Abstract
Chromosome instability (CIN) is found in 85% of colorectal cancers. Defects in mitotic processes are implicated in high CIN and may be critical events in colorectal tumorigenesis. Shugoshin-1 (SGO1) aids in the maintenance of chromosome cohesion and prevents premature chromosome separation and CIN. In addition, integrity of the centrosome may be compromised due to the deficiency of Cohesin and Sgo1 through the disengagement of centrioles. We report here the generation and characterization of SGO1-mutant mice and show that haploinsufficiency of SGO1 leads to enhanced colonic tumorigenesis. Complete disruption of SGO1 results in embryonic lethality, whereas SGO1+/− mice are viable and fertile. Haploinsufficiency of SGO1 results in genomic instability manifested as missegregation of chromosomes and formation of extra centrosomal foci in both murine embryonic fibroblasts and adult bone marrow cells. Enhanced CIN observed in SGO1-deficient mice resulted in an increase in formation of aberrant crypt foci (ACF) and accelerated development of tumors after exposure to azoxymethane (AOM), a colon carcinogen. Together, these results suggest that haploinsufficiency of SGO1 causes enhanced CIN, colonic preneoplastic lesions and tumorigenesis in mice. SGO1 is essential for the suppression of CIN and tumor formation.
Key words: SGO1, mouse genetics, chromosomal instability, centrosome, colon cancer
Since its discovery several years ago, Shugoshin 1 (SGO1) has emerged as a crucial regulator of the cell cycle.1–6 At cellular and molecular levels, SGO1 functions as a protector of centromeric cohesion of sister chromatids in higher eukaryotes.5–7 Depletion of SGO1 by small interfering RNA (siRNA) leads to premature sister chromatid separation.5–8 During mitosis, SGO1 localizes to centromeres in a manner that appears to be dependent on Bub1, Aurora B and survivin.7–13 SGO1 works in concert with protein phosphatase 2A (PP2A) to protect centromeric cohesion during mitosis and meiosis.14,15 It is implicated in microtubule dynamics and required for tension generation at the kinetochore.2,6 In addition to the function of SGO1 in centromeres, sSGO1, a major splice variant of SGO1, has an important function in centrosome dynamics through mediating centriole cohesion.16 A recent study supports the centrosomal function of Sgo1 in further detail.17 Importantly, both cohesin and Sgo1 are shown to be involved in engagement of centrioles and thus in centrosomal integrity.17 Given the importance of centromeric cohesion and centrosome dynamics in the maintenance of chromosomal stability during cell division, it is conceivable that deregulated function of SGO1 would lead to major chromosomal instability.
Chromosomal instability has long been appreciated as a driving force for tumorigenesis, since aneuploidy is prevalent in the majority of solid tumors.18–20 However, several recent studies show that in certain physiological contexts, chromosomal instability induced by deregulated checkpoint genes is not always associated with tumor development in a straightforward manner.21–24 For example, haploinsufficiency of CENP-E, a spindle checkpoint component, results in enhanced aneuploidy formation and a modest increase in spontaneous tumors in spleen and lung. However, CENP-E+/− mice develop fewer tumors when these mice are challenged with carcinogens or in a genetically susceptible background, suggesting that chromosomal instability may suppress tumorigenesis in a context-dependent manner.22
Defects in the chromosome cohesion system and SGO1 may play a critical role in genomic instability, and cancers in human colon. Barber et al. attempted to identify genes involved in CIN in human colon cancer with a tumor DNA sequencing approach. They identified 11 somatic mutations distributed among five genes in a part that included 132 colorectal cancers. All but one of these 11 mutations were in the homologs of yeast genes that regulate sister chromatid cohesion, strongly suggesting a critical relationship between chromosome cohesion and CIN in colon cancer.25 Consistently, SGO1 downregulation is implicated in human colon cancer. Among 46 colorectal cancer cases, hSGO1 mRNA expression was decreased in the tumor tissue in comparison with the corresponding normal tissue (p = 0.032).26 However, direct evidence linking SGO1 to colonic tumor development was lacking. Furthermore, no genetic studies have been reported in mouse models with regard to functions of SGO1 in the maintenance of chromosomal stability and acceleration or suppression of tumor development.
To determine the physiological function of SGO1, we have generated SGO1 haploinsufficient (+/−) mutant mice. Mouse embryonic fibroblasts (MEFs) from SGO1+/− animals were found to contain lower levels of SGO1 than MEFs from wild-type embryos. SGO1 deficiency resulted in increased number of spindle centrosomal foci, enhanced chromosome missegregation and formation of micronuclei at an enhanced rate. Moreover, SGO1+/− animals were prone to higher preneoplastic lesions and rapid development of colonic tumors after exposure to a colon carcinogen.
Results
Ablation of SGO1 results in embryonic lethality.
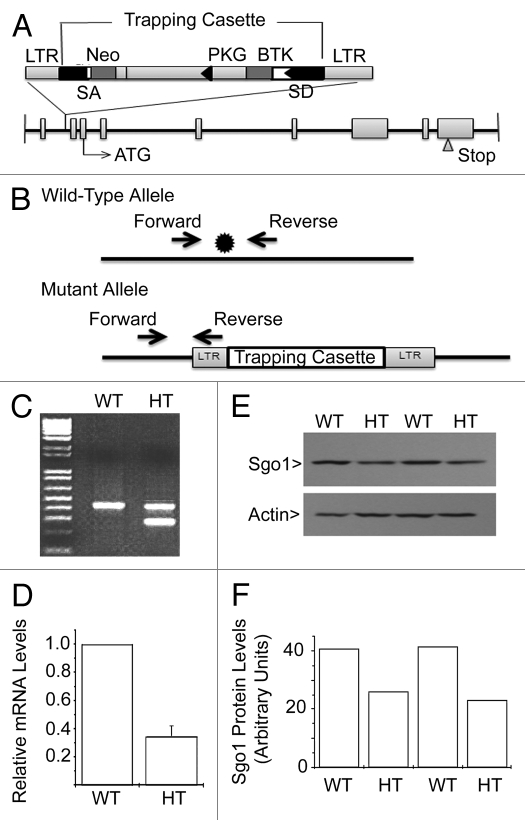
The coding region of the mouse SGO1 gene spans more than 20 kilobase pairs (kbp) with 8 introns ranging in size from 130 bp (intron 3) to 6,116 bp (intron 2). Mouse ES cells with a targeted disruption of the SGO1 locus were obtained by a gene-trapping method.27 The targeting retroviral vector, which contained a neomycin resistance cassette, was inserted between exons 1 and 2 of SGO1 (Fig. 1A and B). Two independent 129/Sv-derived ES cell lines were injected into C57/BL6 blastocysts, and the resulting chimeric mice were backcrossed to wild-type (WT) C57/BL6 animals. Tail tissues from offspring of backcrossed mice were digested to extract DNA and PCR was used to identify the genotypes. DNA samples from WT mice showed a band of 423 bp whereas DNA from heterozygous (HT) mice contained an additional band of 280 bp (Fig. 1C).
