Abstract
West Africa is characterized by a migration history spanning more than 150,000 years. Climate changes but also political circumstances were responsible for several early but also recent population movements that shaped the West African mitochondrial landscape. The aim of the study was to establish a Ghanaian mtDNA dataset for forensic purposes and to investigate the diversity of the Ghanaian population sample with respect to surrounding populations. We sequenced full mitochondrial control regions of 193 Akan people from Ghana and excluded two apparently close maternally related individuals due to preceding kinship testing. The remaining dataset comprising 191 sequences was applied as etalon for quasi-median network analysis and was subsequently combined with 99 additional control region sequences from surrounding West African countries. All sequences were incorporated into the EMPOP database enriching the severely underrepresented African mtDNA pool. For phylogeographic considerations, the Ghanaian haplotypes were compared to those of 19 neighboring populations comprising a total number of 6198 HVS1 haplotypes. We found extensive genetic admixture between the Ghanaian lineages and those from adjacent populations diminishing with geographical distance. The extent of genetic admixture reflects the long but also recent history of migration waves within West Africa mainly caused by changing environmental conditions. Also, evidence for potential socio-economical influences such as trade routes is provided by the occurrence of U6b and U6d sequences found in Dubai but also in Tunisia leading to the African West Coast via Mauritania and Senegal but also via Niger, Nigeria to Cameroon.
Keywords: MtDNA, Sequencing, Control region, EMPOP, Africa, Forensic science, Ghana
1. Introduction
The so-called “Empire of Ghana”, also known as “Wagadou Empire”, flourished from 750 to 1076 AD and covered an area that spread from Mauritania to Senegal including the western parts of Mali that became rich by trading salt and gold. Besides its name, however, it is in no ways related to modern Ghana [1]. Under British rule, the area was named “Gold coast” due to the vast natural resources. With the declaration of independence in 1957, the former colony was renamed into the “Republic of Ghana”. The area of modern Ghana was reported to be first populated approximately 150,000 years before present (ybp) [2]. The settlers belonged to the Sango or Sangoan culture but with the beginning of a period of heavy dryness 25,000–15,000 ybp those people moved away from the plains [2]. Archaeological evidence from that region dates further human presence to 5800 ybp. The Kintapo culture arose during a short but dry phase from about 4000 to 2700 ybp subsiding on livestock breeding and forest land cultivation [3]. With continuous disappearance of the rain forest and the introduction of oil palm trees, a transition to farming and cultivation of forestland took place. Nevertheless, people left the region due to ongoing dryness [3]. Today's autochthonous population is considered to be descendant of groups that came from the North and Northeast immigrating the areas of the savannah between the 9th and 10th century. Between the 11th and the 15th century major migration waves took place around the Volta basin. Most present-day Ghanaians are considered descendants from several of these migrant groups, the first of which probably came down to the Volta River basin [2,4,5]. Based on language and culture, historical geographers and cultural anthropologists have classified the indigenous people of Ghana into five major groupings; Akan, Ewe, Mole-Dagbani, Guan and Ga-Adangbe [4]. The Akan ethnic group is most dominant comprising 44% of Ghana's population [6]. Whereas the Akan people occupy nearly the whole South and West of the Black Volta River and are reported to have migrated from the North occupying the forest and coastal areas as early as in the 13th century, there are no empirical data to trace such and the current representation of 3 West Africa on the EMPOP database is low with very little forensic mtDNA reports from that region of Africa. In this study we therefore investigated the matrilineal genetic diversity based on complete mitochondrial control region (CR) sequences from 191 Akan people and compared them to those from neighboring regions in order to infer phylogeographic patterns to help enrich the body of forensic mtDNA population data and also to contribute to the current EMPOP database.
2. Materials and methods
2.1. Samples and DNA extraction
A set of 193 samples was randomly selected from a group of 5100 apparently unrelated, 18–55 years old residents of a rural area in the Asante Akim North District, Ashanti Region, Ghana. All belonged to the Akan ethnic group. Donors gave their informed consent before samples were taken. They were then anonymized for analysis and transferred to Hamburg, Germany, for the forensic and laboratory data analysis. The study was approved by the Committee on Human Research Publication and Ethics, School of Medical Sciences, Kwame Nkrumah University of Science and Technology, Kumasi, Ghana. Total DNA was extracted from peripheral blood using standard protocols [7].
2.2. MtDNA sequencing and haplogroup assignment
Full mitochondrial control regions were amplified and sequenced according to EMPOP recommendations [8] resulting in sequences with a common reading frame from position 16024 to 576. Sequences were aligned to the rCRS using Sequencher V4.8 (GeneCodes, Ann Arbor, MI) following updated nomenclature guidelines for mtDNA [9]. According to our in-house data quality management process, the resulting consensus sequences were inspected independently by two different analysts and reviewed by a third person. Haplogroup affiliations were assigned according to the mitochondrial phylogenetic tree [10], build 11. Haplotypes are provided for forensic searches under accession numbers EMP00003, EMP00046, EMP00081 and EMP00082 in the EMPOP database (www.empop.org; [11]) and in supplementary file 1.
