Abstract
AGC kinases, including the three Akt (protein kinase B) isoforms, protein kinase A (PKA) and all protein kinase C (PKC) isoforms, require activation loop phosphorylation (threonine 308 in Akt1) as well as phosphorylation of a C-terminal residue (serine 473 in Akt1) for catalytic activity and phosphorylation of downstream targets. Conversely, phosphatases reverse these phosphorylations. Virtually all cellular processes are affected by AGC kinases, a circumstance that has led to intense scrutiny of the molecular mechanisms that regulate phosphorylation of these kinases. Here, we review a new layer of control of phosphorylation in Akt, PKA and PKC pointing to ATP binding pocket occupancy as a means to decelerate dephosphorylation of these and, potentially, other kinases. This additional level of kinase regulation opens the door to search for new functional motifs for the rational design of non-ATP-competitive kinase inhibitors that discriminate within and between protein kinase families.
Key words: inhibitors hijacking kinase activation, activation loop phosphorylation, dephosphorylation, phosphatase resistance, PKA, PKB, PKC
Introduction
Protein kinases modify protein function by attaching phosphate groups to specific amino acids. They have been implicated in affecting many aspects of metabolism and cell fate and play key roles in the pathogenesis of human diseases, including metabolic disorders, degenerative diseases and cancer. The human “kinome” of 518 protein kinases has been broadly divided into nine functional groups. Among the best known kinase families are tyrosine kinases (TK) and serine/threonine protein kinases, including map kinases, Ca2+/calmodulin-dependent protein kinases (CaMK) and the AGC kinases.1
Members of AGC kinases, including protein kinase A (PKA), Akt/Protein kinase B, protein kinase C (PKC) and protein kinase G (PKG) represent vital molecular sensors and signaling intermediaries that coordinate cellular responses to signals emanating from the intracellular milieu and the extracellular environment. For example, cellular calcium activates protein kinase C; intracellular levels of cyclic AMP and cyclic GMP activate protein kinase A (PKA) and protein kinase G (PKG), respectively. In addition, membrane-bound second messengers, i.e., diacylglycerol and D3-phosphorylated phosphatidylinosites, activate protein kinase C kinases and Akt/PKB kinases, respectively. In consideration of the importance of AGC protein kinases in regulating cell fate, the molecular mechanisms that control the phosphorylation of these kinases have been studied extensively.
Akt Kinase Allosteric Conformations Regulate Akt Activation Loop Phosphorylation
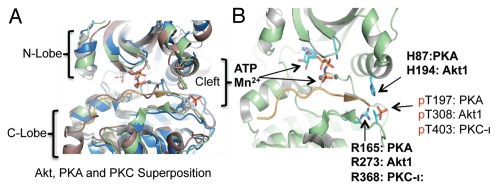
Most of this previous work focused on molecular mechanisms controlling kinase activation. Analyses of the X-ray crystallography structures of PKA provided critical insights into dynamic changes in the structure of the kinase domain of AGC kinases associated with their activation.2,3 In general terms, the catalytic subunits of protein kinases are defined by two lobes, a smaller N-terminal lobe and a larger C-terminal lobe (Fig. 1A). These two lobes border a deep cleft that contains both the ATP/Mg2+ acceptor site and a groove that accommodates binding of the kinase substrate(s). In many kinases, access of ATP and substrate to the active site cleft is controlled by the “activation” loop, which, by way of phosphorylation, undergoes marked conformational changes.5 In the unphosphorylated state, the activation loop is typically disordered and serves to sterically hinder access of both nucleotide and substrate to the catalytic cleft. Upon phosphorylation, it moves away from the catalytic center and adopts a conformation that allows ATP and substrate binding, and results in a “closed” conformation of the N- and C-lobes. Activation loop phosphorylations of Akt1, PKA and PKCι occur at threonine 308, threonine 197 and threonine 403, respectively (Fig. 1B).6,7
Figure 1.