Figure 1.
Generation of SGO1 mutant mice. (A) Schematic representation of knockout construct. Disruption of the SGO1 locus was obtained by a gene trap method. LTR, viral long-terminal repeat; SA, splice acceptor; NEO, neomycin; pA, poly adenylation sequence. (B) Structure of the wild-type and mutant SGO1 loci. *denotes the gene trap cassette insertion site. (C) Representative genotyping result by PCR. WT, wild type; HT, heterozygous. PCR products were run on 1% agarose gels. The WT allele generates a 423 bp PCR product, while the mutant allele generates a 280 bp product. (D) Relative mRNA level of SGO1 from mice tail tissue. mRNA levels were measured by quantitative-RT-PCR. The average tissue mRNA level of WT mice was arbitrarily set to 1, and the tissue mRNA level of HT mice was divided by the WT average level (n = 3/group). (E) Western blot analysis of SGO1 levels in tail tissue. (F) The signals shown in the western blot film were quantified by densitometry.
The first year of breeding with crosses among SGO1+/− mice yielded more than 200 live animals. Genotyping of these animals revealed the absence of SGO1−/− animals, suggesting embryonic lethality of SGO1-null mice. The number of SGO1+/− mice was about 2-fold that of WT mice (SGO1+/+) (Table 1), which is consistent with the prediction by the Mendelian segregation rule. These results are also in agreement with previously published mouse genetic studies regarding the role of spindle checkpoint components in supporting mouse development.24,28,29 Quantitative PCR revealed that compared with WT MEFs, SGO1+/− MEFs contained a lower level of SGO1 mRNA (Fig. 1D). Consistent with the mRNA expression, SGO1 protein level was also lower in SGO1+/− cells than that in WT cells (Fig. 1E and F). Combined, these results indicate that we have successfully obtained mice with a specific disruption at the SGO1 genomic locus.
Table 1.
Viability of SGO1 mutant mice
| SGO1+/− X SGO1+/− | |||
| Genotype of F1 | SGO1+/+ | SGO1+/− | SGO1−/− |
| Observed frequency | 32.5% (26) | 67.5% (54) | 0 |
| Expected frequency | 25% | 50% | 25% |
SGO1+/− mice were crossed and genotype of the F1 was tested. Among total 80 F1 mice, 26 were SGO1+/+ mice, 54 were SGO1+/− mice but there was no SGO1−/− mice, indicating that SGO1−/− is lethal and did not allow live birth.
Haploinsufficency of SGO1 results in missegregation of chromosomes and formation of extra microtubule organization centers.
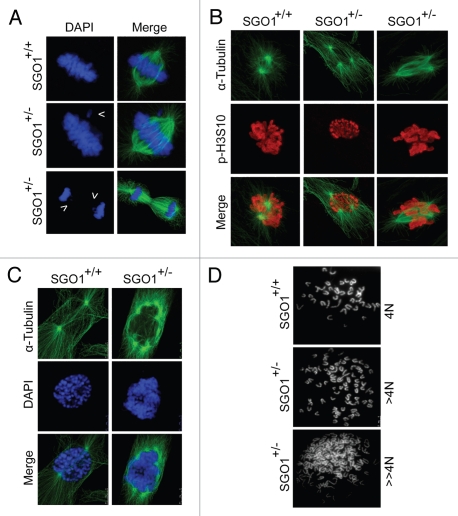
Cell-based studies show that depletion or inactivation of SGO1 induces chromosome missegregation and cell death in longer term.5,10,11,30 Given the importance of SGO1 in mediating sister chromatid cohesion, we reasoned that its haploinsufficiency (i.e., a decrease in expression) could induce mitotic defects. We first examined paired MEFs prepared from littermates. We found that misaligned chromosomes were frequently present in SGO1+/− metaphase cells (Fig. 2A, arrow in the middle part and Fig. S1). We also observed an increased frequency of the anaphase bridge or lagging chromosomes during anaphase in SGO1+/− MEFs (Fig. 2A, arrows in lower part and Fig. S1). The phenotype of the presence of misaligned or lagging chromosomes in metaphase and anaphase was not as dramatic as shown in cells with a complete depletion of SGO1.5,6,16,26 In this study, the rate was ∼12% in lagging chromosome and ∼25% in anaphase bridge (in wild type, ∼2% and ∼1%, respectively) (Fig. S1). Typically, there were a few chromosomes located at one side of the metaphase plate.
Figure 2.
SGO+/− MEFs exhibit enhanced chromosome missegregation. (A) Paired MEFs with indicated genotypes were stained with DAPI (blue) and antibody to α-tubulin (green). Arrows show the misaligned chromosome (middle part) or the lagging or missegregated chromosomes (lower part). (B) Paired MEFs with indicated genotypes were stained with antibodies to α-tubulin (green) and p-H3S10 (red). (C) Paired MEFs with indicated genotypes were stained with DAPI (blue) and antibody to α-tubulin (green). (D) Paired MEFs were subjected to chromosome spread analysis as described in Materials and Methods. Representative images are shown.
Since sSGO1, the major spliced form of SGO1, localizes to the centrosome and spindle poles and it also has a function in centrosomal dynamics during the cell cycle,1,5,16 we examined the microtubule nucleation number in paired MEFs. We observed that, whereas SGO1+/+ MEFs typically exhibited two spindle poles in mitotic cells, SGO1+/− MEFs frequently contain more than two microtubule organization centers (Fig. 2B and C). Some SGO1+/− cells contained more than four microtubule organization centers (Fig. 2C), which usually was correlated with polyploid DNA content. Indeed, chromosome spread analysis revealed that, whereas wildtype mitotic MEFs contained a normal number of chromosomes (40 pairs of chromatids), a significant number of SGO1+/− mitotic MEFs were polyploid or aneuploid, with the chromosome number exceeding 40 pairs (Fig. 2D).
