Abstract
The K antigenic capsular polysaccharide forms a structural layer, the capsule, on the surfaces of Escherichia coli cells. The capsule provides an important protective covering that helps protect encapsulated bacteria from host immune defenses. The assembly and translocation of the capsule requires proteins in the inner and outer membranes. The inner membrane protein Wzc is a tyrosine autokinase that plays an essential role in what is believed to be a coordinated biosynthesis and secretion process. Mutants lacking Wzc can form K antigen oligosaccharides but are unable to polymerize high molecular weight capsular polymers. Wzc homologs have been identified in exopolymer biosynthesis systems in many different Gram-negative and -positive bacteria. Using single particle averaging on cryo-negatively stained samples, we have produced the first three-dimensional structure of this type of membrane protein in its phosphorylated state at ~14 Å resolution. Perfluoro-octanoate-PAGE analysis of detergent-solubilized oligomeric Wzc and symmetry analysis of the transmission electron microscopy data clearly demonstrated that Wzc forms a tetrameric complex with C4 rotational symmetry. Viewed from the top of the complex, the oligomer is square with a diameter of ~100 Å and can be divided into four separate densities. From the side, Wzc is ~110 Å high and has a distinctive appearance similar to an extracted molar tooth. The upper “crown” region is ~55 Å high and forms a continuous ring of density. Four unconnected “roots” (~65 Å high) emerge from the underside of the crown. We propose that the crown is formed by protein-protein contacts from the four Wzc periplasmic domains, while each root represents an individual cytoplasmic tyrosine autokinase domain.
Escherichia coli produces ~80 structurally and immunochemically distinct capsular polysaccharides (1). These major surface structures define the K antigen in serotyping analyses, and they form a surface structure known as the capsule. The capsule is a continuous and coherent surface layer, and its primary role is to provide a protective barrier and maintain the cell in a hydrated state. It is also an important virulence determinant that enables pathogenic bacteria to evade or counteract the nonspecific host defense during the early (preimmune) phase of infection by interfering with the action of complement and phagocytes (1). Capsular polysaccharides vary in monosaccharide composition, glycosidic linkages, and substitution with side branch glycoses and non-carbohydrate residues. Based on structural, genetic, and biochemical criteria, the capsules of E. coli have been classified into four groups (2), and in terms of biosynthesis, there are two distinct pathways. Groups 1 and 4 capsules use a Wzy-dependent system, whereas assembly of groups 2 and 3 follows an ATP-binding cassette transporter-dependent process. Both biosynthesis mechanisms are widely represented in capsules and other glycoconjugates in a variety of bacterial species. The E. coli system therefore serves as a paradigm for this process in a broad range of bacteria, including significant pathogens of plants, livestock, and humans (2-4).
The current working model for the Wzy-dependent assembly pathway is based on dissection of the system by biochemical and genetic means with some interpretation of corresponding reactions in lipopolysaccharide O antigen synthesis (3, 4). In this model, individual repeat unit oligosaccharides are assembled on undecaprenol diphosphate lipid carriers at the cytoplasmic face of the inner membrane, which provide a pool of subunits for assembly. These intermediates are then transported across the inner membrane in what is proposed to be a “flippase”-type reaction by a process involving the Wzx protein. Once the oligosaccharide moiety is transferred to the periplasm, it forms the substrate for the polymerization machinery. The characteristic component of this process is the putative polymerase Wzy, an integral inner membrane protein. These initial steps in the Wzy-dependent pathway are conserved in groups 1 and 4 E. coli capsules and in lipopolysaccharide O antigens, capsules, and exopolysaccharides from a variety of bacteria. Notably, these early reactions are found in some Gram-positive bacteria, including staphylococci and streptococci (4). The terminal stages in assembly of these polymers on bacterial cell surfaces differ according to the nature of the glycoconjugate involved and the structure of the cell envelope (i.e. Gram-negative versus Gram-positive). The terminal stages in group 1 capsule assembly require the outer membrane protein Wza, a member of the outer membrane auxiliary family (5). The presence of Wza is essential for the assembly of the prototype group 1 capsule structure on the surface of E. coli serotype K30 (6, 7). Wza forms stable oligomeric “donut”-shaped complexes with a molecular mass of ~300 kDa, and a preliminary study of Wza, based on small two-dimensional crystalline areas, revealed ring-like complexes with an average outer diameter of ~90–110 Å and a central stained region of ~20–35 Å in diameter (20). A three-dimensional structure for Wza, calculated using single particle analysis of cryo-negatively stained data (9), revealed that Wza forms an octamer with an unusual rotational organization. Oligomeric Wza appears to be formed by a novel tetrameric ring of dimerically associated Wza (i.e. Wza is a tetramer of dimers). Viewed from the side, the oligomer has two distinctive layers with a cavity in the center of the complex (9). Wza is thought to provide a capsule translocon for export of capsular polysaccharide across the outer membrane, and it certainly has structural features in common with members of the secretin superfamily of proteins (e.g. PilQ, pIV, PulD, XcpQ, and YscC (10-14)). These secretins translocate a variety of proteins and pili through the outer membrane. However, Wza shares no sequence homology with them (6, 7). The remaining outer membrane component Wzi is required for surface association of the capsule (15), and the means of capsule attachment is currently unclear.
