Summary
Sen1 of S. cerevisiae is a known component of the NRD complex implicated in transcription termination of nonpolyadenylated as well as some polyadenylated RNA polymerase II transcripts. We now show that Sen1 helicase possesses a wider function by restricting the occurrence of RNA:DNA hybrids that may naturally form during transcription, when nascent RNA hybridizes to DNA prior to its packaging into RNA protein complexes. These hybrids displace the nontranscribed strand and create R loop structures. Loss of Sen1 results in transient R loop accumulation and so elicits transcription-associated recombination. SEN1 genetically interacts with DNA repair genes, suggesting that R loop resolution requires proteins involved in homologous recombination. Based on these findings, we propose that R loop formation is a frequent event during transcription and a key function of Sen1 is to prevent their accumulation and associated genome instability.
Graphical Abstract
Highlights
► Nascent RNA forms hybrids with underwound DNA upstream of elongating Pol II ► Single-stranded DNA so formed is prone to damage which results in genome instability ► Sen1 helicase acts to remove R loops by resolving RNA:DNA hybrids ► Sen1 function in Pol II elongation and termination may relate to R loop resolution
Introduction
In S. cerevisiae nascent transcripts formed by RNA polymerase II (Pol II) on protein-coding genes are immediately processed, packaged, and exported to the cytoplasm (Luna et al., 2008; Moore and Proudfoot, 2009). Messenger RNA (mRNA) packaging protects transcripts from degradation, but also the DNA template from invasion of nascent RNA into the DNA duplex behind elongating Pol II (Aguilera and Gómez-González, 2008). The resulting RNA:DNA hybrid exposes single stranded (ss) nontemplate DNA, a structure referred to as an R loop. R loop formation has been associated with increased occurrence of transcription-associated mutation (TAM) or recombination (TAR), presumably because both induced and spontaneous lesions are more likely to occur on ssDNA. Thus, deletion of genes encoding the THO (Thp2, Hpr1, Mft1, and Tho2) and THSC or TREX-2 (Thp1, Sac3, Sus1, and Cdc31) complexes required for mRNP formation in S. cerevisiae—or, similarly, the splicing factor ASF/SF2 in metazoans—increase levels of R loop formation and consequently TAM and TAR (Chávez et al., 2000; Fischer et al., 2002; Gallardo and Aguilera, 2001; González-Aguilera et al., 2008; Huertas and Aguilera, 2003; Li and Manley, 2005). R loop formation in these mutants may also be connected to Pol II stalling, consequently interfering with processive elongation (Mason and Struhl, 2005; Rondón et al., 2003) and RNA processing (Libri et al., 2002; Rougemaille et al., 2008). Similarly, DNA replication may be compromised when replication forks encounter R loops or a stalled Pol II (Wellinger et al., 2006).
Although little is known about R loop resolution in yeast, in mammals their formation and resolution play a productive role in the stimulation of class switch recombination (CSR) and somatic hypermutation (SHM) in clonally expanding B cells (Yu et al., 2003). Both processes are initiated by activation induced deaminase (AID) (Muramatsu et al., 1999). Double-strand breaks (DSBs) subsequently trigger CSR via nonhomologous end joining (NHEJ) (Yu and Lieber, 2003). Although S. cerevisiae does not express AID, ectopically expressed AID can recognize ssDNA in R loops as a substrate when expressed in mRNA packaging mutants (Gómez-González and Aguilera, 2007; González-Aguilera et al., 2008).
Many events during transcription are orchestrated by proteins binding to the carboxy-terminal domain (CTD) of the Pol II largest subunit. CTD consists in yeast of 26 hepta-peptide repeats (YSPTSPS) that are dynamically modified during transcription. In particular, serine phosphorylation occurs during early (ser5-, 7-P) and late (ser2-P) elongation phases to allow stage specific binding of elongation and RNA processing factors (Kim et al., 2009; Komarnitsky et al., 2000). Transcription termination is also directed by different CTD-bound proteins that recognize specific sequences on the emerging nascent RNA. For protein-coding genes, this requires polyA (pA) site recognition by a ser2-P CTD bound multicomponent cleavage and polyadenylation complex (CF IA/B and CPF), as well as degradation of the downstream RNA by Rat1 exonuclease (Gross and Moore, 2001; Kim et al., 2004b; Meinhart and Cramer, 2004).
Termination of many noncoding RNAs requires an additional component, the NRD complex (Sen1, Nab3, and Nrd1), in which Nrd1 is bound to ser5-P CTD (Steinmetz et al., 2001; Vasiljeva et al., 2008). NRD-dependent termination also requires recognition of frequent short RNA sequences by Nrd1 and Nab3 (GUAA/G and UCUU respectively) (Carroll et al., 2007), although the exact sequence and NRD component requirements may vary for different terminators (Kuehner and Brow, 2008). Nrd1 ser5-P CTD specificity confines this termination pathway to transcriptional stages in which ser5-P CTD prevails and leaves promoter distal Nrd1/Nab3 binding sites unrecognized (Arigo et al., 2006a; Gudipati et al., 2008). Furthermore, as Nrd1 interacts with the exosome, NRD-terminated RNA is either degraded to protein protected stable transcripts (e.g., snoRNAs) or completely, as is the case with cryptic unstable transcripts (CUTs) (Arigo et al., 2006b; Thiebaut et al., 2006; Vasiljeva and Buratowski, 2006). Recent genome-wide transcription profiling studies reveal the wide extent of CUTs produced by Pol II and terminated by NRD. This further emphasizes the biological importance of NRD-dependent termination (Neil et al., 2009; Xu et al., 2009). Importantly, both termination pathways can substitute for each other and so provide mutual fail-safe termination mechanisms (Kim et al., 2006; Rasmussen and Culbertson, 1998). Thus, NRD termination is also important to rescue polymerases that fail to terminate at a polyA signal, especially on highly transcribed genes. Interestingly, these genes show a particular requirement for Sen1 (Rondón et al., 2009).
SEN1 codes for a 240 kDa superfamily I helicase (DeMarini et al., 1992), and its S. pombe homolog possesses 3′-5′ nucleic acid unwinding activity (Kim et al., 1999). The essential C terminus contains the helicase domain, a nuclear localization sequence (NLS), and a domain necessary for interaction with the Glc7 phosphatase component of CPF (Nedea et al., 2008; Ursic et al., 1995; Winey and Culbertson, 1988). The Sen1 975 N-terminal amino acids, although dispensable for growth, interact with Pol II, RNase III endonuclease Rnt1, and the nucleotide excision repair endonuclease Rad2 (Ursic et al., 2004). Mutation of the Sen1 helicase domain results in direct and indirect pleiotropic defects in transcript processing and termination, leading to a perturbed genome-wide profile of Pol II and defective Pol I transcription termination (Kawauchi et al., 2008; Rasmussen and Culbertson, 1998; Steinmetz et al., 2001, 2006; Ursic et al., 1997). Although the severe character of these phenotypes may be explicable by the limiting presence of Sen1 in NRD (as it is only present at 125 copies/cell) (Ghaemmaghami et al., 2003), they have not been clearly attributed to a molecular function of Sen1. Employing the temperature sensitive sen1-1 mutant (helicase domain G1747D), we set out to characterize the molecular role of Sen1 in transcription termination. We now identify broad functions for Sen1 during Pol II transcription in reducing R loop formation and consequent prevention of transcription-associated genome instability.