Comparison of ATP-regulated phosphatase resistant structures of AGC kinases. (A) Superposition of Akt2-Mn-AMPPNP-Gsk3 structure (color green, PDB code: 1o6k5), PKA-Mn-ATP-PKI structure (color brown, PDB code: 1cdk31) and PKC-ι-ATP (color blue, PDB code: 3a8w4). For clarity, only AMPPNP/Mn2+ and pThr from the 1o6k structure are shown. (B) Structural representation of the protected activation loop conformation in the presence of ATP in the three AGC kinases. Under these conditions, the phosphorylated activation loop shown is further stabilized by histidine (H194 in Akt1, H87 in PKA and not conserved in PKC) and arginine (R273 in Akt1, R165 in PKA, R368 in PKC-ι). The structure is modeled on active human Akt2 crystal structures bound to ATP analog AMPPNP and Mn2+ (PDB 1o6k). However, ATP and Mn2+ and corresponding Akt1 amino acid residues are indicated for clarity.
Phosphorylation of the Akt/PKB activation loop is further constrained by close apposition of the N-terminal pleckstrin homology (PH) domain concealing threonine 308.8,9 This constraint is relieved by membrane translocation and binding to PtdIns(3,4,5)P3 lipids which unmask the unphosphorylated activation loop (threonine 308 in Akt1). Thus, the transition of the inactive Akt kinase to the active conformation requires at least two sequential steps affecting the PH domain first and the activation loop second. Of note, the active conformation adopted by Akt kinase is homologous not only to that of the other AGC kinases, but also to other protein kinase groups, including map kinases, Ca2+/calmodulin-dependent protein kinases and tyrosine kinases.2,6
Intra-Molecular Regulation of Akt Activation Loop Dephosphorylation by ATP Binding
Very recently, a novel control mechanism has come to light that targets activation loop phosphorylation at Akt threonine 308.10 In contrast to the mechanisms discussed above, this mechanism controls dephosphorylation of the activation loop by the type 2 phosphatase (PP2A). Biochemical studies and molecular modeling indicated that occupancy of the Akt nucleotide-binding pocket by ATP markedly decelerated Akt dephosphorylation. This phenomenon was observed in cells and, importantly, was replicated in vitro using a cell-free dephosphorylation assay using purified PP2A and Akt proteins. It became apparent that not only ATP, but also ATP analogs, including non-hydrolysable ATP-γS and ATP-competitive Akt inhibitors (A-443654 and GSK690693), similarly inhibited Akt dephosphorylation. Interestingly, ATP and several ATP “mimics” not only decelerated Akt1 dephosphorylation on threonine 308, but also on serine 473. Analysis of the three-dimensional (3D) structures of Akt bound to either ATP or A-443654 6,11 further suggested that this conformation is enabled by direct interaction of phosphorylated threonine 308, with at least two other residues (histidine 194 and arginine 273) of the ATP binding pocket (Fig. 1B). Mutational analysis confirmed that arginine 273 (arginine 274 in Akt2) and histidine 194 in Akt1 were required to inhibit threonine 308 dephosphorylation.10
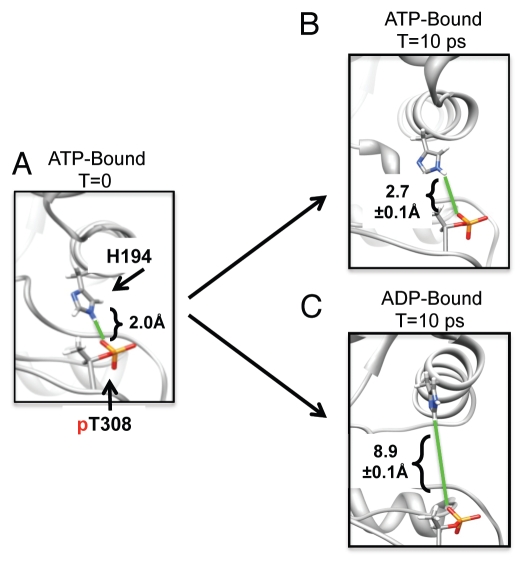
The product of ATP hydrolysis, ADP and the ADP-βS analog, were markedly less effective in blocking dephosphorylation.10 All-atom molecular modeling and molecular dynamics (MD) simulations of ATP vs. ADP binding in Akt kinase revealed that the histidine 194/phospho-threonine 308 interaction is consistently more stable in the ATP-bound state than in the ADP-bound state (Fig. 2). These observations suggested that conformational change in the histidine 194/phosphothreonine 308 interaction upon ATP hydrolysis directly contributes to greater solvent exposure of the phosphorylated threonine 308 and, consequently, less protection of dephosphorylation by ADP. Experimental hydrogen-deuterium exchange studies of PKA have also shown that nucleotide binding dramatically affects the conformational dynamics of the C-helix and other regions outside of the binding site.12 In this context, the results from MD simulations suggest that occupancy of the ATP-binding pocket of Akt kinases stabilizes a “closed” conformational state of the Akt kinase domain that restricts phosphatase access to threonine 308 (and serine 473), thereby sustaining Akt phosphorylation. The possibility that ATP-bound states may exhibit less conformational flexibility than ADP-bound states should not be surprising in the context of known conformational changes in the catalytic cycle of the enzyme. The first conformational change after ATP binding is presumed to orient ATP and active site residues for catalysis (into a more rigid conformational state), while the second conformational change after catalysis presumably relaxes this complex (into a slightly less rigid conformational state) that allows release of product, ADP and bound Mg2+.13,14
Figure 2.