The polymerization and assembly of group 1 capsular polymer appears to be a coupled process in that mutants defective in late steps do not accumulate intracellular or periplasmic polymer (6, 16, 20). This would suggest a connection between the inner and outer membranes, and Wza can, in fact, be cross-linked to an integral inner membrane component, Wzc in vivo (20). Analysis of the primary sequence of the highly conserved Wzc homolog from E. coli K12 predicts a bitopic protein with large cytoplasmic and periplasmic domains (17), and no structural studies have been performed in detail on any member of the Wzc family. In Gram-positive bacteria, functional homologs of Wzc are composed of two polypeptides, one representing the periplasmic component and the other the cytoplasmic C-terminal domain (4, 18). The C-terminal cytoplasmic domain of Wzc from E. coli K30 contains tyrosine autokinase activity with seven tyrosine residues in the last 17 residues (ASSYYRYGHNHYGYSYYDKK721) (16, 19). In the prototype system from E. coli K30, capsule assembly is dependent on the activity of Wzc and its cognate cytoplasmic phosphotyrosine phosphatase, Wzb (6, 16, 19). Analysis of different wzc mutants with defects in the Walker box or in the C-terminal tyrosine-rich domain indicated that both are essential for capsule assembly and that the phosphorylation process occurs via trans-phosphorylation requiring the interaction of Wzc monomers (16). No single tyrosine residue is essential or required for function in capsule assembly; it appears that the load of tyrosine phosphorylation rather than precise location is important for function (19). The requirement for the Wzb phosphatase is consistent with a cycling of phosphorylation during export (16). The absence of Wzc does not prevent the cell from making K30 antigen oligosaccharides, but high molecular weight capsular polymers are absent (6, 16, 19, 20). Thus, Wzc may provide a key role in regulating capsule formation and coupling Wza to the inner membrane.
Structural studies of the protein apparatus employed in carbohydrate export remain in their infancy, yet these molecular machines are very important in understanding the biology of capsular polysaccharide biosynthesis and the development of possible new anti-microbial drugs directed against this essential feature in many pathogens. To further understand this complex system, we have determined the three-dimensional structure of the inner membrane protein Wzc to a resolution of 14 Å. This is the first structural data available on this important class of integral inner membrane protein.
MATERIALS AND METHODS
Wzc Purification
The open reading frame of the gene encoding E. coli Wzc with an N-terminal His tag (His6-Wzc) was cloned in the expression vector pWQ305 (20) (pBAD24-derivative (21)) and transformed into E. coli CWG281 (16) cells. 10-liter agitated fermenter batches of LB medium (containing 100 μg/m ampicillin) were used to grow bacteria expressing His6-Wzc. Expression of Wzc was induced at A600 = 0.5 with 0.02% (w/v) l-arabinose, and the cells were grown for a further 4 h at 37 °C. The cells were then collected by centrifugation at 6,000 × g (4 °C) for 15 min, and the cell pellet was resuspended in 20 mm sodium phosphate, pH 7.0, and 1 mm phenylmethylsulfonyl fluoride. Bacterial cells were disrupted using a French press (110 megapascals), and the crude extract was fractionated by ultracentrifugation (85,000 × g). Cell envelopes were resuspended in solubilizing buffer (20 mm sodium phosphate, pH 7.0, 500 mm NaCl, and 0.1% (w/v) dodecyl maltoside (DDM)4) and incubated at 4 °C overnight. Cell debris was removed by centrifugation at 85,000 × g for 1 h at 4 °C, and the resulting supernatant was loaded onto a 5-ml Hi-Trap chelating HP column (Amersham Biosciences) equilibrated with solubilizing buffer containing 0.008% (w/v) DDM and eluted with a step gradient using the same buffer containing 1 m imidazole. The sample was then dialyzed against 20 mm Tris-base, pH 7.0, 300 mm NaCl, and 0.008% (w/v) DDM and loaded onto a Superdex 200 size exclusion column (Pharmacia). The His6-Wzc was then eluted using 1.2× column volumes of the same buffer, with one major peak at ~350 kDa. The sample was dialyzed overnight in 20 mm Tris-base, pH 7.5, 100 mm NaCl, and 0.008% (w/v) DDM. Matrix-assisted laser desorption ionization time-of-flight showed that the Wzc protein was purified in a fully phosphorylated state with all C-terminal Tyr residues modified (data not shown).
