Rapamycin was first identified as a potent antifungal metabolite produced by a bacteria strain isolated from a soil sample on Easter Island. The later identification of a Ser/Thr kinase termed target of rapamycin (TOR) led to the discovery of a complicated signaling network that governs cell growth, proliferation and survival in response to nutrients, growth factors and hormones. Rapamycin has already been used clinically as an immunosuppressant in organ transplantation recipients. Much broader interest has been attracted to the TOR signaling because of the potential to develop TOR-based therapeutics for cancer, diabetes, neurodegenerative diseases and aging. Recently, a cardioprotective function of rapamycin has been shown to reverse cardiomyopathy.1–3 However, genetic manipulations of TOR in rodents have not yet supported a therapeutic function of TOR signaling inhibition.4 In contrast, a detrimental effect on cardiac function was actually suggested in a conditional knockout study of TOR in mouse.5
Through the use of a transposon-based mutagenesis screen in zebrafish, we identified a zeberafish TOR mutant, ztorxu015.6 Homozygous ztor mutants exhibited smaller cardiomyocyte cell size, and most died at 10 d post-fertilization, demonstrating a critical role of TOR in cardiomyocyte growth and larval development. To elucidate functions of TOR signaling in cardiomyopathies, we utilized the first two adult zebrafish models of cardiomyopathy that were induced by either chronic anemia or doxorubicin (DOX) stress.6,7 In contrast to previous studies in rodents, we studied ztor heterozygous fish, in which TOR protein reduced to 65% of that in normal wild type. We found that partial reduction of TOR exerted no overt phenotypes in heterozygous ztor fish. Importantly, heterozygous ztor preserved cardiac functions and improved fish survival significantly in both adult fish models of cardiomyopathies. Detailed studies uncovered dynamic activities of TOR signaling during pathogenesis of these two different types of cardiomyopathies. Intriguingly, we found that although TOR inhibition by rapamycin treatment protected fish against late-onset cardiomyopathy induced by low-dose DOX, it deteriorated high-dose DOX-induced acute cardiotoxicity. Together, our genetic studies in the zebrafish models suggested that dose- and stage-dependent functions are key attributes to consider when developing TOR-based therapeutics for cardiomyopathies.
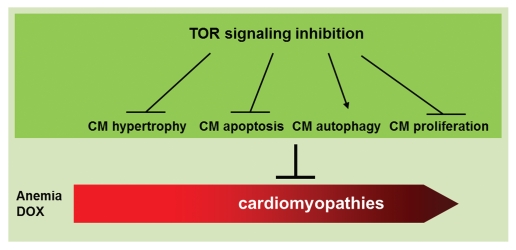
At the cellular level, our zebrafish studies uncovered four effects of TOR signaling inhibition. First, TOR signaling inhibition exerts an anti-hypertrophy function on cardiomyocytes in both the anemia model and the DOX model. This is consistent with a critical role for TOR in cell size control. Second, TOR signaling inhibition exerts an anti-apoptosis function. This effect would be highly beneficial in the DOX model, in which a high level of apoptosis contributes to the pathogenesis, but might be less so in the anemia model, in which no significantly activated apoptosis was detected. Third, TOR signaling inhibition exerts a pro-autophagy effect. The TOR-autophagy signaling sub pathways warrant more detailed studies, because of the recently proposed concept that cardiomyopathies are proteinopathies caused by the expression of aggregationprone proteins, while autophagy helps to eliminate these proteins.8 Fourth, TOR signaling inhibition exerts an anti-proliferative effect in both models. Given our recent data suggesting that activated proliferation might play a favorable role in attenuating cardiomyopathy,9 the precise nature of this anti-proliferative effect of TOR signaling inhibition on cardiomyopathies remains to be clarified. The relatively high cardiomyocyte proliferation capacity in adult zebrafish, especially in the anemia model, makes zebrafish an efficient model to further elucidate this point.7
In summary, by studying a heterozygous ztor mutant in zebrafish, we provided the first genetic evidence to support the therapeutic benefit of TOR signaling inhibition on cardiomyopathies. Its effects on two cardiomyopathy models further suggest that TOR signaling might be one of the common pathological pathways in cardiomyopathies of different etiology. Dose- and stage-specific effects of TOR signaling inhibition might be able to explain the discrepancy between rapamycin treatment and genetic studies of TOR in rodents. Therefore, our data strongly suggest that rapamycin should be further pursued as a candidate therapeutic compound for cardiomyopathy/heart failure. Further investigations are also warranted to discern distinct functions of different cellular events on the pathogenesis, as well as to define TOR downstream sub-pathways that confer this cardioprotective effect, which will lead to the development of compounds of better therapeutic benefit with fewer side effects.
Figure 1.
Cardiomyopathies are progressive diseases that are caused by different etiology. Two adult zebrafish models of cardiomyopathies have been generated, both of which can be attenuated by TOR signaling inhibition. Four cellular changes in cardiomyocytes (CM) are affected by TOR signaling inhibition, whose contribution needs to be discerned to develop better therapeutics for cardiomyopathies.
Comment on: Ding Y, et al. Circ Res. 2011;109:658–669. doi: 10.1161/CIRCRESAHA.111.248260.
References
- 1.Marin TM, et al. J Clin Invest. 2011;121:1026–1043. doi: 10.1172/JCI44972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McMullen JR, et al. Circulation. 2004;109:3050–3055. doi: 10.1161/01.CIR.0000130641.08705.45. [DOI] [PubMed] [Google Scholar]
- 3.Shioi T, et al. Circulation. 2003;107:1664–1670. doi: 10.1161/01.CIR.0000057979.36322.88. [DOI] [PubMed] [Google Scholar]
- 4.Shen WH, et al. J Biol Chem. 2008;283:13842–13849. doi: 10.1074/jbc.M801510200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang D, et al. J Clin Invest. 2010;120:2805–2816. doi: 10.1172/JCI43008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ding Y, et al. Circ Res. 2011;109:658–669. doi: 10.1161/CIRCRESAHA.111.248260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun X, et al. PLoS One. 2009;4:6596. doi: 10.1371/journal.pone.0006596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rothermel BA, et al. Circ Res. 2008;103:1363–1369. doi: 10.1161/CIRCRESAHA.108.186551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoage T, et al. Biochem Biophys Res Commun. 2011;415:490–496. doi: 10.1016/j.bbrc.2011.10.100. [DOI] [PMC free article] [PubMed] [Google Scholar]