Destabilization of intra-molecular protection of the phosphorylated Akt activation loop by ATP hydrolysis as suggested by molecular dynamics simulation. (A) ATP complexes with Mn2+ ions were modeled into the crystal structure of Akt kinase bound to AMP-PNP (1o6k). H194 and pT308 interactions were measured as 2 Å (green line) by the distance of a hydrogen bond from the E2 hydrogen to the phosphate oxygen. (B) Molecular dynamics simulation of H194 and pT308 interaction distance (green line) after 10 ps in ATP-bound Akt kinase. The first 1 ps of the simulation was for equilibration, and the ensemble average distance was calculated over 1–10 ps. (C) After hydrolysis of ATP to ADP, MD simulation shows that H194 and pT308 interaction distance (green line) increases by 3-fold after 10 ps.
A Mechanistic Explanation for “Paradoxical” Akt Hyperphosphorylation
Pharmacological inhibition of Akt activity with certain ATP-competitive inhibitors (A-443654 and GSK690693) is accompanied by robust “paradoxical” hyperphosphorylation of the Akt activation loop (threonine 308) in different tumor cells and in animals.15–18 Inhibitor-induced Akt hyperphosphorylation occurs while Akt substrate phosphorylation is inhibited and, thus, was initially thought to represent a homeostatic feedback mechanism to compensate for Akt signal loss.17 However, this view was challenged by several subsequent studies. First, directly inhibiting the Akt targets TSC1/2-mTORC1-S6K failed to inhibit Akt hyperphosphorylation by A443654.15 Second, cell-based chemical genetic studies demonstrated that binding of synthetic chemicals to the ATP binding site can induce Akt hyperphosphorylation.19 However, prior work did not illuminate the mechanism of drug-induced hyperphosphorylation. By use of the in vitro reconstitution assay, we unequivocally show that occupancy of the ATP binding site by the ATP competitors A-443654 and GSK690693 restrains phosphatase access to threonine 308 of Akt1 (T. Chan unpublished data and ref. 10). Dephosphorylation resistance provides a simple intra-molecular mechanism for the Akt hyperphosphorylation induced by ATP competitive inhibitors.
ATP Binding Inhibits Dephosphorylation of the PKA and PKC Activation Loops
Analysis of the three-dimensional (3D) structures of phosphorylated activation loop of Akt, PKA and PKC-ι when bound to ATP analogs (Fig. 1A; PDB 1o6k, 1cdk and 3a8w, respectively) shows that these activation loops are in almost identical protected conformations through direct contact with a conserved arginine (Fig. 1B).2,3 Interestingly, the contacting histidine is conserved in Akt and PKA but not in PKC. However, in contrast to Akt/PKB, phosphorylation of PKA and PKC activation loops occurs during the synthesis of the enzymes, not during kinase activation.20–22 The pre-activation loop phosphorylated or “primed” PKA and PKC kinases are folded into larger inactive structures. Phosphorylation events outside the activation loop or binding to second messengers, such as cAMP, diacylglycerol or calcium unleashes the catalytic activities of “primed” PKA and PKC enzymes.