Perfluoro-octanoic Acid (PFO)-PAGE of Oligomeric Wzc
PFO-PAGE was performed using a variation on previously reported methods (22). 10-μl Wzc samples (50–600 μg/ml) were mixed 1:1 with PFO sample buffer (100 mm Tris, pH 7.5, 8% (w/v) Na-PFO, and 20% (v/v) glycerol) and incubated at 4 °C for 10 min. Samples were then loaded onto 7.5% slab gels containing 375 mm Tris, pH 7.5, and run in a buffer containing 25 mm Tris, pH 7.5, 200 mm glycerol, and 8% (w/v) Na-PFO for 2 h (100 V) at 4 °C. All gels and solutions were precooled to 4 °C for several h before use.
Cryo-negative Staining of Wzc
Carbon-coated copper grids (No. 400) were glow-discharged and placed shiny side down on the surface of a 3-μl droplet of sample containing 50 μg/ml His6-Wzc for 2 min and then immediately placed on a 10-μl droplet of freshly prepared 12% (w/v) ammonium molybdate tetrahydrate (pH 6.8) containing 1% (w/v) trehalose for several seconds. Grids where then briefly blotted onto double-layered Whatman 50 filter paper, and data were immediately recorded in Oxford system cryo-stage at ~100 K. Images for each oligomeric complex were recorded and processed as described in Table 1.
TABLE 1. Cryo-electron microscopy and image analysis information.
| Microscope | Philips CM200 FEG | ||
| Mode | TEM low dose recording at 100K | ||
| Operating voltage | 200 kV | ||
| Micrograph film | Kodak SO-163 | ||
| Scanner | UMAX Photolook 3000–256 greyscale | ||
| Spot size | 6 | ||
| Electron dose (electron/Å2) | <15 | ||
| Calibrated magnification | 37.6× | ||
| Processing Å/pixel | 3.39 | ||
| Sample | Wzc | Wzc Ni-NTA-gold | |
| Defocus range (μm) | −1.8 to −2.6 | −1.2 to −2.0 | |
| Selection box size (Å) | 164 | 164 | |
| Total particles selected | 5,515 | 1,750 | |
| Particle classes | 124 | 62 | |
| Reconstruction mask radius (Å) | 130 | 130 | |
| Spatial resolution of volume at FSC = 0.5 (Å) | 14 | 22 | |
Image Processing and Structure Calculation from Single Particle Data
Particles were interactively selected using BOXER software and contrast-normalized. The contrast transfer function (CTF) for each micrograph was determined using the program CTFIT (23), and corrections for amplitudes and phases for particles in each dataset were applied using a structure factor dataset to flip phases (kindly provided by Dr. Steve Ludtke, National Center for Macromolecular Imaging). A set of reference-free class averages were then generated with the complex orientated in multiple particle positions. By following previously described strategies (13), a preliminary three-dimensional model was determined from class averages that represented distinct views of the Wzc complex. The relative orientations of the characteristic views were determined using a Fourier common lines routine, and the resulting averages were combined to generate the preliminary three-dimensional model. The three-dimensional structure was subsequently refined using C4 symmetry, using eight rounds of iterative projection matching, with each refinement assessed by examining convergence by comparison of the Fourier shell correlation of the three-dimensional models generated from each iteration. The final three-dimensional volume was fully converged after six rounds of iterative refinement, and resolution was determined by Fourier shell correlation analysis as previously described (13).
Ni-NTA Nanogold Labeling of His6-Wzc
10 μl of N-terminal His6-Wzc (50 μg/ml) was incubated with 3.3 μl (1.8 nm) of Ni-NTA-nanogold (Nanoprobes) for 18 h at 4 °C. Samples were then centrifuged at 13,000 rpm in a bench-top centrifuge for 10 min before cryo-negative staining as described above.
Homology Modeling
The crystal structure of chain A of the septum site-determining protein MinD (complexed with Mg-ADP) of the hyperthermophile Pyrococcus horikoshii was used for fitting to the Wzc structure using residues 15–171 (156 amino acids out of 243 total) of the MinD chain A, a region that shares 24% identity and 42% similarity with a continuous region of the Wzc amino acid sequence and which corresponds to ~61% of its predicted cytoplasmic domain. The BLAST score was 2e-06 for the overall comparison, and examination showed that many of the identical residues are involved at the nucleotide-binding pocket (data not shown).