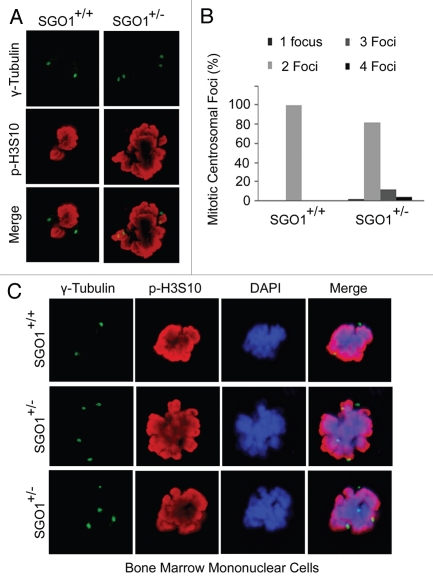
To investigate whether extra microtubule organization centers observed in SGO1+/− MEFs were centrosome-based, we stained MEFs with an antibody to γ-tubulin, a centrosome/centriole marker. We observed that in comparison with wild-type MEFs, SGO1+/− MEFs frequently exhibited an abnormal number of centrosomal foci during mitosis (Fig. 3A and B). To confirm that extra centrosomal foci also occurred in cell types other than MEFs in SGO1+/− mice, we obtained mouse bone marrow cells derived from paired littermates at 8 weeks of age and stained these cells with antibodies to γ-tubulin and phospho-H3S10. Fluorescent microscopy revealed that a significant fraction of SGO1+/− bone marrow cells contained more than two γ-tubulin foci during mitosis (Fig. 3C). These observations are consistent with the notion that SGO1 also functions in mediating centriole cohesion during the cell cycle.16,17 An alternative interpretation of the cytology data is that the increase in centrosome number is an indirect effect of increased polyploidy in SGO1+/− cells.
Figure 3.
SGO+/− MEFs exhibit extra centrosomal foci. (A) Paired MEFs were stained with the antibodies to γ-tubulin (green) and p-H3S10 (red). DNA was stained with DAPI (blue). (B) Paired MEFs were stained with the antibody to γ-tubulin (green). Centrosomal numbers in mitotic cells were counted for each type of MEFs (n = 50 for each). (C) Bone marrow cells from mice of each genotype were stained with the antibodies to γ-tubulin (green) and p-H3S10 (red). DNA was stained with DAPI (blue). Representative images are shown.
SGO1+/− mice show higher acute cell death with AOM treatment.
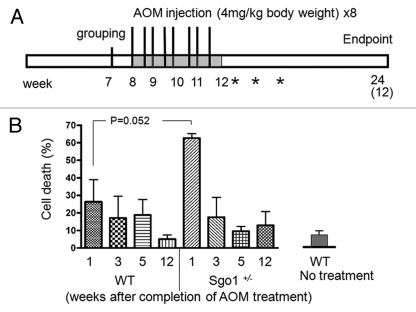
We investigated whether SGO1+/− mice were more susceptible to tumorigenesis after treatment with AOM, a colon carcinogen that creates DNA adducts and damage and initiates/facilitates carcinogenesis (Fig. 4A).31–35 Defects in chromosome cohesion are implicated in chromosome instability and colonic tumorigenesis.25,26 However, mutations linked to chromosome instability have not always been linked to elevated tumorigenesis. Such mutations can be oncogenic in one organ and function as tumor suppressors in another.22,23,36 We hypothesized that the context-dependent response may be due to activation of cellular responses such as cell death or senescence, which may serve to antagonize tumorigenesis.37 To test this hypothesis and to characterize the response of SGO1+/− mice to AOM treatments over time, we monitored DNA content in colonic mucosa as a marker for cell death with FACS analysis. One week after completion of AOM treatment, the percent of cells with sub-G1 DNA content (i.e., the cell death rate) in colonic mucosa was high both in WT and in SGO1+/− mice due to an acute response to AOM. However, the percent of cells with sub-G1 DNA content was significantly higher in SGO1+/− mice (∼60%) than in WT (∼25%) (Fig. 4B). After three weeks, the acute cell death subsided, and the difference between wild-type and SGO1+/− mice showed no statistical significance (Fig. 4B). Cell death in WT colonic mucosa kept decreasing at five and 12 weeks after AOM treatment, suggesting an ongoing recovery processes. Colonic cell death rate in untreated WT was less than 10%, and in WT samples 12 weeks after AOM, the cell death rate returned to the basal level. In the SGO1+/− background, the sub-G1 content as revealed by FACS analysis remained rather variable 5 and 12 weeks post AOM treatment.
Figure 4.
SGO1+/− mice show enhanced acute colonic cell death with AOM treatments. (A) Schematic presentation of mouse tumorigenesis assay. At 7 weeks of age, mice were grouped, and starting at 8 weeks of age, they were given s.c. azoxymethane (AOM, 4 mg/kg body weight) two times weekly for 4 weeks. Asterisks indicate timing for FACS sample harvest to assess acute response to AOM on 1, 3 and 5 weeks after completion of AOM treatments. Twelve weeks after completion of AOM treatment, all remaining mice were sacrificed, and their colons were removed for FACS, immunoblotting and ACF/adenoma counting. (B) Colonic mucosa in SGO1+/− mice show a higher acute cell death rate in response to AOM treatments. Cell death rate (i.e., percentage of cells with sub-G1 DNA) is estimated with FACS analysis of colonic mucosa in AOM-treated wild-type and SGO1+/− mice. Cell death rate in untreated wild-type is less than 10%.
SGO1+/− mice show more frequent colonic ACF and tumors.
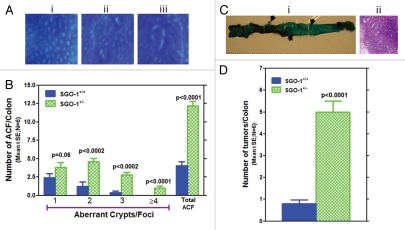
Twelve weeks after completion of AOM treatment (endpoint), all mice were sacrificed, and colonic ACF and tumors were counted. SGO1+/− mice showed significantly more ACF and multicrypt ACF (Fig. 4C). The frequency of ACF was approximately three times higher in SGO1+/− mice (n = 6, p < 0.0001). Frequency of colonic tumors were increased in SGO1+/− mice by 5-fold (n = 6, p < 0.0001). Most of these colonic tumors were adenomas (Fig. 5C and D).
Figure 5.
SGO1+/− mice show enhanced colonic tumorigenesis with AOM treatments. (A) Examples of precancerous lesions (ACF): (i) Normal-looking colonic crypts (control), (ii) ACF with single crypt (upper right) and with two crypts (lower left), (iii) Large ACF with four crypts. (B) SGO1−/+ mice are more susceptible to the development of precancerous legions (ACF) of various sizes. Size of ACF, indicated by number of crypts, is larger in SGO1−/+ mice (green bars) than in wild type (blue bars). (C) (i) Methylene blue-stained colon from SGO1+/−. Arrows indicate adenoma-like masses of cells, differentially stained with Methylene blue. (ii) Hematoxylin-stained colonic tissue sections from the adenoma. (D) Frequency of colon tumors is higher in Sgo1+/− mice. Unit is incidence/animal. p-value is calculated with Student t-test.
SGO1+/− mice differentially express tumorigenesis-relevant markers p53, Bcl-2, IL-6 and COX2.
