Abstract
Cardiac remodeling and hypertrophy are the pathological consequences of cardiovascular disease and are correlated with its associated mortality. Activity of the transcription factor NF-κB is increased in the diseased heart; however, our present understanding of how the individual subunits contribute to cardiovascular disease is limited. We assign a new role for the c-Rel subunit as a stimulator of cardiac hypertrophy and fibrosis. We discovered that c-Rel-deficient mice have smaller hearts at birth, as well as during adulthood, and are protected from developing cardiac hypertrophy and fibrosis after chronic angiotensin infusion. Results of both gene expression and cross-linked chromatin immunoprecipitation assay analyses identified transcriptional activators of hypertrophy, myocyte enhancer family, Gata4, and Tbx proteins as Rel gene targets. We suggest that the p50 subunit could limit the prohypertrophic actions of c-Rel in the normal heart, because p50 overexpression in H9c2 cells repressed c-Rel levels and the absence of cardiac p50 was associated with increases in both c-Rel levels and cardiac hypertrophy. We report for the first time that c-Rel is highly expressed and confined to the nuclei of diseased adult human hearts but is restricted to the cytoplasm of normal cardiac tissues. We conclude that c-Rel-dependent signaling is critical for both cardiac remodeling and hypertrophy. Targeting its activities could offer a novel therapeutic strategy to limit the effects of cardiac disease.
Cardiovascular disease underpins the development of cardiac hypertrophy and heart failure and is the primary cause of death in the developed world.1 During periods of acute physical and metabolic stress, the heart employs hemodynamic coping mechanisms, including increasing stroke volume and heart rate to meet the increased demand. In response to prolonged stress, the heart undergoes a physiological compensatory mechanism whereby it becomes enlarged (ie, cardiac hypertrophy). This process is governed by a series of biochemical and molecular changes in the heart, including cardiac remodeling and the reactivation of a group of genes collectively known as the fetal gene program.2–4 Recent discoveries in both animal models and the clinic suggest that cardiac hypertrophy is a dynamic process that may be reversible.5–7 Nonetheless, despite considerable research efforts, the complex signaling events regulating development and reversion of cardiac hypertrophy are not fully understood.
Nuclear factor-κB (NF-κB) is a pleiotropic transcription factor that, in addition to playing fundamental roles in immunity, also regulates the expression of genes controlling cell survival and fate.8 NF-κB activity is elevated during cardiovascular disease, and its signaling is strongly implicated in the development of cardiac remodeling (fibrosis), hypertrophy, and heart failure.9–13 The NF-κB/Rel family comprises five members: RelA (p65), NF-κB1 (p105/p50), NF-κB2 (p100/p52), c-Rel, and RelB. These are divided into two classes. Class I subunits are synthesized as precursors p105 and p100; these proteins are then proteolytically processed, yielding the p50 and p52 subunits, respectively. The full-length proteins contain ankyrin repeat domains and can act as inhibitory κB proteins. RelA (p65), c-Rel, and RelB proteins comprise the class II subunits. Importantly, only class II subunits contain a transactivation domain in the C-terminus allowing them to interact with the transcriptional machinery. The five subunits combine either as a homodimer or as heterodimers that bind to a decameric DNA consensus sequence known as the κB site to modulate gene transcription.8 NF-κB is activated via two pathways, the canonical (classical) and noncanonical pathway. Canonical signaling uses the RelA, p50, and c-Rel subunits, whereas activation of the noncanonical pathway is mediated by RelB and p100/p52. Clinical studies have linked canonical NF-κB signaling with susceptibility and progression of cardiac disease, in that increased nuclear RelA has been observed in failing human hearts,14,15 whereas NFKB1 gene polymorphisms are associated with an increased susceptibility to developing dilated cardiomyopathy.16,17
Global inhibition of this pathway using either pharmacological NF-κB inhibitors, transgenic mice, or expression of dominant negative IκB under the control of a cardiac specific promoter in rodent models of heart disease is cardioprotective.18–21 These data highlight NF-κB as a potential therapeutic target. However, recent data generated from studies using cardiac-specific Nemo (or IKKγ), a regulatory subunit of NF-κB,22 or IKKβ knockout mice,23 the upstream kinases regulating canonical NF-κB signaling, revealed that activation of RelA is critical for promoting myocyte survival and cardiac homeostasis. This suggests that long-term pan-blockade of IKK/RelA-dependent NF-κB signaling in the diseased heart is likely to be detrimental. However, NF-κB biology is complex, with multiple levels of control (including regulatory stimuli, activating kinases, post-translational modifications, and subunit composition).24–27 Each subunit is functionally distinct and has discrete biochemical characteristics. Growing evidence and knowledge now suggest that the Rel subunit is important in study of NF-κB signaling in the context of disease.28–33 Given that the RelA subunit is critical for cardiac homeostasis and therefore unlikely to be a good therapeutic target, we decided to investigate the roles of the two other canonical NF-κB subunits, c-Rel and NF-κB/p50, in cardiac disease. These may provide more selective therapeutic targets, leaving the cardioprotective actions of RelA signaling intact.
In the present study, we began by comparing cardiac expression of c-Rel in normal versus end-stage cardiomyopathic human hearts. We show for the first time that c-Rel was found in the nucleus of diseased but not normal hearts. We discovered that gene deletion of the c-Rel subunit in mice protects against development of stress-induced cardiac hypertrophy and fibrosis. Finally, we suggest that the p50 subunit may act to antagonize the prohypertrophic affects of c-Rel. With this report, we unravel some of the complexities of NF-κB system within heart pathophysiology and identify a new role for the c-Rel subunit as a positive regulator of cardiac hypertrophy. We suggest that the disease-related shift in balance from p50 toward c-Rel-dependent signaling has a positive influence on the expression of genes controlling the adaptive response of the stressed heart and that, unlike RelA, the c-Rel subunit may prove to be a good therapeutic target for the treatment of cardiovascular disease.
Materials and Methods
Mice
Experiments were performed on either mixed C57Bl/6;129PF2/J Nfkb1−/− and F2 hybrid Nfkb1+/+ wild-type (WT) control mice (Jackson Laboratory, Bar Harbor, ME) or genetically modified mice lacking Nfkb1 or Rel on a pure C57Bl/6 background and C57Bl/6 WT controls.34,35 Rel-deficient mice were a kind gift of Hsiou Chi-Liou (Weill Cornell Medical College). Mice were housed in pathogen-free conditions. Age-matched, 12-week-old or newborn (day 1) male Nfkb1−/−, Rel−/−, and WT control mice were weighed, then euthanized by cervical dislocation; the hearts were removed and weighed.
Blood Pressure Measurements
WT, Nfkb1−/−, and Rel−/− mice were anesthetized by intraperitoneal injection of a water solution with a fentanyl and fluanisone combination (Hypnorm; VetaPharma, Leeds, UK) and midazolam at the following doses, per gram of body weight: 3.8 μg fentanyl, 120 μg fluanisone, and 60 μg midazolam. A 1.4-French Millar Mikro-Tip pressure-volume catheter (ADInstruments, Oxford, UK) was introduced into the right carotid artery and placed in the ascending aorta. Systolic and diastolic pressures were recorded using PowerLab Chart5 software and analyzed using Millar PVAN data analysis software version 3 (ADInstruments). Data are reported as means ± SEM of five mice per group.
Angiotensin Infusion
Age-matched adult WT or Rel−/− mice were anesthetized with isoflurane, and Alzert 2004 mini-osmotic pumps (Durect, Cupertino, CA) containing either 0.9% sterile saline (vehicle) or angiotensin (700 μg/kg per day) were placed in the flank of the mouse. After 4 weeks of infusion, mice were weighed, then euthanized by cervical dislocation; the hearts were removed and weighed. All data are reported as means ± SEM of six WT and five Rel−/− mice per group.
Cell Culture
The rat embryonic cardiomyocyte cell line H9c2 (a kind gift from Deborah Henderson, Newcastle, UK) was cultured on plastic in Dulbecco's modified Eagle's medium, supplemented with 100 units/mL penicillin, 100 μg/mL streptomycin, 2 mmol/L l-glutamine, and 10% fetal calf serum, and was maintained at 37°C in an atmosphere of 5% CO2.
Human Left Ventricular Myocardium
Human left ventricular (LV) tissue was obtained from male patients undergoing cardiac transplantation for end-stage heart failure. Normal nondonor suitable human LV tissue was obtained from healthy male individuals involved in road traffic accidents. Tissue was formalin-fixed, processed, and embedded in paraffin blocks.