2.3. Identification of maternally closely related individuals using STR markers
All identical mitochondrial haplotypes (disregarding length variants around positions 309, 16193 and 573) were tested for accidental double analysis of the same individual or close maternal relationship between individuals (monozygotic twins, full or half siblings as well as mother/child) by STR analysis using PowerPlex ESX17 System (Promega Corporation, Madison, WI, USA). Relevant scenarios were evaluated using “Familias”, a freeware to compute probabilities and likelihoods for paternity and identification cases [13,14]. Likelihood ratios were calculated for each putative related versus unrelated sample pair. Cut-off values for exclusion scenarios were applied according to Evett [15], and modified by Bodner et al. [12].
2.4. Post laboratory data inspection
The error-rate in mtDNA analysis is generally still unacceptably high as manifested by transcriptional, clerical and interpretational errors in the literature. The visualization of datasets in form of quasi-median networks has proven a powerful tool to unmask data idiosyncrasies that could represent such errors [16]. We have recently updated network tools for investigating West Eurasian mtDNA CR sequences [17] and present here an etalon file representative of West African CR sequences in conjunction with an adapted filter file. In addition to the speedy filter presented in [11] the following transitions/transversions were filtered: A95C, G185A, G185T, C186A, A189G, A189C, G247A, G316A, A357G, G513A, C16114A, C16114T, C16168T, C16184T, A16230G and A16399G.
2.5. Diversity indices and population genetic analysis
Within our Ghana sample set the random match probability was calculated as the sum of squared CR frequencies, disregarding length variants at positions 16193, 309 and 573. This sample set was compared with 6007 published mtDNA HVS1 sequences from 19 surrounding populations [18–38]. All sequences were aligned to the greatest common range comprising positions 16030–16365. Length variants at position 16193 were disregarded. Intra- as well as interpopulation comparisons were performed based on the number of pairwise differences between sequences, molecular diversity indices and the molecular variance (AMOVA). Calculations were performed using the Arlequin software package (http://cmpg.unibe.ch/software/arlequin35/; version 3.5.1.2). In order to detect the underlying interpopulation structure we applied a multidimensional scaling (MDS) approach which uses a function minimization algorithm to maximize the goodness-of-fit. We used average population pairwise differences for calculating the multidimensional scaling plot (Supplementary Fig. 1).
3. Results and discussion
3.1. Identification of maternally closely related individuals
Identical haplotypes were further investigated with nuclear markers (STRs) to test for accidental double analysis of the same individual or presence of closely maternally related individuals following the concept outlined in [12]. Twins and double analysis of the same person were excluded due to the absence of identical STR genotypes. Mother/child relationships were excluded by direct comparison of the relevant STR genotypes. Likelihood ratios (LR) for full and half sibling constellations versus unrelated were calculated for 79 sample pairs that shared identical mtDNA haplotypes. In 75 cases LR values did not suggest close maternal relationship (LR < 10). In two sample pairs LR values indicated the presence of siblings (LRs of 1.5 × 106 and 3.3 × 104, respectively). We therefore excluded one of the identical haplotypes of each pair for further analysis. In two other cases LR values did not indicate the presence of closely maternally related individuals (LRs of 129 and 236, respectively) but potentially more distant relationships. We therefore decided to retain these haplotypes in our dataset resulting in a final number of 191 mtDNA haplotypes.
3.2. Observed CR haplotypes and diversity indices
In the Ghanaian population sample of 191 individuals we observed a total of 127 different haplotypes of which 96 were unique (disregarding length variants around positions 16193, 309 and 573) (Table 1). The most common mitochondrial CR haplotype represented the haplogroup (hg) L1b sequence motif 16126C-16187T-16189C-16223T-16264T-16270T-16278T-16293G-16311C-16519C-73G-146C-152C-182T-185T-195C-247A-263G-(309.1C)-315.1C-357G-523DEL-524DEL with nine (4.7%) observations followed by the L1c sequence motif 16129A-16182C-16183C-16189C-16213A-16215G-16223T-16278T-16291T-16294T-16311C-16355T-16360T-16519C-73G-151T-152C-182T-186A-189C-247A-263G-(309.1C)-315.1C-316A-523DEL-524DEL with eight (4.2%) observations reflecting the low diversity within the L1 haplotypes in the Akan population. The random match probability (disregarding length variants at positions 16193, 309 and 573) was 1.3%. Hence, the power of discrimination by CR was calculated as 98.7%.
Table 1.