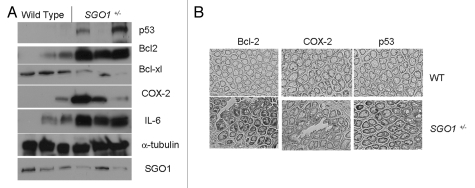
We tested expression of select biomarkers in wild-type and in SGO1+/− mice normal-looking colonic mucosa via immunoblotting (Fig. 6A). Although there is variation among animals, colonic mucosa from SGO1+/− mice showed an increase in COX2, Bcl-2, IL-6 and p53 expression and a decrease in Bcl-xL. Interestingly, there seems to be a correlation between the amount of p53, Bcl-2, IL-6 and Sgo1, suggesting a link between expression of p53, Bcl-2, IL-6 and Sgo1.
Figure 6.
Differential marker protein expressions in wild-type and SGO1+/− mice. (A) Extracts of colonic mucosa from three wild-type and three SGO1+/− mice were subjected to immunoblotting. All samples are from the endpoint (12 weeks after completion of AOM treatment). α-tubulin used as loading control. (B) Immunohistochemistry of endpoint samples for the overexpressed proteins (Bcl-2, left column; COX2, middle column; p53, right column) counterstained with Hematoxylin. Upper part: normal-looking colonic crypts from wild type. Lower part: normal-looking colonic crypts from SGO+/− mice.
The overall crypt structures in SGO1 mice were indistinguishable from that in wild type by histological observation alone. We investigated via immunohistochemistry whether these overexpressed proteins (i.e., Bcl-2, COX2 and p53) localize in particular cell populations or whether they are evenly expressed in the normal-looking colonic mucosal tissues (Fig. 6B and S3). In WT, only limited immunohistochemistry signals were observed, and the proteins were sporadically expressed in a few cells in each crypt (Fig. 6B, upper row). In SGO1+/− mice, the expression was still in a limited number of clustered cells in each crypt, but the number of stained cells was larger and the signal was more intense (Fig. 6B, lower row).
Discussion
In this report, we described the characterization of SGO1-haploinsufficent mice and provided evidence that the SGO1 defect can significantly enhance colonic tumorigenesis triggered by AOM treatment. Complete ablation of SGO1 results in embryonic lethality, indicating that it is essential for animal development. This embryonic lethality is also in agreement with the role of other mitotic genes involved in regulating spindle checkpoint and the metaphase to anaphase transition. Mice with SGO1 haploinsufficiency develop normally and are completely fertile. However, SGO1+/− cells frequently contain extra microtubulenucleation centers or spindle poles (centrosomes), which are accompanied by an elevated chromosome missegregation, suggesting enhanced chromosome instability during cell division.
The increase in centrosome number can be interpreted as a result of (1) over-replication of centrosome or (2) premature separation of duplicated centrosomes.38,39 A recent publication demonstrated the novel function of cohesin-Sgo1 to engage centrioles, paired core components of centrosomes.17 Thus, loss of Sgo1 function would lead to premature centriolecentrosome separation. Our observation agrees with the newly identified function of cohesin-Sgo1, which supports possibility of enhanced centrosomes.
We demonstrated that Sgo1 haploinsufficiency in mice resulted in two phenotypes at the cellular level, namely, chromosome cohesion defect and centrosome number defect, both of which would lead to enhanced chromosome instability. There is no evidence at present on centrosome number increase in colon cancer, but there is ample evidence for centrosome-mediated aneuploidial defects being implicated in enhanced carcinogenesis.38,39 Centrosome number defect can lead to multipolar mitosis, which is a major source of subsequent aneuploidy in daughter cells.40,41 Other mutations that lead to centrosome number defect can be carcinogenic. Knockout of p53 leads to centrosome number defect, and p53-knockout mice are prone to carcinogenesis.42,43
Previous reports indicated that, rather than simply being oncogenic, high chromosome instability from defects in mitotic machinery can be both oncogenic and tumor suppressive in an environment-dependent manner.20,22,36 In our AOM-induced tumorigenesis assay, the SGO1+/− mice showed a significant increase in size and frequency of ACF, which are precursor lesions of colonic tumors,31–35 and an increased number of colonic tumors at the endpoint, indicating that the SGO1 defect can be an enhancing factor for colonic tumorigenesis. This observation is reminiscent of our previous results on colonic tumorigenesis with BubR1 haploinsufficiency mice, which show a spindle checkpoint defect and high mitotic chromosome instability.37,44 Similar pro-tumorigenic effects of other mitotic defect models have been observed.19,36,45,46 The pro-tumorigenic effect can be explained at least in part, since chromosome instability can accelerate loss of heterozygosity of tumor suppressor(s).47,48 Detailed tumor analysis in molecular level requires more abundant samples from later endpoints and will be addressed in an upcoming study.
We investigated the duality of aneuploidy in promoting or suppressing carcinogenesis. The underlying mechanism by which aneuploidy suppresses tumor development is unclear. We hypothesized that cell death as a result of CIN can eliminate these cells at an accelerated rate. To investigate possible dual responses and mechanism(s), we tested whether cellular responses that serve to prevent cancer (e.g., elevated cell death or senescence) are activated in SGO1+/− mice. Indeed SGO1+/− mice showed an elevated rate of acute cell death, suggesting that SGO1 colonic cells may be inherently prone to death upon AOM treatment. Our observation is a first demonstration that enhanced cell death is operating in high CIN model animals after direct carcinogen challenge, which supports a hypothesis that tumor promotion or suppression is, indeed, a result of balance between enhancement of cell proliferation and elimination.
We identified differential expression of several markers for colonic tumorigenesis in SGO1+/− mice. COX2, p53 and Bcl-2 overexpression have been reported in human cancers,49–51 and their usefulness as prognostic or staging markers has been recognized. However, it was unexpected that these markers are expressed in normal-looking tissues (Fig. 6). Expression of the inflammation markers COX2 and IL-6 also correlated with SGO1 reduction in this study. Human colon cancer patients have higher serum IL-6 levels than do healthy controls, and higher IL-6 levels are associated with increasing tumor stages and tumor size as well as with metastasis and decreased survival.52 COX2 has been established as a major target for colon cancer chemoprevention with specific inhibitors.53–55 However, when in the process of tumorigenesis, the marker expressions start was ambiguous. Our results place the timing of p53 and Bcl-2 expression quite early in normal-looking mucosa. The localization pattern of the marker-positive cells was different from that of known colon cancer stem cell markers (e.g., CD133 and LGR5).56 This new observation invites next stage investigation, including whether the p53 and Bcl-2 are functional forms or not.
