are present in banana flowers [11].The chloroform, water and ethanol extract of Musa sapientum flowers were found to exhibit hypoglycaemic activity in alloxan diabetic rat [12]. Intraperitoneal administration of prunin (naringenin 7-O-β-D-glucoside) produces a significant hypoglycemic effect in diabetic rats. [13]. Chronic treatment with hesperitin and naringenin was found to lower the blood glucose level of db/db mice [14]. Banana flower extracts are able to promote glucose uptake into the cells, which could be beneficial in diabetes mellitus. It can be hypothesized that consumption of nutraceutical-rich extract of banana flower could replace some amount of insulin being taken for diabetes mellitus [15]. Although insulin has become one of the most important therapeutic agents known to medicine, there is a continuing effort to find insulin substitutes, secretagogues, or sensitizers from synthetic or plant sources for the treatment of diabetes mellitus [16]. In the present study we have attempted to dock the banana flower flavonoids with insulin receptor tyrosine kinase to understand the interactions. This insilico approach can be further investigated to generate more effective and potential insulin receptor tyrosine kinase activators through ligand based drug designing approaches.
Methodology
Preparation of Protein and ligands:
The three dimensional structure of Insulin Receptor of Homosapiens was obtained from Protein Database (PDB: ID 1IR3) [17] .The list of Flavonoids and other compounds present in banana flower were obtained from USDA database [18]. The CID files of the ligands were obtained from NCBI Pubchem [19]. The list of the ligands present in the banana flower initially used are Leucocyaniin (CID155206), Cyanidin (CID128861), Malvidin (CID159287), Pelargonidin(CID440832), Peonidin(CID441773), Hesperetin (CID 72281), Naringenin (CID 932), Hesperetin Triacetate (CID 457809), Hesperetin Dihydrochalcone ( CID 147608 ), Naringenin Pelargonidin ( CID 439246), Naringenin Flavanone (CID 25244584), Hesperetin (Anion) (CID 49859576). The energy minimization of the prepared ligand was carried out with Swiss-PDB Viewer V.4.02. [20] Molinspiration [21] an online tool, was used to perform QSAR studies in order to identify important molecular properties (logP, polar surface area, number of hydrogen bond donors and acceptors and others), as well as prediction of bioactivity score for the most important drug targets. Various properties of ligands such as logP, molecular weight, H bond donors, H bond acceptors, number of atoms were obtained. Lipinski’s Rule of Five was then applied to select probable ligands. Those compounds that had more than one violation (i.e Leucocyanidin) were eliminated. Active site analysis of the Insulin Receptor was carried out using Swiss PDB Viewer (SPDBV) V.4.02 and from the PDB ligand Explorer.
Molecular Docking Studies
Binding mode and selectivity of Insulin receptor tyrosine kinase with individual flavonoids, was studied by docking softwares like Autodock 4.0 [22], Autodock VINA [23]. Autodock 4.0 uses Monte Carlo simulated annealing and Lamarckian genetic algorithm (LGA) to create a set of possible conformations. LGA is used as a global optimizer and energy minimization as a local search method. Possible orientations are evaluated with AMBER force field model in conjunction with free energy scoring functions and a large set of protein-Ligand complexes with known protein-Ligand constants. The newest version 4 contains side chain flexibility. Hydrogen atoms, Kollman charges were added. The grid was centered in the active site region which involves all functional amino acid residues. Grid maps were generated using the Autogrid Program. Docking was performed using the Lamarckian genetic algorithm. In the present study docking was performed by creating an initial population of 150 individuals, 5 random torsions to each of the 150 individuals, Lamarckian Genetic Algorithm (LGA), was implemented with a maximum of 2500000 energy evaluations. AutoDock Vina is a new opensource program for drug discovery, molecular docking and virtual screening, offering multi-core capability, high performance and enhanced accuracy and ease of use. AutoDock Vina significantly improves the average accuracy of the binding mode predictions compared to AutoDock 4.0.
Discussion
We analyzed the interactions of the banana flower flavonoids with insulin receptor tyrosine kinase and observed that these compounds can activate the kinase domain since the phosphorylated tyrosines were bound to the residues located in the activated loop of the protein.
Structural Aspects and Interactions of IR Tyrosine kinase:
Insulin receptor is a tyrosine protein kinase that undergoes autophosphorylation upon insulin binding. The activated receptor phosphorylates substrate proteins on multiple Tyr residues to propagate insulin signal transduction and insulin action [24]. Three important structural regions are identified in the intracellular part of the beta subunit. They are juxtamembrane (JM) region, the kinase region, and the carboxyl-terminal (CT) region. The JM region (Arg940-Leu987) contains one autophosphorylation site; the CT region (Leu1245- Ser1343) contains two autophosphorylation sites. The intracellular portion of the insulin receptor, contain functional groups that bind to the effector molecules. Insulin receptor substrates (IRS-1, -2, -3 and -4) are the targets for insulin receptor kinase. The JM region serves as a binding site for IRS-1. Phosphorylation of the juxtamembrane of the insulin receptor mediates insulin's metabolic functions. The catalytic loop is formed by residues 1130-1137 and the activation loop comprises of residues 1150-1171 [25]. Extensive studies on activated and inactive IR tyrosine kinases indicate that in the activated state the activation loop is tris-phosphorylated and moves out of the active site. The phosphorylated tyrosines become hydrogenbonded to residues in the activation loop [26].