Histology, Immunohistochemistry, and Image Analysis
Sirius Red and H&E staining was performed as described previously.31 Photomicrographs were taken using a Leica DMR and JVC camera system (Leica Microsystems, Wetzlar, Germany). Densitometry and cell areas were calculated using the Leica Qwin image analysis system in at least six ×20 fields in the left ventricle. Immunohistochemical staining for mouse α-sarcomeric actin, laminin, and c-Rel was performed as follows. Slides were deparaffinized, and then citric saline antigen retrieval was performed. For c-Rel, a combination of citric saline and trypsin antigen retrieval was performed. Endogenous peroxidase activity was blocked using hydrogen peroxide, and further inhibition of nonspecific binding was achieved using an avidin/biotin blocking kit (Vector Laboratories, Peterborough, UK) followed by incubation with 20% swine serum (Dako, Ely, UK). Antibody specific to α-sarcomeric actin or laminin (Abcam, Cambridge, UK) was diluted 1:250 in PBS and c-Rel (Santa Cruz Biotechnology, Santa Cruz, CA) diluted 1:50 in PBS with 20% swine serum, then applied to the slides and incubated overnight at 4°C. Slides were washed and amplification of the antigen was achieved using a quick kit (Vector Laboratories) for α-sarcomeric actin or biotinylated swine anti-rabbit diluted 1:200 (Dako), followed by ABC complex (Vector Laboratories) for c-Rel and laminin. Slides were then washed and presence of α-sarcomeric actin, laminin, or c-Rel was visualized with diaminobenzidine staining. Slides were then counterstained with Mayer's hematoxylin, dehydrated, cleared in Clearene solvent (Leica Microsystems), and mounted in Pertex mounting medium (Leica Microsystems).
Total RNA Isolation and cDNA Synthesis
Total RNA was isolated from 10 to 15 μg of heart tissue or ∼5 × 106 H9c2 cells using a total RNA purification kit (Qiagen, Valencia, CA: Crawley, UK). Heart tissue was homogenized using a sterile pestle in Qiagen RLT lysis buffer, then 0.01% v/v proteinase K was added to the homogenate, followed by incubation at 55°C for 10 minutes. RNA was then isolated according to the manufacturer's instructions. RNA was treated with 1 μL DNase (Promega, Madison, WI) for 30 minutes at 37°C and first-strand cDNA was produced via incubation with random hexamer primer [p(dN)6] and 100 units MMLV reverse transcriptase, as described previously.31
SDS-PAGE and Immunoblotting
Murine heart tissue was lysed in Laemmli buffer, heated at 95°C for 10 minutes, sonicated, and centrifuged for 30 minutes at 16,000 × g. Total protein (10 to 30 μg) was fractionated by 9% SDS-PAGE and transferred to nitrocellulose. Blots were blocked with 0.1% TBS/Tween 20 containing 3% nonfat dried milk before overnight incubation with primary antibodies; c-Rel (rabbit, 1:1000; Santa Cruz Biotechnology), α-sarcomeric actin (mouse, 1:1000; Abcam), p105/p50 (rabbit, 1:1000; Abcam), or GAPDH (rabbit, 1:2000; Abcam). Membranes were washed in TBS/Tween 20 and incubated with either mouse anti-rabbit horseradish peroxidase conjugate (1:2000; Cell Signaling Technology, Danvers, MA) or goat anti-rabbit horseradish peroxidase conjugate (1:5000; Sigma-Aldrich, St. Louis, MO) for 2 hours. Blots were washed and antigen detected by enzymatic chemiluminescence (Amersham; GE Healthcare, Little Chalfont, UK).
SYBR Green Quantitative RT-PCR
Primers amplifying the mouse and rat target gene sequences of interest were designed using OLIGO software version 4.0 (Molecular Biology Insights, Cascade, CO) (Table 1). Gata4-specific primers were as described previously.36 Relative level of transcriptional difference between knockout and control mice was calculated as [1/(2A)] ×100, where A is the difference between mean wild-type CT and mean knockout CT after the GAPDH value has been deducted from the target gene for each animal.
Table 1.
SYBR Green Quantitative RT-PCR Primers
| Gene | Forward primer | Reverse primer |
|---|---|---|
| Rat/mouse Gapdh | 5′-GCACAGTCAAGGCCGAGAAT-3′ | 5′-GCCTTCTCCATGGTGGTGAA-3′ |
| mouse RelA | 5′-GCCCAGACCGCAGTATCC-3′ | 5′-GTCCCGCACTGTCACCTG-3′ |
| mouse RelB | 5′-CTGGCTCCCTGAAGAACC-3′ | 5′-CGCTCTCCTTGTTGATTC-3′ |
| mouse Rel | 5′-CTCTGCCTCCCATTGTTTCTA-3′ | 5′-GGCTTCCCAGTCATTCAACAC-3′ |
| mouse p52 | 5′-CTATCTGGTGATTGTGGA-3′ | 5′-GGCCGGTCCCTCATAGTT-3′ |
| mouse Mef 2A | 5′-TACAAATCACACGCATAA-3′ | 5′-GAAAATGATGAGTGCTAT-3′ |
| mouse Mef 2B | 5′-GCAACGCCTCTTCCAGTA-3′ | 5′-CAAGGAGCAGCGGGTAGG-3′ |
| mouse Mef 2C | 5′-ATGAGCGTAACAGACAGG-3′ | 5′-TTGGTGCTGTTGAAGATG-3′ |
| mouse Mef 2D | 5′-ATGGGGAGGAAAAAGATT-3′ | 5′-GGAGTGGTTGAAGATGAT-3′ |
| mouse Nkx2-5 | 5′-GCCAACAGCAACTTCGTGAACT-3′ | 5′-ACTCTGCACGGTGTTCAAGTCC-3′ |
| mouse Tbx-5 | 5′-CTGGCCTTAATCCCAAAACGA-3′ | 5′-GCCAGTTACGGACCATTTGTATC-3′ |
| mouse Tbx-20 | 5′-GAGCAGCTCCTCAACAGATGG-3′ | 5′-CCGTGGCTGGTACTTATGCATT-3′ |
| mouse Foxm1b | 5′-CCTGTCTCCTCCACTCCTA-3′ | 5′-ACTGGGCTGAAATCTAACC-3′ |
| mouse ANP | 5′-GCTCCTTCTCCATCACCCTG-3′ | 5′-ACCGGCATCTTCTCCTCCA-3′ |
| mouse BNP | 5′-TATCTGTCACCGCTGGGAGG-3′ | 5′-TTGTGAGGCCTTGGTCCTTC-3′ |
| rat Nkx2-5 | 5′-AGCGGCAGGACCAGACTC-3′ | 5′-CGTAGGCGGGAGCGTAGG-3′ |
| rat TBx5 | 5′-GGTCCGTAACTGGTAAAG-3′ | 5′-ATTTTCGTCTGCTTTCAC-3′ |
| rat TBx20 | 5′-CAGATGGTGTCCTTTGAA-3′ | 5′-CCGTGTGGTCTTTCTTCT-3′ |
| rat Mef 2A | 5′-CAACATTAGCAGAGTCCA-3′ | 5′-TATTCGCACCAGTATTTC-3′ |
| rat Mef 2B | 5′-AGGGGGACTGGGCACATC-3′ | 5′-GTGGTTAGGAAGGCGAAGC-3′ |
| rat Mef 2C | 5′-GCAGACGATTCAGTAGGT-3′ | 5′-CCTTTGTTTTCTTTCTTG-3′ |
| rat Mef 2D | 5′-TGTCCCCAAGTCGTGAAC-3′ | 5′-GCCGCATCCTCTTCACAG-3′ |
| rat GATA4 | 5′-GCAGGGCGGAGGCAGTGG-3′ | 5′-AGGCGGTGGGGTGTAAGC-3′ |
| rat ANP | 5′-ACCTGGAGGAGAAGATGCCG-3′ | 5′-TGTTGCAGCCTAGTCCGCTC-3′ |
| rat BNP | 5′-GTGCTGCCCCAGATGATT-3′ | 5′-GGTCTATCTTCTGCCCAAAG-3′ |
| rat Rel | 5′-TCGGTGTGTAAAGAAAAA-3′ | 5′-TCCATAGACTCGCTGACT-3′ |
| rat p50 | 5′-TGGTCACAAATGGAAAAA-3′ | 5′-ATAGGCAAGGTCAGAATG-3′ |
| rat RelB | 5′-AGCGAAAGCGGGGACTGC-3′ | 5′-GATGGTGGGAGGAACAGG-3′ |
| rat RelA | 5′-TCACCAAAGACCCACCTC-3′ | 5′-AGGGGTTATTGTTGGTCT-3′ |
| rat p52 | 5′-ATGCGGAGAATGAAGAGC-3′ | 5′-GCATCCCCCAGTGACAGC-3′ |
The annealing temperature was 55°C for all primers.
Plasmid DNA, Cell Transfection, and Reporter Assays
Overexpression plasmids encoding respiratory syncytial virus β galactosidase (RSV-β-gal), RSV-p50, and RSV-c-Rel, described previously,37 were a gift from Neil D. Perkins (Newcastle University, UK). Transfection of H9c2 cells with 3 μg of plasmid DNA was achieved using linear polyethylenimine (m.w., ∼25,000; Polysciences, Warrington, PA; Eppelheim, Germany) or Effectene (Qiagen) reagent, as described previously.37,38 An 828-bp sequence from the Gata4 promoter was amplified from rat genomic DNA using primers rat Gata4 PR Forward 5′-TTAGCTAGCTCAAACTCCTCTAATCTCAGCAA-3′ and rat Gata4 PR Reverse 5′-AACGATATCTGAAGTTGGATTACTGGAACAAT-3′. The forward primer contains a recognition site for NheI at the 5′ end and the reverse primer contains a cut site for EcoRV at the 3′ end. The PCR product was digested with NheI and EcoRV, purified, and then cloned into an NheI-EcoRV digested pGL4.17 vector (Promega) using T4 DNA ligase. The cloned promoter sequence was then verified using sequencing. H9c2 cells were cotransfected with 1 μg Gata4-luciferase and 3 μg of either RSV-β-gal, RSV-p50, or RSV-c-Rel and 10 ng of Renilla (pRLTK) vector using an Effectene transfection kit (Qiagen). Cells were lysed in passive lysis buffer 48 hours after transfection, and luciferase and Renilla activity was measured using a Stop & Glo kit (Promega).