We observed that these marker expressions coincided in colonic mucosal tissues in SGO1+/− mice. Whether these expressions are linked with direct causal relationships in mice, or whether they are independently or coincidentally associated with Sgo1 defect, awaits further investigation. Our current scenario is that (1) the whole chromosome instability is detected by the p53 system, leading to accumulation of p53,40,57,58 and (2) the chromosome instability triggers an inflammatory response with elevated COX2 and IL6 expression, possibly because of increased cell death. (3) COX2 expression triggers overexpression of Bcl-2, reduction of Bcl-xl and additional chromosome instability, as observed in transgenic mice conditionally expressing COX2 under a mammary gland specific promoter59 and in the breast cancer cell line MCF7 with induced COX2 expression.60 Bcl-2 is usually considered to be an anti-apoptotic factor, and its expression is associated with cell survival. However, once p53 and Bcl-2 are overexpressed simultaneously, they can form a complex that directly triggers mitochondrial cell death.61 Also, formation of the p53-Bcl-2 complex depletes the Bcl-2-Bax complex, which also triggers cell death.62 As a whole, the p53-COX2-Bcl-2 expression may create a pro cell-death condition, and the expressed markers may function to antagonize chromosome cohesion defect-mediated tumorigenesis. This hypothetical scenario should be tested in subsequent in vitro experiments. Systematic characterization of the expression of various markers and elucidation of their relevance to chromosomal instability also requires further study.
The increase in COX2 in SGO1 haploinsufficient mice was also unexpected. Our result is the first in vivo demonstration that an SGO1 defect facilitates COX2 overexpression in the colon. It also suggests that at least a part of carcinogenic activity may be exerted or aided by the COX2 expression, in which case COX2 inhibitors may prevent cancers enhanced by an SGO1 defect. Testing COX2 inhibitors on SGO1+/− mice may be helpful to assess to what extent the COX2 overexpression is involved in tumorigenesis in SGO1+/− mice.
Material and Methods
Generation of SGO1-deficient mice.
The SGO1-mutant mice were generated as in text in collaboration with Lexicon Genetics. Mouse embryonic stem (ES) cells with a targeted disruption of the SGO1 locus were obtained by a gene-trapping method.27 The targeting retroviral vector, which contained a neomycin resistance cassette, was inserted between exons 1 and 2 of SGO1. The nucleotide sequence surrounding the insertion site (*) was determined as 5′-TGT ACG CCT CCA ATG AAT ATT TAC* TAT AAG ATG AAT AAT TGA TAC TTT. Two independent 129/Sv-derived ES cell lines were injected into C57/BL6 blastocysts, and the resulting chimeric mice were backcrossed to wild-type C57/BL6 animals.
Genotyping.
Genomic DNA was recovered from mouse tail or MEFs. The nucleotide sequences of the PCR primers were as follows: forward primer, 5′-GAA AAG TAA GTC TGC TTA TGG CTC A-3′; reverse primer, 5′-CAG GTG TTG TAG AAT AAT CCA AGC-3′; and reverse primer long-terminal repeat (LTR), 5′-ATA AAC CCT CTT GCA GTT GCA TC-3′. PCR products were fractionated by electrophoresis on a 1% agarose gel and detected by ethidium bromide staining.
Immunoblotting for MEFs.
MEFs were derived from embryonic day 13.5 (E13.5) embryos (produced from SGO1+/− intercrosses). Bone marrow stromal cells were collected by flushing the tibia of 8-week-old littermates with respective genotypes and isolated as described by Kopen et al.63 Cells were cultured under 5% CO2 in dishes containing Dulbecco's minimum essential medium (DMEM) supplemented with 15% fetal bovine serum (FBS) and antibiotics (100 µg/mL penicillin, 50 µg/mL streptomycin sulfate). Total cellular proteins were extracted from mouse tail tissues and subjected to immunoblotting with antibodies against SGO1 (developed in the W. Dai lab16,64) and β-actin (Cell Signaling Technology, 4970). Specific signals were detected with horseradish peroxidase-conjugated secondary antibodies (Cell Signaling Technology, 7074) and enhanced chemiluminescence reagents (Thermo Scientific, 32132).
Real-time quantitative RT-PCR and statistical analysis.
Total RNA was isolated from the mouse tail tissue with the TRIzol Plus RNA purification system (Invitrogen Life Technologies, 12183555). RNA was reverse transcribed into first strand cDNA (Invitrogen Life Technologies, 11752-050) before quantitative polymerase chain reaction (qPCR) conducted by Applied Biosystem Life Technologies 7300 real-time PCR system with SYBR® Green PCR Master Mix (Applied Biosystem Life Technologies, 4309155) according to the manufacturer's instructions. The murine SGO1-specific and β-actin-specific primer sequences (sense/antisense) were used as follows: β-actin forward primer: 5′-GGC ATA GAG GTC TT-3′, β-actin reverse primer: 5′-CAC AGG CAT TGT ATG GAC TC-3′, SGO1 forward primer 1: 5′-ATG GCT AAG GAA AGG TGT CAG-3′, SGO1 reverse primer 1: 5′-CTG CGT TTA GTC AGA GCC ACT-3′, SGO1 forward primer 2: 5′-CCA AAG TGA GAG AAG CAC AGG-3′, SGO1 reverse primer 2: 5′-CTG TGT TTG CTT GGT TCT TCT-3′. The specificity of the amplification product was determined from a melting curve analysis. Standard curves were generated for each gene by preparing serial dilutions of the respective cDNA gene template of known quantities. Relative quantities of SGO1 mRNA were obtained by normalizing its signal to that of β-actin. All data were expressed as means ± SD. The difference between groups was analyzed using Student t-test.
Chromosome spread and fluorescence microscopy.
Cells fixed in methanol or 4% paraformaldehyde (PFA) were treated with 0.1% Triton X-100 on ice and then washed three times with PBS. After blocking with 2.0% BSA in PBS for 15 min, cells were incubated for 1 h with antibodies to γ-tubulin, α-tubulin, (Sigma, T5326, F2168, respectively), or phosphor H3 serine 10 (Santa Cruz Technology, sc-8656-R), washed with PBS, and then incubated with appropriate secondary antibodies conjugated with Rhodamine-Red-X or FITC (Jackson Immuno Research). Finally, cells were stained with 4′,6-diamidino-2-phenylindole (DAPI, 1 µg/ml, Fluka).
MEFs seeded for 24 h were treated with nocodazol (50 ng/ml) for 16 h to arrest cells in pro-metaphase. Cells detached from the culture plates by shaking were incubated in 75 mM KCl for 20 min at 37°C. These cells were then fixed in three changes of methanol/acetic acid (3:1), and the fixed cell pellets were used for slide spreads. Slides were air-dried for at least 2 d at 37°C before examination. For MEFs of each genotype, at least 50 metaphase spreads were examined. Fluorescence microscopy was performed on a Nikon microscope, and images were captured using a digital camera (Optronics). For confocal imaging, a Leica TCS SP5 (Leica Microsystems) was utilized. For determination of mitotic index, 20 fields of stained MEFs of each genotype were chosen randomly and the percentages were determined by positive staining of phosphor-H3Ser10 vs. 4,6-diamidino-2-phenylindole (DAPI).