Diversity indices for 6198 published HVS1 mtDNA sequences including 191 Ghanaian samples from this study. HVS1 motives span from 16030 to 16365, length variation around position 16193 was disregarded. The comparison was performed among 20 populations (abbreviated according to the international three letter code).
| Population | GHAa | GHA | CAF | CMR | EGY | ETH | GAB |
|---|---|---|---|---|---|---|---|
| N | 191 | 191 | 56 | 1131 | 312 | 185 | 946 |
| Reference | This study | This study | [37] | [18,19,20,21,37,38] | [22,23] | [19,24,33] | [37] |
| Nr. of haplotypes | 127 | 99 | 11 | 393 | 204 | 105 | 264 |
| Nr. of unique haplotypes | 96 | 69 | 3 | 252 | 150 | 64 | 144 |
| Haplotype diversity | 0.987 | 0.98 | 0.81 | 0.98 | 0.99 | 0.99 | 0.98 |
| Mean pairwise differences | 15.55 ± 6.97 | 7.79 ± 3.64 | 6.01 ± 2.90 | 9.85 ± 4.51 | 7.03 ± 3.30 | 9.96 ± 4.57 | 9.68 ± ± 4.44 |
| Population | GHA | BFA | GNB | KEN | LBY | MAR | MLI |
|---|---|---|---|---|---|---|---|
| N | 238 | 137 | 373 | 178 | 129 | 56 | 147 |
| Reference | [20] | [18,19,34] | [25,33] | [19,26,27,33] | [27] | [28] | [19,34,36] |
| Nr. of haplotypes | 137 | 38 | 183 | 140 | 19 | 42 | 98 |
| Nr. of unique haplotypes | 101 | 17 | 120 | 116 | 5 | 37 | 76 |
| Haplotype diversity | 0.98 | 0.95 | 0.98 | 0.99 | 0.67 | 0.95 | 0.98 |
| Mean pairwise differences | 7.00 ± 3.30 | 6.57 ± 3.12 | 7.79 ± 3.64 | 9.56 ± 4.40 | 4.12 ± 2.06 | 6.34 ± 3.05 | 6.72 ± ± 3.19 |
| Population | MRT | NER | NGA | SEN | SLE | TCD | TUN |
|---|---|---|---|---|---|---|---|
| N | 64 | 75 | 1302 | 268 | 277 | 69 | 64 |
| Reference | [36] | [19,34] | [19,20,35] | [30] | [31] | [18,33] | [28] |
| Nr. of haplotypes | 43 | 53 | 449 | 165 | 151 | 40 | 49 |
| Nr. of unique haplotypes | 33 | 42 | 278 | 125 | 98 | 30 | 41 |
| Haplotype diversity | 0.96 | 0.97 | 0.99 | 0.99 | 0.99 | 0.95 | 0.97 |
| Mean pairwise differences | 6.41 ± 3.07 | 7.62 ± 3.56 | 8.43 ± 3.90 | 6.75 ± 3.19 | 7.90 ± 3.69 | 7.04 ± 3.35 | 7.51 ± 3.55 |
Diversity indices of 191 mtDNA sequences from Ghana based on complete control region haplotypes spanning positions 16024-576 (length variation around positions 16193, 309 and 573 was disregarded).
3.3. Point heteroplasmy
We found nine point heteroplasmic transitions and one point heteroplasmic transversion in a total of ten samples, out of which six (16189Y, 16093Y, 215R, 64Y, 16309R, 228R) coincide with typical heteroplasmic hotspots [39]. In two instances (16286Y and 16525R) we found heteroplasmy that was not observed earlier [39]. In addition, four of these six heteroplasmic mutations affected evolutionary fast sites [40,41]. The observed point heteroplasmy at position 215 confirms earlier reports [39] that explain the evolutionary stability of this position with regulatory functions [42]. A mixture containing mitochondrial populations carrying either a C or a T at position 16093 occurred twice in our dataset. The transversion 16182M was also observed by [39] at a frequency of 0.06%.
3.4. Haplogroup composition and distribution
The 191 haplotypes from Ghana were assigned to 45 distinct CR haplogroups (Table 2) according to the mitochondrial phylogeny [10], build 11. The overwhelming majority (98.4%) belonged to the three Africa-specific haplogroups L1–L3. The occurrence of L1–L3 branches in West Africa are documented by Salas et al. [43], Veeramah et al. [20] and González et al. [36] with which our data are in general accordance. However, within L1 (26% of all sequences in our sample set) our results indicate a dominance of L1b lineages (54%) in Ghanaians, whereas the L1c cluster constituted the majority in [43]. Recent findings of Veeramah et al. display generally lower levels of L1 lineages in Ghana, in total 16%, but with similar ratios between L1b (50%) and L1c (32%) as compared to our dataset. A similar observation accounts for L3e and L3f subclusters, which were most frequent (66.2%) among Ghanaian L3 lineages in our study which is also in accordance with observations of Veeramah et al. [20] whereas L3b and L3d were the dominant L3 haplogroups in [43]. Contrary to Veeramah et al. reporting the absence of L3e1 haplotypes in their Ghanaian samples while being present in neighboring regions we detected at least low levels (2%) of L3e1 haplotypes in our data. Haplogroup distribution of West African population of Mauritania [36] showed significantly higher proportions of non L lineages. Approximately 55% of their sequences had Euroasiatic provenance. This dominant Euroasiatic influence which cannot be recognized in regions south of Mauritania is explained by the fact that the Mauritanian samples derived from Berber/Arabic individuals from Northern regions. Mauritania constitutes an important barrier to southwards gene flow as three quarters of the region is desert or semidesert.