RESULTS AND DISCUSSION
Transmission Electron Microscopy (TEM) of Cryo-negatively Stained Wzc Samples
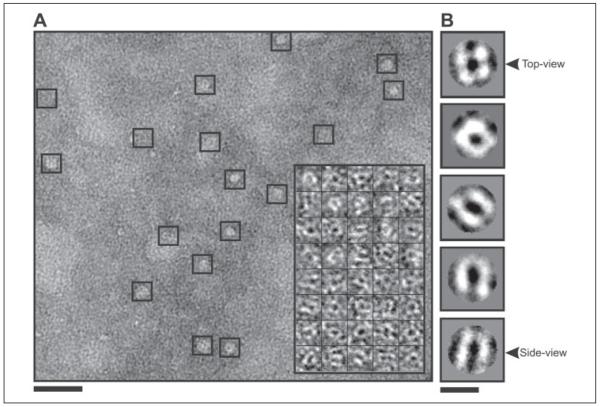
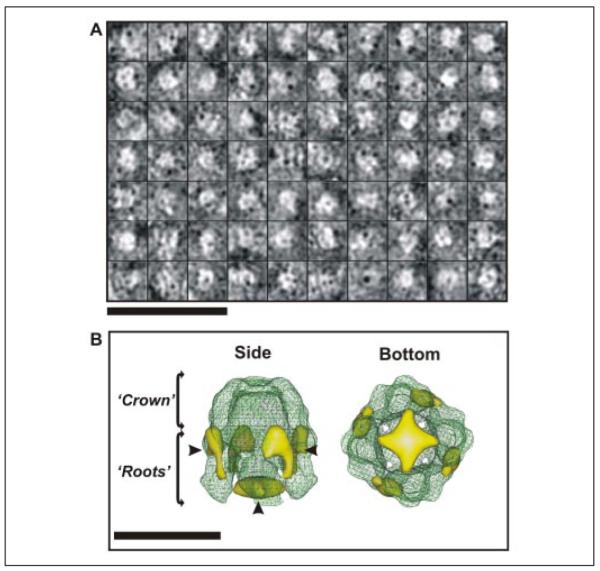
Fig. 1A shows an area of a micrograph of cryo-negatively stained, DDM-solubilized His6-Wzc. As can be seen, the population of complexes was homogeneous, well dispersed, and uniform in size. The inset boxes in Fig. 1A show a magnified and low pass-filtered montage of particles selected for analysis in this study. Two types of high contrast projections could be easily observed from the bulk of the data. One class of particle was between 90 and 100 Å in width with a square appearance and a large stained center. The other class of particles was slightly larger (~110 × 100 Å) and had a two-layered rounded rectangular appearance with a stain-filled line running along the center of the particle. Analysis of the total selected particles (n = 5515), using reference-free two-dimensional projection classification, identified many additional views of the Wzc particles, some of which are displayed in Fig. 1B. The most obvious class was the particle top view, which had an evident 4-fold rotational symmetry axis. The rounded rectangular particles had a strong bilateral symmetry and represent the side view of the Wzc oligomer (Fig. 1B). The other class averages represented various intermediate orientations between these two archetypal projections (Fig. 1B, central panels). The reference-free classification shows that the particles were positioned within the preservative stain layer in multiple orientations and so were suitable for Euler angle determination using cross-common line projection matching (23).
FIGURE 1. Cryo-negatively stained His6-Wzc.
A, shows DDM-solubilized His6-Wzc preparations used for cryo-negative staining. Individual Wzc complexes are shown in boxes. The image was recorded at ~2 μm defocus and was CTF-corrected and contrast-enhanced for presentation. Scale bar = 500 Å. The inset box shows a representative montage of CTF corrected raw Wzc particles, which have been low pass-filtered to 20 Å resolution for clarity. Each individual box is 164 Å2. B illustrates a selection of unsymmetrized two-dimensional projection averages calculated from the particle data-set using reference free alignment in EMAN (23). Each class average contains 80–100 particles and the circular masks had radii of 65 Å. Scale bar = 100 Å.
Identification of C4 Rotational Symmetry in Tetrameric Wzc Complexes
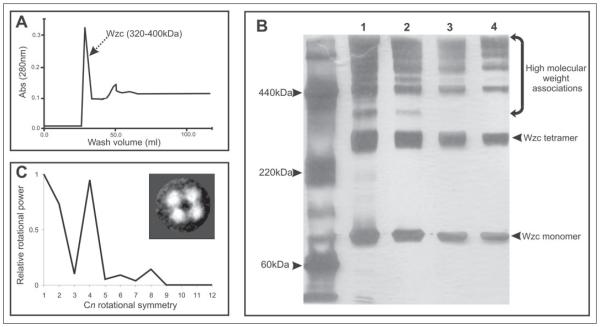
In addition to the 4-fold organization of His6-Wzc observed on TEM “raw” data, several other analyses independently demonstrated that Wzc has a tetrameric organization. First, purification of Wzc using size exclusion chromatography produced one sample peak, which eluted at 350–400 kDa (Fig. 2A). The nature of these Wzc oligomers was further characterized using analytical PFO-PAGE. PFO is a “mild” detergent that preserves high affinity interactions in many membrane protein complexes (22). Separation of the Wzc population clearly identified two main bands that migrated at ~90 kDa and ~350 kDa (Fig. 2B). Given that the Wzc monomer is 82 kDa, the bands are interpreted as monomeric and tetrameric Wzc species. The formation of the tetramer is not an artifact of the protein concentration and is observed across a wide range of protein concentrations (Fig. 2B, lanes 1–4). Several bands were observed with molecular masses in excess of 500 kDa; however no larger oligomer complexes were observed by TEM (Fig. 1A). These bands may therefore represent transient oligomer-oligomer contacts in PFO or correspond to the higher molecular mass aggregates we occasionally observed.