Mouse tumorigenesis assay.
All mice were housed in the OUHSC pathogen-free rodent barrier facility. Experiments were performed under compliance of IACUC guideline and permission. At 7 weeks of age, mice [20 wild type, 20 SGO1+/− (10 male and 10 female each)] were grouped, and starting at 8 weeks of age, they were given s.c. azoxymethane (AOM, 4 mg/kg body weight) two times weekly for 4 weeks.37 One, three and five weeks after completion of AOM treatment, three male mice from both groups were sacrificed by CO2 euthanasia to harvest colonic mucosa to assess acute response to AOM with FACS analysis. Twelve weeks after completion of AOM treatment, all remaining mice were killed (10 wild type, 10 SGO1), and their colons were removed. Colons from each group were opened longitudinally and flushed with PBS and fixed flat between two pieces of filter paper in 10% buffered formalin for ACF/tumor analysis. The ACF were stained with Methylene Blue solution and counted according to our standard procedure,64 and colonic tumors were histopathologically evaluated. Normal-looking mucosal tissues of the rest of the colons from each group were scraped and snapfrozen for immunoblots and for FACS analysis.
Immunoblot analysis of colonic tissues.
Immunoblots for colonic mucosal tissues were performed as previously described in reference 65. The following antibodies were used: anti-Bcl-2 (Santa Cruz Biotechnology, SC-492), anti-Bcl-xL (SC-8392), anti-p53 (SC-6243), anti-Bax (SC-526), anti-caspase 3 (SC-7148), anti-COX2 (SC-1745), anti-α-tubulin (SC-8035) and anti-SGO1 (Novus Biologicals, NBP1-44080). Immune complexes were detected with appropriate secondary antibodies conjugated with Horseradish peroxidase (Sigma) and with chemiluminescence reagents (Thermo Scientific).
Fluorescence activated cell sorting (FACS) analysis.
Colonic mucosal tissue was rinsed with PBS and scraped, and cells were suspended in PBS, immediately fixed with ice-cold ethanol (final 80–90%). The cells were resuspended and rehydrated in PBS with 20 µM propidium iodide, 50 mM sodium citrate and 0.1 mg/ml RNaseA for 16 h at room temperature. Cells were filtered through Nylon mesh and subjected to FACS analysis with a FACS caliber cell sorter (OUHSC). For each sample, 20,000 to 50,000 cells were analyzed. The data were analyzed with Summit 4.3 software (Dako Colorado, Inc.).
Immunohistochemistry.
Colonic tissues were fixed with 10% buffered formalin and embedded in paraffin. The Histostain SP kit (Invitrogen) was used with aforementioned primary antibodies at 1.0 µg/ml.
Acknowledgements
We thank Dr. Julie Sando for editorial and scientific suggestions. We also thank Dr. Stan Lightfoot for assessments of histo-pathological abnormalities.
Abbreviations
- SGO1
shugoshin 1
- CIN
chromosome instability
- AOM
azoxymethane
- siRNA
small interfering RNA
- MEFs
mouse embryonic fibroblasts
- FACS
fluorescence activated cell sorter
- ACF
aberrant crypt foci
- WT
wild type
- COX2
cyclooxygenase-2
Conflict of Interest
There are no competing financial interests in relation to this work.
Grant Support
This study was supported in part by a US Public Service Award to W.D. (CA090658), and in part by HIEHS center fund and Superfund grants (ES000260 and ES010344). This work is also supported in part by US Public Service Award (CA102947) and Kerley-Cade chair Research endowment (OUHSC) to C.V.R.
Supplementary Material
References
- 1.Wang X, Dai W. Shugoshin, a guardian for sister chromatid segregation. Exp Cell Res. 2005;310:1–9. doi: 10.1016/j.yexcr.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 2.Indjeian VB, Stern BM, Murray AW. The centromeric protein Sgo1 is required to sense lack of tension on mitotic chromosomes. Science. 2005;307:130–133. doi: 10.1126/science.1101366. [DOI] [PubMed] [Google Scholar]
- 3.Kitajima TS, Kawashima SA, Watanabe Y. The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature. 2004;427:510–517. doi: 10.1038/nature02312. [DOI] [PubMed] [Google Scholar]
- 4.Dudas A, Ahmad S, Gregan J. Sgo1 is required for cosegregation of sister chromatids during achiasmate meiosis I. Cell Cycle. 2011;10:951–955. doi: 10.4161/cc.10.6.15032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McGuinness BE, Hirota T, Kudo NR, Peters JM, Nasmyth K. Shugoshin prevents dissociation of cohesin from centromeres during mitosis in vertebrate cells. PLoS Biol. 2005;3:86. doi: 10.1371/journal.pbio.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salic A, Waters JC, Mitchison TJ. Vertebrate shugoshin links sister centromere cohesion and kinetochore microtubule stability in mitosis. Cell. 2004;118:567–578. doi: 10.1016/j.cell.2004.08.016. [DOI] [PubMed] [Google Scholar]
- 7.Watanabe Y, Kitajima TS. Shugoshin protects cohesin complexes at centromeres. Philos Trans R Soc Lond B Biol Sci. 2005;360:515–521. doi: 10.1098/rstb.2004.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rivera T, Losada A. Shugoshin and PP2A, shared duties at the centromere. Bioessays. 2006;28:775–779. doi: 10.1002/bies.20448. [DOI] [PubMed] [Google Scholar]
- 9.Gregan J, Spirek M, Rumpf C. Solving the shugoshin puzzle. Trends Genet. 2008;24:205–207. doi: 10.1016/j.tig.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang Z, Sun Y, Harley SE, Zou H, Yu H. Human Bub1 protects centromeric sister-chromatid cohesion through Shugoshin during mitosis. Proc Natl Acad Sci USA. 2004;101:18012–18017. doi: 10.1073/pnas.0408600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawashima SA, Yamagishi Y, Honda T, Ishiguro K, Watanabe Y. Phosphorylation of H2A by Bub1 prevents chromosomal instability through localizing shugoshin. Science. 2010;327:172–177. doi: 10.1126/science.1180189. [DOI] [PubMed] [Google Scholar]
- 12.Kawashima SA, Tsukahara T, Langegger M, Hauf S, Kitajima TS, Watanabe Y. Shugoshin enables tensiongenerating attachment of kinetochores by loading Aurora to centromeres. Genes Dev. 2007;21:420–435. doi: 10.1101/gad.1497307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kitajima TS, Hauf S, Ohsugi M, Yamamoto T, Watanabe Y. Human Bub1 defines the persistent cohesion site along the mitotic chromosome by affecting Shugoshin localization. Curr Biol. 2005;15:353–359. doi: 10.1016/j.cub.2004.12.044. [DOI] [PubMed] [Google Scholar]