Results
Role of Sen1 Helicase Domain in Transcription Termination
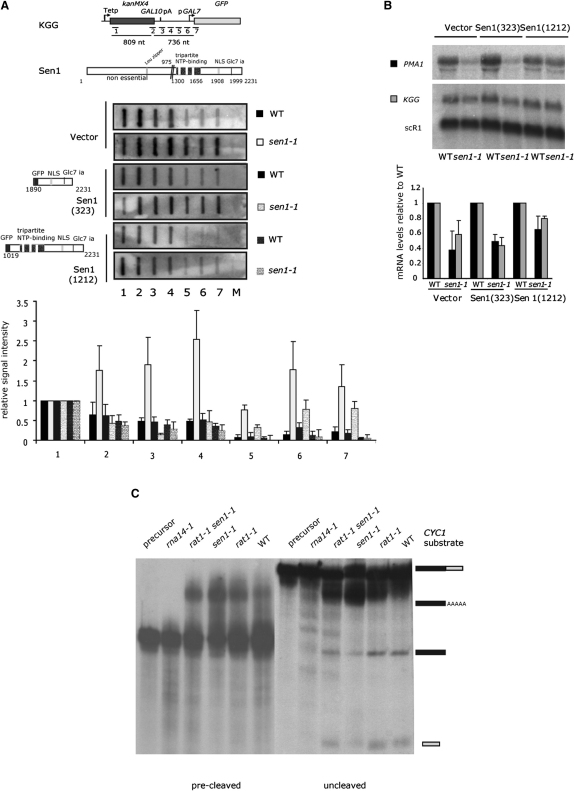
Mutation of the Sen1 helicase domain results in genome-wide transcription termination defects of noncoding RNAs, but also of some protein coding genes (Steinmetz et al., 2006). Thus, when tested by transcription run on (TRO) experiments with the plasmid gene construct KGG (Figure 1A), with the KanMX4 gene terminated by the weak GAL10 pA signal (Morillon et al., 2003), sen1-1 mutants grown for 150 min at nonpermissive temperature (37°C) show a strong termination defect (Figure 1A, upper panels) (Rondón et al., 2009). This indicates either a requirement for Sen1 in Rat-dependent termination or that some transcripts over the weak GAL10 pA signal are terminated by the NRD failsafe termination mechanism.
Figure 1.
Sen1 Helicase Is Required for Transcription Termination but Not Transcript 3′ Processing
(A) Top: pKGG and positions of ssM13 probes (1–7). Domain structure of Sen1. Middle: representative TRO filters of WT and sen1-1 cells cotransformed with KGG and empty vector, Sen1(323), or Sen1(1212) constructs. Transformants were grown for 150 min at 37°C before TRO. Bottom: quantification based on four repeat experiments.
(B) RNA isolated from the inocules used for TRO analysis probed for KGG and endogenous PMA1 mRNA, as well as Pol III transcript scR1. Bottom: quantification of four repeat experiments.
(C) In vitro cleavage and polyadenylation assays performed with extracts from WT and mutant cells grown for 150 or 90 min (rna14-1) at 37°C with CYC1 3′ pA as substrate. Positions of uncleaved, polyadenylated, cleaved, and 3′ end cleavage product are indicated.
All error bars represent the standard deviation (SD). See also Figure S1.
To determine whether Sen1 protein-protein interactions or its helicase function are required for transcription termination, we repeated TRO analysis in WT and sen1-1 cells transformed with additional Sen1 expression constructs. Transcribed from an ACT1 promoter, these either contained the NLS and the Glc7 interaction domain [Sen1(323)] or additionally the C-terminal helicase domain [Sen1(1212)] (Nedea et al., 2008). As shown in Figure 1A, Sen1(1212) but not Sen1(323) rescued the sen1-1 termination defect, implying that the sen1-1 termination defect is caused by loss of helicase function and not Glc7 mediated recruitment of CPF. We also examined steady-state mRNA produced from the KGG construct (Figure 1B). mRNA levels were reduced in sen1-1 cells and partially complemented by coexpression of Sen1(1212), but not Sen1(323). Similarly, coexpression of Sen1(1212) restored wild-type levels of endogenous PMA1 mRNA, also previously shown to display mild termination defects in sen1-1 (Kawauchi et al., 2008).
The above results indicate that the Sen1 helicase domain is required both for efficient Pol II termination and mRNA accumulation. As these effects could be attributed to defective 3′ end processing, we employed an in vitro cleavage and polyadenylation assay using CYC1 3′ flanking RNA as the pA substrate (Figure 1C). sen1-1 shows no defects in RNA 3′ end processing. Confirmation of this result is provided by reverse transcription analysis of ACT1 pA usage, in which sen1-1 showed WT pA selection (Figure S1 available online). In contrast, a CF IA mutant strain, rna14-1, showed the expected defect in both in vitro 3′ end processing and in vivo pA selection (Figure 1C and Figure S1). Finally, like sen1-1, the rat1-1 termination mutant (or both combined) had no effect on mRNA 3′ end formation but stabilized the 3′ end cleavage product, indicative of loss of exonuclease “torpedo” function (Kim et al., 2004b; Luo et al., 2006). Overall, these combined analyses show that the Sen1 helicase is dispensable for 3′ transcript processing but is required to promote transcriptional termination.
Mitotic Recombination Is Increased in sen1-1
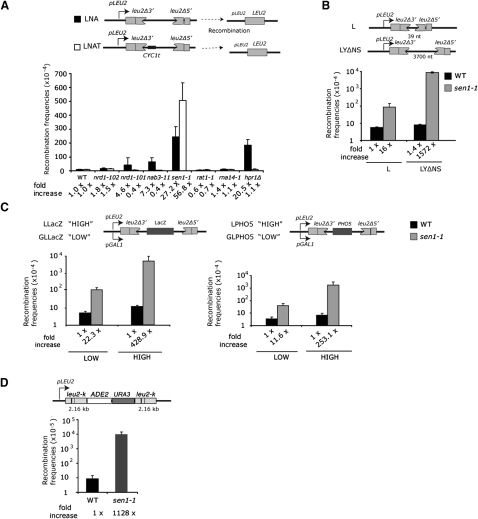
Since S. pombe Sen1 can use RNA:DNA hybrids as an in vitro substrate (Kim et al., 1999), we considered the possibility that Sen1 may remove RNA:DNA hybrids formed by nascent RNA and the template strand. Such hybrids were previously shown to form in THO mutants, causing increased rates of transcription associated mitotic recombination (Huertas and Aguilera, 2003). RNA:DNA hybrids may also be naturally encountered in transcribed regions downstream of pA signals, where THO is undetectable on chromatin (Kim et al., 2004a; Luna et al., 2005). We therefore tested whether sequences downstream of a pA signal elicit TAR in sen1-1. We employed a plasmid borne recombination substrate that carries two truncated regions of LEU2 overlapping by 600 nt of homologous sequence (LNA). Lack of THO elicits TAR in LNA and consequent restoration of LEU2, as previously shown (Figure 2A) (Prado et al., 1997). In contrast, when transcription between both repeats is terminated by insertion of the CYC1 38nt pA signal (CYC1t, LNAT), recombination levels in the hpr1Δ strain were reduced to background WT levels, presumably because Pol II termination restricts R loop formation. Similar analysis of LNA and LNAT transformed rat1-1 and rna14-1 showed no detectible increase in recombination, confirming that defects in CPF/Rat1 dependent transcription termination per se do not promote recombination (Luna et al., 2005). In marked contrast, sen1-1 transformed with either LNA or LNAT showed high levels of recombination, suggesting that RNA:DNA hybrids may form throughout the mRNA coding region irrespective of the CYC1t. This lack of CYC1t suppression reiterates the sen1-1 CYC1 termination defect previously reported (Kawauchi et al., 2008; Steinmetz et al., 2006). Moreover, the fact that CYC1t (in LNAT) further stimulated recombination may reflect an increase in R loop formation downstream of pA signals.
Figure 2.
sen1-1 Are Hyperrecombinogenic
(A) Recombination substrates LNA and LNAT. Transformants were grown for 3–4 days at 30°C. Recombination generates a functional LEU2, allowing selection of recombinants on leu-deficient plates. Quantification of recombinants formed from six colonies of four to six transformants is presented.
(B) Recombinants formed in WT and sen1-1 transformed with L and LYΔNS containing homologous repeats separated by 39 or 3900 nt.