While the mechanisms of kinase activation may differ between Akt/PKB and PKA or PKC, regulation of activation loop dephopsphorylation appears to be a shared mechanism between these three AGC kinases. In support of this notion, Taylor and colleagues observed that the activation loop (T197) of PKA resists dephosphorylation by the highly active lambda protein phosphatase in vitro.23 Furthermore, mutating the highly conserved cysteine (C199) near the activation loop enhanced dephosphorylation by phosphatases. Similarly, Srivastava and colleagues showed that binding of ATP/Mg2+ to PKC protected the enzyme from dephosphorylation by PP2A in vitro.24 Finally, Parker and colleagues used chemical genetics to show that binding of synthetic chemicals to PKCε nucleotide pocket alone is sufficient to induce phosphorylation of three “priming” sites in PKCε, the activation loop (T556), the turn motif (T710) and the hydrophobic motif (S729).25
Thus, in all three AGC kinases (PKA, PKB and PKC), occupancy of the nucleotide-binding pocket by ATP or ATP-competitive inhibitors induces activation loop hyperphosphorylation in cells by inhibiting phosphatase access to the activation loop, rather than by autophosphorylation or by an extrinsic feedback loop. Intriguingly, modifying cysteine residues of PKA in vivo by the alkylation agent N-ethylmaleimide (NEM) enhances activation loop dephosphorylation of both PKA and PKC.23 This finding opens a perspective on dephosphorylation kinetics controlled by ATP loading as they relate to metabolic states of cells.
Conclusion
Phosphorylation of the highly conserved activation loop in AGC kinases is widely accepted as a necessary step in the transition of protein kinases to the catalytically active “on” conformation and ATP loading. Based on the results described here, steady-state phosphorylation of the activation loop may be further regulated by an intra-molecular mechanism that controls dephosphorylation kinetics of the activation loop in AGC kinases.
Since both nucleotide binding pockets and activation loops of kinases are highly conserved, it seems possible that occupancy of the nucleotide-binding pocket protects the activation loop from dephosphorylation in other kinase groups and families. For example, inhibitors of Map2k (MEK) have been found to induce activation loop hyperphosphorylation.26–28 At first approximation, the effects of MEK inhibitors seem to be distinct from inhibitor-induced hyperphosphorylation in AGC kinases, because these inhibitors (U0126, PD098059, PD184161, Cl-1040 and AZD6244) are considered allosteric and non-ATP-competitive.26–28 As in the case of Akt, drug-induced hyperphosphorylation of MEK kinases is thought to represent a homeostatic feedback mechanism to compensate for signal loss.29 However, the MEK inhibitors listed, above all, bind to a hydrophobic pocket adjacent to the ATP binding site30 that is similar to the space occupied by γ-phosphate/Mg2+ in Akt and critical for ATP to support the phosphatase-resistant conformation. The importance of ATP γ-phosphate/Mg2+ in regulating Akt1 dephosphorylation is further supported by the findings that ADP does not protect against dephosphorylation, and intra-molecular interactions of the phosphorylated activation loop were more stable in the ATP-bound state than in the ADP-bound state (Fig. 2).10 However, all Mek1/Mek2 crystal structures available to date have been generated without activation loop phosphorylation.30,31
Collectively, these observations raise the intriguing possibility that allosteric control of phosphatase access to activation loop residues and the associated control of dephosphorylation kinetics is a broad mechanism shared across different kinase families. It further suggests that structural features that regulate overall kinase conformation and are located outside the canonical ATP binding pocket (e.g., region surrounding pSer473 in Akt1) are amenable to the design of novel “allosteric” kinase inhibitors with improved selectivity profiles.
References