Table 2.
Haplogroup frequencies of 191 samples from Ghana based on full mtDNA control region sequences (16024-576); haplogroup nomenclature according to phylotree [10], build 11.
| Haplogroup | N | Frequency (%) |
|---|---|---|
| L0a1b1 | 1 | 0.52 |
| L1b* | 1 | 0.52 |
| L1b1* | 18 | 9.42 |
| L1b1a | 8 | 4.19 |
| L1c1* | 1 | 0.52 |
| L1c1d | 1 | 0.52 |
| L1c3* | 2 | 1.05 |
| L1c3a | 15 | 7.85 |
| L1c3b1 | 4 | 2.09 |
| L2a1′2* | 1 | 0.52 |
| L2a1* | 32 | 16.75 |
| L2a1a1 | 1 | 0.52 |
| L2a1a2 | 5 | 2.62 |
| L2a1a3 | 6 | 3.14 |
| L2a1c1 | 2 | 1.05 |
| L2a1c2 | 4 | 2.09 |
| L2a1c5 | 1 | 0.52 |
| L2a1h | 3 | 1.57 |
| L2a1i | 3 | 1.57 |
| L2a1l | 1 | 0.52 |
| L2b* | 2 | 1.05 |
| L2b1* | 1 | 0.52 |
| L2b1a | 1 | 0.52 |
| L2b2 | 5 | 2.62 |
| L2c3 | 1 | 0.52 |
| L2e | 1 | 0.52 |
| L3b* | 10 | 5.24 |
| L3b1b | 3 | 1.57 |
| L3b2 | 2 | 1.05 |
| L3d* | 2 | 1.05 |
| L3d1c | 2 | 1.05 |
| L3d1d | 2 | 1.05 |
| L3d4 | 1 | 0.52 |
| L3e1 | 4 | 2.09 |
| L3e2 | 2 | 1.05 |
| L3e2a1 | 10 | 5.24 |
| L3e2b* | 14 | 7.33 |
| L3e2b3 | 1 | 0.52 |
| L3e3 | 3 | 1.57 |
| L3e4 | 1 | 0.52 |
| L3f1b* | 9 | 4.71 |
| L3f1b1 | 1 | 0.52 |
| L3h1b1a1 | 1 | 0.52 |
| U6a* | 1 | 0.52 |
| U6a3b | 1 | 0.52 |
We were able to dissect the L1 haplotypes in three L1b and five L1c lineages whereof L1b1 (67% of L1b lineages) and L1c3a (65% of L1c lineages) were most frequent. Low diversity in haplogroup L1b based on HVS1 and 2 sequences was also observed by [37] as six out of nine known L1b subclades can only be characterized based on coding region mutations. Contrarily to our study, L1c sequences displayed high diversity values in [37]. Only 0.7% remained further undissolved, while 44.6% were assigned to L1c1a1, 28.5% to L1c1a2, 2.1% to L1c1, 6.8% to L1c1b, 4.6% to L1c2 and 6.2% to L1c3.
Our study confirms the weak signal of haplogroup L0a as well as U6a presence in West Africa [20,36,42]. Within our sample set we found only one L0a1b1 (0.5%) and two U6a (1%) lineages (Table 2). These haplogroups show striking regional differences. Haplogroup L0a reaches a frequency of up to 25% in East Africa and is dispersed at low frequency north of the Sahara [23,27,43], while haplogroup U6a arrives at frequencies up to 45% in North Africa as a re-immigration signal from the Near East [43]. Very interesting, in that respect is the finding, that we only found U6a lineages that have been reported to be dispersed from the North Maghrib region including the Atlas Mountains and the coastal plain of Morocco, Algeria, Tunisia, and Libya to the East including Syria and Ethiopia as well as to the Canary Islands in the west [44]. In contrast, within our Ghanaian population sample we did not find U6b or U6d lineages which have been reported to be distributed only sporadically in the North but found more frequently in western regions such as Senegal and Nigeria [44]. It has been proposed that the occurrence of U6b in diverse western African countries reflects either a wider distribution in the past or more recent gene flow from yet unsampled regions [44]. Our U6b and U6d haplogroup screening among 6198 mtDNA sequences from states neighboring Ghana rather indicates potential migration or trade routes from North Africa to the Western coast, as evidenced by the occurrence of U6b and U6d lineages in neighboring countries Mauritania and Senegal. A second intercontinental route reveals the migration from North Africa via Niger, Nigeria to the Cameroon evidenced by U6b and U6d signals within those countries. U6b lineages have also been detected in Saudi Arabia [38], hence these findings are in agreement with reported trade routes from the North/Near East including the introduction of the camel in that area [4]. The absence of M1 lineages in our Ghanaian population goes along with previous findings that M1 is most frequent in East Africa with two appreciable gradients one diminishing to the South and a second to the West [29].