Cross-Linked Chromatin Immunoprecipitation Assay
A cross-linked chromatin immunoprecipitation (ChIP) assay was performed using 100 μg cross-linked chromatin prepared from H9c2 cells as described previously.38 In each ChIP reaction, 10 μg of antibody raised against c-Rel (Santa Cruz Biotechnology), p50 (Abcam) or appropriate irrelevant antibody control was used. The 3500-bp sequence upstream of ATG for each gene was taken from the ENSEMBL database, and putative transcription factor binding sites were predicted in silico using Promo (available via the ALGGEN server, Polytechnic University of Catalonia, Barcelona, Spain) Transfac 8.3 and TFSearch software (see Supplemental Table S1 at http://ajp.amjpathol.org). ChIP primers for rat brain natriuretic peptide (BNP), rat atrial natriuretic peptide (ANP), rat Gata4, rat Mef2A promoters, and Tgfβ1 exon 1-intron 1 boundaries were specific. The sequences were as follows. Rat BNP site 1 Forward 5′-CTATACAAGGCCTGCGGTTT-3′ and reverse 5′-TGCCTCTGCTTTATCCTG-3′; rat ANP forward 5′-GAGAGGAGCTGGACCATGAG-3′ and reverse 5′-CCCAGCATCCACATAAAAGC-3′; rat Mef2A forward 5′-TCTGCCCTGACATTCCCTAC-3′ and reverse 5′-AGAGCTCCCAGGAACTACCC-3′; rat Gata4 sites 1 to 4 forward 5′-AGAGGGGCTTTTCGGTAGAA-3′ and reverse 5′-GAAAGTCCGAAGCAGTGTCC-3′; rat Gata4 site 5 forward 5′-GTGCTCAGTCCTTCCTCTGG-3′ and reverse 5′-TGGAATACCACCCGGTAAGA-3′; and rat Tgfβ1 exon 1-intron 1 forward 5′-GACCTGCTGGCAATAGCTTC-3′ and reverse 5′-ACGGGAGTGGGAGCAGAA-3′. The annealing temperature was 55°C. Primers recognizing the PPARγ exon 1-intron 1 boundaries have been described previously.39 Each PCR reaction was performed in triplicate, and the analysis was repeated in at least three independent ChIP experiments. A signal intensity value for each sample was calculated from the mean of the experiments.
Statistical Analysis
P values were calculated using a two-tailed unpaired Student's t-test or one-way analysis of variance with Tukey's post hoc test or Bonferroni's test for individual subgroup comparison if required. In all tests, P < 0.05 was used as the criterion for statistical significance.
Ethics
All animal experiments were approved by the local ethical review committee and were conducted under a United Kingdom Home Office license. All human LV tissue was collected by a protocol approved by the Papworth (Cambridge) Hospital Tissue Bank review board and the Cambridgeshire Research Ethics Committee (Reference: 06/Q0104/64). Written consent was obtained from every individual according to the Papworth Tissue Bank protocol.
Results
c-Rel Positively Regulates Cardiac Growth in Mouse and Human
The canonical or classical NF-κB signaling pathway uses three of the five family members: RelA, c-Rel, and p50. Of these, only RelA and c-Rel contain a transactivation domain.26 The p50 subunit lacks a transactivation domain, and therefore p50 homo-dimers are unable to stimulate gene transcription unless complexed with a coactivator.27,32 Previous studies have reported an increase in RelA activation in the diseased heart,14,15 but studies using genetically modified mice suggest that therapeutic targeting of this subunit or its upstream kinases could be detrimental, given that RelA signaling plays a vital role in preventing cardiomyocyte apoptosis in response to cardiac damage.22,23 However, the role of the other transactivating subunit, c-Rel, in cardiac pathology has not been characterized previously.
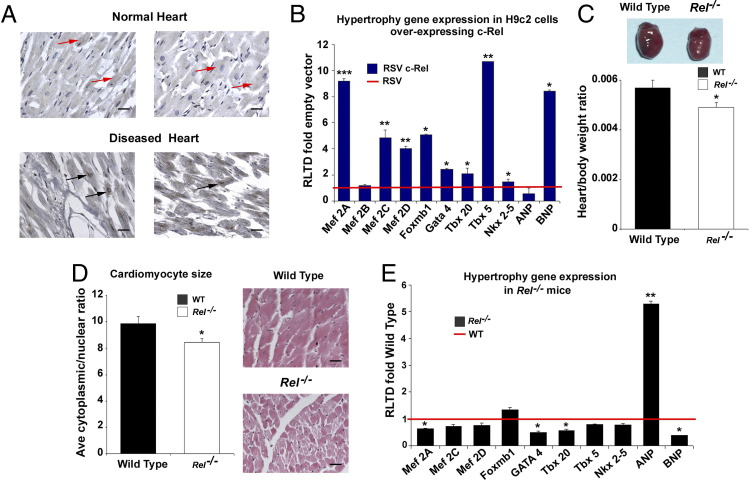
To ascertain whether c-Rel-dependent signaling plays a role in the pathogenesis of cardiac disease, we performed immunohistochemical staining for c-Rel in LV tissue from normal and diseased human hearts (Figure 1A). c-Rel was confined primarily to the nucleus in cardiomyocytes in patients with end-stage ischemic or idiopathic dilated cardiomyopathy. Conversely, in normal, nondiseased heart tissue we observed comparatively lower levels of c-Rel, restricted to the cytoplasm. We hypothesize that the nuclear localization of c-Rel in the diseased heart alters the transcriptional activity of target genes and contributes to disease pathogenesis. To identify candidate prohypertrophic target genes under the control of c-Rel, we overexpressed c-Rel in the neonatal rat cardiac cell line H9c2. These experiments revealed that c-Rel induces multiple genes that promote cardiac hypertrophy, including transcriptional stimulators of hypertrophy (myocyte enhancer factor proteins Mef2A, Mef2C, and Mef2D, Gata4, and Tbx5) and the cardioprotective factor BNP (Figure 1B). Levels of Foxm1b, a transcription factor reported to regulate lung and heart cell size and promote liver regeneration40,41 were also elevated when c-Rel was overexpressed. These data led us to hypothesize that cardiac deletion of c-Rel would result in a reduction in heart size.
Figure 1.
c-Rel regulates cardiac growth in mouse and human. A: Human LV tissue from normal and diseased hearts was stained for c-Rel protein. Black arrows indicate nuclear staining; red arrows indicate cytoplasmic staining. Photomicrographs are representative of five normal and five diseased human hearts. H9c2 cells were transiently transfected with RSV-β-gal (control plasmid) or RSV-c-Rel. B: Quantification of mRNA showed that c-Rel positively regulates cardiac expression of transcriptional activators of hypertrophy and hypertrophic markers. Relative level of transcriptional difference RLTD was expressed as mean fold change ± SEM, compared with RSV, of five independent transfections. C: Representative images of hearts isolated from adult WT (left) and Rel−/− mice (right) show decreased heart size in the knockout mice. Cardiac hypotrophy is further highlighted by a decrease in heart/body weight ratio in mice lacking Rel, compared with WT. Results are expressed as mean ratio change in Rel−/− mice compared with WT ± SEM; n = 20 mice/group. D: Cardiomyocyte size is reduced in Rel−/− mice, compared with controls. Photomicrographs show heart sections from WT and Rel−/− mice, stained with hematoxylin and eosin. Original magnification, ×400. Scale bars: 100 μm. Image analysis was used to calculate mean cytoplasmic/nuclear area ratios. Results are expressed as mean ratio change compared with WT ± SEM; n = 5 mice/group. E: Eight of the 10 genes measured were down-regulated in Rel−/− mice, compared with WT. Data are expressed as mean RLTD fold change ± SEM, relative to WT; n = 5 mice/genotype. All P values were calculated using a one-way analysis of variance or an unpaired two-tailed Student's t-test. *P = 0.05, **P = 0.01, and ***P = 0.001.