- 1.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 2.Huse M, Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109:275–282. doi: 10.1016/S0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- 3.Kannan N, Haste N, Taylor SS, Neuwald AF. The hallmark of AGC kinase functional divergence is its C-terminal tail, a cis-acting regulatory module. Proc Natl Acad Sci USA. 2007;104:1272–1277. doi: 10.1073/pnas.0610251104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takimura T, Kamata K, Fukasawa K, Ohsawa H, Komatani H, Yoshizumi T, et al. Structures of the PKC-iota kinase domain in its ATP-bound and apo forms reveal defined structures of residues 533–551 in the C-terminal tail and their roles in ATP binding. Acta Crystallogr D Biol Crystallogr. 2010;66:577–583. doi: 10.1107/S0907444910005639. [DOI] [PubMed] [Google Scholar]
- 5.Nolen B, Taylor S, Ghosh G. Regulation of protein kinases; controlling activity through activation segment conformation. Mol Cell. 2004;15:661–675. doi: 10.1016/j.molcel.2004.08.024. [DOI] [PubMed] [Google Scholar]
- 6.Yang J, Cron P, Good VM, Thompson V, Hemmings BA, Barford D. Crystal structure of an activated Akt/protein kinase B ternary complex with GSK3-peptide and AMP-PNP. Nat Struct Biol. 2002;9:940–944. doi: 10.1038/nsb870. [DOI] [PubMed] [Google Scholar]
- 7.Yang J, Cron P, Thompson V, Good VM, Hess D, Hemmings BA, et al. Molecular mechanism for the regulation of protein kinase B/Akt by hydrophobic motif phosphorylation. Mol Cell. 2002;9:1227–1240. doi: 10.1016/S1097-2765(02)00550-6. [DOI] [PubMed] [Google Scholar]
- 8.Calleja V, Laguerre M, Larijani B. 3-D structure and dynamics of protein kinase B-new mechanism for the allosteric regulation of an AGC kinase. J Chem Biol. 2009;2:11–25. doi: 10.1007/s12154-009-0016-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu WI, Voegtli WC, Sturgis HL, Dizon FP, Vigers GP, Brandhuber BJ. Crystal structure of human AKT1 with an allosteric inhibitor reveals a new mode of kinase inhibition. PLoS One. 2010;5:12913. doi: 10.1371/journal.pone.0012913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan TO, Zhang J, Rodeck U, Pascal JM, Armen RS, Spring M, et al. Resistance of Akt kinases to dephosphorylation through ATP-dependent conformational plasticity. Proc Natl Acad Sci USA. 2011;108:1120–1127. doi: 10.1073/pnas.1109879108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davies TG, Verdonk ML, Graham B, Saalau-Bethell S, Hamlett CC, McHardy T, et al. A structural comparison of inhibitor binding to PKB, PKA and PKA-PKB chimera. J Mol Biol. 2007;367:882–894. doi: 10.1016/j.jmb.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 12.Andersen MD, Shaffer J, Jennings PA, Adams JA. Structural characterization of protein kinase A as a function of nucleotide binding. Hydrogen-deuterium exchange studies using matrix-assisted laser desorption ionization-time of flight mass spectrometry detection. J Biol Chem. 2001;276:14204–14211. doi: 10.1074/jbc.M011543200. [DOI] [PubMed] [Google Scholar]
- 13.Shaffer J, Adams JA. Detection of conformational changes along the kinetic pathway of protein kinase A using a catalytic trapping technique. Biochemistry. 1999;38:12072–12079. doi: 10.1021/bi991109q. [DOI] [PubMed] [Google Scholar]
- 14.Khavrutskii IV, Grant B, Taylor SS, McCammon JA. A transition path ensemble study reveals a linchpin role for Mg(2+) during rate-limiting ADP release from protein kinase A. Biochemistry. 2009;48:11532–11545. doi: 10.1021/bi901475g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han EK, Leverson JD, McGonigal T, Shah OJ, Woods KW, Hunter T, et al. Akt inhibitor A-443654 induces rapid Akt Ser-473 phosphorylation independent of mTORC1 inhibition. Oncogene. 2007;26:5655–5661. doi: 10.1038/sj.onc.1210343. [DOI] [PubMed] [Google Scholar]
- 16.Levy DS, Kahana JA, Kumar R. AKT inhibitor, GSK690693, induces growth inhibition and apoptosis in acute lymphoblastic leukemia cell lines. Blood. 2009;113:1723–1729. doi: 10.1182/blood-2008-02-137737. [DOI] [PubMed] [Google Scholar]
- 17.Luo Y, Shoemaker AR, Liu X, Woods KW, Thomas SA, de Jong R, et al. Potent and selective inhibitors of Akt kinases slow the progress of tumors in vivo. Mol Cancer Ther. 2005;4:977–986. doi: 10.1158/1535-7163.MCT-05-0005. [DOI] [PubMed] [Google Scholar]
