Abstract
Two frequently applied genetic Bacteroidetes markers for total fecal pollution (AllBac and BacUni) were found in high numbers in pristine soil samples of two alpine catchment areas casting doubt on their value as fecal indicators. This finding underlines the necessity to evaluate assays locally and against non-intestinal samples before application.
Keywords: Water quality, Fecal pollution indicators, Genetic markers, Bacteroidetes, AllBac, BacUni
The contamination of water by fecal pollution has enormous impacts on a global scale since fecal material frequently contains intestinal pathogens in significant numbers. The reliability of traditional indicators for fecal pollution (e.g. E. coli) has been brought into question in various studies during the last 20 years (e.g. Byappanahalli and Fujioka, 1998; Byappanahalli et al., 2006; Fujioka et al., 1999). State-of-the-art microbial hazard and risk assessment approaches increasingly demand comprehensive pollution analysis including the quantification of total microbial fecal pollution and a reliable identification of its major contributing sources (i.e. microbial source tracking — MST) (Farnleitner et al., 2011; Field and Samadpour, 2007). Numerous MST markers have recently been developed targeting source-specific abundant intestinal microbial populations (e.g. the phylum Bacteroidetes) (Santo Domingo et al., 2007). In addition genetic markers for total fecal pollution were proposed (e.g. Kildare et al., 2007; Layton et al., 2006; Okabe et al., 2007) and applied (Bae and Wuertz, 2009; Dick et al., 2010; Schriewer et al., 2010; Silkie and Nelson, 2009; Stapleton et al., 2009; Wuertz et al., 2011) as reference parameters relating source-specific pollution to a level of total fecal pollution. Like any fecal indicator parameter, these genetic markers should be absent in extra-intestinal habitats where no fecal pollution has occurred. While most MST markers were tested for specificity using non-target fecal samples, studies investigating the specificity of markers for total fecal pollution against non-intestinal samples are very rare and cast considerable doubt on their reliability (Dick and Field, 2004; van der Wielen and Medema, 2010).
The aim of this study was to evaluate the reliability of two genetic Bacteroidetes quantitative PCR (qPCR) assays for total fecal pollution, AllBac (Layton et al., 2006) and BacUni (Kildare et al., 2007), by investigating their occurrence and concentration in pristine soil and fecal samples both collected from the same extensively studied alpine catchments. In addition we compared the amplicon sequences found with the two assays in soil and fecal samples, respectively.
Sampling and analysis: 42 soil and 20 fecal samples were concurrently collected in different vegetation zones (woodland, krummholz, alpine grassland) and at different altitudes (800–1800 m above sea level) in two well characterized alpine karst spring catchment areas in Eastern Austria. Detailed quantitative pollution source surveys (pollution source profiles) for the catchments showed that ruminant animals (cattle, red deer, roe deer, chamois) are by far the most important sources of fecal pollution (Farnleitner et al., 2011; Reischer et al., 2011). Soil sampling was restricted to sites with i) no visible signs of fecal pollution and ii) no indication of recent animal activity. Finally a sample was only considered “pristine” if it was negative for the MST marker BacR which is highly specific and sensitive for local ruminant fecal pollution sources (Reischer et al., 2006). Soil samples were taken from a depth of approximately 10 cm below the surface. The AllBac (Layton et al., 2006) and BacUni (Kildare et al., 2007) qPCR assays targeting fecal Bacteroidetes were implemented on our qPCR platforms, optimized for stringent marker detection and applied on the soil and fecal DNA. Marker copy numbers were quantified using plasmid standard dilution series and expressed as marker equivalents (ME) per g wet weight of soil or feces (Reischer et al., 2006). For a deeper comparison between the populations detected in soil and fecal samples, qPCR products of the two assays were cloned and sequenced to determine similarities to sequences of known origin in databases (Altschul et al., 1990; Cole et al., 2009). For a supplementary internal comparison between the sequences recovered from soil and fecal samples, the relatedness of the detected populations was determined by calculating the UniFrac metric (Lozupone and Knight, 2005; Lozupone et al., 2007), a phylogeny-based distance measure for the between-sample-diversity of bacterial communities. For methodical details refer to the supplementary material.