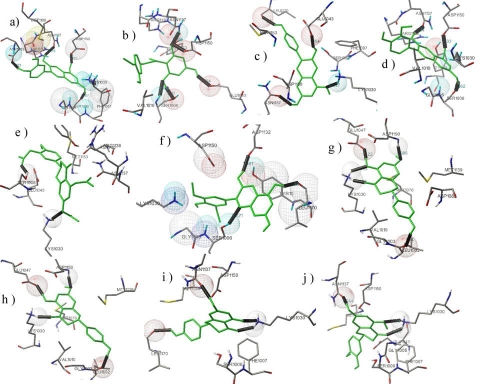
The molecular properties of ligands such as logP, molecular weight, H bond donors, H bond acceptors, number of atoms were obtained using Molinspiration. This tool also gives the analysis with respect to number of violations ie deviation from Lipinski's rule. Lipinski's Rule of Five was then applied to select probable ligands. Those compounds that had more than one violation (i.e Leucocyanidin) were eliminated Table 1 (see supplementary material). Active site analysis of the Insulin Receptor was carried out using Swiss PDB Viewer (SPDBV) V.4.02 and from the PDB ligand Explorer. The active site consists of residues: SER 1006, LYS 1030, GLU 1077, ASP 1083, ASN 1137, and ASP 1150, MET 1079. Docking results indicated that majority of the compounds bind to insulin receptor tyrosine kinase Table 2 (see supplementary material). The binding modes and interactions of each ligand with the functional residues of insulin receptor tyrosine kinase were analyzed in detail by visually inspecting the docked complexes using PMV 1.5.2.From (Figure 1) it is evident that all the ligands interact with most of the residues in the binding pocket. Cyanidin binding energy value from Autodock 4.0 was found to be -5.18 kcal/mol, Autodock Vina it was found to be - 7.9kcal/mol. This compound was found to interact with MET 1139, ASN 1137, ARG 1136, ASP 1083, ASP 1150, LYS 1030, SER 1006 and PHE 1007. Hesperetin Dihydrochalcone binding energy value from Autodock 4.0 was found to be -4.78 kcal/mol, Autodock Vina it was found to be -7.0 kcal/mol. This compound was found to interact with ARG 1136, ASN 1137, ASP 1150 and SER 1006. Pelargonidin binding energy from Autodock 4.0 was -4.93 kcal/mol, Autodock Vina it was found to be -7.7 kcal/mol. This compound was found to interact with ASN 1137, LYS 1030 and SER 1006. Peonidin binding energy from Autodock 4.0 was found to be -4.80 kcal/mol, Autodock Vina it was found to be -7.7 kcal/mol. This compound was found to interact with ASN 1137, LYS 1030 and ASP 1150.
Figure 1.
(a)Cyanidin interaction; (b)Hesperetin Dihydrochalcone interaction; (c)Pelargonidin Interaction; (d) Peonidin Interaction; (e)Hesperitin Triacetate interaction; (f)Malvidin Interaction; (g)Naringenin Interaction; (h)Naringenin Pelargonidin interaction; (i) Naringenin Flavanone interaction; (j)Hesperetin Interaction. Ligands represented in green are bound to tyrosine kinase visualized by PMV 1.5.2. Hydrogen bonds are indicated by black solid lines
Hesperetin triacetate binding energy value from Autodock 4.0 was found to be -6.08 kcal/mol, Autodock Vina it was found to be -8.2 kcal/mol. This compound was found to interact with SER 1006, ASN 1137, and LYS 1030. Malvidin binding energy from Autodock 4.0 was found to be -5.26 kcal/mol, Autodock Vina it was found to be -7.3 kcal/mol. This compound was found to interact with ASP 1150, LYS 1030 and SER 1006. Naringenin binding energy value from Autodock 4.0 was found to be -6.02 kcal/mol, Autodock Vina it was found to be -8.1 kcal/mol. This compound interacted with ASP 1150, ASP 1083, and LYS 1030. Naringenin pelargonidin binding energy value from Autodock 4.0 was found to be -6.02 kcal/mol, Autodock Vina it was found to be -8.1 kcal/mol. This compound was found to interact with ASP 1150 and LYS 1030 .Naringenin flavanone binding energy value from Autodock 4.0 was found to be -5.82 kcal/mol, Autodock Vina it was found to be -8.0 kcal/mol. This compound was found to interact with ASN 1137, ASP 1150, LYS 1030 and SER 1006. Hesperetin binding energy value from Autodock 4.0 was found to be -5.78 kcal/mol, Autodock Vina it was found to be -8.4 kcal/mol. This compound was found to interact with ASN 1137, ASP 1150 and LYS 1030 and SER 1006.