FIGURE 2. Oligomeric nature of His6-Wzc.
The elution profile of His6-Wzc oligomers on a Superdex S200 gel filtration column is shown in A. Oligomers of His6-Wzc were evident in samples analyzed by PFO-PAGE using a 7.5% silver-stained slab gel (B). Lanes 1–4 contain different concentrations of His6-Wzc (600, 300, 100, and 50 μg/ml, respectively). The migration of markers bovine serum albumin (65 kDa), catalase (220 kDa), and ferratin (440 kDa) are indicated on the left of the gel. C shows the rotational power spectrum of the His6-Wzc oligomer. The inset box shows the unsymmetrized class average used for analysis (n = 112). Abs, absorbance.
To quantify the obvious 4-fold symmetry apparent in the reference-free two-dimensional projection classification of Wzc, rotational power analysis was applied to the single particle data (Fig. 2C). This eliminated the possibility of a quasisymmetrical C4 (2–2) arrangement, because strong correlation peaks were only observed for C1, C2, and C4 rotational symmetries, with the strongest signal originating from the C4 comparison. Three-dimensional structure calculations on the Wzc data set were therefore performed using C4 symmetry.
Three-dimensional Structure of the Wzc Complex
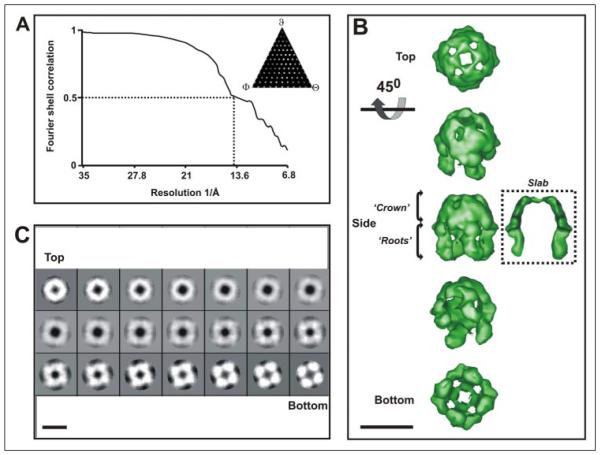
A three-dimensional structure was calculated from the Wzc single particle data set with C4 symmetry imposed. A molecular envelope of Wzc thresholded at 1 σ above the mean density would accommodate a molecular mass of ~350 kDa, assuming a protein packing density of 0.73 g/cm3 (24). This data is consistent with a protein complex of this size (a Wzc tetramer of ~320 kDa with additional associated detergent). The final resolution of the volume was estimated to be 14 Å using the Fourier shell correlation = 0.5 criteria routinely used for cryo-TEM reconstructions (25) (Fig. 3A). The iterative structure refinement converged after six rounds, and matrix vector analysis of the asymmetric triangle (Fig. 3A, inset) revealed no significant gaps in data sampling through three-dimensional space. Fig. 3B shows a surface rendering of the Wzc oligomer density map. Viewed from the top, at a threshold of 2 σ above the mean density, the complex is virtually sealed with small holes around the uppermost surface. The top view of the complex is rounded square, with a square edge of 100 Å and a corner-corner distance of 123 Å. The side view of Wzc shows it is slightly longer (~110 Å high) and has a distinctive appearance similar to an extracted molar tooth with distinctive domed “crown” and separate “root” regions. The upper crown is ~55 Å high and forms a ring of connected density (~20–25 Å thick) around the periphery of the complex. From the underside of the crown, four unconnected roots ~65 Å high emerge. To put the size of the Wzc structure in an in vivo context, the thickness of the E. coli inner membrane measured by cryo-TEM methods in frozen hydrated sections is ~60 Å (26).
FIGURE 3. Three-dimensional structure of His6-Wzc.
A shows the Fourier shell correlation plot calculated from the final iteration of the volume at 1σ above the mean density and a threshold of 1. Data were calculated with C4 symmetry applied using EMAN (23). The inset contains the asymmetric triangle and shows the distribution of the class averages used to calculate the three-dimensional structure between Euler angles φ, ψ, and θ. B shows iso-surface renderings of the volume displayed at a threshold to accommodate 350 kDa of mass. Volume was low pass-filtered to 14 Å resolution, and a high pass Fermi filter (radii and temperature factor = 0.05) was applied to sharpen high resolution frequencies. Scale bar = 100 Å. In C, projections representing 3.4 Å thick slices through the height of the volume (left to right) displayed at 1 σ above the mean density. Scale bar = 100 Å.