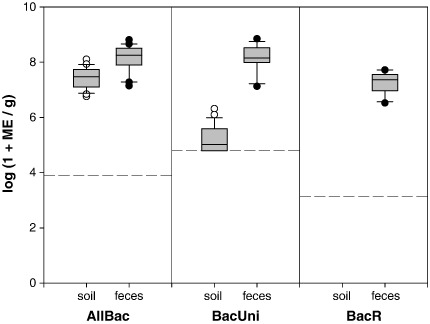
Marker detection in fecal and soil samples: The ruminant-specific BacR marker was not detectable in 29 of 42 soil samples which were hereafter considered “pristine”. Only these “pristine” samples were included in further analysis. All fecal samples found during the investigation were from ruminant animals confirming the assessment of the previous pollution source profile (Reischer et al., 2011). In all of these fecal samples BacR was present at similar levels (median 2.3 × 107 ME g− 1)(Fig. 1). The AllBac marker was found in all fecal samples, too (median 1.8 × 108 ME g− 1). AllBac could also be detected in all pristine soil samples at levels not even one order of magnitude lower than in feces and with little variation between the 29 samples (range from 5.7 × 106 to 1.3 × 108 ME g− 1, median 3.0 × 107 ME g− 1). The BacUni marker showed levels very similar to AllBac in ruminant feces (median 1.4 × 108 ME g− 1) while levels in soil were lower but still detectable in 76% of all samples (median 1.0 × 105 ME g− 1)(Fig. 1).
Fig. 1.
Concentrations of AllBac and BacUni markers for total fecal pollution in pristine soil samples (n = 29) and fecal samples (n = 20) in marker equivalents (ME) per g wet weight. BacR marker concentrations are given as a reference of fecal pollution levels. BacR was not detected in the 29 soil samples which were selected based on this property. Boxes, 25th and 75th percentile; lines within the boxes, median; whiskers, 10th and 90th percentile, respectively; data is log+ 1-transformed; dashed line represents assay limit of detection under the given circumstances.
The surprisingly high levels of the markers in soil motivated us to clone and sequence the qPCR amplicons of a subset of the soil and fecal samples in order to get an impression of the populations detected by the assays (two sequences per sample and assay). Comparison with the GenBank database showed that the sequences amplified from fecal samples showed high similarities with sequences derived from vertebrate fecal samples (average percentage of best hits from fecal origin: 64% for AllBac and 88% for BacUni, see Supplementary Table 1). In soil samples on the other hand only an average of 21% of the best hits for the AllBac and an average of 15% for BacUni sequences showed highest sequence identities with sequences found in vertebrate animal feces. As an additional supporting analysis and visualization of the phylogenetic relatedness of the recovered sequences we applied cluster analysis on the UniFrac distance matrix. Sequence communities from the two assays were clearly distinct (Supplementary Fig. 1) and most of the resulting clusters were exclusively containing either fecal or soil communities.
Applicability of markers for total fecal pollution: The assays tested in this study are the two most widely applied genetic Bacteroidetes markers for total fecal pollution (Bae and Wuertz, 2009; Dick et al., 2010; Silkie and Nelson, 2009; Stapleton et al., 2009; Wuertz et al., 2011). However, the achieved results call the applicability of the proposed assays for the studied Austrian catchment areas into question. Considering their concentrations in pristine soils, neither of them seems to be highly specific for vertebrate intestinal Bacteroidetes populations at all. The quantitative level of BacUni marker in pristine soils is lower in comparison to AllBac. Nevertheless the sequences detected with this assay in soils were more closely related to sequences not derived from vertebrate feces. In general the retrieved sequence information suggests that the assays appear to detect autochthonous, non-intestinal Bacteroidetes populations in soils that are for the most part distinct from the intended fecal target populations. These findings are in accordance with the recently reported unaccountably high concentrations of the AllBac marker in ground and drinking water in the Netherlands (van der Wielen and Medema, 2010). In fact, members of the phylum Bacteroidetes are, in addition to their dominant role in intestinal microbiota, also well known as abundant members of soil microbiota (Lauber et al., 2009; Roesch et al., 2007). The broad application of the tested assays combined with the apparent lack of investigations of their fecal indication performance is even more surprising when considering how vividly and controversially the occurrence of conventional fecal indicators (such as E. coli) in non-intestinal habitats such as soils and sediments has been discussed in recent years (Byappanahalli et al., 2006; Fujioka et al., 1999). The only qPCR assay specifically addressing this issue was published by Dick and Field (2004) who actually amended their original Bacteroidetes assay primers in an addendum in proof in order to exclude non-intestinal targets. Unfortunately these primers also seemed to have performed poorly in the recent Dutch study (van der Wielen and Medema, 2010) and do not seem to be widely used (Wuertz et al., 2011). Scientific literature also lacks information about the potential persistence of fecal Bacteroidetes populations in soil or other extra-intestinal environments. Our results underline the need to develop improved molecular markers for total fecal pollution detection. It should be stated that most proposed assays have been based on a fragmentary puzzle of sequence information which was available at the time of development. To improve assay design more sequence data are necessary, especially about extra-intestinal habitats such as soil or sediment. This study emphasizes the need to test the specificity and sensitivity of qPCR-based assays for total fecal pollution on the local level and especially against non-intestinal environmental samples. Although there is a strong demand and pressure for marker-based detection techniques for total fecal pollution in water quality monitoring and risk assessment, currently none of the tested assays seems to meet one of the most basic requirements, which is being indicative of vertebrate fecal material.