(C) As in (B), with the LLacZ and LPHO5 substrates under control of either LEU2 or glucose-repressed GAL1 promoters to stimulate high or low expression.
(D) Diagram and recombination quantification of chromosomal construct crossed into the WT and sen1-1.
All error bars represent the SD. See also Figure S2.
To determine whether this recombination phenotype was specific to Sen1, we similarly tested other NRD complex components. Although recombination levels were somewhat increased in nab3 and nrd1 CTD-interacting domain mutants (but not the RNA binding domain mutant nrd1-102) transformed with LNA, they were reduced to background levels in LNAT (Figure 2A). This suggests that these NRD mutants still recognize the CYC1t. Why these NRD mutations elicit some recombination is unclear at this point, but may reflect alteration in mRNP biogenesis. The fact that CYC1t abrogates recombination in NRD mutants but stimulates recombination in sen1-1 clearly separates Sen1 function from Nab3 and Nrd1 and argues that Sen1 plays a distinct role outside the NRD complex.
sen1-1 Hyperrecombination Depends on Transcription
Hyperrecombination in THO and THSC/TREX-2 mutants shows clear transcription dependence, as it increases with greater transcript length and transcription rate but decreases when the R loop-forming RNA is removed either by RNase H activity or ribozyme directed RNA cleavage (González-Aguilera et al., 2008; Huertas and Aguilera, 2003). To test whether sen1-1 shows a similar transcription-dependent recombination phenotype, we analyzed sen1-1 recombination levels for various direct-repeat recombination substrates. As shown in Figures 2B and 2C, levels of recombination in sen1-1 correlate with the length and transcriptional rate of the gene. Thus, TAR, although not abolished, was significantly decreased in two different recombination substrates when transcription from a GAL1 promoter was glucose repressed. Finally, we verified that recombination was also stimulated between direct repeats in a chromosomal context (Figure 2D). These observations suggest that there is a correlation between transcriptional activity and sen1-1 recombination levels.
RNA:DNA Hybrids Form in sen1-1
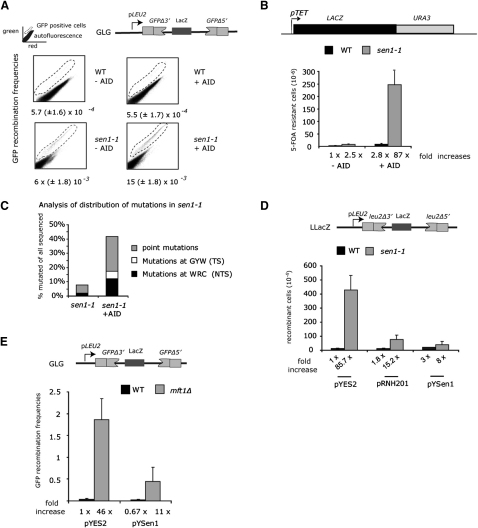
Evidence for RNA:DNA hybrid accumulation in THO mutants derives from expression of the human AID in yeast which was shown to cause a 25-fold increase in TAR (Gómez-González and Aguilera, 2007). Employing 5′ and 3′ truncated overlapping GFP repeats and intervening LacZ sequence as a recombination substrate, FACS analysis of GFP-positive cells showed that AID expressed in sen1-1 also stimulates recombination albeit only 2.5-fold (P[Wilcoxon-rank-sum test] = 0.017; Figure 3A). As discussed below, this moderate but significant stimulation of TAR by AID could reflect the nature of RNA:DNA hybrids formed in sen1-1 or be due to the fact that recombination levels in sen1-1 cells that did not express AID were already very high.
Figure 3.
sen1-1 Cells Form a Substrate for AID and RNase H
(A) AID overexpression increases sen1-1 TAR. pGLG recombinants forming GFP were counted after 12–16 hr growth at 30°C by FACS. AID coexpression increases GFP-positive cells in both WT and sen1-1 strains (p = 0.018, Wilcoxon test).
(B) Coexpression of AID and pLAUR-induced mutations within URA3 in pLAUR were scored as 5-FOA resistant.
(C) URA3 sequence from mutants was amplified and sequenced. The frequency of point mutations on either strand is depicted graphically (see Figure S3 for more detail).
(D) Effect of galactose-induced expression from plasmids pRNH201 (coding RNase H RNH201), pYsen1 (aa 1281–2231 of Sen1 cloned into pYES2), or pYES2 alone on the recombination frequencies in WT and sen1-1 cells produced by the LLacZ system. Note that double selection and growth on galactose reduces the sen1-1 viability and therefore recombination frequencies as compared to data in Figure 2C.
(E) Recombination frequency analysis in the THO-complex mutant mft1Δ with the GLG recombination substrate and AID to increase recombination rates. Overexpression of Sen1 helicase reduces recombination frequency.
All error bars represent the SD. See also Figure S3.
AID C to U deamination preferentially occurs in a WRC (or GYW on the opposite strand) sequence (Pham et al., 2003). In regions of R loop formation, AID has access to the nontranscribed strand (NTS), although some mutations occur on the transcribed strand (TS) (Gómez-González and Aguilera, 2007). In an attempt to analyze the nature of AID-induced mutations in sen1-1, we transformed WT and sen1-1 cells (both ura3–) with plasmid-encoded LACZ::URA3 chimeric gene (pLAUR, Figure 3B) and selected AID-induced ura3 mutations on 5-FOA (5-fluoorotic acid). Although mutation rates were very low in WT and sen1-1 cells grown at semipermissive temperature, AID expression in sen1-1 significantly increased mutation rates (Figure 3B). DNA sequencing of 57 5-FOA-resistant transformants revealed that 42% carried a point mutation in URA3, of which 71% had a point mutation within an AID sequence motif. Of these mutations 70% occurred on C (12; Fisher's test; p < 0.003) or the NTS and only 30% (five; p < 0.05) occur on G or the TS (Figure 3C and Figure S3). The small amount of WT transformants sequenced showed a distribution as earlier reported (Gómez-González and Aguilera, 2007). Since AID expression not only increased the amount of point mutations within the transcribed URA3 in sen1-1 but also preferentially acted on the NTS, this suggests that R loops are formed during transcription in sen1-1 cells and displace the nontranscribed DNA strand.
We next tested whether the high recombination levels seen in sen1-1 were sensitive to RNase H overexpression as previously observed with THO and THSC/TREX2 mutants and indicative of increased R loop formation (González-Aguilera et al., 2008; Huertas and Aguilera, 2003). Thus, RNase H overexpression (from pRNH201) reduced recombination rates of pLLacZ in sen1-1 by 5.6-fold (Figure 3D). Furthermore, the sen1-1 recombination phenotype correlated with deficient helicase activity and could be rescued by high copy expression of pYsen1, encoding for the Sen1 helicase domain. To exclude the possibility that increased recombination levels in sen1-1 could be a consequence of the sen1-1 transcription termination defect, rather than the ability of Sen1 to remove R loops, we tested the capability of Sen1 helicase domain to suppress the hyperrecombination phenotype of the THO mutant mft1Δ, which displays no transcription termination defect but increased levels of R loops. As shown in Figure 3E, overexpression of the Sen1 helicase domain in AID- and pGLG-transformed mft1Δ cells caused a substantial reduction in the number of GFP-recombinant cells. We conclude that Sen1 enzymatic activity is able to directly restrict cotranscriptionally formed R loops.