Macrohaplogroups L4–L6 were absent in our Ghanaian dataset. Based on available data haplogroup L4 seems more abundant within East Africa [44] but only scarcely present in West Africa (Nigeria and Ghana [20]). Haplogroup L5 also seems to show higher frequencies in East Africa [24,26,45,46] with traces in the Congo [36] as well as in Cameroon [21] while haplogroup L6 seems to be more frequent north of the equator [20,24].
3.5. Population comparisons
We compared HVS1 sequences with a common range of 16030–16365 of 20 African populations comprising samples from Ghana (GHA) whereof 191 were from this study, Central African Republic (CAF), Cameroon (CMR), Egypt (EGY), Ethiopia (ETH), Gabon (GAB), Burkina Faso (BFA), Guinea-Bissau (GNB), Kenya (KEN), Libyan Arab Jamahiriya (LBY), Morocco (MAR), Mali (MLI), Mauritania (MRT), Niger (NER), Nigeria (NGA), Senegal (SEN), Sierra Leone (SLE), Chad (TCD) and Tunisia (TUN) (Table 1).
Intrapopulation diversity values (expressed as average number of pairwise differences) ranged from 9.96 ± 4.57 (Ethiopia) to 4.12 ± 2.06 (Libyan Arab Jamahiriya).
The great majority of the observed populations varied significantly in terms of their genetic structure (Supplementary Table 1(3)). AMOVA was used to test for significant variation in the mtDNA structure among the populations (Supplementary Table 1). The vast majority of the observed variance (92.7%) within the 20 populations was attributable to differences within populations, only 7.3% represented differences among populations (Supplementary Table 1 (1)). An MDS plot was calculated to depict the dissimilarities between the observed populations as distance matrix based on average population pairwise differences (Supplementary Fig. 1). The MDS plot clearly evidenced that our Ghana sequences clustered together with those populations being geographically closest (Sierra Leone, Guinea Bissau, Mali, Niger, Nigeria, Burkina Faso) which could reflect recent maternal gene flow and admixture among those areas. Also, geographically more distant populations clustered together such as Tunisia, Morocco, Libyan Arab Jamahiriya and Egypt as well as Ethiopia and Kenya, but also Senegal and Mauritania.
3.6. Network analysis
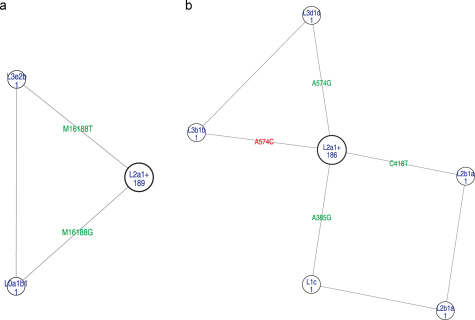
The passage of the 191 Ghanaian CR sequences through the modified filter resulted in simple quasi-median network torsi for both HVS1 (16024-16569) and HVS2 (1-576) sequences (Fig. 1). The addition of 99 West African CR sequences only slightly increased the complexity of both network torsi (Supplementary Fig. 2a and b). The expected star-like shaped torsi even remained for larger data sets including high-quality literature data (up to a total size of 450, data not shown) which demonstrates the robustness of the etalon and the adapted filter. We filtered both speedy (e.g. transitions at 16384, 16399) as well as relatively stable mutations (e.g. transitions at 357, 16320), as those would otherwise introduce reticulations and parallel structures that make the network difficult to read. Similar to earlier experience [17] we expect this etalon to serve as probative means to evaluate new population data from West Africa.
Fig. 1.
Quasi-median network torsi of: (a) HVS1 (16024-16569) and (b) HVS2 (1-576) of 191 Ghanaian sequences after passage through the adapted EMPOP speedy filter [11].
4. Conclusions
Here we present the first mtDNA data set providing full control region sequences from Ghana which is of high forensic importance due to the actual underrepresentation of African mtDNA data in general. So far, EMPOP holds only 377 African sequences that meet the quality criterion “forensic data” (see www.empop.org; “concept”) representing 100 from Kenya in East Africa and 277 from Egypt in North Africa. Other mtDNA databases show comparable lack of African mtDNA data. In addition we present a West African etalon dataset that can be used in conjunction with an adapted filter for graphical representation and quality control of new West African mtDNA sequences by means of quasi-median network analysis.