Membrane Orientation of the Wzc Complex in the Cytoplasmic Membrane
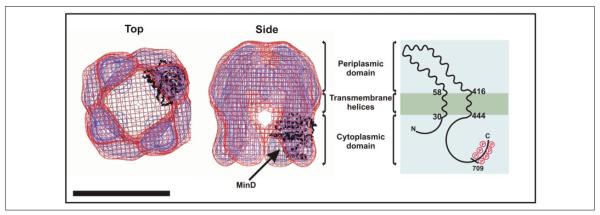
Wzc is a bitopic inner membrane protein, and the periplasmic location of the domain flanked by the two transmembrane segments has been established by Wzc-PhoA fusion experiments (17). The C terminus must be cytoplasmic to facilitate tyrosine phosphorylation, and as a consequence, the N terminus must also be located in the cytoplasm. To investigate the membrane orientation of the Wzc three-dimensional structure in a quantitative fashion, the N-terminal His6 tag was exploited for Ni-NTA-nanogold labeling; this label is a highly specific His6 reagent that is pH neutral and non-denaturing. Using Ni-NTA-nanogold to label oligomeric complexes may also aid single particle averaging, because the prominent electron density acts as a “target” for two-dimensional alignment algorithms (www.nanoprobes.com). Fig. 4A shows a montage of cryo-negatively stained Wzc particles, which have been incubated with Ni-NTA-nanogold. These Wzc complexes were essentially identical in appearance to those observed without nanogold (Fig. 1A), but multiple gold densities can be easily observed bound to the proteins. With a smaller gold-labeled data set, we were able to generate a second three-dimensional structure at ~22 Å with additional bound gold density contained within the volume (Fig. 4B). Nanogold (shown in gold) was found in two locations: on the upper half of the root regions and at the bottom of the cavity formed by the roots underneath the crown. As this second Ni-NTA-nanogold location is not physically connected to the protein envelope, it may also represent nonspecific binding or trapped gold particles. However, the location of gold in either position suggests that the roots contain the N terminus and hence are in the cytoplasm, whereas the crown is in the periplasm. Given the location of the N terminus on a root, the alternatively flipped orientation is inconsistent with relative observed volumes of the protein domains. We therefore predict that the root contains the tyrosine kinase domain and is cytoplasmic. For further confirmation, the structure of MinD (an ATP-binding protein) from P. horikoshii was manually fitted into an individual root volume (Fig. 5) (see “Materials and Methods”). There is a good volume match between the MinD model and molecular envelope of Wzc, and this is consistent with our experimentally determined location of the two domains. It should be appreciated that, at 14 Å resolution, the MinD model can only act as an appropriately sized globular volume, and precise orientation of the model within the root domain is not possible. The overall appearance of the structure further suggests this orientation; the roots are globular and compact as would be expected for a soluble kinase-like domain.
FIGURE 4. Three-dimensional location of the cytoplasmic N terminus of His6-Wzc identified by Ni-NTA-nanogold labeling.
A shows a montage of cryo-negatively stained Wzc particles labeled with Ni-NTA-nanogold (median diameter = 18 Å) used to calculate a three-dimensional volume. The montage has been CTF-corrected and low pass-filtered to 22 Å resolution to highlight the nanogold density bound to Wzc. Scale bar = 500 Å. Wire frame representations of the His6-Wzc volume with Ni-NTA-nanogold are illustrated in B. The volume was low pass-filtered to 22 Å resolution, and the green mesh shows Wzc at 1σ above the mean density. The locations of the bound nanogold are shown in gold surface at −3.5 σ below the mean density. Scale bar = 100 Å.
FIGURE 5. Orientation of the His6-Wzc tetramer in the cytoplasmic membrane.
The figure illustrates modeling of MinD fragment into the root densities of the Wzc three-dimensional structure. The volume is displayed at 1 σ (red mesh) and 1.5 σ (blue mesh) above the mean density, and the fitted MinD α-polypeptide chain is shown in black. Scale bar = 100 Å. The schematic on the far right shows the predicted topology of Wzc approximately matched with the corresponding domain distribution within the three-dimensional structure.
Functional Significance
The E. coli K12 and K30 Wzc homologs share sequence (65% identity; 90% similarity) and topological conservation, and they appear to perform similar functions (27). The K12 homolog has been reported to form higher molecular weight aggregates, such as trimers and hexamers after cross-linking with dithiobis(succinimidyl propionate) (17). However, the subsequent finding that Wzc can be cross-linked to Wza (20) complicates these interpretations. The data described above give unambiguous evidence of the existence of Wzc tetramers. Oligomerization of Wzc is mostly driven by contacts with the periplasmic portions of the protein. Interestingly, this domain contains sequences with a high probability of forming coiled-coil structures (28), which are often implicated in protein-protein interactions in a variety of different systems (29). Although we see no interaction between the roots, such an interaction is predicted from the capacity of the protein to trans-phosphorylate (16); adjacent Wzc complexes could be involved in this but seem less likely. The possible role of additional proteins and cellular factors in mediating conformational changes that would facilitate interactions between the roots and phosphorylation are beyond the scope of this study.