Conclusions
The docked conformations of all the flavonoids indicate that the phosphorylated tyrosines namely PTR -1158, PTR -1162 and PTR – 1163 remain in the activation loop .PTR -1158 was bound to ASP 1156, PTR-1162 was hydrogen bonded to THR 1160 ,ARG 1164 and PTR-1163 was bound to GLY 1166 all of which are a part of the activation loop. This structural orientation clearly suggests that insulin receptor tyrosine kinase in its active state. Hence it can be concluded that theses flavonoids can be considered as activators of Insulin Receptor Tyrosine kinase. Based on Autodock binding energies it can be hypothesized that Hesperitin triacetate,Naringenin,Naringenin pelargonidin and naringinen flavonone are potent activators of IR tyrosine kinase.
Supplementary material
Acknowledgments
We wish to thank the management of Sreenidhi Institute of Science and Technology for providing the necessary facilities to carryout this work.
Footnotes
Citation:Ganugapati et al, Bioinformation 8(5): 216-220 (2012)
References
- 1.ID Goldfine. Endocr Rev. 1987;8:235. doi: 10.1210/edrv-8-3-235. [DOI] [PubMed] [Google Scholar]
- 2.CR Kahn, MF White. J Clin Invest. 1988;82:1551. [Google Scholar]
- 3.CR Kahn, et al. Diabetes. 1994;43:1066. doi: 10.2337/diab.43.8.1066. [DOI] [PubMed] [Google Scholar]
- 4.MF White, et al. Recent Prog Horm Res. 1998;53:119. [PubMed] [Google Scholar]
- 5.RS Thies, et al. Diabetes. 1990;39:250. [Google Scholar]
- 6.A Virkamaki, et al. J Clin Invest. 1999;103:931. doi: 10.1172/JCI6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DE Moller, JS Flier. N Engl J Med. 1991;325:938. doi: 10.1056/NEJM199109263251307. [DOI] [PubMed] [Google Scholar]
- 8.JA Ross, CM Kasum. Annu Rev Nutr. 2002;22:19. doi: 10.1146/annurev.nutr.22.111401.144957. [DOI] [PubMed] [Google Scholar]
- 9.Brahmachari Goutam, et al. Medicinal Chemistry. 2011;661:187. [Google Scholar]
- 10.L Pari, J Umamaheswari. Phytother Res. 2000;14:136. doi: 10.1002/(sici)1099-1573(200003)14:2<136::aid-ptr607>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 11.GPP Lima, et al. Int J Food Sci Technol. 2008;43:1838. [Google Scholar]
- 12.Dhanabal, et al. J Herbal Pharmacother. 2005;5:7. [PubMed] [Google Scholar]
- 13.JS Choi, et al. Planta Med. 1991;57:208. doi: 10.1055/s-2006-960075. [DOI] [PubMed] [Google Scholar]
- 14.UJ Jung, et al. J Nutr. 2004;134:2499. doi: 10.1093/jn/134.10.2499. [DOI] [PubMed] [Google Scholar]
- 15.JJ Bhaskar, et al. J Physiol Biochem. 2011;67:415. doi: 10.1007/s13105-011-0091-5. [DOI] [PubMed] [Google Scholar]
- 16.PN Pushparaj, et al. Life Sci. 2001;70:535. [Google Scholar]
- 17. http://www.rcsb.org/pdb/home/home.do.
- 18. www.nal.usda.gov/fnic/foodcomp.
- 19. http://pubchem.ncbi.nlm.nih.gov/
- 20.N Guex, MC Peitsch. Protein Data Bank Quaterly Newsletter. 1996;77:7. [Google Scholar]
- 21. www.molinspiration.com.
- 22.GM Morris, et al. J Comput Chem. 2009;30:2785. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O Trott, AJ Olson. J Comput Chem. 2010;31:455. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.B Cheatham, CR Kahn. Endocr Rev. 1995;16:117. doi: 10.1210/edrv-16-2-117. [DOI] [PubMed] [Google Scholar]
- 25.K Paz, et al. J Biol Chem. 1996;271:6998. doi: 10.1074/jbc.271.12.6998. [DOI] [PubMed] [Google Scholar]
- 26.P De Meyts, J Whittaker. Nat Rev Drug Discov. 2002;1:769. doi: 10.1038/nrd917. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.