Acknowledgements
We would like to thank Christian Timmann, Rolf D. Horstmann (Dept. of Molecular Medicine) and Bernhard Nocht (Institute for Tropical Medicine, Hamburg, Germany) for the collection of samples and Daniela Niederwieser (Institute of Legal Medicine, Innsbruck Medical University) for valuable technical assistance. This study received support by the Austrian Science Fund (FWF): project L397.
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.fsigen.2011.05.011.
Appendix A. Supplementary data
References
- 1.Munson P.J. Archaeology and the prehistoric origins of the Ghana empire. J. Afr. Hist. 1980;21:457–466. [Google Scholar]
- 2.Shillington K. 2nd ed. Palgrave Macmillan; New York: 2005. History of Africa. [Google Scholar]
- 3.Casey J., Sawatzky R., Godfrey-Smith D.I., Quickert N., Wollstonecroft M., Hawkins A. Report of investigations at the Birimi site in Northern Ghana. Nyame Akuma. 1997;48:32–38. [Google Scholar]
- 4.La Verle B. Claitor's Law Books and Publishing Division; 1994. Ghana: a Country Study. [Google Scholar]
- 5.Gocking R.S. Greenwood Press; London: 2005. The History of Ghana. [Google Scholar]
- 6.The World Factbook, Washington, DC, Central Intelligence Agency, https://www.cia.gov/library/publications/the-world-factbook/index.html, 2009.
- 7.Miller S.A., Dykes D.D., Polesky H.F. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parson W., Bandelt H.J. Extended guidelines of mtDNA typing of population data in forensic science. Forensic Sci. Int. Genet. 2007;1:13–19. doi: 10.1016/j.fsigen.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 9.Bandelt H.-J., Parson W. Consistent treatment of length variants in the human mtDNA control region: a reappraisal. Int. J. Legal Med. 2007;122:11–21. doi: 10.1007/s00414-006-0151-5. [DOI] [PubMed] [Google Scholar]
- 10.van Oven M., Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum. Mutat. 2008;1039:e386–e394. doi: 10.1002/humu.20921. [DOI] [PubMed] [Google Scholar]
- 11.Parson W., Dür A. EMPOP – a forensic mtDNA database. Forensic Sci. Int. Genet. 2007;1:88–92. doi: 10.1016/j.fsigen.2007.01.018. [DOI] [PubMed] [Google Scholar]
- 12.Bodner M., Irwin J.A., Coble M.D., Parson W. Inspecting close maternal relatedness: towards better mtDNA population samples in forensic databases. Forensic Sci. Int. Genet. 2011;5:138–141. doi: 10.1016/j.fsigen.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Egeland T., Mostad P.F., Mevåg B., Stenersen M. Beyond traditional paternity and identification cases. Selecting the most probable pedigree. Forensic Sci. Int. 2000;110:47–59. doi: 10.1016/s0379-0738(00)00147-x. [DOI] [PubMed] [Google Scholar]
- 14.Drábek J. Validation of software for calculating the likelihood ratio for parentage and kinship. Forensic Sci. Int. Genet. 2009;3:112–118. doi: 10.1016/j.fsigen.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 15.Evett I.W., Weir B.S. Sinauer Associates; Sunderland, MA: 1998. Interpreting DNA Evidence. [Google Scholar]
- 16.Bandelt H.J., Dür A. Translating DNA data tables into quasi-median networks for parsimony analysis and error detection. Mol. Phylogenet. Evol. 2007;42:256–271. doi: 10.1016/j.ympev.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 17.Zimmermann B., Röck A., Huber G., Krämer T., Schneider P.M., Parson W. Application of a west Eurasian-specific filter for quasi-median network analysis: sharpening the blade for mtDNA error detection. Forensic Sci. Int. Genet. 2011;5:133–137. doi: 10.1016/j.fsigen.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Černý V., Hájek M., Bromová M., Čmejla R., Diallo I., Brdička R. MtDNA of Fulani nomads and their genetic relationships to neighboring sedentary populations. Hum. Biol. 2006;78:9–27. doi: 10.1353/hub.2006.0024. [DOI] [PubMed] [Google Scholar]
- 19.Watson E., Bauer K., Aman R., Weiss G., von Haeseler A., Pääbo S. mtDNA sequence diversity in Africa. Am. J. Hum. Genet. 1996;59:437–444. [PMC free article] [PubMed] [Google Scholar]