Cell envelope-spanning multienzyme complexes have been identified in other assembly systems, and some of these contain intracellular membrane-associated proteins with ATPase motifs. For example, the type II protein-secretion pathway in Vibrio cholerae requires the cytoplasmic EpsE ATPase for formation of a subcomplex with membrane proteins EpsL and EpsM (30). This may serve to couple energy to protein export through the trans-envelope type II complex (31, 32). EpsE is proposed to form a hexameric ring-like structure, and it represents a larger hexameric ATPase family involved in protein secretion (reviewed in Ref. 31). In the type IV secretion system of Agrobacterium tumefaciens, VirB4 is one of at least three ATPases associated with the inner membrane (31). In a comparable manner, Wzc may be involved in the assembly of a functional capsule assembly complex or in providing energy for capsule export. Wzc is not related in sequence or domain organization to these proteins, and the wzc mutants so far studied provide no additional insight into such possibilities. Although the structure reported here is from a fully phosphorylated Wzc derivative, phosphorylation is not essential for oligomer formation per se (data not shown), and its role could be confined to mediating additional interactions in a larger biosynthetic complex or dictating specific functions within the complex.
Interaction of Wzc with Wza
Experimental data clearly indicate that Wza and Wzc interact (20). Recent evidence implicates the periplasmic domain in these interactions, and there is an element of sequence specificity in the recognition of Wza homologs by their cognate Wzc proteins (27). These interactions are essential for function in capsule assembly. The oligomeric structure of Wza is an octamer (9, 20), whereas Wzc is a tetramer. Despite the difference in organization, it is striking that both Wza and Wzc oligomers share 4-fold rotational symmetry and furthermore have very similar widths. This is very suggestive that a specific complex forms between the two oligomers during carbohydrate export. If such a direct complex exists, it would span the periplasmic space. Analogous membrane-spanning complexes are predicted and modeled for several systems (e.g. the type I hemolysin export system requires the assembly of the HlyABD-TolC complex (33) and the AcrAB-TolC drug efflux pump from E. coli (34)). In these systems, structural details are largely absent, although a cryo-TEM reconstruction has been performed on the “needle” complex of the type III protein-secretion system (8). However, the differences in the process and proteins involved in type III secretion and those involved in capsular polysaccharide export prevent any extrapolation from the type III protein structural studies. Rather, further structural studies will be required to characterize the putative Wza-Wzc system.
Cryo-electron microscopy provides a unique methodology to investigate the structure and interactions of the capsule export system proteins. After establishing the “building blocks” of this biosynthetic machine, it is anticipated that the interactions of larger assemblies will be amenable to study in the future. A structural understanding of the mechanisms of capsule export would not only provide a rationale for the development of new therapeutic strategies but could also illuminate and inform Gram-negative secretion processes in general.
Acknowledgments
We thank Prof. Per Bullough and Dr. Peiyi Wang for excellent cryo-electron microscopy facilities and support and Dr. Jeremy Derrick for useful discussions.
Footnotes
The abbreviations used are: DDM, dodecyl maltoside; PFO, perfluoro-octanoic acid; CTF, contrast transfer function; TEM, transmission electron microscopy; Ni-NTA, nickel-nitrilotriacetic acid.
REFERENCES
- 1.Jann K, Jann B. In: Escherichia coli: Mechanisms of Virulence. Sussman M, editor. Cambridge University Press; Cambridge, UK: 1997. pp. 113–143. [Google Scholar]
- 2.Whitfield C, Roberts IS. Mol. Microbiol. 1999;31:1307–1319. doi: 10.1046/j.1365-2958.1999.01276.x. [DOI] [PubMed] [Google Scholar]
- 3.Raetz CRH, Whitfield C. Annu. Rev. Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Whitfield C, Paiment A. Carbohydr. Res. 2003;338:2491–2502. doi: 10.1016/j.carres.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 5.Paulsen IT, Beness AM, Saier MH., Jr. Microbiology. 1997;143:2685–2699. doi: 10.1099/00221287-143-8-2685. [DOI] [PubMed] [Google Scholar]