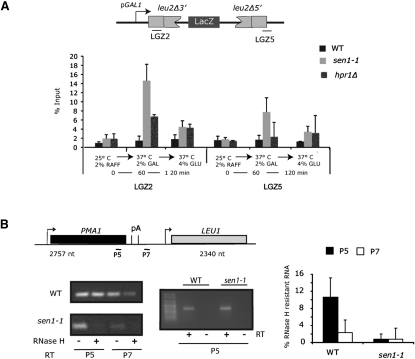
To obtain independent evidence for the existence of R loops formed in sen1-1, we employed both DNA (DIP; Figure 4A) and chromatin immunoprecipitation analysis (ChIP; Figure S4A) with the RNA:DNA hybrid-specific antibody (S9.6) (Hu et al., 2006). Yeast transformed with pGLLacZ displayed hybrid signal over the recombining LEU2 sequence in sen1-1 but not WT cells when shifted for 1 hr to nonpermissive temperature and induced by galactose. Hybrid signal then diminished to WT background levels when transcription was repressed by glucose addition to the medium (Figure 4A). Notably, the untranscribed origin region of pGLLacZ gave background signals (Figure S4B). Furthermore, where DIP signals were detected on LGZ2 and LGZ5 in sen1-1 grown in galactose, these signals were sensitive to RNase H digestion prior to immunoprecipitation (Figure S4C). Overall, these data demonstrate that R loop formation is highly dynamic, closely following transcriptional activation and repression of pGLLacZ. Parallel experiments with the THO mutant strain hpr1Δ gave significant but lower levels of hybrid signal over the 5′ positioned LEU sequence (LGZ2) but not over LGZ5 (Figure 4A). The 3′ LGZ5 probe spans the LEU2 pA signal and so will lack transcripts in hpr1Δ due to Pol II termination. sen1-1 in contrast is termination defective so that read-through transcripts in this strain will still elicit R loop formation.
Figure 4.
R Loops Form in sen1-1 Cells
(A) DIP analysis on pGLLacZ in sen1-1, hpr1Δ, and WT cells with antibody against RNA:DNA hybrids (S9.6). Coimmunoprecipitated DNA was detected by real-time qPCR. Inocules were grown in raffinose, induced with galactose for 1 hr at 37°C, and successively repressed at 37°C by addition of 4% glucose.
(B) Reverse transcription (RT) of RNA isolated from genomic DNA preparations after treatment with RNase H. The levels of RNase H resistant RNA were measured after RT with P5 and P7 primers followed by PCR with P5 amplicon (as shown on the gene map). Signals obtained were free from DNA contamination based on minus RT controls. Left: representative gel. Right: quantification of RT normalized triplicate repeats by real-time qRT-PCR, further normalized to P5 amplicon signal obtained from non-RNase H-digested samples.
All error bars represent the SD. See also Figure S4.
The S9.6 antibody was also employed in a regular ChIP analysis of endogenous PMA1 and compared to Pol II ChIP profiles. RNA:DNA hybrid signal was detected over PMA1 (Figure S4A), although general signal intensities were lower than those obtained by DIP analysis of the highly expressed pGLLacZ. When normalized to the gene 5′ end in WT and sen1-1 cells, signals were detected wherever Pol II was present on chromatin (Kawauchi et al., 2008). Of note, in chromatin isolated from rna14-1 that shows no recombination phenotype, little hybrid could be detected downstream of the pA signals, even through Pol II is still present at these positions. This may indicate that while hybrid is degraded downstream of the pA signal in presence of Sen1 and Rat1, it is stabilized in rat1-1 sen1-1 and to a lesser extent in sen1-1. A modest accumulation of hybrid signal was observed over the pA signal compared to the gene body (P6 versus P5) in all tested strains and may indicate a region that is particularly prone to form RNA:DNA hybrids.
In view of the only modest RNA:DNA hybrid accumulation observed over PMA1, we developed an independent assay for R loop formation over this same gene (Figure 4B). Genomic DNA with associated nascent transcripts was isolated from WT and sen1-1 cells, shifted for 1 hr to 37°C, and subjected to RNase H digestion. Subsequently, DNA was digested with DNase I and nascent RNA that survived this treatment was detected by RT-PCR with RT primers either within PMA1 (P5) or downstream of the pA signal (P7). As shown in gel fractionation of these amplified DNAs, sen1-1 chromatin associated RNA is highly sensitive to RNase H treatment. Quantitative PCR (qPCR) values (ratio of RNase H sensitive to total signal) show that although in sen1-1 chromatin associated RNA is nearly fully RNase H sensitive, in WT RNase H sensitivity increases downstream of the PMA1 pA signal. This suggests that in sen1-1 a higher fraction of RNA forms R loop structures with genomic DNA, whereas in the WT R loops are particularly prone to forming downstream of the pA site.
From these combined analyses, we conclude that R loops accumulate in sen1-1 in a transcription-dependent manner. Furthermore, they imply that WT-transcribed and subsequently packaged RNA still forms some level of RNA:DNA hybrids. The amount of RNA in R loop conformation appears to differ throughout the transcribed PMA1 gene, with a particular prevalence over the pA site.
SEN1 Genetic Interaction with Homologous Recombination Genes
Experiments presented so far suggest that R loops formed in sen1-1 elicit TAR. However, to substantiate this conclusion and further exclude the possibility that sen1-1 TAR is an indirect consequence of its transcription termination defect, we employed genomic analysis. A comparison of RNA steady-state levels of sen1-1 to WT cells grown for 150 min at 30°C was performed by hybridization to strand-specific tiling arrays (data available at http://www.ebi.ac.uk/arrayexpress/ with accession number E-TABM-863) (David et al., 2006). This revealed that among the transcripts whose expression was significantly changed (adjusted p value of < 0.01), stable unannotated transcripts were overrepresented (19% stable unannotated transcripts versus 7% for open reading frames [ORFs]), confirming a Sen1 role in the termination of some of these genes (Figure S5A and Table S4). Importantly, THO or THSC genes were absent among the 347 ORFs that were significantly changed, excluding the possibility that R loops are formed in sen1-1 as an indirect consequence of alteration in THO or THSC/TREX-2 expression. We next considered whether the 347 significantly changed ORF transcript levels correspond to DNA damage repair and cell-cycle (DDCC) genes. DDCC genes were underrepresented, and among those changed, no clear trend was observed (Figure S5B and Table S4), with some mildly upregulated (i.e., REC104, POL4, SMC5, SCM4, and TOP2) and others downregulated (i.e., DIA2, MMS2, and NEJ1).
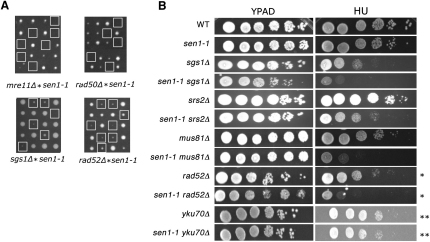
To define which DNA repair mechanism was induced by R loops formed in sen1-1, we searched for synthetic genetic interactions of sen1-1 with mutants of candidate genes involved in either NHEJ or homologous recombination (HR). At permissive temperature (25°C) or in presence of replicative stress, we observed genetic interaction with various factors involved in HR but not in NHEJ (Figure 5). Thus, sen1-1 (but not nrd1-102; Figure S5C) causes synthetic lethality with rad50 and mre11 deletion mutants, both found in a complex with Xrs2 and involved in initial recognition and ss resection at a DSB (Figure 5A). The critical need for HR in sen1-1 cells is demonstrated by the phenotypes of double mutants of sen1-1 and rad52Δ, sgs1Δ, srs2Δ, or mus81Δ. These all showed synthetic defects or displayed increased sensitivity to growth in hydroxyurea (HU). In contrast, sen1-1 yku70Δ double mutants showed neither growth defects nor increased sensitivity to HU, suggesting that NHEJ is not required for cell survival of sen1-1 cells (Figure 5B). Taken together, these data suggest that proteins involved in HR but not NHEJ are important to maintain sen1-1 viability.
Figure 5.
sen1-1 Shows Synthetic Genetic Interaction with DNA Damage Repair Genes
(A) Synthetic interactions between sen1-1 and MRX gene mutations: mre11Δ, rad50Δ. Also shown are synthetic interactions between sen1-1 and HR gene mutants sgs1Δ and rad52Δ. White boxes indicate spores that carry both mutations.