To test this idea, we isolated hearts from normal, nonstressed adult Rel−/− and WT mice. Rel-deficient mice are viable and apparently normal, apart from some modest immune defects, attenuated responses to liver injury, and certain bacterial infections. We observed that the Rel−/− hearts appeared to be smaller than the WT controls (Figure 1C). This was verified by a significant reduction in heart/body weight ratio in Rel knockout mice, compared with WT controls. Blood pressure did not differ significantly between Rel knockout and WT mice (see Supplemental Table S2 at http://ajp.amjpathol.org), suggesting that the cardiac phenotype is not a consequence of hemodynamic changes. The reduction in overall heart size and heart/body weight ratio was accompanied by a decrease in the cytoplasmic/nuclear ratio of cardiomyocytes in Rel-deficient hearts (Figure 1D). To determine whether the reduction in heart size is a feature of development or a postnatal phenotype, we measured the heart/body weight ratio in newborn mice. This ratio was also significantly reduced in Rel−/− newborn mice, confirming that c-Rel can control cardiac growth during development as well as in adult life (see Supplemental Figure S1A at http://ajp.amjpathol.org).
To determine whether c-Rel controls genes that regulate cardiomyocyte size, we next measured expression of hypertrophy-associated genes in both genotypes. The transcription factors Mef2A, Mef2C, Mef2D, Gata4, Tbx5, and Tbx20 were expressed at reduced levels in Rel-deficient heart (Figure 1E). Of note, all of these genes were significantly up-regulated by overexpression of Rel (Figure 1B). BNP was expressed at reduced levels in Rel−/− hearts (Figure 1E); by contrast, ANP levels were elevated, which may indicate that a compensatory mechanism operates to ensure appropriate physiological levels of this natriuretic peptide.
Gene Deletion of c-Rel Protects Against Angiotensin-Induced Cardiac Hypertrophy and Fibrosis
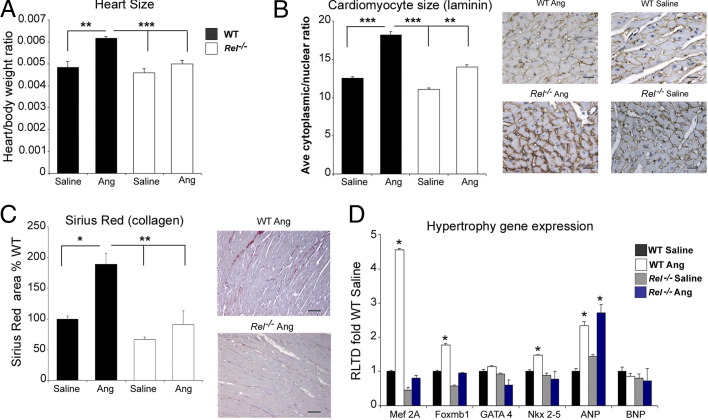
Our data show that c-Rel stimulates cardiac growth under normal physiological conditions and that it is confined to the nucleus in human diseased hearts. We therefore asked whether c-Rel-dependent gene transcription is required for the development of cardiac hypertrophy. To test this hypothesis, we subjected both Rel−/− and WT mice to 4 weeks of angiotensin or saline (vehicle) infusion via a mini-osmotic pump. Prolonged exposure to angiotensin causes hypertension and chronic pressure overload, triggering cardiac remodeling, which in turn results in cardiac hypertrophy and fibrosis. Calculation of heart/body weight ratio confirmed the reduction in heart size in saline-infused Rel−/− mice, compared with the WT (Figure 2A). Chronic angiotensin infusion caused a 27% increase in heart/body weight ratio in WT mice. In Rel−/− mice, however, the ratio was increased by 9%, only a third of that observed in WT animals. This cardioprotective effect was further supported by a statistically significant reduction in the cytoplasmic/nuclear area ratio of LV cardiomyocytes in Rel−/− mice, compared with WT mice, treated with angiotensin, as shown by laminin immunostaining (Figure 2B; see also Supplemental Figure S1B at http://ajp.amjpathol.org).
Figure 2.
Rel knockout mice are protected from angiotensin-induced cardiac hypertrophy and fibrosis. A: The heart/body weight ratio was calculated in Rel−/− and WT mice after 4 weeks of saline vehicle or angiotensin (Ang) infusion. B: Mean cytoplasmic/nuclear area ratio was calculated using image analysis of laminin-stained hearts. Data are expressed as mean percentage change, compared with WT. Cardiomyocyte size was increased in WT mice after angiotensin, indicative of a hypertrophic response. By contrast, cardiomyocytes of angiotensin-treated Rel−/− mice were smaller than in the WT, suggesting a cardioprotective effect. Representative photomicrographs show heart sections from angiotensin-infused WT and Rel−/− mice immunostained with anti-laminin antibodies. Cardiac remodeling and fibrosis was reduced in angiotensin-infused Rel−/− mice, compared with WT. Original magnification, ×400. Scale bars: 100 μm. C: Densitometric analysis of Sirius Red-stained heart tissues showed a statistically significant increase in collagen deposition in WT mice after angiotensin infusion; however, less collagen was observed in both the saline and angiotensin-infused Rel−/− mice. Representative photomicrographs show Sirius Red-stained heart sections from angiotensin-treated WT and Rel−/− mice. Data are expressed as mean ratio or percentage change ± SEM; n = 5 (Rel−/−) and n = 6 (WT) mice/group. D: Relative mRNA levels of hypertrophy-associated genes were determined in Rel−/− and WT mice after angiotensin infusion. Angiotensin infusion induces cardiac expression of transcriptional regulators of hypertrophy; however, this response is attenuated in Rel−/− mice. The RLTD was calculated between WT and Rel−/− mice and expressed as a mean fold change ± SEM, relative to WT; n = 4 mice/genotype. All P values were calculated using a one-way analysis of variance. *P = 0.05, **P = 0.01, and ***P = 0.001.
Cardiac remodeling and fibrosis are features of this model. Morphometric analysis of Sirius Red staining for collagen revealed that mice lacking Rel develop significantly less fibrosis than control mice (Figure 2C). We also noted a failure to up-regulate expression of key prohypertrophic transcription factors, including Mef2A and Nkx2-5, and a suppression of basal Gata4 levels in chronic angiotensin-treated Rel−/− mice. The inability to induce Mef2A expression after angiotensin treatment in these mice is the likely explanation for the observed increase in levels of the cardioprotective peptide ANP; Naya et al42 reported that ANP expression is elevated in Mef2A-deficient mice. Up-regulation of ANP is a normal physiological response to protect the heart from damage caused by pressure overload. Elevated expression of ANP could be another mechanism conveying protection to Rel-deficient mice from chronic angiotensin infusion-induced hypertrophy (Figure 2D).
c-Rel Directly Binds Hypertrophy-Related Gene Promoters
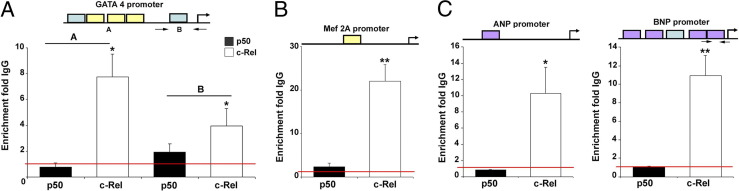
To determine whether c-Rel directly regulates genes that promote cardiac hypertrophy or whether the effects observed in vitro and in vivo are indirect, we performed ChIP assays. With this technique, we were able to demonstrate a physical interaction between Rel and κB regions of the Gata4 (Figure 3A) and Mef2A (Figure 3B) gene promoters, providing further evidence for c-Rel as a transcriptional regulator of prohypertrophic genes. To determine whether c-Rel or p50 can transactivate the Gata4 promoter, we cotransfected H9c2 cells with a Gata4-luciferase construct and either RSV-β-gal, RSV-p50, or RSV-c-Rel. Overexpression of c-Rel increased activity of the Gata4-luciferase reporter construct by twofold, whereas overexpression of p50 was without effect, suggesting that c-Rel but not p50 is a critical transcriptional regulator of the Gata4 gene (see Supplemental Figure S2A at http://ajp.amjpathol.org).
Figure 3.
c-Rel binds to hypertrophy-related gene promoters. Cross-linked ChIP analysis revealed binding of c-Rel to the promoters of transcriptional activators of hypertrophy (A and B) and to cardioprotective factors (C). In the promoter schematics, blue boxes indicate NF-κB-c-Rel consensus sequences, yellow boxes indicate preferential c-Rel consensus sequences, and purple boxes indicate NF-κB consensus sequences. H9c2 cells were formalin-fixed. Chromatin was then isolated and sheared by sonication, and 100 μg of chromatin was incubated with an anti-c-Rel antibody, anti-p50 antibody, or irrelevant IgG isotype control. Immunoprecipitation reactions were performed, proteins were digested, and cross-links were reversed before purification of genomic DNA and qRT-PCR amplification of promoters of interest. Binding was normalized to total input genomic DNA and is expressed as fold IgG control. Data are representative of at least three separate experiments. All P values were calculated using one way analysis of variance. *P = 0.05, **P = 0.01.