- 6.Drummelsmith J, Whitfield C. Mol. Microbiol. 1999;31:1321–1332. doi: 10.1046/j.1365-2958.1999.01277.x. [DOI] [PubMed] [Google Scholar]
- 7.Drummelsmith J, Whitfield C. EMBO J. 2000;19:57–66. doi: 10.1093/emboj/19.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marlovits TC, Kubori T, Sukhan A, Thomas DR, Galan JE, Unger VM. Science. 2004;306:1040–1042. doi: 10.1126/science.1102610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beis K, Collins RF, Ford RC, Kamis AB, Whitfield C, Naismith JH. J. Biol. Chem. 2004;279:28227–28232. doi: 10.1074/jbc.M402913200. [DOI] [PubMed] [Google Scholar]
- 10.Bitter W. Arch. Microbiol. 2003;179:307–314. doi: 10.1007/s00203-003-0541-8. [DOI] [PubMed] [Google Scholar]
- 11.Bitter W, Koster M, Latjinhouwers M, de Cock H, Tommassen J. Mol. Microbiol. 1998;27:209–219. doi: 10.1046/j.1365-2958.1998.00677.x. [DOI] [PubMed] [Google Scholar]
- 12.Collins RF, Ford RC, Kitmitto A, Olsen R, Tonjum T, Derrick JP. J. Bacteriol. 2003;183:3825–3832. doi: 10.1128/JB.183.13.3825-3832.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collins RF, Frye SA, Kitmitto A, Ford RC, Tonjum T, Derrick JP. J. Biol. Chem. 2004;279:39751–39756. doi: 10.1074/jbc.M405971200. [DOI] [PubMed] [Google Scholar]
- 14.Nouwen N, Stahleberg H, Pugsley AP, Engel A. EMBO J. 2000;19:2229–2236. doi: 10.1093/emboj/19.10.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rahn A, Beis K, Naismith JH, Whitfield C. J. Bacteriol. 2003;185:5882–5890. doi: 10.1128/JB.185.19.5882-5890.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wugeditsch T, Paiment A, Hocking J, Drummelsmith J, Forrester C, Whit-field C. J. Biol. Chem. 2001;276:2361–2371. doi: 10.1074/jbc.M009092200. [DOI] [PubMed] [Google Scholar]
- 17.Doublet P, Grangeasse C, Obadia B, Vaganay E, Cozzone AJ. J. Biol. Chem. 2002;277:37339–37348. doi: 10.1074/jbc.M204465200. [DOI] [PubMed] [Google Scholar]
- 18.Cozzone AJ, Grangeasse C, Doublet P, Duclos B. Arch. Microbiol. 2004;181:170–181. doi: 10.1007/s00203-003-0640-6. [DOI] [PubMed] [Google Scholar]
- 19.Paiment A, Hocking J, Whitfield C. J. Bacteriol. 2002;184:6437–6447. doi: 10.1128/JB.184.23.6437-6447.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nesper J, Hill CMD, Paiment A, Harauz G, Beis K, Naismith JH, Whitfield C. J. Biol. Chem. 2003;278:49763–49772. doi: 10.1074/jbc.M308775200. [DOI] [PubMed] [Google Scholar]
- 21.Guzman LM, Belin D, Carson MJ, Beckwith J. J. Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramjeesingh M, H L, Huan LJ, Garami E, Bear CE. Biochem. J. 1999;342:119–123. [PMC free article] [PubMed] [Google Scholar]
- 23.Ludtke SJ, Baldwin PR, Chiu W. J. Struct. Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 24.Harpaz Y, Gerstein M, Chothia C. Structure (Camb.) 1994;2:641–649. doi: 10.1016/s0969-2126(00)00065-4. [DOI] [PubMed] [Google Scholar]
- 25.Frank J. Three-dimensional Electron Microscopy of Macromolecular Assem-blies. Plenum Press; San Diego, CA: 1996. [Google Scholar]
- 26.Matias VR, Al-Amoudi A, Dubochet J, Beveridge TJ. J. Bacteriol. 2003;185:6110–6118. doi: 10.1128/JB.185.20.6112-6118.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reid AN, Whitfield C. J. Bacteriol. 2005;187:5470–5481. doi: 10.1128/JB.187.15.5470-5481.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morona R, Van Den Bosch L, Daniels C. Microbiology (Read.) 2000;146:1–4. doi: 10.1099/00221287-146-1-1. [DOI] [PubMed] [Google Scholar]
- 29.Burkhard R. J. Struct. Biol. 2001;134:204–218. doi: 10.1006/jsbi.2001.4336. [DOI] [PubMed] [Google Scholar]
- 30.Sandkvist M. Mol. Microbiol. 2001;40:271–283. doi: 10.1046/j.1365-2958.2001.02403.x. [DOI] [PubMed] [Google Scholar]
- 31.Camberg JL, Sandkvist M. J. Bacteriol. 2005;187:249–256. doi: 10.1128/JB.187.1.249-256.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robien MA, Krumm BE, Sandkvist M, Hol WG. J. Mol. Biol. 2003;333:657–674. doi: 10.1016/j.jmb.2003.07.015. [DOI] [PubMed] [Google Scholar]
- 33.Thanabalu T, Koronakis E, Hughes C, Koronakis V. EMBO J. 1998;17:6487–6496. doi: 10.1093/emboj/17.22.6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nikaido H, Zgurskaya HI. J. Mol. Microbiol. Biotechnol. 2001;3:215–218. [PubMed] [Google Scholar]