- 20.Veeramah K.R., Connell B.A., Pour N.A., Powell A., Plaster C.A., Zeitlyn D., Mendell N.R., Weale M.E., Bradman N., Thomas M.G. Little genetic differentiation as assessed by uniparental markers in the presence of substantial language variation in peoples of the Cross River region of Nigeria. BMC Evol. Biol. 2010;10:92. doi: 10.1186/1471-2148-10-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coia V., Destro-Bisol G., Verginelli F., Battaggia C., Boschi I., Cruciani F., Spedini G., Comas D., Calafell F. Brief communication: mtDNA variation in North Cameroon: lack of Asian lineages and implications for back migration from Asia to sub-Saharan Africa. Am. J. Phys. Anthropol. 2005;128:678–681. doi: 10.1002/ajpa.20138. [DOI] [PubMed] [Google Scholar]
- 22.Kujanová M., Pereira L., Fernandes V., Pereira J.B., Černý V. Near eastern neolithic genetic input in a small oasis of the Egyptian Western Desert. Am. J. Phys. Anthropol. 2009;140:336–346. doi: 10.1002/ajpa.21078. [DOI] [PubMed] [Google Scholar]
- 23.Saunier J.L., Irwin J.A., Strouss K.M., Ragab H., Sturk K.A., Parsons T.J. Mitochondrial control region sequences from an Egyptian population sample. Forensic Sci. Int. Genet. 2009;3:e97–103. doi: 10.1016/j.fsigen.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 24.Poloni E.S., Naciri Y., Bucho R., Niba R., Kervaire B., Excoffier L., Langaney A., Sanchez-Mazas A. Genetic evidence for complexity in ethnic differentiation and history in East Africa. Ann. Hum. Genet. 2009;73:582–600. doi: 10.1111/j.1469-1809.2009.00541.x. [DOI] [PubMed] [Google Scholar]
- 25.Rosa A., Brehm A., Kivisild T., Metspalu E., Villems R. MtDNA profile of West Africa Guineans: towards a better understanding of the Senegambia region. Ann. Hum. Genet. 2004;68:340–352. doi: 10.1046/j.1529-8817.2004.00100.x. [DOI] [PubMed] [Google Scholar]
- 26.Brandstätter A., Peterson C.T., Irwin J.A., Mpoke S., Koech D.K., Parson W., Parsons T.J. Mitochondrial DNA control region sequences from Nairobi (Kenya): inferring phylogenetic parameters for the establishment of a forensic database. Int. J. Legal Med. 2004;118:294–306. doi: 10.1007/s00414-004-0466-z. [DOI] [PubMed] [Google Scholar]
- 27.Ottoni C., Martínez-Labarga C., Loogväli E.L., Pennarun E., Achilli A., De A.F., Trucchi E., Contini I., Biondi G., Rickards O. First genetic insight into Libyan Tuaregs: a maternal perspective. Ann. Hum. Genet. 2009;73:438–448. doi: 10.1111/j.1469-1809.2009.00526.x. [DOI] [PubMed] [Google Scholar]
- 28.Turchi C., Buscemi L., Giacchino E., Onofri V., Fendt L., Parson W., Tagliabracci A. Polymorphisms of mtDNA control region in Tunisian and Moroccan populations: an enrichment of forensic mtDNA databases with Northern Africa data. Forensic Sci. Int. Genet. 2009;3:166–172. doi: 10.1016/j.fsigen.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 29.González A.M., Larruga J.M., Abu-Amero K.K., Shi Y., Pestano J., Cabrera V.M. Mitochondrial lineage M1 traces an early human backflow to Africa. BMC Genomics. 2007;8:223. doi: 10.1186/1471-2164-8-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rando J.C., Pinto F., González A.M., Hernández M., Larruga J.M., Cabrera V.M., Bandelt H.-J. Mitochondrial DNA analysis of northwest African populations reveals genetic exchanges with European, near-eastern, and sub-Saharan populations. Ann. Hum. Genet. 1998;62:531–550. doi: 10.1046/j.1469-1809.1998.6260531.x. [DOI] [PubMed] [Google Scholar]
- 31.Jackson B.A., Wilson J.L., Kirbah S., Sidney S.S., Rosenberger J., Bassie L., Alie J.A., McLean D.C., Garvey W.T., Ely B. Mitochondrial DNA genetic diversity among four ethnic groups in Sierra Leone. Am. J. Phys. Anthropol. 2005;128:156–163. doi: 10.1002/ajpa.20040. [DOI] [PubMed] [Google Scholar]
- 32.Diegoli T.M., Irwin J.A., Just R.S., Saunier J.L., O’Callaghan J.E., Parsons T.J. Mitochondrial control region sequences from an African American population sample. Forensic Sci. Int. Genet. 2009;4:e45–e52. doi: 10.1016/j.fsigen.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 33.Černý V., Fernandes V., Costa M.D., Hájek M., Mulligan C.J., Pereira L. Migration of Chadic speaking pastoralists within Africa based on population structure of Chad Basin and phylogeography of mitochondrial L3f haplogroup. BMC Evol. Biol. 2009;9:63. doi: 10.1186/1471-2148-9-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pereira L., Černý V., Cerezo M., Silva N.M., Hájek M., Vašíková A., Kujanová M., Brdička R., Salas A. Linking the sub-Saharan and West Eurasian gene pools: maternal and paternal heritage of the Tuareg nomads from the African Sahel. Eur. J. Hum. Genet. 2010:1–9. doi: 10.1038/ejhg.2010.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vigilant L., Stoneking M., Harpending H., Hawkes K., Wilson A.C. African populations and the evolution of human mitochondrial DNA. Science. 1991;27:1503–1507. doi: 10.1126/science.1840702. [DOI] [PubMed] [Google Scholar]