Acknowledgments
This study was financed by the Austrian Science Fund (FWF) project P22032 granted to G.H.R as well as the FWF DKplus “Vienna Doctoral Programme on Water Resource Systems” (W1219-N22) granted to A.H.F. G.H.R. is a recipient of an APART Fellowship of the Austrian Academy of Sciences. The study was also supported within a research cooperation with the Vienna Waterworks (Groundwater Resource Systems Vienna). Technical equipment was funded by the Innovative Project HOAL of the Vienna University of Technology. Our thanks go to Hannes Müller for his work on cloning.
Footnotes
Supplementary data to this article can be found online at doi:10.1016/j.mimet.2012.01.009.
Appendix A. Supplementary data
Supplementary material is available online and contains detailed methods, Supplementary Table 1, Supplementary Fig. 1 and a list of all investigated sequences determined in this study.
Supplementary material.
References
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bae S., Wuertz S. Rapid decay of host-specific fecal Bacteroidales cells in seawater as measured by quantitative PCR with propidium monoazide. Water Res. 2009;43:4850–4859. doi: 10.1016/j.watres.2009.06.053. [DOI] [PubMed] [Google Scholar]
- Byappanahalli M.N., Fujioka R.S. Evidence that tropical soil environment can support the growth of Escherichia coli. Water Sci. Technol. 1998;38:171–174. [Google Scholar]
- Byappanahalli M.N., Whitman R.L., Shively D.A., Sadowsky M.J., Ishii S. Population structure, persistence, and seasonality of autochthonous Escherichia coli in temperate, coastal forest soil from a Great Lakes watershed. Environ. Microbiol. 2006;8:504–513. doi: 10.1111/j.1462-2920.2005.00916.x. [DOI] [PubMed] [Google Scholar]
- Cole J.R., Wang Q., Cardenas E., Fish J., Chai B., Farris R.J., Kulam-Syed-Mohideen A.S., McGarrell D.M., Marsh T., Garrity G.M., Tiedje J.M. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick L.K., Field K.G. Rapid estimation of numbers of fecal Bacteroidetes by use of a quantitative PCR assay for 16S rRNA genes. Appl. Environ. Microbiol. 2004;70:5695–5697. doi: 10.1128/AEM.70.9.5695-5697.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick L.K., Stelzer E.A., Bertke E.E., Fong D.L., Stoeckel D.M. Relative decay of Bacteroidales microbial source tracking markers and cultivated Escherichia coli in freshwater microcosms. Appl. Environ. Microbiol. 2010;76:3255–3262. doi: 10.1128/AEM.02636-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnleitner A.H., Reischer G.H., Stadler H., Kollanur D., Sommer R., Zerobin W., Barella M., Truesdale J.A., Casarez E.A., Di Giovanni G.D. Agricultural and rural watersheds. In: Hagedorn C., Blanch A.R., Harwood V.J., editors. Microbial Source Tracking: Methods, Applications, and Case Studies. Springer; New York, USA: 2011. pp. 399–433. [Google Scholar]
- Field K.G., Samadpour M. Fecal source tracking, the indicator paradigm, and managing water quality. Water Res. 2007;41:3517–3538. doi: 10.1016/j.watres.2007.06.056. [DOI] [PubMed] [Google Scholar]
- Fujioka R., Sian-Denton C., Borja M., Castro J., Morphew K. Soil: the environmental source of Escherichia coli and enterococci in Guam's streams. J. Appl. Microbiol. 1999;85:83S–89S. doi: 10.1111/j.1365-2672.1998.tb05286.x. [DOI] [PubMed] [Google Scholar]
- Kildare B.J., Leutenegger C.M., McSwain B.S., Bambic D.G., Rajal V.B., Wuertz S. 16S rRNA-based assays for quantitative detection of universal, human-, cow-, and dog-specific fecal Bacteroidales: a Bayesian approach. Water Res. 2007;41:3701–3715. doi: 10.1016/j.watres.2007.06.037. [DOI] [PubMed] [Google Scholar]