- 14.Riedel CG, Katis VL, Katou Y, Mori S, Itoh T, Helmhart W, et al. Protein phosphatase 2A protects centromeric sister chromatid cohesion during meiosis I. Nature. 2006;441:53–61. doi: 10.1038/nature04664. [DOI] [PubMed] [Google Scholar]
- 15.Kitajima TS, Sakuno T, Ishiguro K, Iemura S, Natsume T, Kawashima SA, et al. Shugoshin collaborates with protein phosphatase 2A to protect cohesin. Nature. 2006;441:46–52. doi: 10.1038/nature04663. [DOI] [PubMed] [Google Scholar]
- 16.Wang X, Yang Y, Duan Q, Jiang N, Huang Y, Darzynkiewicz Z, et al. sSgo1, a major splice variant of Sgo1, functions in centriole cohesion where it is regulated by Plk1. Dev Cell. 2008;14:331–341. doi: 10.1016/j.devcel.2007.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schöckel L, Mockel M, Mayer B, Boos D, Stemmann O. Cleavage of cohesin rings coordinates the separation of centrioles and chromatids. Nat Cell Biol. 2011;13:966–972. doi: 10.1038/ncb2280. [DOI] [PubMed] [Google Scholar]
- 18.Duesberg P, Li R, Fabarius A, Hehlmann R. Aneuploidy and cancer: from correlation to causation. Contrib Microbiol. 2006;13:16–44. doi: 10.1159/000092963. [DOI] [PubMed] [Google Scholar]
- 19.Rao CV, Yamada HY, Yao Y, Dai W. Enhanced genomic instabilities caused by deregulated microtubule dynamics and chromosome segregation: a perspective from genetic studies in mice. Carcinogenesis. 2009;30:1469–1474. doi: 10.1093/carcin/bgp081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weaver BA, Cleveland DW. Aneuploidy: instigator and inhibitor of tumorigenesis. Cancer Res. 2007;67:10103–10105. doi: 10.1158/0008-5472.CAN-07-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weaver BA, Cleveland DW. The aneuploidy paradox in cell growth and tumorigenesis. Cancer Cell. 2008;14:431–433. doi: 10.1016/j.ccr.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weaver BA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell. 2007;11:25–36. doi: 10.1016/j.ccr.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 23.Weaver BA, Silk AD, Cleveland DW. Low rates of aneuploidy promote tumorigenesis while high rates of aneuploidy cause cell death and tumor suppression. Cell Oncol. 2008;30:453. doi: 10.3233/CLO-2008-0445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Q, Liu T, Fang Y, Xie S, Huang X, Mahmood R, et al. BUBR1 deficiency results in abnormal megakaryopoiesis. Blood. 2004;103:1278–1285. doi: 10.1182/blood-2003-06-2158. [DOI] [PubMed] [Google Scholar]
- 25.Barber TD, McManus K, Yuen KW, Reis M, Parmigiani G, Shen D, et al. Chromatid cohesion defects may underlie chromosome instability in human colorectal cancers. Proc Natl Acad Sci USA. 2008;105:3443–3448. doi: 10.1073/pnas.0712384105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iwaizumi M, Shinmura K, Mori H, Yamada H, Suzuki M, Kitayama Y, et al. Human Sgo1 downregulation leads to chromosomal instability in colorectal cancer. Gut. 2009;58:249–260. doi: 10.1136/gut.2008.149468. [DOI] [PubMed] [Google Scholar]
- 27.Zambrowicz BP, Friedrich GA, Buxton EC, Lilleberg SL, Person C, Sands AT. Disruption and sequence identification of 2,000 genes in mouse embryonic stem cells. Nature. 1998;392:608–611. doi: 10.1038/33423. [DOI] [PubMed] [Google Scholar]
- 28.Babu JR, Jeganathan KB, Baker DJ, Wu X, Kang-Decker N, van Deursen JM. Rae1 is an essential mitotic checkpoint regulator that cooperates with Bub3 to prevent chromosome missegregation. J Cell Biol. 2003;160:341–353. doi: 10.1083/jcb.200211048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rohrabaugh SL, Hangoc G, Kelley MR, Broxmeyer HE. Mad2 haploinsufficiency protects hematopoietic progenitor cells subjected to cell cycle stress in vivo and to inhibition of redox function of Ape1/Ref-1 in vitro. Exp Hematol. 2011;39:415–423. doi: 10.1016/j.exphem.2010.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clift D, Bizzari F, Marston AL. Shugoshin prevents cohesin cleavage by PP2A(Cdc55)-dependent inhibition of separase. Genes Dev. 2009;23:766–780. doi: 10.1101/gad.507509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanaka T. Colorectal carcinogenesis: Review of human and experimental animal studies. J Carcinog. 2009;8:5. doi: 10.4103/1477-3163.49014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Der Kraak L, Meunier C, Turbide C, Jothy S, Gaboury L, Marcus V, et al. A two-locus system controls susceptibility to colitis-associated colon cancer in mice. Oncotarget. 2010;1:436–446. doi: 10.18632/oncotarget.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raju J. Azoxymethane-induced rat aberrant crypt foci: relevance in studying chemoprevention of colon cancer. World J Gastroenterol. 2008;14:6632–6635. doi: 10.3748/wjg.14.6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen J, Huang XF. The signal pathways in azoxymethane-induced colon cancer and preventive implications. Cancer Biol Ther. 2009;8:1313–1317. doi: 10.4161/cbt.8.14.8983. [DOI] [PubMed] [Google Scholar]
- 35.Fenoglio-Preiser CM, Noffsinger A. Aberrant crypt foci: A review. Toxicol Pathol. 1999;27:632–642. doi: 10.1177/019262339902700604. [DOI] [PubMed] [Google Scholar]
- 36.Ricke RM, van Ree JH, van Deursen JM. Whole chromosome instability and cancer: a complex relationship. Trends Genet. 2008;24:457–466. doi: 10.1016/j.tig.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dai W, Wang Q, Liu T, Swamy M, Fang Y, Xie S, et al. Slippage of mitotic arrest and enhanced tumor development in mice with BubR1 haploinsufficiency. Cancer Res. 2004;64:440–445. doi: 10.1158/0008-5472.CAN-03-3119. [DOI] [PubMed] [Google Scholar]
- 38.Zyss D, Gergely F. Centrosome function in cancer: guilty or innocent? Trends Cell Biol. 2009;19:334–346. doi: 10.1016/j.tcb.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 39.Fukasawa K. Aberrant activation of cell cycle regulators, centrosome amplification and mitotic defects. Horm Cancer. 2011;2:104–112. doi: 10.1007/s12672-010-0060-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi Q, King RW. Chromosome nondisjunction yields tetraploid rather than aneuploid cells in human cell lines. Nature. 2005;437:1038–1042. doi: 10.1038/nature03958. [DOI] [PubMed] [Google Scholar]