- 36.González A.M., Cabrera V.M., Larruga J.M., Tounkara A., Noums G., Thomas B.N., Moulds J.M. Mitochondrial DNA variation in Mauritania and Mali and their genetic relationship to other Western Africa populations. Ann. Hum. Genet. 2006;70:631–657. doi: 10.1111/j.1469-1809.2006.00259.x. [DOI] [PubMed] [Google Scholar]
- 37.Quintana-Murci L., Quach H., Harmant C., Luca F., Massonnet B., Patin E., Sica L., Mouguiama-Daouda P., Comas D., Tzur S., Balanovsky O., Kidd K.K., Kidd J.R., van der Veen L., Hombert J.M., Gessain A., Verdu P., Froment A., Bahuchet S., Heyer E., Dausset J., Salas A., Behar D.M. Maternal traces of deep common ancestry and asymmetric gene flow between Pygmy hunter-gatherers and Bantu-speaking farmers. Proc. Natl. Acad. Sci. U.S.A. 2008;105:1596–1601. doi: 10.1073/pnas.0711467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tishkoff S.A., Gonder M.K., Henn B.M., Mortensen H., Knight A., Gignoux C., Fernandopulle N., Lema G., Nyambo T.B., Ramakrishnan U., Reed F.A., Mountain J.L. History of click-speaking populations of Africa inferred from mtDNA and Y chromosome genetic variation. Mol. Biol. Evol. 2007;24:2180–2195. doi: 10.1093/molbev/msm155. [DOI] [PubMed] [Google Scholar]
- 39.Irwin J.A., Saunier J.L., Niederstätter H., Strouss K.M., Sturk K.A., Diegoli T.M., Brandstätter A., Parson W., Parsons T.J. Investigation of heteroplasmy in the human mitochondrial DNA control region: a synthesis of observations from more than 5000 global population samples. J. Mol. Evol. 2009;68:516–527. doi: 10.1007/s00239-009-9227-4. [DOI] [PubMed] [Google Scholar]
- 40.Bandelt H.-J., Kong Q.P., Richards M., Macaulay V. Estimation of mutation rates and coalescence times: some caveats. In: Bandelt H.-J., Macaulay V., Richards M., editors. Human Mitochondrial DNA and the Evolution of Homo Sapiens. Springer-Verlag; Berlin, Germany: 2006. pp. 47–90. [Google Scholar]
- 41.Bandelt H.-J., Quintana-Murci L., Salas A., Macaulay V. The fingerprint of phantom mutations in mitochondrial DNA data. Am. J. Hum. Genet. 2002;71:1150–1160. doi: 10.1086/344397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Santos C., Sierra B., Álvarez L., Ramos A., Fernández E., Nogués R., Aluja M.P. Frequency and pattern of heteroplasmy in the control region of human mitochondrial DNA. J. Mol. Evol. 2008;67:191–200. doi: 10.1007/s00239-008-9138-9. [DOI] [PubMed] [Google Scholar]
- 43.Salas A., Richards M., De la Fe T., Lareu M.V., Sobrino B., Sánchez-Diz P., Macaulay V., Carracedo Á. The making of the African mtDNA landscape. Am. J. Hum. Genet. 2002;71:1082–1111. doi: 10.1086/344348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maca-Meyer N., González A.M., Pestano J., Flores C., Larruga J.M., Cabrera V.M. Mitochondrial DNA transit between West Asia and North Africa inferred from U6 phylogeography. BMC Genetics. 2003;4:15. doi: 10.1186/1471-2156-4-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Castrì L., Tofanelli S., Garagnani P., Bini C., Fosella X., Pelotti S., Paoli G., Pettener D., Luiselli D. mtDNA variability in two Bantu-speaking populations (Shona and Hutu) from Eastern Africa: implications for peopling and migration patterns in sub-Saharan Africa. Am. J. Phys. Anthropol. 2009;140:302–311. doi: 10.1002/ajpa.21070. [DOI] [PubMed] [Google Scholar]
- 46.Pereira L., Macaulay V., Torroni A., Scozzari R., Prata M.J., Amorim A. Prehistoric and historic traces in the mtDNA of Mozambique: insights into the Bantu expansions and the slave trade. Ann. Hum. Genet. 2001;65:439–458. doi: 10.1017/S0003480001008855. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.