(B) Analysis of HU sensitivity of double mutants grown at 25°C. Growth was compared on YPAD plates ± 50 mM HU (10 mM ∗ or 100 mM ∗∗ as indicated).
DNA Damage Foci in sen1-1 Nuclei
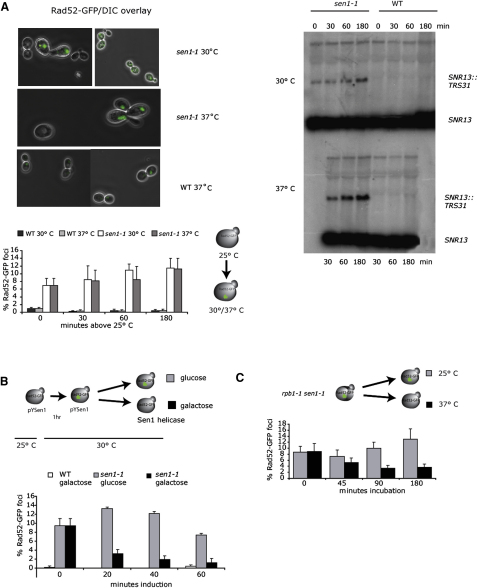
In cells that accumulate DNA damage, factors involved in DNA repair are rapidly recruited to the damage site (Lisby et al., 2001). Consistent with the role of R loops in DNA damage as seen in sen1-1, we observed that many sen1-1 cells display an accumulation of GFP-tagged Rad52 (encoded on pWJ144; Figure 6A). This percentage of sen1-1 cells forming foci increased from 8% at 25°C to 13% when cultures were shifted to semipermissive (30°C) or nonpermissive (37°C) temperature for 3 hr. In contrast, only about 1% of WT cells formed foci under these conditions. Importantly, steady-state RNA analysis from the same cell populations showed that Rad52 foci formation correlated with accumulation of a bicistronic transcript from the SNR13 snoRNA gene, which results from termination at the next available pA when NRD-dependent termination is defective (Rasmussen and Culbertson, 1998). As shown in Figure 6A (right panels), the SNR13-TRS31 transcript is already present in sen1-1 at 25°C but increases over time at 30°C and to a greater extent at 37°C. These results connect the various sen1-1 phenotypes observed in this work and support the assumption that they are caused by mutation of Sen1 helicase domain in sen1-1.
Figure 6.
DNA Repair Foci in sen1-1 Nuclei
(A) Time course of WT and sen1-1 grown at log phase and shifted to 30° or 37°C. At indicated time points aliquots were spotted on microscope slides and foci-containing cells scored based on 300 cells. Representative pictures (top left) and quantification of 3-5 repeats are shown (bottom left). Simultaneously isolated RNA was analyzed by Northern Blot against SNR13 to show accumulation of bi-cistronic SNR13-TRS31 transcript in sen1-1.
(B) WT and sen1-1 cells transformed with pWJ144 and pYSen1 grown in raffinose and shifted to 30°C for 1hr, before Sen1 helicase fragment expression was induced or repressed by addition of 2% galactose or 2% glucose to the medium respectively. Only WT in galactose shown and foci containing cells scored as in A.
(C) sen1-1 rpb1-1 cells transformed with pWJ144 were grown in logarithmic phase at 25°C and then shifted to 37°C. Shutoff of transcription results in decrease of Rad52 foci, either by reduced accumulation or Rad52 turnover.
All error bars represent the SD.
Using this assay as an indicator of ongoing HR in sen1-1, we reinvestigated the role of Sen1 helicase in preventing or resolving transcription dependent R loops. WT and sen1-1 transformed with Rad52-GFP and pYSen1, which encodes the Sen1 helicase domain transcribed from the GAL1 promoter, were grown in raffinose as a neutral carbon source for 1 hr at 30°C. After addition of glucose or galactose to respectively repress or activate Sen1 helicase expression, the amount of Rad52 foci forming cells was counted. Expression of the Sen1 helicase domain rapidly reduced the number of sen1-1 nuclei displaying Rad52 foci to almost WT levels (Figure 6B). Employing a sen1-1 rpb1-1 double mutant, which allows rapid transcription shutdown at the nonpermissive temperature due to a mutation in the Pol II largest subunit (rpb1-1) (Ursic et al., 2004) (Figure 6C), we could also correlate the formation of Rad52 foci to transcriptional activity. When grown at 37°C, Rad52-GFP transformed sen1-1 rpb1-1 cells displayed a time-dependent decrease in Rad52 foci as compared to growth at 25°C. Since sen1-1 cells accumulate Rad52 foci when shifted to 37°C, these data indicate that upon transcription shutdown, Rad52 foci disappear either as DNA damage is immediately repaired or the cells die.
Discussion
The deleterious effects of transcription on genome integrity have been suggested by various observations (Aguilera, 2002). Whenever the integrated process of transcript processing, packaging, and export in eukaryotes is disrupted, genome instability can be observed (Baaklini et al., 2004; Broccoli et al., 2004; Jimeno et al., 2002; Luna et al., 2005). This has been shown to derive from R loops, which preferentially form when mRNP biogenesis is disrupted. Our data show that Sen1 helicase plays a pivotal role in the prevention of genome instability by recombination. A large fraction of this instability is transcription dependent and linked to the formation of R loop structures. The exact nature of these structures remains to be established, but we show their accumulation can still occur with normal mRNP biogenesis. If these structures are not removed by either Sen1 helicase or RNase H directed degradation, they can exert a deleterious effect on genome stability, as is further illustrated by SEN1 genetic interaction with HR genes. The occurrence of Rad52 foci, as a marker for ongoing recombination, shows that recombination is related to transcription, as well as to the presence of a functional Sen1 helicase domain. In summary, we suggest that R loop formation is more frequent than hitherto anticipated and requires active removal by helicases such as Sen1.
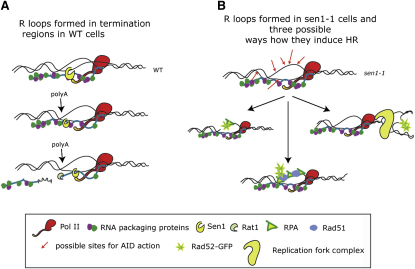
We suggest that as soon as the nascent transcript emerges from the polymerase body, mRNA packaging and R loop formation occur in kinetic competition (Figure 7). A fragile equilibrium between protective mRNA packaging and the hiding of specific recognition sequences is likely to exist (Bucheli and Buratowski, 2005; Bucheli et al., 2007). Therefore, RNA packaging is likely to be incomplete so leaving some transcript available for R loop formation. For pervasive CUT transcription, termination depends on NRD and by inference on Sen1 (Arigo et al., 2006b). If R loops formed in sen1-1 extend to many CUT loci, then their accumulation, even if transient, would cover substantial regions of the genome. In both CUTs and mRNA coding genes, R loops could interfere with DNA replication, induce ssDNA breaks, or be recognized as recombination intermediates. Any of these possibilities could explain the essential need for DSB sensing proteins in sen1-1 (Figure 5). However, the different genetic interactions of sen1-1 and hpr1 with HR or S phase checkpoint genes suggest structural and functional differences of the replication/recombinogenic intermediates that are formed (Gómez-González et al., 2009). Alternatively, these differences may hint at a transcription-independent role of Sen1 in DNA damage repair that is yet to be uncovered.
Figure 7.
Cotranscriptional Functions of Sen1
(A) Model for Sen1 cotranscriptional function especially in termination regions.
(B) Model for R loop accumulation in sen1-1 showing three ways they may elicit HR: processing of nicks in ssDNA, ssDNA recognition, and collapse of colliding replication forks.