To rule out nonspecific interaction of the c-Rel antibody with the transcriptional machinery, we performed ChIP analysis on the exon 1-intron 1 boundaries of Tgfb1 and Pparg, two control genes that lack c-Rel binding sites. We did not observe an interaction of p50 or c-Rel with either gene, suggesting the interactions with Gata4, Mef2A, and the natriuretic peptides are specific (Supplemental Figure S2B at http://ajp.amjpathol.org). This identifies c-Rel but not p50 as the critical NF-κB subunit regulating the Mef2A promoter. These data are consistent with our observations in the angiotensin infusion model, in which Rel gene-deficient mice fail to induce expression of Mef2A in response to angiotensin. c-Rel was also recruited to the gene promoters of the cardioprotective peptides ANP and BNP (Figure 3C). These results verify that c-Rel not only promotes transcription of hypertrophy-associated genes but also directly binds their promoters.
The p50 Subunit of NF-κB Represses c-Rel Expression and Limits Cardiac Hypertrophy and Fibrosis
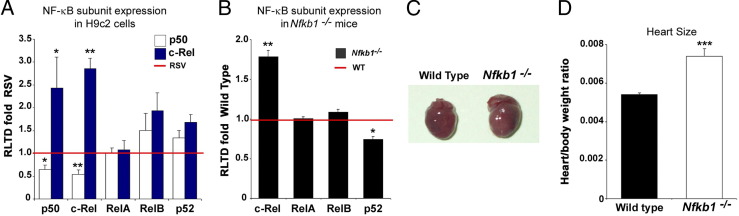
Regulation of c-Rel at the transcriptional level is poorly defined. To determine whether overexpression of either p50 or c-Rel affects endogenous expression of the five NF-κB subunits, we overexpressed p50 and c-Rel in H9c2 cells and measured each family member at the RNA level. Our data revealed that p50 can repress the expression of c-Rel, whereas overexpression of c-Rel leads to its own positive regulation (Figure 4A). These data suggest that a function of p50 in the normal heart may be to limit expression of c-Rel and limit its influence on cardiac physiology. To investigate this further, we compared NF-κB subunit expression in the hearts of WT and Nfkb1 (p50) knockout mice. Our data confirmed that c-Rel is repressed by p50 (Figure 4B; see also Supplemental Figure S3A at http://ajp.amjpathol.org), given that mice lacking this subunit have elevated cardiac levels of c-Rel. We then predicted that hearts lacking p50 may be enlarged, because the repressive effects of p50 have been lost.
Figure 4.
p50 suppresses the prohypertrophic effects of c-Rel. H9c2 cells were transiently transfected with expression plasmids (control RSV-β-gal, RSV-p50, or RSV-c-Rel). RNA was isolated, and cDNA was generated and used as a template for qRT-PCR. The RLTD was calculated and expressed as mean fold change ± SEM, relative to RSV, of five independent transfections. A: Gene expression levels of the individual NF-κB subunits revealed that overexpression of p50 repressed expression of c-Rel. B: RNA was isolated from the hearts of adult WT and Nfkb1−/− mice, and mRNA levels of the NF-κB subunits RelA, NF-κB2, c-Rel, and RelB were quantified using qRT-PCR. c-Rel was the only NF-κB subunit to be up-regulated in Nfkb1−/− mice. The RLTD between WT and Nfkb1−/− mice was calculated and expressed as a mean fold change ± SEM relative to WT; n = 5 mice/genotype. *P = 0.05, **P = 0.01. C: Representative photographs of hearts isolated from adult WT and Nfkb1−/− mice show increased heart size in Nfkb1−/− mice. D: The heart/body weight ratio was calculated in Nfkb1−/− and WT control mice on a pure C57Bl/6 background; data are expressed as mean percentage change ± SEM, compared with WT; n = 17 mice/genotype. An increase in heart/body weight ratio was observed in male Nfkb1−/− mice, compared with WT controls, in both backgrounds. ***P < 0.001.
To test this idea, we isolated hearts from Nfkb1 (p50−/−) gene knockout and WT mice. Hearts from Nfkb1−/− mice were indeed larger than the WT controls, as confirmed by heart/body weight ratios (Figure 4, C and D). This prohypertrophic phenotype was conserved in two different mouse genetic backgrounds (Figure 4D; see also Supplemental Figure S3B at http://ajp.amjpathol.org). Hypertrophy of Nfkb1−/−hearts was confirmed by a statistically significant increase in the number of sarcomeric units and cardiac sarcomeric actin levels (Figure 5A; see also Supplemental Figure S3A at http://ajp.amjpathol.org), as well as by an increase in the cytoplasmic/nuclear ratio (Figure 5B; see also Supplemental Figure S3C at http://ajp.amjpathol.org). There was no difference in blood pressure between the two genotypes, suggesting that the spontaneous development of cardiac hypertrophy in Nfkb1−/− mice is not driven by pre-load or after-load effects on the heart (see Supplemental Table S1 at http://ajp.amjpathol.org). For newborn mice, however, we found no evidence of difference in heart/body ratio, suggesting that hypertrophy develops with adulthood (see Supplemental Figure S3D at http://ajp.amjpathol.org).
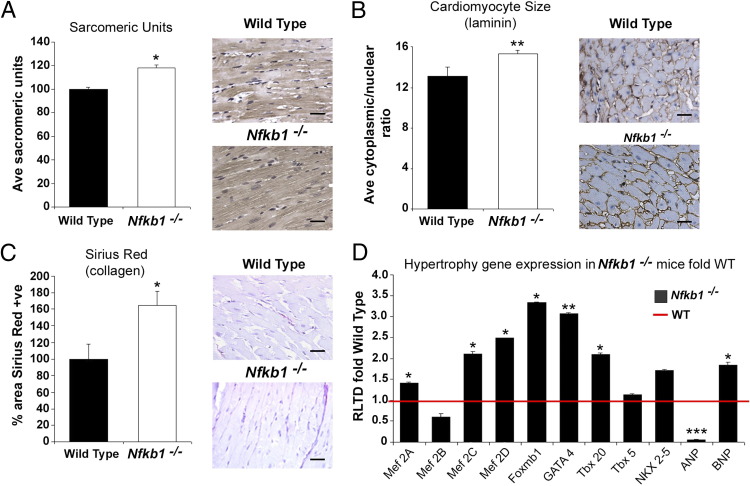
Figure 5.
Features of cardiac hypertrophy and fibrosis are observed in Nfkb1 knockout mice. A: Heart sections were stained for α-sarcomeric actin and sarcomeric units were counted. The mean number (Ave) of sarcomeric units in Nfkb1−/− mice was significantly increased, compared with WT. *P = <0.05. B: Representative photomicrographs of laminin-stained WT and Nfkb1−/− mouse hearts. Mean cardiomyocyte cytoplasmic/nuclear area ratios were calculated using image analysis software. The ratio was significantly increased in Nfkb1−/− hearts, compared with WT. **P < 0.01. C: Representative photomicrographs of Sirius Red-stained heart sections from WT and Nfkb1−/− mice. Densitometric analysis revealed a statistically significant increase in collagen deposition (red fibers) in Nfkb1−/− mice. *P = <0.05. Data are expressed as means ± SEM of eight WT and six Nfkb1−/− mice. +ve, positive. D: Relative mRNA levels of the transcriptional regulators of hypertrophy myocyte enhancer factor 2 (Mef2) A, C, and D (but not Mef2B), Gata4, and Foxm1b were elevated in Nfkb1−/− mice, compared with WT. Deletion of Nfkb1 was associated with an increase in gene expression of Nkx2-5 and of Tbx20. Cardiac mRNA levels of the cardioprotective proteins brain natriuretic peptide (BNP) and atrial natriuretic peptide (ANP) were quantified. BNP expression was increased in the Nfkb1−/− mice, compared with WT; however, expression of ANP was barely detectable in Nfkb1−/− mice. RLTD was calculated between WT and Nfkb1−/− mice and expressed as mean fold change ± SEM, relative to WT; n = 5 mice/genotype. Original magnification, ×400. Scale bars: 100 μm.
Cardiac remodeling and fibrosis are common features of heart damage and disease. Sirius Red staining revealed spontaneous development of fibrosis in the hearts of 12-week-old Nfkb1−/− mice (Figure 5C). We next sought to determine whether transcription factors that drive cardiac hypertrophy are up-regulated in the hearts of Nfkb1−/− mice. Levels of cardiac gene expression of Mef2 isoforms 2A, 2C and 2D were increased in Nfkb1−/− mice, compared with controls, and all were down-regulated in Rel-deficient mice. Gata4 is a key transcriptional regulator linked to the development of cardiac hypertrophy, and its expression was also elevated in these mice (Figure 5D). We also observed an increased expression of Foxm1b in Nfkb1−/− mice (Figure 5D), which may be highly relevant, given the role previously suggested for this factor in organ hypertrophy. Consistent with these data, markers of cardiac hypertrophy Nkx2-5 and TBx20 were also elevated in Nfkb1−/− hearts.
The vasoactive hormones BNP and ANP are produced by cardiomyocytes and are cardioprotective.43 BNP levels were modestly elevated in Nfkb1−/− heart; by contrast expression of ANP was markedly suppressed. Of note, genes for Mef2A, Gata4, and the natriuretic peptide BNP were not only elevated in the Nfkb1−/− hearts, but were also identified as c-Rel target genes in the ChIP analysis.