- 18.Rhodes N, Heerding DA, Duckett DR, Eberwein DJ, Knick VB, Lansing TJ, et al. Characterization of an Akt kinase inhibitor with potent pharmacodynamic and antitumor activity. Cancer Res. 2008;68:2366–2374. doi: 10.1158/0008-5472.CAN-07-5783. [DOI] [PubMed] [Google Scholar]
- 19.Okuzumi T, Fiedler D, Zhang C, Gray DC, Aizenstein B, Hoffman R, et al. Inhibitor hijacking of Akt activation. Nat Chem Biol. 2009;5:484–493. doi: 10.1038/nchembio.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taylor SS, Buechler JA, Yonemoto W. cAMP-dependent protein kinase: framework for a diverse family of regulatory enzymes. Annu Rev Biochem. 1990;59:971–1005. doi: 10.1146/annurev.bi.59.070190.004543. [DOI] [PubMed] [Google Scholar]
- 21.Chan TO, Rittenhouse SE, Tsichlis PN. AKT/PKB and other D3 phosphoinositide-regulated kinases: kinase activation by phosphoinositide-dependent phosphorylation. Annu Rev Biochem. 1999;68:965–1014. doi: 10.1146/annurev.biochem.68.1.965. [DOI] [PubMed] [Google Scholar]
- 22.Parekh DB, Ziegler W, Parker PJ. Multiple pathways control protein kinase C phosphorylation. EMBO J. 2000;19:496–503. doi: 10.1093/emboj/19.4.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Humphries KM, Deal MS, Taylor SS. Enhanced dephosphorylation of cAMP-dependent protein kinase by oxidation and thiol modification. J Biol Chem. 2005;280:2750–2758. doi: 10.1074/jbc.M410242200. [DOI] [PubMed] [Google Scholar]
- 24.Srivastava J, Goris J, Dilworth SM, Parker PJ. Dephosphorylation of PKCdelta by protein phosphatase 2Ac and its inhibition by nucleotides. FEBS Lett. 2002;516:265–269. doi: 10.1016/S0014-5793(02)02500-0. [DOI] [PubMed] [Google Scholar]
- 25.Cameron AJ, Escribano C, Saurin AT, Kostelecky B, Parker PJ. PKC maturation is promoted by nucleotide pocket occupation independently of intrinsic kinase Activity. Nat Struct Mol Biol. 2009;16:624–630. doi: 10.1038/nsmb.1606. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y, Van Becelaere K, Jiang P, Przybranowski S, Omer C, Sebolt-Leopold J. A role for K-ras in conferring resistance to the MEK inhibitor, CI-1040. Neoplasia. 2005;7:336–347. doi: 10.1593/neo.04532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huynh H, Soo KC, Chow PK, Tran E. Targeted inhibition of the extracellular signal-regulated kinase kinase pathway with AZD6244 (ARRY-142886) in the treatment of hepatocellular carcinoma. Mol Cancer Ther. 2007;6:138–146. doi: 10.1158/1535-7163.MCT-06-0436. [DOI] [PubMed] [Google Scholar]
- 28.Yip-Schneider MT, Klein PJ, Wentz SC, Zeni A, Menze A, Schmidt CM. Resistance to mitogen-activated protein kinase kinase (MEK) inhibitors correlates with upregulation of the MEK/extracellular signal-regulated kinase pathway in hepatocellular carcinoma cells. J Pharmacol Exp Ther. 2009;329:1063–1070. doi: 10.1124/jpet.108.147306. [DOI] [PubMed] [Google Scholar]
- 29.Friday BB, Yu C, Dy GK, Smith PD, Wang L, Thibodeau SN, et al. BRAF V600E disrupts AZD6244-induced abrogation of negative feedback pathways between extracellular signal-regulated kinase And Raf proteins. Cancer Res. 2008;68:6145–6153. doi: 10.1158/0008-5472.CAN-08-1430. [DOI] [PubMed] [Google Scholar]
- 30.Ohren JF, Chen H, Pavlovsky A, Whitehead C, Zhang E, Kuffa P, et al. Structures of human MAP kinase kinase 1 (MEK1) and MEK2 describe novel noncompetitive kinase inhibition. Nat Struct Mol Biol. 2004;11:1192–1197. doi: 10.1038/nsmb859. [DOI] [PubMed] [Google Scholar]
- 31.Fischmann TO, Smith CK, Mayhood TW, Myers JE, Reichert P, Mannarino A, et al. Crystal structures of MEK1 binary and ternary complexes with nucleotides and inhibitors. Biochemistry. 2009;48:2661–2674. doi: 10.1021/bi801898e. [DOI] [PubMed] [Google Scholar]
- 32.Bossemeyer D, Engh RA, Kinzel V, Ponstingl H, Huber R. Phosphotransferase and substrate binding mechanism of the cAMP-dependent protein kinase catalytic subunit from porcine heart as deduced from the 2.0 A structure of the complex with Mn2+ adenylyl imidodiphosphate and inhibitor peptide PKI(5-24) EMBO J. 1993;12:849–859. doi: 10.1002/j.1460-2075.1993.tb05725.x. [DOI] [PMC free article] [PubMed] [Google Scholar]