How may R loop accumulation in sen1-1 be related to its transcription termination defect? R loops were originally hypothesized to slow down transcription elongation, thereby enhancing termination (Proudfoot, 1989). This would give time for the Rat1 5′-3′ exonuclease “torpedo” to catch up with Pol II but would require R loop resolution by an enzymatic activity such as Sen1 prior to degradation. Based on observations made on THO mutants, R loops have been suggested to interfere with transcription elongation (Huertas and Aguilera, 2003; Mason and Struhl, 2005). Employing sen1-1, in which transcript processing is normal, we predict that reduced steady-state RNA accumulation is due to reduced transcript elongation. Furthermore, the data presented here support the view that R loops preferentially form in termination regions. Thus, we employed the LNA/LNAT recombination substrates, anticipating that even though the CYC1 pA would not elicit termination (Kawauchi et al., 2008), it should serve as a 3′ processing signal (Figure 1C), promoting disassembly of THO and consequent R loop formation (Kim et al., 2004a). Compared to LNA, sen1-1 recombination levels increased 2-fold in LNAT. This demonstrates for sen1-1 in contrast to THO mutants, that RNA cleavage in the context of a pA is not sufficient to relieve recombination. To reiterate this point, Figure S2 shows that in a ribozyme containing substrate, recombination levels in sen1-1 are reduced similar to hpr1Δ (Huertas and Aguilera, 2003). As both ribozyme cleaved ends are unprotected they are likely to be degraded and so reduce R loop forming substrate. However, this appears not to be the case if RNA in sen1-1 cells is cleaved at a pA, possibly as RNA downstream to the polyA signal may be less packaged and so temporarily protected from degradation by R loop formation. Although these studies require a more detailed biochemical analysis, we predict from these initial results that R loops may play a role in transcriptional termination.
In summary the molecular and genetic effects of Sen1 inactivation presented here reveal that Sen1 acts to protect the heavily transcribed genome from R loop-mediated DNA damage. Of note, mutations in the helicase domain of the human SEN1 gene ortholog SETX (encoding Senataxin) cause the neurodegenerative diseases, Ataxia with Oculomotor Apraxia Type II (AOAII), and juvenile amyotrophic lateral sclerosis (ALS4). Like sen1-1 these SETX mutants show defects in transcription, RNA processing, and DNA damage repair (Moreira et al., 2004; Suraweera et al., 2007, 2009). It remains to be established whether the tendency of transcription to induce R loop formation is a general feature of all eukaryotic genomes. It is possible that a range of dedicated helicases act to resolve these potentially harmful structures.
Experimental Procedures
Yeast Cultivation and Genetic and Cell Biology Methods
Yeast strains, plasmids, and primers are listed in Tables S1–S3. Standard methods are detailed in the Supplemental Experimental Procedures. In outline, genetic crossing of single mutants (sen1-1 and nrd1-102) with HR and NHEJ mutants employed standard conditions. Recombination frequencies were scored by counting of LEU+ cells or by FACS analysis of GFP+ cells. GFP-Rad52 nuclear foci were detected by epifluorescent microscopy.
Transcription Run On Analysis
The transcription run on (TRO) method and probes for pKGG are as described (Morillon et al., 2003; Rondón et al., 2009).
Northern Blot Analysis
For RNA isolation, strains were grown in minimal (selective) media at indicated temperatures. After acidic phenol RNA isolation, RNA (15 μg) was separated by 1% formaldehyde agarose gel electrophoresis. RNA immobilized on nitrocellulose membranes was detected with random primed probes.
Chromatin and DNA Immunoprecipitation
ChIP employed real-time qPCR as previously described (Rondón et al., 2009). S9.6 purified antibody was employed for immunoprecipitation and was a kind gift from Stephen Leppla (Hu et al., 2006) and Antonin Morillon (Institut Curie, Paris). For DIP analysis, sonicated, deproteinized chromatin was immunoprecipitated with S9.6 antibody and amplified by qPCR as further detailed in the Supplemental Experimental Procedures (Liu et al., 2005). RNase H sensitivity was measured by treatment with RNase H prior to immunoprecipitation.
RT Analysis of Genomic DNA-Associated RNA
Sequential RNase H (NEB, 2 hr at 37°C) and DNase I (Roche, 4 hr at 37°C) digestion of genomic DNA (5 μg) isolated from logarithmic phase cells cultivated for 1 hr at 37°C with yeast breaking buffer (2% [v/v] Triton X-100, 1% [w/v] SDS, 100 mM NaCl, 10 mM Tris [pH 8.0], and 1 mM EDTA [pH 8.0]), phenol, and glass beads. Isolated RNA was reverse transcribed (Invitrogen Superscript III) according to the manufacturer.
Further Standard Experimental Procedures
These are presented in the Supplemental Experimental Procedures and include in vitro 3′ end processing, 3′end RACE, and microarray analysis.
Acknowledgments
Thanks to our lab colleagues, especially Monika Gullerova and Jurgi Camblong, Julien Gagneur, Sandra Clauder-Muenster, and Charles Giradot (submission to ArrayExpress). Funding was from the Wellcome Trust to N.J.P., the Spanish Ministry of Science and Innovation (BFU2006-05260 and Consolider Ingenio 2010 CSD2007-0015) and Junta de Andalucía (BIO102 and CV2549) to A.A., and a joint ESF grant to N.J.P. and A.A. (NuRNASu). H.E.M. was recipient of a B.I.F. studentship and a European Molecular Biology Organization (EMBO) short-term fellowship. B.G. received a predoctoral FPU training grant from the Spanish Ministry of Science and Innovation. P.G. was supported by a long-term EMBO fellowship.
Published: January 6, 2011
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, five figures, and four tables and can be found with this article online at doi:10.1016/j.molcel.2010.12.007.
Contributor Information
Andrés Aguilera, Email: aguilo@us.es.
Nick J. Proudfoot, Email: nicholas.proudfoot@path.ox.ac.uk.