Discussion
Activation of NF-κB is strongly linked with the development of cardiac growth and hypertrophy, and blockade of this pathway has been proposed as a promising approach for prevention of adverse cardiac remodeling.18–20 This idea is supported by clinical observations and in rodent models of heart disease. Clinically, increased nuclear translocation of the RelA subunit has been observed in end-stage dilated cardiomyopathy. Furthermore, Santos et al16 recently reported a polymorphism in the Nfkb1 gene that leads to reduced gene activation, which is associated with an increase in onset and progression of cardiac remodeling, cardiac deterioration, and heart failure in patients with dilated cardiomyopathy. In rodent models of cardiac hypertrophy involving angiotensin II, isoproterenol, and aortic-banding, significant cardioprotective effects were observed either in mice expressing an NF-κB super-repressor or in rodents treated with pan NF-κB or IKK inhibitors.44,45 Other studies, however, have indicated that NF-κB signaling is cardioprotective in heart disease, particularly those linked to the RelA subunit, because inhibiting the actions of this subunit is likely to severely affect cardiomyocyte homeostasis and survival.22,23,27,46 Hence, long-term pan-blockade of NF-κB signaling could be detrimental. An alternative approach would be to target one or more of the other four NF-κB subunits. Here, we have described for the first time a pivotal role for c-Rel as a stimulator of cardiac hypertrophy and remodeling.
Our data suggest that c-Rel is predominantly found in the cytoplasm of cardiomyocytes in the normal human heart, whereas its localization is primarily nuclear in the diseased human heart. Mice lacking c-Rel develop with reduced heart size and are protected from angiotensin-induced cardiac hypertrophy and fibrosis. We further suggest that the p50 subunit limits the influence of c-Rel on cardiac physiology by suppressing its expression. The importance of this crosstalk between the two subunits was highlighted by our observation of spontaneous development of cardiac hypertrophy and fibrosis in mice lacking p50. Grumont et al47 identified three putative NF-κB binding sites in the Rel gene promoter. Their electrophoretic mobility shift assay analysis revealed that RelA:p50 heterodimers and p50:p50 homodimers bind the κB1 and κB3 sites; however, these sites exhibit a higher affinity for the p50 homodimer, whereas dimers containing RelA, c-Rel, and p50 are bound to the κB2 site. Expression of c-Rel is regulated at the transcriptional level in immune cells, and control of c-Rel has been shown to vary greatly depending on stimulus, cell type and cell function.48 Future investigation into the mechanism by which p50 regulates c-Rel expression in the normal and diseased heart is therefore warranted, given that targeting the crosstalk between p50 and c-Rel may reveal therapeutic opportunities.
Alternatively, the interaction of c-Rel with its target hypertrophy-related genes (Mef2a and Gata4) may offer strategies to inhibit cardiac remodeling. Activation of Mef2A-dependent transcription is required for cardiac development and growth and for initiating the phenotypic reprogramming of myocytes that results in cardiac remodeling and hypertrophy2,42,49,50 Mef2A appears to be a direct target of c-Rel, and therefore a plausible explanation for the reduction in Rel knockout heart size at birth and in adulthood could be insufficient production of Mef2A (although this would require further investigation). Furthermore, deficiency of c-Rel leads to a failure to induce Mef2A expression after chronic angiotensin infusion, which was associated with protection from cardiac fibrosis and enlargement. Mef2A expression was also induced in cardiac cells overexpressing c-Rel and in the hearts of mice lacking p50. Defining the nature of the c-Rel-containing NF-κB complexes assembled at the promoters of Mef2a and other c-Rel target genes identified in the present study will be an important step toward improving our understanding of the transcriptional mechanisms that regulate stress-induced remodeling of the heart. Moreover, the regulatory events that lead to nuclear accumulation of c-Rel in the diseased human heart will be of great interest and could be a potential source of therapeutic targets.
Footnotes
Supported by a British Heart Foundation project grant (PG/08/051/25141, FS/07/021, to D.A.M. and P.A.T.); grants from the Medical Research Council (G0900535), Wellcome Trust, and the European Union FP7 Programme (D.A.M. and F.O.); and by a Ph.D. studentship from the Gerald Kerkut Charitable Trust (S.G.-P.).
S.G.-P. and N.F. contributed equally to the present work.
Supplemental material for this article can be found at http://ajp.amjpathol.org or at doi: 10.1016/j.ajpath.2011.11.007.
Supplementary data
Rel−/− mice are born with smaller hearts and do not develop cardiac hypertrophy in response to pressure overload. A: Heart/body weight ratio was decreased in newborn (day 1) mice lacking c-Rel (Rel−/− mice), compared with WT mice. Data are expressed as means ± SEM; n £ 20 mice/group B: The cytoplasmic/nuclear ratio was calculated for WT and Rel null mice, with and without angiotensin (Ang) infusion. Data are expressed as means ± SEM; n = 5 (Rel−/−) and n = 6 (WT) mice/group. Representative photomicrographs show H&E staining of hearts from WT and c-Rel knockout mice 4 weeks after angiotensin infusion. Original magnification, ×400. Scale bars: 100 μm. All P values were calculated using a one-way analysis of variance or an unpaired two-tailed Student's t-test. **P = 0.01; ***P = 0.001.
Gata4 promoter activity is induced by c-Rel but not p50. A: The region of the Gata4 promoter containing one NF-κB/c-Rel site (blue box) and three c-Rel sites (yellow boxes) was cloned upstream of the luciferase gene and cotransfected into H9c2 cells with either RSV-β-gal, RSV-p50, or RSV-c-Rel. Overexpression of c-Rel increased Gata4 promoter activity twofold, whereas overexpression of p50 was without effect. Data are expressed as mean luciferase units ± SEM, normalized to Renilla, of four independent transfections. **P < 0.01. B: Recruitment of c-Rel and p50 to the exon 1-intron 1 boundary of the rat Tgfβ1 gene, which spans the ATG, or the exon 1-intron 1 boundary of the Pparγ gene was measured. These regions do not contain either a p50- or c-Rel-binding site and therefore act as a negative control for the ChIP analysis (Figure 3). Binding of c-Rel and p50 to these genes was not detected, suggesting that the antibodies are not nonspecifically interacting with the basal transcriptional machinery. Binding was normalized to total input genomic DNA and expressed as fold-IgG control (red line). Data are representative of three separate experiments.
Cardiac hypertrophy develops spontaneously in nfkb1−/− mice during adulthood and is associated with increased expression of c-Rel. A: Western blot analysis of whole heart lysates isolated from adult WT or nfkb1−/− mice shows that total cardiac c-Rel levels are increased in mice lacking the nfkb1 gene. Deletion of p105/p50 is associated with a modest increase in a-sarcomeric actin levels. As expected, nfkb1−/− mice do not express either the p105 or the p50 product of the nfkb1 gene. B: Heart/body weight ratio was calculated in nfkb1−/− and WT mice on a mixed C57Bl/6;129PF2/J F2 genetic background. Data are expressed as the mean ratio change ± SEM, compared with WT mice; n = 8 mice/genotype. An increase in heart/body weight ratio was observed in male Nfkb1−/− mice, compared with WT controls, and statistical significance was achieved. **P < 0.01. C: Heart/body weight ratio was increased in nfkb1−/− mice, compared with WT control mice on a pure C57Bl/6 background. Data are expressed as the mean ratio change compared with WT mice ± SEM; n = 17 mice/genotype. ***P < 0.001. Data are representative of three individual mice per genotype. D: There was no difference in cytoplasmic/nuclear ratios in male newborn (day 1) WT and nfkb1−/− mice. Data are expressed as the mean ratio change, ± SEM, compared with WT mice, for 11 WT and 15 nfkb1−/− mice.