- Lauber C.L., Hamady M., Knight R., Fierer N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 2009;75:5111–5120. doi: 10.1128/AEM.00335-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layton A., McKay L., Williams D., Garrett V., Gentry R., Sayler G. Development of Bacteroides 16S rRNA gene TaqMan-based real-time PCR assays for estimation of total, human, and bovine fecal pollution in water. Appl. Environ. Microbiol. 2006;72:4214–4224. doi: 10.1128/AEM.01036-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C., Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C.A., Hamady M., Kelley S.T., Knight R. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 2007;73:1576–1585. doi: 10.1128/AEM.01996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe S., Okayama N., Savichtcheva O., Ito T. Quantification of host-specific Bacteroides-Prevotella 16S rRNA genetic markers for assessment of fecal pollution in freshwater. Appl. Microbiol. Biotechnol. 2007;74:890–901. doi: 10.1007/s00253-006-0714-x. [DOI] [PubMed] [Google Scholar]
- Reischer G.H., Kasper D.C., Steinborn R., Mach R.L., Farnleitner A.H. Quantitative PCR method for sensitive detection of ruminant fecal pollution in freshwater and evaluation of this method in alpine karstic regions. Appl. Environ. Microbiol. 2006;72:5610–5614. doi: 10.1128/AEM.00364-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reischer G.H., Kollanur D., Vierheilig J., Wehrspaun C., Mach R.L., Sommer R., Stadler H., Farnleitner A.H. Hypothesis-driven approach for the identification of fecal pollution sources in water resources. Environ. Sci. Technol. 2011;45:4038–4045. doi: 10.1021/es103659s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesch L.F., Fulthorpe R.R., Riva A., Casella G., Hadwin A.K.M., Kent A.D., Daroub S.H., Camargo F.A.O., Farmerie W.G., Triplett E.W. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 2007;1:283–290. doi: 10.1038/ismej.2007.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santo Domingo J.W., Bambic D.G., Edge T.A., Wuertz S. Quo vadis source tracking? Towards a strategic framework for environmental monitoring of fecal pollution. Water Res. 2007;41:3539–3552. doi: 10.1016/j.watres.2007.06.001. [DOI] [PubMed] [Google Scholar]
- Schriewer A., Miller W.A., Byrne B.A., Miller M.A., Oates S., Conrad P.A., Hardin D., Yang H.-H., Chouicha N., Melli A., Jessup D., Dominik C., Wuertz S. Presence of Bacteroidales as a predictor of pathogens in surface waters of the central California coast. Appl. Environ. Microbiol. 2010;76:5802–5814. doi: 10.1128/AEM.00635-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silkie S.S., Nelson K.L. Concentrations of host-specific and generic fecal markers measured by quantitative PCR in raw sewage and fresh animal feces. Water Res. 2009;43:4860–4871. doi: 10.1016/j.watres.2009.08.017. [DOI] [PubMed] [Google Scholar]
- Stapleton C., Kay D., Wyer M., Davies C., Watkins J., Kay C., McDonald A., Porter J., Gawler A. Evaluating the operational utility of a Bacteroidales quantitative PCR-based MST approach in determining the source of faecal indicator organisms at a UK bathing water. Water Res. 2009;43:4888–4899. doi: 10.1016/j.watres.2009.09.015. [DOI] [PubMed] [Google Scholar]
- van der Wielen P.W.J.J., Medema G. Unsuitability of quantitative Bacteroidales 16S rRNA gene assays for discerning fecal contamination of drinking water. Appl. Environ. Microbiol. 2010;76:4876–4881. doi: 10.1128/AEM.03090-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuertz S., Wang D., Reischer G.H., Farnleitner A.H. Library-independent source tracking methods. In: Hagedorn C., Blanch A.R., Harwood V.J., editors. Microbial Source Tracking: Methods, Applications, and Case Studies. Springer; New York, USA: 2011. pp. 61–113. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.