Supplemental Information
References
- Aguilera A. The connection between transcription and genomic instability. EMBO J. 2002;21:195–201. doi: 10.1093/emboj/21.3.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera A., Gómez-González B. Genome instability: a mechanistic view of its causes and consequences. Nat. Rev. Genet. 2008;9:204–217. doi: 10.1038/nrg2268. [DOI] [PubMed] [Google Scholar]
- Arigo J.T., Carroll K.L., Ames J.M., Corden J.L. Regulation of yeast NRD1 expression by premature transcription termination. Mol. Cell. 2006;21:641–651. doi: 10.1016/j.molcel.2006.02.005. [DOI] [PubMed] [Google Scholar]
- Arigo J.T., Eyler D.E., Carroll K.L., Corden J.L. Termination of cryptic unstable transcripts is directed by yeast RNA-binding proteins Nrd1 and Nab3. Mol. Cell. 2006;23:841–851. doi: 10.1016/j.molcel.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Baaklini I., Hraiky C., Rallu F., Tse-Dinh Y.C., Drolet M. RNase HI overproduction is required for efficient full-length RNA synthesis in the absence of topoisomerase I in Escherichia coli. Mol. Microbiol. 2004;54:198–211. doi: 10.1111/j.1365-2958.2004.04258.x. [DOI] [PubMed] [Google Scholar]
- Broccoli S., Rallu F., Sanscartier P., Cerritelli S.M., Crouch R.J., Drolet M. Effects of RNA polymerase modifications on transcription-induced negative supercoiling and associated R-loop formation. Mol. Microbiol. 2004;52:1769–1779. doi: 10.1111/j.1365-2958.2004.04092.x. [DOI] [PubMed] [Google Scholar]
- Bucheli M.E., Buratowski S. Npl3 is an antagonist of mRNA 3′ end formation by RNA polymerase II. EMBO J. 2005;24:2150–2160. doi: 10.1038/sj.emboj.7600687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucheli M.E., He X., Kaplan C.D., Moore C.L., Buratowski S. Polyadenylation site choice in yeast is affected by competition between Npl3 and polyadenylation factor CFI. RNA. 2007;13:1756–1764. doi: 10.1261/rna.607207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll K.L., Ghirlando R., Ames J.M., Corden J.L. Interaction of yeast RNA-binding proteins Nrd1 and Nab3 with RNA polymerase II terminator elements. RNA. 2007;13:361–373. doi: 10.1261/rna.338407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chávez S., Beilharz T., Rondón A.G., Erdjument-Bromage H., Tempst P., Svejstrup J.Q., Lithgow T., Aguilera A. A protein complex containing Tho2, Hpr1, Mft1 and a novel protein, Thp2, connects transcription elongation with mitotic recombination in Saccharomyces cerevisiae. EMBO J. 2000;19:5824–5834. doi: 10.1093/emboj/19.21.5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David L., Huber W., Granovskaia M., Toedling J., Palm C.J., Bofkin L., Jones T., Davis R.W., Steinmetz L.M. A high-resolution map of transcription in the yeast genome. Proc. Natl. Acad. Sci. USA. 2006;103:5320–5325. doi: 10.1073/pnas.0601091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMarini D.J., Winey M., Ursic D., Webb F., Culbertson M.R. SEN1, a positive effector of tRNA-splicing endonuclease in Saccharomyces cerevisiae. Mol. Cell. Biol. 1992;12:2154–2164. doi: 10.1128/mcb.12.5.2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer T., Strässer K., Rácz A., Rodriguez-Navarro S., Oppizzi M., Ihrig P., Lechner J., Hurt E. The mRNA export machinery requires the novel Sac3p-Thp1p complex to dock at the nucleoplasmic entrance of the nuclear pores. EMBO J. 2002;21:5843–5852. doi: 10.1093/emboj/cdf590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo M., Aguilera A. A new hyperrecombination mutation identifies a novel yeast gene, THP1, connecting transcription elongation with mitotic recombination. Genetics. 2001;157:79–89. doi: 10.1093/genetics/157.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S., Huh W.K., Bower K., Howson R.W., Belle A., Dephoure N., O'Shea E.K., Weissman J.S. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Gómez-González B., Aguilera A. Activation-induced cytidine deaminase action is strongly stimulated by mutations of the THO complex. Proc. Natl. Acad. Sci. USA. 2007;104:8409–8414. doi: 10.1073/pnas.0702836104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-González B., Felipe-Abrio I., Aguilera A. The S-phase checkpoint is required to respond to R-loops accumulated in THO mutants. Mol. Cell. Biol. 2009;29:5203–5213. doi: 10.1128/MCB.00402-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Aguilera C., Tous C., Gómez-González B., Huertas P., Luna R., Aguilera A. The THP1-SAC3-SUS1-CDC31 complex works in transcription elongation-mRNA export preventing RNA-mediated genome instability. Mol. Biol. Cell. 2008;19:4310–4318. doi: 10.1091/mbc.E08-04-0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross S., Moore C.L. Rna15 interaction with the A-rich yeast polyadenylation signal is an essential step in mRNA 3′-end formation. Mol. Cell. Biol. 2001;21:8045–8055. doi: 10.1128/MCB.21.23.8045-8055.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudipati R.K., Villa T., Boulay J., Libri D. Phosphorylation of the RNA polymerase II C-terminal domain dictates transcription termination choice. Nat. Struct. Mol. Biol. 2008;15:786–794. doi: 10.1038/nsmb.1460. [DOI] [PubMed] [Google Scholar]
- Hu Z., Zhang A., Storz G., Gottesman S., Leppla S.H. An antibody-based microarray assay for small RNA detection. Nucleic Acids Res. 2006;34:e52. doi: 10.1093/nar/gkl142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P., Aguilera A. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol. Cell. 2003;12:711–721. doi: 10.1016/j.molcel.2003.08.010. [DOI] [PubMed] [Google Scholar]
- Jimeno S., Rondón A.G., Luna R., Aguilera A. The yeast THO complex and mRNA export factors link RNA metabolism with transcription and genome instability. EMBO J. 2002;21:3526–3535. doi: 10.1093/emboj/cdf335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawauchi J., Mischo H., Braglia P., Rondon A., Proudfoot N.J. Budding yeast RNA polymerases I and II employ parallel mechanisms of transcriptional termination. Genes Dev. 2008;22:1082–1092. doi: 10.1101/gad.463408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.D., Choe J., Seo Y.S. The sen1(+) gene of Schizosaccharomyces pombe, a homologue of budding yeast SEN1, encodes an RNA and DNA helicase. Biochemistry. 1999;38:14697–14710. doi: 10.1021/bi991470c. [DOI] [PubMed] [Google Scholar]
- Kim M., Ahn S.H., Krogan N.J., Greenblatt J.F., Buratowski S. Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO J. 2004;23:354–364. doi: 10.1038/sj.emboj.7600053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Krogan N.J., Vasiljeva L., Rando O.J., Nedea E., Greenblatt J.F., Buratowski S. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature. 2004;432:517–522. doi: 10.1038/nature03041. [DOI] [PubMed] [Google Scholar]
- Kim M., Vasiljeva L., Rando O.J., Zhelkovsky A., Moore C., Buratowski S. Distinct pathways for snoRNA and mRNA termination. Mol. Cell. 2006;24:723–734. doi: 10.1016/j.molcel.2006.11.011. [DOI] [PubMed] [Google Scholar]
- Kim M., Suh H., Cho E.J., Buratowski S. Phosphorylation of the yeast Rpb1 C-terminal domain at serines 2, 5, and 7. J. Biol. Chem. 2009;284:26421–26426. doi: 10.1074/jbc.M109.028993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarnitsky P., Cho E.J., Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehner J.N., Brow D.A. Regulation of a eukaryotic gene by GTP-dependent start site selection and transcription attenuation. Mol. Cell. 2008;31:201–211. doi: 10.1016/j.molcel.2008.05.018. [DOI] [PubMed] [Google Scholar]
- Li X., Manley J.L. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005;122:365–378. doi: 10.1016/j.cell.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Libri D., Dower K., Boulay J., Thomsen R., Rosbash M., Jensen T.H. Interactions between mRNA export commitment, 3′-end quality control, and nuclear degradation. Mol. Cell. Biol. 2002;22:8254–8266. doi: 10.1128/MCB.22.23.8254-8266.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M., Rothstein R., Mortensen U.H. Rad52 forms DNA repair and recombination centers during S phase. Proc. Natl. Acad. Sci. USA. 2001;98:8276–8282. doi: 10.1073/pnas.121006298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Noll D.M., Lieb J.D., Clarke N.D. DIP-chip: rapid and accurate determination of DNA-binding specificity. Genome Res. 2005;15:421–427. doi: 10.1101/gr.3256505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna R., Jimeno S., Marín M., Huertas P., García-Rubio M., Aguilera A. Interdependence between transcription and mRNP processing and export, and its impact on genetic stability. Mol. Cell. 2005;18:711–722. doi: 10.1016/j.molcel.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Luna R., Gaillard H., González-Aguilera C., Aguilera A. Biogenesis of mRNPs: integrating different processes in the eukaryotic nucleus. Chromosoma. 2008;117:319–331. doi: 10.1007/s00412-008-0158-4. [DOI] [PubMed] [Google Scholar]