References
- 1.Franco M., Cooper R.S., Bilal U., Fuster V. Challenges and opportunities for cardiovascular disease prevention. Am J Med. 2011;124:95–102. doi: 10.1016/j.amjmed.2010.08.015. [DOI] [PubMed] [Google Scholar]
- 2.Barry S.P., Davidson S.M., Townsend P.A. Molecular regulation of cardiac hypertrophy. Int J Biochem Cell Biol. 2008;40:2023–2039. doi: 10.1016/j.biocel.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 3.Chien K.R. Stress pathways and heart failure. Cell. 1999;98:555–558. doi: 10.1016/s0092-8674(00)80043-4. [DOI] [PubMed] [Google Scholar]
- 4.Frey N., Olson E.N. Cardiac hypertrophy: the good, the bad, and the ugly. Annu Rev Physiol. 2003;65:45–79. doi: 10.1146/annurev.physiol.65.092101.142243. [DOI] [PubMed] [Google Scholar]
- 5.Chevalier B., Callens-el Amrani F., Heymes C., Swynghedauw B. Molecular basis of regression of cardiac hypertrophy. Am J Cardiol. 1994;73:10C–17C. doi: 10.1016/0002-9149(94)90618-1. [DOI] [PubMed] [Google Scholar]
- 6.Perrino C., Rockman H.A. Reversal of cardiac remodeling by modulation of adrenergic receptors: a new frontier in heart failure. Curr Opin Cardiol. 2007;22:443–449. doi: 10.1097/HCO.0b013e3282294d72. [DOI] [PubMed] [Google Scholar]
- 7.Wohlschlaeger J., Schmitz K.J., Schmid C., Schmid K.W., Keul P., Takeda A., Weis S., Levkau B., Baba H.A. Reverse remodeling following insertion of left ventricular assist devices (LVAD): a review of the morphological and molecular changes. Cardiovasc Res. 2005;68:376–386. doi: 10.1016/j.cardiores.2005.06.030. [DOI] [PubMed] [Google Scholar]
- 8.Hayden M.S., Ghosh S. Signaling to NF-kappaB. Genes Dev. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- 9.Gupta S., Sen S. Role of the NF-kappaB signaling cascade and NF-kappaB-targeted genes in failing human hearts. J Mol Med (Berl) 2005;83:993–1004. doi: 10.1007/s00109-005-0691-z. [DOI] [PubMed] [Google Scholar]
- 10.Li Y., Ha T., Gao X., Kelley J., Williams D.L., Browder I.W., Kao R.L., Li C. NF-kappaB activation is required for the development of cardiac hypertrophy in vivo. Am J Physiol Heart Circ Physiol. 2004;287:H1712–H1720. doi: 10.1152/ajpheart.00124.2004. [DOI] [PubMed] [Google Scholar]
- 11.Van der Heiden K., Cuhlmann S., Luong le A., Zakkar M., Evans P.C. Role of nuclear factor kappaB in cardiovascular health and disease. Clin Sci (Lond) 2010;118:593–605. doi: 10.1042/CS20090557. [DOI] [PubMed] [Google Scholar]
- 12.Zelarayan L., Renger A., Noack C., Zafiriou M.P., Gehrke C., van der Nagel R., Dietz R., de Windt L., Bergmann M.W. NF-kappaB activation is required for adaptive cardiac hypertrophy. Cardiovasc Res. 2009;84:416–424. doi: 10.1093/cvr/cvp237. [DOI] [PubMed] [Google Scholar]
- 13.Gordon J.W., Shaw J.A., Kirshenbaum L.A. Multiple facets of NF-kappaB in the heart: to be or not to NF-kappaB. Circ Res. 2011;108:1122–1132. doi: 10.1161/CIRCRESAHA.110.226928. [DOI] [PubMed] [Google Scholar]
- 14.Frantz S., Fraccarollo D., Wagner H., Behr T.M., Jung P., Angermann C.E., Ertl G., Bauersachs J. Sustained activation of nuclear factor kappa B and activator protein 1 in chronic heart failure. Cardiovasc Res. 2003;57:749–756. doi: 10.1016/s0008-6363(02)00723-x. [DOI] [PubMed] [Google Scholar]
- 15.Saito T., Giaid A. Cyclooxygenase-2 and nuclear factor-kappaB in myocardium of end stage human heart failure. Congest Heart Fail. 1999;5:222–227. [PubMed] [Google Scholar]
- 16.Santos D.G., Resende M.F., Mill J.G., Mansur A.J., Krieger J.E., Pereira A.C. Nuclear factor (NF) kappaB polymorphism is associated with heart function in patients with heart failure. BMC Med Genet. 2010;11:89. doi: 10.1186/1471-2350-11-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou B., Rao L., Peng Y., Wang Y., Li Y., Gao L., Chen Y., Xue H., Song Y., Liao M., Zhang L. Functional polymorphism of the NFKB1 gene promoter is related to the risk of dilated cardiomyopathy. BMC Med Genet. 2009;10:47. doi: 10.1186/1471-2350-10-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gupta S., Young D., Sen S. Inhibition of NF-kappaB induces regression of cardiac hypertrophy, independent of blood pressure control, in spontaneously hypertensive rats. Am J Physiol Heart Circ Physiol. 2005;289:H20–H29. doi: 10.1152/ajpheart.00082.2005. [DOI] [PubMed] [Google Scholar]
- 19.Kawano S., Kubota T., Monden Y., Kawamura N., Tsutsui H., Takeshita A., Sunagawa K. Blockade of NF-kappaB ameliorates myocardial hypertrophy in response to chronic infusion of angiotensin II [Erratum appeared in Cardiovasc Res 2006;69:556] Cardiovasc Res. 2005;67:689–698. doi: 10.1016/j.cardiores.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 20.Kawano S., Kubota T., Monden Y., Tsutsumi T., Inoue T., Kawamura N., Tsutsui H., Sunagawa K. Blockade of NF-kappaB improves cardiac function and survival after myocardial infarction. Am J Physiol Heart Circ Physiol. 2006;291:H1337–H1344. doi: 10.1152/ajpheart.01175.2005. [DOI] [PubMed] [Google Scholar]
- 21.Timmers L., van Keulen J.K., Hoefer I.E., Meijs M.F., van Middelaar B., den Ouden K., van Echteld C.J., Pasterkamp G., de Kleijn D.P. Targeted deletion of nuclear factor kappaB p50 enhances cardiac remodeling and dysfunction following myocardial infarction. Circ Res. 2009;104:699–706. doi: 10.1161/CIRCRESAHA.108.189746. [DOI] [PubMed] [Google Scholar]
- 22.Kratsios P., Huth M., Temmerman L., Salimova E., Al Banchaabouchi M., Sgoifo A., Manghi M., Suzuki K., Rosenthal N., Mourkioti F. Antioxidant amelioration of dilated cardiomyopathy caused by conditional deletion of NEMO/IKKgamma in cardiomyocytes. Circ Res. 2010;106:133–144. doi: 10.1161/CIRCRESAHA.109.202200. [DOI] [PubMed] [Google Scholar]
- 23.Hikoso S., Yamaguchi O., Nakano Y., Takeda T., Omiya S., Mizote I., Taneike M., Oka T., Tamai T., Oyabu J., Uno Y., Matsumura Y., Nishida K., Suzuki K., Kogo M., Hori M., Otsu K. The I{kappa}B kinase {beta}/nuclear factor {kappa}B signaling pathway protects the heart from hemodynamic stress mediated by the regulation of manganese superoxide dismutase expression. Circ Res. 2009;105:70–79. doi: 10.1161/CIRCRESAHA.108.193318. [DOI] [PubMed] [Google Scholar]
- 24.Chakraborty J.B., Mann D.A. NF-kappaB signalling: embracing complexity to achieve translation. J Hepatol. 2010;52:285–291. doi: 10.1016/j.jhep.2009.10.030. [DOI] [PubMed] [Google Scholar]
- 25.Leung T.H., Hoffmann A., Baltimore D. One nucleotide in a kappaB site can determine cofactor specificity for NF-kappaB dimers. Cell. 2004;118:453–464. doi: 10.1016/j.cell.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 26.Perkins N.D. Post-translational modifications regulating the activity and function of the nuclear factor kappa B pathway. Oncogene. 2006;25:6717–6730. doi: 10.1038/sj.onc.1209937. [DOI] [PubMed] [Google Scholar]
- 27.Zhong H., May M.J., Jimi E., Ghosh S. The phosphorylation status of nuclear NF-kappa B determines its association with CBP/p300 or HDAC-1. Mol Cell. 2002;9:625–636. doi: 10.1016/s1097-2765(02)00477-x. [DOI] [PubMed] [Google Scholar]
- 28.Campbell I.K., Gerondakis S., O'Donnell K., Wicks I.P. Distinct roles for the NF-kappaB1 (p50) and c-Rel transcription factors in inflammatory arthritis. J Clin Invest. 2000;105:1799–1806. doi: 10.1172/JCI8298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Campbell S.J., Anthony D.C., Oakley F., Carlsen H., Elsharkawy A.M., Blomhoff R., Mann D.A. Hepatic nuclear factor kappa B regulates neutrophil recruitment to the injured brain. J Neuropathol Exp Neurol. 2008;67:223–230. doi: 10.1097/NEN.0b013e3181654957. [DOI] [PubMed] [Google Scholar]
- 30.Gerondakis S., Grumont R., Gugasyan R., Wong L., Isomura I., Ho W., Banerjee A. Unravelling the complexities of the NF-kappaB signalling pathway using mouse knockout and transgenic models. Oncogene. 2006;25:6781–6799. doi: 10.1038/sj.onc.1209944. [DOI] [PubMed] [Google Scholar]
- 31.Gieling R.G., Elsharkawy A.M., Caamano J.H., Cowie D.E., Wright M.C., Ebrahimkhani M.R., Burt A.D., Mann J., Raychaudhuri P., Liou H.C., Oakley F., Mann D.A. The c-Rel subunit of nuclear factor-kappaB regulates murine liver inflammation, wound-healing, and hepatocyte proliferation. Hepatology. 2010;51:922–931. doi: 10.1002/hep.23385. [DOI] [PubMed] [Google Scholar]