- 41.Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fukasawa K, Choi T, Kuriyama R, Rulong S, Vande Woude GF. Abnormal centrosome amplification in the absence of p53. Science. 1996;271:1744–1747. doi: 10.1126/science.271.5256.1744. [DOI] [PubMed] [Google Scholar]
- 43.Fukasawa K. Centrosome amplification, chromosome instability and cancer development. Cancer Lett. 2005;230:6–19. doi: 10.1016/j.canlet.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 44.Rao CV, Yang YM, Swamy MV, Liu T, Fang Y, Mahmood R, et al. Colonic tumorigenesis in BubR1+/− ApcMin/+ compound mutant mice is linked to premature separation of sister chromatids and enhanced genomic instability. Proc Natl Acad Sci USA. 2005;102:4365–4370. doi: 10.1073/pnas.0407822102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Foijer F, Draviam VM, Sorger PK. Studying chromosome instability in the mouse. Biochim Biophys Acta. 2008;1786:73–82. doi: 10.1016/j.bbcan.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schvartzman JM, Sotillo R, Benezra R. Mitotic chromosomal instability and cancer: mouse modelling of the human disease. Nat Rev Cancer. 2010;10:102–115. doi: 10.1038/nrc2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baker DJ, Jin F, Jeganathan KB, van Deursen JM. Whole chromosome instability caused by Bub1 insufficiency drives tumorigenesis through tumor suppressor gene loss of heterozygosity. Cancer Cell. 2009;16:475–486. doi: 10.1016/j.ccr.2009.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baker DJ, van Deursen JM. Chromosome missegregation causes colon cancer by APC loss of heterozygosity. Cell Cycle. 2010;9:1711–1716. doi: 10.4161/cc.9.9.11314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang D, Dubois RN. The role of COX2 in intestinal inflammation and colorectal cancer. Oncogene. 2010;29:781–788. doi: 10.1038/onc.2009.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kerns BJ, Jordan PA, Moore MB, Humphrey PA, Berchuck A, Kohler MF, et al. p53 overexpression in formalin-fixed, paraffin-embedded tissue detected by immunohistochemistry. J Histochem Cytochem. 1992;40:1047–1051. doi: 10.1177/40.7.1607637. [DOI] [PubMed] [Google Scholar]
- 51.Basu A, Haldar S. The relationship between BcI2, Bax and p53: consequences for cell cycle progression and cell death. Mol Hum Reprod. 1998;4:1099–1109. doi: 10.1093/molehr/4.12.1099. [DOI] [PubMed] [Google Scholar]
- 52.Knupfer H, Preiss R. Serum interleukin-6 levels in colorectal cancer patients—a summary of published results. Int J Colorectal Dis. 2010;25:135–140. doi: 10.1007/s00384-009-0818-8. [DOI] [PubMed] [Google Scholar]
- 53.Reddy BS, Rao CV. Novel approaches for colon cancer prevention by cyclooxygenase-2 inhibitors. J Environ Pathol Toxicol Oncol. 2002;21:155–164. [PubMed] [Google Scholar]
- 54.Rostom A, Dubé C, Lewin G, Tsertsvadze A, Barrowman N, Code C, et al. US Preventive Services Task Force. Nonsteroidal anti-inflammatory drugs and cyclooxygenase-2 inhibitors for primary prevention of colorectal cancer: a systematic review prepared for the US Preventive Services Task Force. Ann Intern Med. 2007;146:376–389. doi: 10.7326/0003-4819-146-5-200703060-00010. [DOI] [PubMed] [Google Scholar]
- 55.Samoha S, Arber N. Cyclooxygenase-2 inhibition prevents colorectal cancer: from the bench to the bed side. Oncology. 2005;69:33–37. doi: 10.1159/000086630. [DOI] [PubMed] [Google Scholar]
- 56.Kemper K, Grandela C, Medema JP. Molecular identification and targeting of colorectal cancer stem cells. Oncotarget. 2010;1:387–395. doi: 10.18632/oncotarget.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Duensing A, Duensing S. Guilt by association? p53 and the development of aneuploidy in cancer. Biochem Biophys Res Commun. 2005;331:694–700. doi: 10.1016/j.bbrc.2005.03.157. [DOI] [PubMed] [Google Scholar]
- 58.Ho CC, Hau PM, Marxer M, Poon RY. The requirement of p53 for maintaining chromosomal stability during tetraploidization. Oncotarget. 2010;1:583–595. doi: 10.18632/oncotarget.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu CH, Chang SH, Narko K, Trifan OC, Wu MT, Smith E, et al. Overexpression of cyclooxygenase-2 is sufficient to induce tumorigenesis in transgenic mice. J Biol Chem. 2001;276:18563–18569. doi: 10.1074/jbc.M010787200. [DOI] [PubMed] [Google Scholar]
- 60.Singh B, Cook KR, Vincent L, Hall CS, Berry JA, Multani AS, et al. Cyclooxygenase-2 induces genomic instability, BCL2 expression, doxorubicin resistance and altered cancer-initiating cell phenotype in MCF7 breast cancer cells. J Surg Res. 2008;147:240–246. doi: 10.1016/j.jss.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 61.Mihara M, Erster S, Zaika A, Petrenko O, Chittenden T, Pancoska P, et al. p53 has a direct apoptogenic role at the mitochondria. Mol Cell. 2003;11:577–590. doi: 10.1016/S1097-2765(03)00050-9. [DOI] [PubMed] [Google Scholar]
- 62.Deng X, Gao F, Flagg T, Anderson J, May WS. Bcl2's flexible loop domain regulates p53 binding and survival. Mol Cell Biol. 2006;26:4421–4434. doi: 10.1128/MCB.01647-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kopen GC, Prockop DJ, Phinney DG. Marrow stromal cells migrate throughout forebrain and cerebellum, and they differentiate into astrocytes after injection into neonatal mouse brains. Proc Natl Acad Sci USA. 1999;96:10711–10716. doi: 10.1073/pnas.96.19.10711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang X, Yang Y, Dai W. Differential subcellular localizations of two human Sgo1 isoforms: implications in regulation of sister chromatid cohesion and microtubule dynamics. Cell Cycle. 2006;5:635–640. doi: 10.4161/cc.5.6.2547. [DOI] [PubMed] [Google Scholar]
- 65.Yamada HY, Rao CV. BRD8 is a potential chemosensitizing target for spindle poisons in colorectal cancer therapy. Int J Oncol. 2009;35:1101–1109. doi: 10.3892/ijo_00000425. [DOI] [PubMed] [Google Scholar]
- 66.Rao CV, Rivenson A, Katiwalla M, Kelloff GJ, Reddy BS. Chemopreventive effect of oltipraz during different stages of experimental colon carcinogenesis induced by azoxymethane in male F344 rats. Cancer Res. 1993;53:2502–2506. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.