- Luo W., Johnson A.W., Bentley D.L. The role of Rat1 in coupling mRNA 3′-end processing to transcription termination: implications for a unified allosteric-torpedo model. Genes Dev. 2006;20:954–965. doi: 10.1101/gad.1409106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason P.B., Struhl K. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol. Cell. 2005;17:831–840. doi: 10.1016/j.molcel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- Meinhart A., Cramer P. Recognition of RNA polymerase II carboxy-terminal domain by 3′-RNA-processing factors. Nature. 2004;430:223–226. doi: 10.1038/nature02679. [DOI] [PubMed] [Google Scholar]
- Moore M.J., Proudfoot N.J. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- Moreira M.C., Klur S., Watanabe M., Németh A.H., Le Ber I., Moniz J.C., Tranchant C., Aubourg P., Tazir M., Schöls L. Senataxin, the ortholog of a yeast RNA helicase, is mutant in ataxia-ocular apraxia 2. Nat. Genet. 2004;36:225–227. doi: 10.1038/ng1303. [DOI] [PubMed] [Google Scholar]
- Morillon A., Karabetsou N., O'Sullivan J., Kent N., Proudfoot N., Mellor J. Isw1 chromatin remodeling ATPase coordinates transcription elongation and termination by RNA polymerase II. Cell. 2003;115:425–435. doi: 10.1016/s0092-8674(03)00880-8. [DOI] [PubMed] [Google Scholar]
- Muramatsu M., Sankaranand V.S., Anant S., Sugai M., Kinoshita K., Davidson N.O., Honjo T. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J. Biol. Chem. 1999;274:18470–18476. doi: 10.1074/jbc.274.26.18470. [DOI] [PubMed] [Google Scholar]
- Nedea E., Nalbant D., Xia D., Theoharis N.T., Suter B., Richardson C.J., Tatchell K., Kislinger T., Greenblatt J.F., Nagy P.L. The Glc7 phosphatase subunit of the cleavage and polyadenylation factor is essential for transcription termination on snoRNA genes. Mol. Cell. 2008;29:577–587. doi: 10.1016/j.molcel.2007.12.031. [DOI] [PubMed] [Google Scholar]
- Neil H., Malabat C., d'Aubenton-Carafa Y., Xu Z., Steinmetz L.M., Jacquier A. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature. 2009;457:1038–1042. doi: 10.1038/nature07747. [DOI] [PubMed] [Google Scholar]
- Pham P., Bransteitter R., Petruska J., Goodman M.F. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature. 2003;424:103–107. doi: 10.1038/nature01760. [DOI] [PubMed] [Google Scholar]
- Prado F., Piruat J.I., Aguilera A. Recombination between DNA repeats in yeast hpr1delta cells is linked to transcription elongation. EMBO J. 1997;16:2826–2835. doi: 10.1093/emboj/16.10.2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N.J. How RNA polymerase II terminates transcription in higher eukaryotes. Trends Biochem. Sci. 1989;14:105–110. doi: 10.1016/0968-0004(89)90132-1. [DOI] [PubMed] [Google Scholar]
- Rasmussen T.P., Culbertson M.R. The putative nucleic acid helicase Sen1p is required for formation and stability of termini and for maximal rates of synthesis and levels of accumulation of small nucleolar RNAs in Saccharomyces cerevisiae. Mol. Cell. Biol. 1998;18:6885–6896. doi: 10.1128/mcb.18.12.6885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondón A.G., Jimeno S., García-Rubio M., Aguilera A. Molecular evidence that the eukaryotic THO/TREX complex is required for efficient transcription elongation. J. Biol. Chem. 2003;278:39037–39043. doi: 10.1074/jbc.M305718200. [DOI] [PubMed] [Google Scholar]
- Rondón A.G., Mischo H.E., Kawauchi J., Proudfoot N.J. Fail-safe transcriptional termination for protein-coding genes in S. cerevisiae. Mol. Cell. 2009;36:88–98. doi: 10.1016/j.molcel.2009.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougemaille M., Dieppois G., Kisseleva-Romanova E., Gudipati R.K., Lemoine S., Blugeon C., Boulay J., Jensen T.H., Stutz F., Devaux F., Libri D. THO/Sub2p functions to coordinate 3′-end processing with gene-nuclear pore association. Cell. 2008;135:308–321. doi: 10.1016/j.cell.2008.08.005. [DOI] [PubMed] [Google Scholar]
- Steinmetz E.J., Conrad N.K., Brow D.A., Corden J.L. RNA-binding protein Nrd1 directs poly(A)-independent 3′-end formation of RNA polymerase II transcripts. Nature. 2001;413:327–331. doi: 10.1038/35095090. [DOI] [PubMed] [Google Scholar]
- Steinmetz E.J., Warren C.L., Kuehner J.N., Panbehi B., Ansari A.Z., Brow D.A. Genome-wide distribution of yeast RNA polymerase II and its control by Sen1 helicase. Mol. Cell. 2006;24:735–746. doi: 10.1016/j.molcel.2006.10.023. [DOI] [PubMed] [Google Scholar]
- Suraweera A., Becherel O.J., Chen P., Rundle N., Woods R., Nakamura J., Gatei M., Criscuolo C., Filla A., Chessa L. Senataxin, defective in ataxia oculomotor apraxia type 2, is involved in the defense against oxidative DNA damage. J. Cell Biol. 2007;177:969–979. doi: 10.1083/jcb.200701042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suraweera A., Lim Y., Woods R., Birrell G.W., Nasim T., Becherel O.J., Lavin M.F. Functional role for senataxin, defective in ataxia oculomotor apraxia type 2, in transcriptional regulation. Hum. Mol. Genet. 2009;18:3384–3396. doi: 10.1093/hmg/ddp278. [DOI] [PubMed] [Google Scholar]
- Thiebaut M., Kisseleva-Romanova E., Rougemaille M., Boulay J., Libri D. Transcription termination and nuclear degradation of cryptic unstable transcripts: a role for the nrd1-nab3 pathway in genome surveillance. Mol. Cell. 2006;23:853–864. doi: 10.1016/j.molcel.2006.07.029. [DOI] [PubMed] [Google Scholar]
- Ursic D., DeMarini D.J., Culbertson M.R. Inactivation of the yeast Sen1 protein affects the localization of nucleolar proteins. Mol. Gen. Genet. 1995;249:571–584. doi: 10.1007/BF00418026. [DOI] [PubMed] [Google Scholar]
- Ursic D., Himmel K.L., Gurley K.A., Webb F., Culbertson M.R. The yeast SEN1 gene is required for the processing of diverse RNA classes. Nucleic Acids Res. 1997;25:4778–4785. doi: 10.1093/nar/25.23.4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursic D., Chinchilla K., Finkel J.S., Culbertson M.R. Multiple protein/protein and protein/RNA interactions suggest roles for yeast DNA/RNA helicase Sen1p in transcription, transcription-coupled DNA repair and RNA processing. Nucleic Acids Res. 2004;32:2441–2452. doi: 10.1093/nar/gkh561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasiljeva L., Buratowski S. Nrd1 interacts with the nuclear exosome for 3′ processing of RNA polymerase II transcripts. Mol. Cell. 2006;21:239–248. doi: 10.1016/j.molcel.2005.11.028. [DOI] [PubMed] [Google Scholar]
- Vasiljeva L., Kim M., Mutschler H., Buratowski S., Meinhart A. The Nrd1-Nab3-Sen1 termination complex interacts with the Ser5-phosphorylated RNA polymerase II C-terminal domain. Nat. Struct. Mol. Biol. 2008;15:795–804. doi: 10.1038/nsmb.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellinger R.E., Prado F., Aguilera A. Replication fork progression is impaired by transcription in hyperrecombinant yeast cells lacking a functional THO complex. Mol. Cell. Biol. 2006;26:3327–3334. doi: 10.1128/MCB.26.8.3327-3334.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M., Culbertson M.R. Mutations affecting the tRNA-splicing endonuclease activity of Saccharomyces cerevisiae. Genetics. 1988;118:609–617. doi: 10.1093/genetics/118.4.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Wei W., Gagneur J., Perocchi F., Clauder-Münster S., Camblong J., Guffanti E., Stutz F., Huber W., Steinmetz L.M. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033–1037. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K., Lieber M.R. Nucleic acid structures and enzymes in the immunoglobulin class switch recombination mechanism. DNA Repair (Amst.) 2003;2:1163–1174. doi: 10.1016/j.dnarep.2003.08.010. [DOI] [PubMed] [Google Scholar]
- Yu K., Chedin F., Hsieh C.L., Wilson T.E., Lieber M.R. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat. Immunol. 2003;4:442–451. doi: 10.1038/ni919. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.