- 32.Pereira S.G., Oakley F. Nuclear factor-kappaB1: regulation and function. Int J Biochem Cell Biol. 2008;40:1425–1430. doi: 10.1016/j.biocel.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y., Rickman B.H., Poutahidis T., Schlieper K., Jackson E.A., Erdman S.E., Fox J.G., Horwitz B.H. c-Rel is essential for the development of innate and T cell-induced colitis. J Immunol. 2008;180:8118–8125. doi: 10.4049/jimmunol.180.12.8118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liou H.C., Jin Z., Tumang J., Andjelic S., Smith K.A., Liou M.L. c-Rel is crucial for lymphocyte proliferation but dispensable for T cell effector function. Int Immunol. 1999;11:361–371. doi: 10.1093/intimm/11.3.361. [DOI] [PubMed] [Google Scholar]
- 35.Sha W.C., Liou H.C., Tuomanen E.I., Baltimore D. Targeted disruption of the p50 subunit of NF-kappa B leads to multifocal defects in immune responses. Cell. 1995;80:321–330. doi: 10.1016/0092-8674(95)90415-8. [DOI] [PubMed] [Google Scholar]
- 36.Nebigil C.G., Hickel P., Messaddeq N., Vonesch J.L., Douchet M.P., Monassier L., Gyorgy K., Matz R., Andriantsitohaina R., Manivet P., Launay J.M., Maroteaux L. Ablation of serotonin 5-HT(2B) receptors in mice leads to abnormal cardiac structure and function. Circulation. 2001;103:2973–2979. doi: 10.1161/01.cir.103.24.2973. [DOI] [PubMed] [Google Scholar]
- 37.Perkins N.D., Schmid R.M., Duckett C.S., Leung K., Rice N.R., Nabel G.J. Distinct combinations of NF-kappa B subunits determine the specificity of transcriptional activation. Proc Natl Acad Sci USA. 1992;89:1529–1533. doi: 10.1073/pnas.89.5.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mann J., Oakley F., Akiboye F., Elsharkawy A., Thorne A.W., Mann D.A. Regulation of myofibroblast transdifferentiation by DNA methylation and MeCP2: implications for wound healing and fibrogenesis. Cell Death Differ. 2007;14:275–285. doi: 10.1038/sj.cdd.4401979. [DOI] [PubMed] [Google Scholar]
- 39.Mann J., Chu D.C., Maxwell A., Oakley F., Zhu N.L., Tsukamoto H., Mann D.A. MeCP2 controls an epigenetic pathway that promotes myofibroblast transdifferentiation and fibrosis. Gastroenterology. 2010;138:705–714. doi: 10.1053/j.gastro.2009.10.002. 714.e1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim I.M., Ramakrishna S., Gusarova G.A., Yoder H.M., Costa R.H., Kalinichenko V.V. The forkhead box m1 transcription factor is essential for embryonic development of pulmonary vasculature. J Biol Chem. 2005;280:22278–22286. doi: 10.1074/jbc.M500936200. [DOI] [PubMed] [Google Scholar]
- 41.Wang X., Bhattacharyya D., Dennewitz M.B., Kalinichenko V.V., Zhou Y., Lepe R., Costa R.H. Rapid hepatocyte nuclear translocation of the forkhead box M1B (FoxM1B) transcription factor caused a transient increase in size of regenerating transgenic hepatocytes. Gene Expr. 2003;11:149–162. doi: 10.3727/000000003108749044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Naya F.J., Black B.L., Wu H., Bassel-Duby R., Richardson J.A., Hill J.A., Olson E.N. Mitochondrial deficiency and cardiac sudden death in mice lacking the MEF2A transcription factor. Nat Med. 2002;8:1303–1309. doi: 10.1038/nm789. [DOI] [PubMed] [Google Scholar]
- 43.Nishikimi T., Maeda N., Matsuoka H. The role of natriuretic peptides in cardioprotection. Cardiovasc Res. 2006;69:318–328. doi: 10.1016/j.cardiores.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 44.Kawamura N., Kubota T., Kawano S., Monden Y., Feldman A.M., Tsutsui H., Takeshita A., Sunagawa K. Blockade of NF-kappaB improves cardiac function and survival without affecting inflammation in TNF-alpha-induced cardiomyopathy. Cardiovasc Res. 2005;66:520–529. doi: 10.1016/j.cardiores.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 45.Zhang J., Ping P., Vondriska T.M., Tang X.L., Wang G.W., Cardwell E.M., Bolli R. Cardioprotection involves activation of NF-kappa B via PKC-dependent tyrosine and serine phosphorylation of I kappa B-alpha. Am J Physiol Heart Circ Physiol. 2003;285:H1753–H1758. doi: 10.1152/ajpheart.00416.2003. [DOI] [PubMed] [Google Scholar]
- 46.Bergmann M.W., Loser P., Dietz R., von Harsdorf R. Effect of NF-kappa B Inhibition on TNF-alpha-induced apoptosis and downstream pathways in cardiomyocytes. J Mol Cell Cardiol. 2001;33:1223–1232. doi: 10.1006/jmcc.2001.1385. [DOI] [PubMed] [Google Scholar]
- 47.Grumont R.J., Richardson I.B., Gaff C., Gerondakis S. rel/NF-kappa B nuclear complexes that bind kB sites in the murine c-rel promoter are required for constitutive c-rel transcription in B-cells. Cell Growth Differ. 1993;4:731–743. [PubMed] [Google Scholar]
- 48.Grumont R.J., Gerondakis S. The murine c-rel proto-oncogene encodes two mRNAs the expression of which is modulated by lymphoid stimuli. Oncogene Res. 1990;5:245–254. [PubMed] [Google Scholar]
- 49.Xu J., Gong N.L., Bodi I., Aronow B.J., Backx P.H., Molkentin J.D. Myocyte enhancer factors 2A and 2C induce dilated cardiomyopathy in transgenic mice. J Biol Chem. 2006;281:9152–9162. doi: 10.1074/jbc.M510217200. [DOI] [PubMed] [Google Scholar]
- 50.Potthoff M.J., Olson E.N. MEF2: a central regulator of diverse developmental programs. Development. 2007;134:4131–4140. doi: 10.1242/dev.008367. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Rel−/− mice are born with smaller hearts and do not develop cardiac hypertrophy in response to pressure overload. A: Heart/body weight ratio was decreased in newborn (day 1) mice lacking c-Rel (Rel−/− mice), compared with WT mice. Data are expressed as means ± SEM; n £ 20 mice/group B: The cytoplasmic/nuclear ratio was calculated for WT and Rel null mice, with and without angiotensin (Ang) infusion. Data are expressed as means ± SEM; n = 5 (Rel−/−) and n = 6 (WT) mice/group. Representative photomicrographs show H&E staining of hearts from WT and c-Rel knockout mice 4 weeks after angiotensin infusion. Original magnification, ×400. Scale bars: 100 μm. All P values were calculated using a one-way analysis of variance or an unpaired two-tailed Student's t-test. **P = 0.01; ***P = 0.001.
Gata4 promoter activity is induced by c-Rel but not p50. A: The region of the Gata4 promoter containing one NF-κB/c-Rel site (blue box) and three c-Rel sites (yellow boxes) was cloned upstream of the luciferase gene and cotransfected into H9c2 cells with either RSV-β-gal, RSV-p50, or RSV-c-Rel. Overexpression of c-Rel increased Gata4 promoter activity twofold, whereas overexpression of p50 was without effect. Data are expressed as mean luciferase units ± SEM, normalized to Renilla, of four independent transfections. **P < 0.01. B: Recruitment of c-Rel and p50 to the exon 1-intron 1 boundary of the rat Tgfβ1 gene, which spans the ATG, or the exon 1-intron 1 boundary of the Pparγ gene was measured. These regions do not contain either a p50- or c-Rel-binding site and therefore act as a negative control for the ChIP analysis (Figure 3). Binding of c-Rel and p50 to these genes was not detected, suggesting that the antibodies are not nonspecifically interacting with the basal transcriptional machinery. Binding was normalized to total input genomic DNA and expressed as fold-IgG control (red line). Data are representative of three separate experiments.
Cardiac hypertrophy develops spontaneously in nfkb1−/− mice during adulthood and is associated with increased expression of c-Rel. A: Western blot analysis of whole heart lysates isolated from adult WT or nfkb1−/− mice shows that total cardiac c-Rel levels are increased in mice lacking the nfkb1 gene. Deletion of p105/p50 is associated with a modest increase in a-sarcomeric actin levels. As expected, nfkb1−/− mice do not express either the p105 or the p50 product of the nfkb1 gene. B: Heart/body weight ratio was calculated in nfkb1−/− and WT mice on a mixed C57Bl/6;129PF2/J F2 genetic background. Data are expressed as the mean ratio change ± SEM, compared with WT mice; n = 8 mice/genotype. An increase in heart/body weight ratio was observed in male Nfkb1−/− mice, compared with WT controls, and statistical significance was achieved. **P < 0.01. C: Heart/body weight ratio was increased in nfkb1−/− mice, compared with WT control mice on a pure C57Bl/6 background. Data are expressed as the mean ratio change compared with WT mice ± SEM; n = 17 mice/genotype. ***P < 0.001. Data are representative of three individual mice per genotype. D: There was no difference in cytoplasmic/nuclear ratios in male newborn (day 1) WT and nfkb1−/− mice. Data are expressed as the mean ratio change, ± SEM, compared with WT mice, for 11 WT and 15 nfkb1−/− mice.